The structure of the homing endonuclease I-SceI from S. cerevisiae has been determined in complex with its target DNA and catalytic cations, revealing a two-metal-ion mechanism.
Keywords: gene targeting, protein–DNA interaction, genetics, I-SceI, homing endonuclease, DNA cleavage, Saccharomyces cerevisiae
Abstract
Homing endonucleases are highly specific DNA-cleaving enzymes that recognize and cleave long stretches of DNA. The engineering of these enzymes provides instruments for genome modification in a wide range of fields, including gene targeting. The homing endonuclease I-SceI from the yeast Saccharomyces cerevisiae has been purified after overexpression in Escherichia coli and its crystal structure has been determined in complex with its target DNA. In order to evaluate the number of ions that are involved in the cleavage process, thus determining the catalytic mechanism, crystallization experiments were performed in the presence of Mn2+, yielding crystals that were suitable for X-ray diffraction analysis. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 80.11, b = 80.57, c = 130.87 Å, α = β = γ = 90°. The self-rotation function and the Matthews coefficient suggested the presence of two protein–DNA complexes in the asymmetric unit. The crystals diffracted to a resolution limit of 2.9 Å using synchrotron radiation. From the anomalous data, it was determined that three cations are involved in catalysis and it was confirmed that I-SceI follows a two-metal-ion DNA-strand cleavage mechanism.
1. Introduction
Homing endonucleases (HEs) recognize long DNA sequences and generate double-strand breaks that can be used to promote gene targeting through homologous recombination. These enzymes, which are also known as meganucleases, are highly sequence-specific enzymes with recognition sites that are 12–45 base pairs in length. Because of the stringency of the recognition site, HEs display a very low frequency of cleavage even in whole genomes and thus are very powerful tools for the manipulation of the genomes of mammalian and plant cells (Choulika et al., 1994 ▸, 1995 ▸; Rouet et al., 1994 ▸). The HEs are grouped into several families, of which the LAGLIDADG family of homing endonucleases (LHEs), which are named after a common amino-acid motif, is the most abundant. Within the LHE family there are two subfamilies: enzymes that contain one copy of the LAGLIDADG motif and those with two LAGLIDADG motifs. The monomeric LHEs have two asymmetric domains that enable them to recognize nonpalindromic DNA targets, whilst homodimeric LHEs recognize palindromic (or nearly palindromic) DNA targets. Information extracted from the structures of different homodimeric members (Chevalier et al., 2001 ▸, 2004 ▸; Molina, Redondo et al., 2015 ▸; Redondo et al., 2014 ▸) indicated that catalysis by the LHEs is based on a cleavage mechanism involving two metal ions per DNA strand and involving three catalytic ions in total, one of which is used in the cleavage of both strands. Notwithstanding, structural analysis of the monomeric LHE I-DmoI in the presence of noncatalytic and catalytic ions (Marcaida et al., 2008 ▸) revealed the presence of only two catalytic cations in the cleaved state, suggesting different catalytic mechanisms for the monomeric and homodimeric LHEs. Nevertheless, a detailed analysis of the catalytic mechanism of I-DmoI, identifying all of its intermediate states, showed that the catalytic mechanism is also based on a two-metal-ion catalytic mechanism, as in its homodimeric counterparts. Thus, three catalytic ions are required during catalysis for monomeric and homodimeric HEs, although in the monomeric I-DmoI the central catalytic ion is substituted by a water molecule when the cleavage reaction is complete (Molina, Stella et al., 2015 ▸).
Apart from I-DmoI, there is no other monomeric LHE deposited in the Protein Data Bank for which the number of catalytic ions has been unequivocally determined by X-ray crystallography using their anomalous signal. To shed light on the possibility of this two-metal-ion mechanism being common to all LHEs, we have solved the crystal structure of another monomeric LHE, I-SceI, in complex with its target DNA and with Mn2+ as the catalytic ion, since it can be directly observed by X-ray crystallography. From the anomalous signal provided by Mn2+, three catalytic ions were located in the active site occupying the canonical catalytic positions (Molina, Stella et al., 2015 ▸) and supporting the idea of a common cleavage mechanism for all LHEs based on a two-metal-ion mechanism, with one of them being involved in the cleavage of both strands. Curiously, the anomalous signal revealed the presence of an extra Mn2+ ion located outside the catalytic centre that was apparently not involved in enzyme catalysis.
2. Materials and methods
2.1. Protein expression and purification
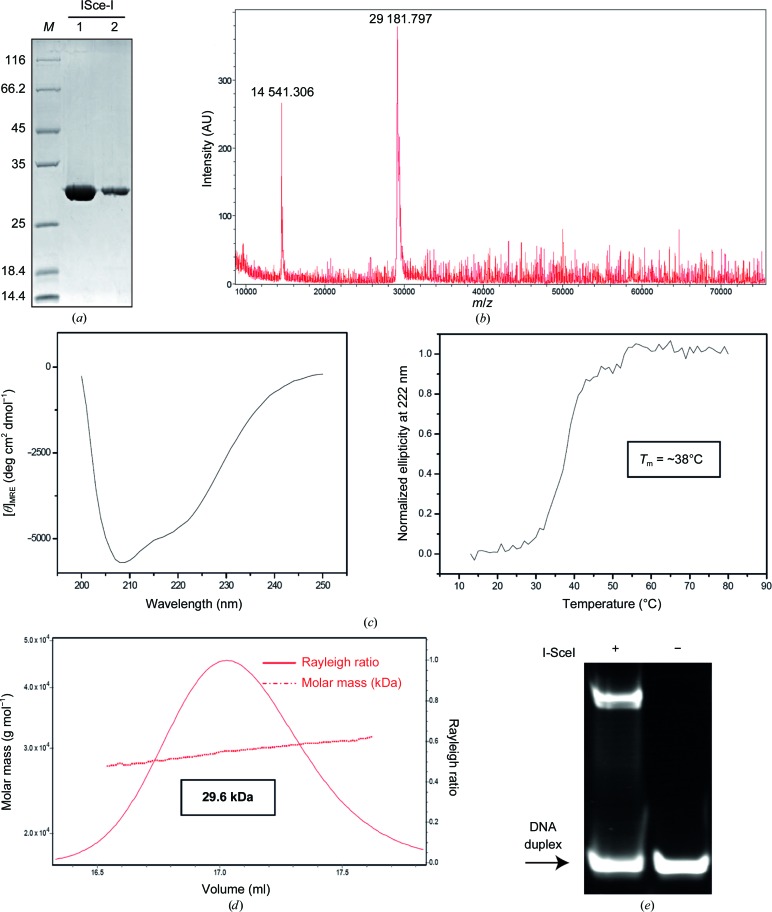
Escherichia coli strain BL21 (DE3) cells were transformed with pET-24d(+) vector containing the gene encoding the I-SceI protein. The polypeptide chain expressed corresponds to UniProt sequence P03882 with an Ala residue inserted after the initial methionine and the additional C-terminal sequence AAALEHHHHHHHH. Cells were grown in ZYP-5052 auto-inducible medium (Studier, 2005 ▸) at 37°C for 2 h and then at 20°C for about 16 h. Cells were harvested by centrifugation (at 9000g for 20 min at 4°C), resuspended in lysis buffer [50 mM sodium phosphate pH 8.0, 300 mM NaCl, 5%(v/v) glycerol] containing protease inhibitors (Complete EDTA-free tablets, Roche), flash-frozen in liquid nitrogen and stored at −80°C. After thawing, solid DNase and lysozyme were added to the suspension and the cells were disrupted by sonication. The cell lysate was ultracentrifuged at 12 000g for 30 min at 4°C. SDS–PAGE showed the protein to predominantly be in the pellet. Nevertheless, the amount of protein in the soluble fraction was sufficient to set up a purification procedure using three steps, all performed at 4°C, using chromatographic equipment and columns from GE Healthcare. Firstly, the supernatant was applied onto a 5 ml HiTrap Chelating HP column loaded with Co2+ ions and equilibrated in 50 mM sodium phosphate pH 8.0, 300 mM NaCl. The column was washed first with the same buffer and then with 32 mM imidazole, and the protein was eluted with 800 mM imidazole. The tubes in the fraction collector contained 20 mM sodium phosphate pH 7.0, 150 mM NaCl to dilute the eluted protein fourfold. This was performed to minimize protein precipitation and to reduce the concentration of imidazole and salt for the next chromatographic step. Protein-rich fractions were pooled and applied onto a 5 ml HiTrap Heparin HP column equilibrated with 20 mM sodium phosphate pH 7.0, 150 mM NaCl. The protein was eluted with a linear gradient from 0 to 50% of 20 mM sodium phosphate pH 7.0, 2 M NaCl in 15 column volumes. Protein-rich fractions were pooled and loaded onto a HiLoad 16/60 200 Superdex column equilibrated with 10 mM HEPES pH 8.0, 5%(v/v) glycerol with 500 mM KCl (the approximate salt concentration in the pool of protein fractions eluted from the heparin column, as estimated from conductivity measurements). This gel-filtration step polished the protein and changed the buffer at the same time, purifying the protein into a buffer more suitable for crystallization trials. The protein eluted as a symmetric peak and was concentrated (using 3 kDa cutoff Centriprep Amicon Ultra devices), flash-frozen in liquid nitrogen and stored at −80°C until use. The protein concentration was determined using the theoretical molar extinction coefficient at 280 nm (49 975 M −1 cm−1) calculated from the amino-acid composition. An overloaded SDS–PAGE stained with SimplyBlue SafeStain (Invitrogen) displayed the protein preparation (Fig. 1 ▸ a). The molar mass measured by mass spectrometry (MALDI) indicated that the initial methionine was not present in the purified protein (Fig. 1 ▸ b). Protein secondary structure and thermal stability were analysed by circular dichroism (CD; Fig. 1 ▸ c). Far-UV CD spectra (250–190 nm) were recorded at 20°C on a Jasco-810 dichrograph equipped with a Peltier thermoelectric temperature controller and previously calibrated with d-10-camphorsulfonic acid. The spectra were acquired in continuous mode with 2 nm bandwidth, 4 s response and a scan speed of 100 nm min−1. We used 0.2 cm path-length quartz cuvettes (Hellma) and a protein sample with a concentration of 2 µM in PBS for CD analysis. Ten scans were accumulated to obtain the final spectrum. Thermal denaturation was induced by heating the sample from 12 to 80°C at 1°C min−1. The change in ellipticity at 222 nm was recorded at 1°C intervals.
Figure 1.
Expression, purification and crystallization of I-SceI. (a) SDS–PAGE gel showing the degree of purity of the protein samples. (b) MALDI of I-SceI showing a major peak at 29 181.797 Da. A small peak at 14 541.306 Da arising from the doubly charged molecule can be also observed. (c) Protein secondary structure (left panel) and thermal stability (right panel) analysed by circular dichroism. (d) SEC-MALLS data of the I-SceI protein preparation. (e) Band-shift assay. The DNA target was incubated with and without I-SceI protein for 1 h on ice. The mixture was separated on a native polyacrylamide gel. The arrow indicates the band corresponding to free DNA target.
2.2. Size-exclusion chromatography with multi-angle laser-light scattering (SEC-MALLS)
This experiment was performed at 25°C using a Superdex 200 10/300 GL column (GE Healthcare) attached to a DAWN HELEOS light-scattering detector and an Optilab rEX differential refractive-index detector (Wyatt Technology; Fig. 1 ▸ d). The column was equilibrated with 20 mM sodium phosphate pH 7.0, 500 mM NaCl, 0.03%(w/v) NaN3 and the SEC-MALLS system was calibrated with a sample of BSA at 1 g l−1 in the same buffer. A 100 µl sample of I-SceI at 0.4 g l−1 in the same buffer was injected and run through the column at a flow rate of 0.5 ml min−1. Data acquisition and analysis were carried out using the ASTRA software (Wyatt). Based on numerous measurements of BSA samples under the same or similar conditions, we estimate that the experimental error in the molar mass is around 5%.
2.3. I-SceI–DNA complex formation
The I-SceI target was purchased from Eurofins MWG Operon and consisted of two strands of sequence 5′-CACGCTAGGGATAACAGGGTAATAC-3′ and 5′-GGTATTACCCTGTTATCCCTAGCGT-3′. The target sequence was chosen according to a previously reported DNA sequence (Moure et al., 2003 ▸). The construct forms a 25 bp duplex with one overhang nucleotide at the 5′ end after annealing by heating and slow cooling. Before complex formation, the protein was dialyzed against 10 mM HEPES pH 8, 100 mM KCl, 5% glycerol, 1.4 mM β-mercaptoethanol. The I-SceI–DNA complex was obtained in the presence of 2 mM MnCl2 by incubation of the meganuclease and the oligonucleotide samples in a 1:1.5 molar ratio (protein:DNA). The mixture was incubated for 60 min on ice and then spun down for 10 min to remove insoluble material. The final concentration of I-SceI in the DNA–protein complex solution was 6 mg ml−1 (as determined by the Bradford assay). A band-shift assay was used to test the binding of I-SceI to the duplex (Fig. 1 ▸ e).
2.4. Crystallization
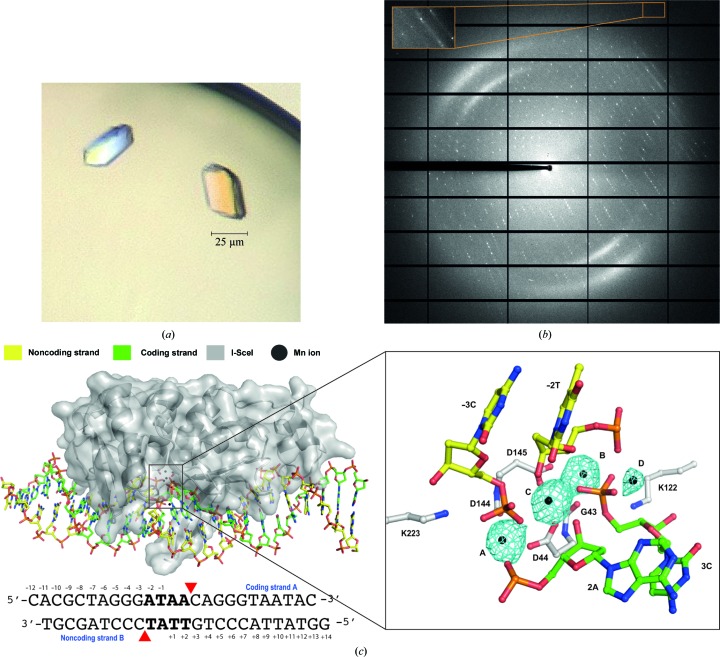
Crystallization screening was performed immediately after complex formation with a Cartesian MicroSys robot (Genomic Solutions). Using the sitting-drop method with drops of 0.2 µl DNA–protein complex sample mixed with an equal volume of well solution, the best diffracting crystals were obtained after 3–5 d in 0.1 M magnesium chloride, 0.1 M HEPES pH 7.0, 15% PEG 4000 (Protein Complex Suite screen from Qiagen, condition No. 24). The dimensions of the largest crystals were 0.025 × 0.025 × 0.015 mm (Fig. 2 ▸ a). These crystals were picked up and immersed into a cryoprotectant solution consisting of 0.1 M magnesium chloride, 0.1 M HEPES pH 7.0, 15% PEG 4000, 20% ethylene glycol and diffracted to 2.9 Å resolution (Fig. 2 ▸ b).
Figure 2.
Crystallization, diffraction experiments and crystal structure solution of the I-SceI–DNA complex in the presence of catalytic ions. (a) I-SceI–DNA complex crystals grown in the presence of 2 mM Mn2+ and (b) the corresponding X-ray diffraction pattern. The inset indicated by an arrow shows the high-resolution reflections at 2.9 Å. (c) Overall structure of I-SceI–DNA (left) and detailed view (right) of its active site in the presence of 2 mM Mn2+. Anomalous difference Fourier maps (cyan), revealing the number and the position of ions, show density contoured at 8σ.
2.5. Data collection, structure solution, model building and refinement
Crystals were removed from the drop and flash-cooled in liquid nitrogen. Diffraction data were recorded using a PILATUS 6M detector on the PXI beamline at the Swiss Light Source (SLS), Villigen, Switzerland. The best data set was collected using the fine φ-slicing method with Δφ = 0.2°, choosing the wavelength of the K absorption edge of Mn (λ = 1.89 Å). Processing and scaling were accomplished using XDS (Kabsch, 2010 ▸) and SCALA from the CCP4 package (Evans, 2006 ▸). Data-collection and refinement statistics are summarized in Table 1 ▸. The protein–DNA complex structure was solved by molecular replacement as implemented in Phaser (McCoy et al., 2007 ▸). The search model was based on PDB entry 1r7m (I-SceI–DNA–Ca2+; Moure et al., 2003 ▸). The structures were then subjected to iterative cycles of model building and refinement with Coot (Emsley et al., 2010 ▸) and PHENIX (Adams et al., 2010 ▸).
Table 1. Data-collection and refinement statistics for native I-SceI–DNA crystals grown in the presence of 2 mM Mn2+ .
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 80.11, b = 80.57, c = 130.87, α = β = γ = 90 |
| Wavelength | 1.89 |
| Resolution (Å) | 43.62–2.90 (3.06–2.90) |
| R merge | 0.10 (0.58) |
| Mean I/σ(I) | 12.1 (2.7) |
| Completeness (%) | 100.0 (99.2) |
| Multiplicity | 8.1 (7.9) |
| Refinement | |
| Resolution (Å) | 42.90–2.90 |
| No. of reflections | 36160 |
| R work/R free | 0.19/0.25 |
| No. of molecules in asymmetric unit | 2 |
| DNA strands cleaved? | Both |
| No. of ions in active sites | 8 |
| No. of non-H atoms | |
| Protein | 3736 |
| DNA | 2003 |
| Ions | 8 |
| Water | 22 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.253 |
| Average B factors (Å2) | |
| Protein | 66.05 |
| DNA | 60.26 |
| Ions | 57.58 |
| Water | 72.13 |
| Ramachandran plot | |
| Favoured (%) | 92.17 |
| Allowed (%) | 7.38 |
| Outliers (%) | 0.45 |
| PDB code | 5a0m |
3. Results and discussion
The I-SceI nuclease was expressed in E. coli and the recombinant protein was purified using three consecutive chromatography steps: the soluble fraction was subjected to a HiTrap Chelating HP affinity purification column loaded with Co2+, a HiTrap Heparin HP affinity purification column and a final polishing step using gel-filtration purification on a HiLoad 16/60 200 Superdex column. Protein purity was confirmed by overloaded SDS–PAGE stained with SimplyBlue SafeStain (Invitrogen; Fig. 1 ▸ a). The recombinant I-SceI construct used in this work is 249 residues in length, with a theoretical molecular mass of 29 307 Da and a pI of 9.24. However, mass spectrometry showed a single-charge peak corresponding to purified protein with a measured molecular mass of 29 182 Da (Fig. 1 ▸ b), indicating that the initial methionine was processed during expression (Hirel et al., 1989 ▸). Protein structure and stability were first checked by circular dichroism and thermal denaturation, demonstrating the proper folding of the protein (Fig. 1 ▸ c). A SEC-MALLS experiment shows that I-SceI behaves as a monomer in solution (Fig. 1 ▸ d), since its measured molar mass is 29.6 kDa. A band-shift assay confirmed the binding of I-SceI to the DNA duplex (Fig. 1 ▸ e).
The crystal of I-SceI in complex with its dsDNA target in the presence of 2 mM Mn2+ (Fig. 2 ▸ a) belonged to space group P212121, with unit-cell parameters a = 80.11, b = 80.57, c = 130.87 Å, α = β = γ = 90°. The Matthews coefficient (V M = 2.1 Å3 Da−1) and the self-rotation function (data not shown) suggested the presence of two protein–DNA complexes in the asymmetric unit and a solvent content of 48%. The diffraction data (Fig. 2 ▸ b) were 100% complete, with a multiplicity of 8.1 and an overall mean I/σ(I) of 12.1. Detailed statistics for the data set are summarized in Table 1 ▸.
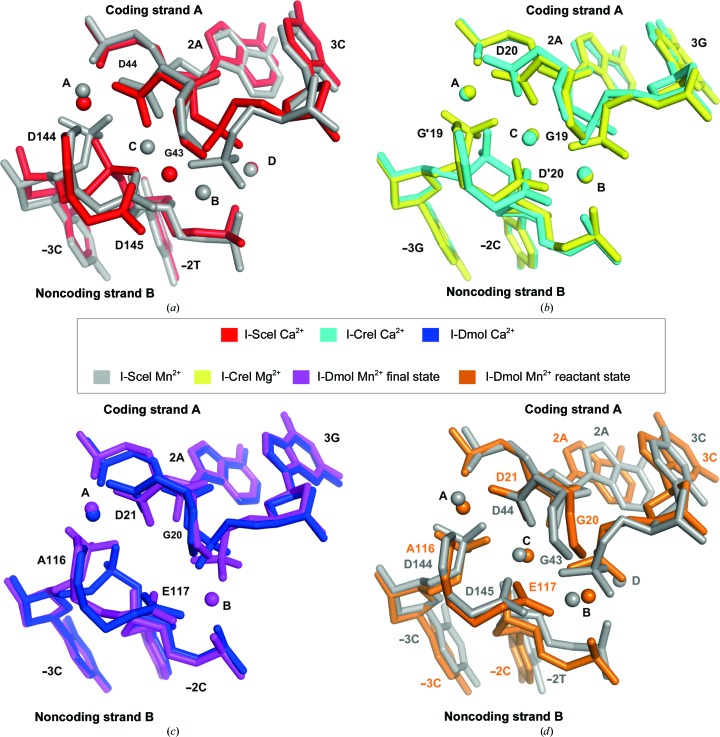
The crystal structure of the protein–DNA complex in the presence of Mn2+ was solved by X-ray crystallography (Fig. 2 ▸ c, left). Unlike Mg2+, the presence of bound Mn2+ ion can be directly observed by X-ray crystallography owing to its strong anomalous signal, thus helping to locate the metal ions. The use of Mn2+ instead of Mg2+ as the catalytic ion did not seem to alter the cleavage reaction, and structures in the presence of Mg2+ showed an identical active-centre organization in other HEs (Molina, Stella et al., 2015 ▸). Despite the use of a higher concentration of Mg2+ than of Mn2+ in crystal growth, since the cleavage reaction is carried out before the crystallization step, with a low diffusion rate, the anomalous signal unambiguously revealed the presence of four Mn2+ ions, three of which were located at the active site (Fig. 2 ▸ c, right). In addition, lower coordination requirements have been reported for Mn2+ than Mg2+ (Yang, 2011 ▸), which would not help in Mn2+ diffusion. As expected, the DNA was cleaved in both strands, while the overall structure was similar to the corresponding noncleaved state described by Moure et al. (2003 ▸) in the presence of Ca2+ as a noncatalytic ion (Cα r.m.s.d. of 0.45 Å). However, on comparing noncleaved and cleaved structures of the I-SceI–DNA complex at the active site, apart from the DNA breaks, the position and the number of noncatalytic and catalytic ions were found to be different (Fig. 3 ▸ a). This result contrasts with what has been reported for the homodimeric homing endonuclease I-CreI, where the number and positions of catalytic and noncatalytic ions in the active site were the same (Fig. 3 ▸ b). Curiously, in other monomeric HEs described, such as I-DmoI, the situation also differed between catalytic and noncatalytic structures (Marcaida et al., 2008 ▸; Fig. 3 ▸ c). On the other hand, on comparing the active site of monomeric I-SceI and I-DmoI in the presence of Mn2+, we found that three out of the four cations were occupying the canonical catalytic positions (Molina, Stella et al., 2015 ▸), suggesting that the fourth position (position D) is not involved in catalysis and is just an artefact of crystallization (Fig. 3 ▸ d). The catalytic mechanism of I-SceI has been proposed to be a sequential mechanism as a consequence of an asymmetric arrangement of the three Ca2+ ions in the active site (Moure et al., 2003 ▸). In this model, the third Ca2+ ion corresponds to the Mn2+ ion at position D in the I-SceI–DNA complex structure shown here, thus indicating that the third catalytic ion at position B was not present in the corresponding calcium-bound structure (Fig. 3 ▸ a). This new structure, together with the information previously reported in the literature, suggests a common catalytic mechanism for both monomeric and homodimeric LADGLIDADG HEs (Molina, Stella et al., 2015 ▸).
Figure 3.
Number and positions of metal ions at the active site in I-SceI–DNA and other LADGLIDADG homing endonuclease protein–DNA complexes. (a) Comparison of the active site of the I-SceI–DNA complex in the presence of noncatalytic ions (2 mM Ca2+; red) and catalytic ions (2 mM Mn2+; grey). (b) Comparison of the active site of the I-CreI–DNA complex in the presence of noncatalytic ions (2 mM Ca2+; cyan) and catalytic ions (2 mM Mg2+; yellow). (c) Comparison of the active site of the I-DmoI–DNA complex in the presence of noncatalytic ions (2 mM Ca2+; blue) and catalytic ions (2 mM Mn2+; purple). (d) Comparison of the active site of the I-SceI–DNA (grey) and I-DmoI–DNA (orange) complexes both in the presence of catalytic ions (2 mM Mn2+). The I-DmoI–DNA complex structure corresponds to the state before cleavage.
These are the first crystals of I-SceI in complex with DNA in the presence of catalytic ions, and the structure shows three metal ions in the final reaction step of a monomeric LHE for the first time. Thanks to this structural analysis, we have reinforced the idea of a common cleavage mechanism for all LHEs based on a two-metal-ion mechanism.
Supplementary Material
PDB reference: I-SceI nuclease complexed with its dsDNA target and three catalytic metal ions, 5a0m
Acknowledgments
We thank the Swiss Light Source staff for their support. This work was supported by Ministerio de Economía y Competitividad (JCI-2011-09308, BFU2011-23815/BMC, CTQ2014-56966-RR), the Fundación Ramón Areces and the Comunidad Autónoma de Madrid (CAM-S2010/BMD-2305).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Chevalier, B., Monnat, R. J. Jr & Stoddard, B. L. (2001). Nature Struct. Biol. 8, 312–316. [DOI] [PubMed]
- Chevalier, B., Sussman, D., Otis, C., Noël, A.-J., Turmel, M., Lemieux, C., Stephens, K., Monnat, R. J. Jr & Stoddard, B. L. (2004). Biochemistry, 43, 14015–14026. [DOI] [PubMed]
- Choulika, A., Perrin, A., Dujon, B. & Nicolas, J.-F. (1994). C. R. Acad. Sci. III, 317, 1013–1019. [PubMed]
- Choulika, A., Perrin, A., Dujon, B. & Nicolas, J.-F. (1995). Mol. Cell. Biol. 15, 1968–1973. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Hirel, P. H., Schmitter, M. J., Dessen, P., Fayat, G. & Blanquet, S. (1989). Proc. Natl Acad. Sci. USA, 86, 8247–8251. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Marcaida, M. J., Prieto, J., Redondo, P., Nadra, A. D., Alibés, A., Serrano, L., Grizot, S., Duchateau, P., Pâques, F., Blanco, F. J. & Montoya, G. (2008). Proc. Natl Acad. Sci. USA, 105, 16888–16893. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Molina, R., Redondo, P., López-Méndez, B., Villate, M., Merino, N., Blanco, F. J., Valton, J., Grizot, S., Duchateau, P., Prieto, J. & Montoya, G. (2015). J. Biol. Chem. 290, 28727–28736. [DOI] [PMC free article] [PubMed]
- Molina, R., Stella, S., Redondo, P., Gomez, H., Marcaida, M. J., Orozco, M., Prieto, J. & Montoya, G. (2015). Nature Struct. Mol. Biol. 22, 65–72. [DOI] [PubMed]
- Moure, C. M., Gimble, F. S. & Quiocho, F. A. (2003). J. Mol. Biol. 334, 685–695. [DOI] [PubMed]
- Redondo, P., Merino, N., Villate, M., Blanco, F. J., Montoya, G. & Molina, R. (2014). Acta Cryst. F70, 256–259. [DOI] [PMC free article] [PubMed]
- Rouet, P., Smih, F. & Jasin, M. (1994). Proc. Natl Acad. Sci. USA, 91, 6064–6068. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Yang, W. (2011). Q. Rev. Biophys. 44, 1–93. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: I-SceI nuclease complexed with its dsDNA target and three catalytic metal ions, 5a0m