Abstract
Pathogenicity islands are capable of excision and insertion within bacterial chromosomes. We describe a protein, Rox, that stimulates excision of the Shigella resistance locus pathogenicity island in Shigella flexneri. Sequence analysis suggests that Rox belongs to a new subfamily of recombination directionality factors, which includes proteins from P4, enterohemorrhagic Escherichia coli, and Yersinia pestis.
A variety of mobile genetic elements, including insertion sequences, transposons, plasmids, bacteriophages, and pathogenicity islands (PAIs), mediate the lateral acquisition of foreign DNA in bacteria. PAIs are large regions of the chromosome that are genetically and functionally distinct units (7). These elements are often incorporated into bacterial chromosomes by site-specific integration at tRNA genes, and some are capable of excision. PAIs often have G+C contents that vary from those of their host bacteria, suggesting that they are acquired by horizontal transfer. Thus far, PAIs have been identified predominantly in gram-negative enteric and uropathogenic organisms, such as Salmonella spp. and Vibrio cholerae, and in a variety of other pathogens, e.g., Escherichia coli, Helicobacter pylori, Yersinia enterocolitica, and Shigella flexneri (9). S. flexneri carries five distinct PAIs: Shigella island 2 (SHI-2), SHI-3, the she PAI, the Shigella resistance locus (SRL) PAI, and the mxi-spa gene cluster on the large virulence plasmid (1, 13, 15, 17, 28, 29).
The SRL PAI is a 67-kb island first identified in S. flexneri 2a YSH6000 (13). The element was discovered due to its carriage of a locus mediating resistance to ampicillin, streptomycin, chloramphenicol, and tetracycline, the SRL. The SRL PAI also encodes a functional ferric citrate transport system, a number of insertion sequences, and a large number of phage-like genes. Turner et al. have shown that the SRL PAI can undergo precise insertion and deletion in an integrase-dependent manner (26, 27).
The importance of genetic acquisition and loss was highlighted by the recent sequencing of the S. flexneri 2a genome, which showed that the major differences between S. flexneri and E. coli arose primarily through the loss or acquisition of DNA, including PAIs and bacteriophages (8). However, very little is known about the movement of PAIs into and out of bacterial genomes. Several PAIs, including the SRL and she PAIs, are capable of spontaneous deletion from the chromosomes in which they normally reside (18, 27). Like many PAIs, the SRL PAI encodes an integrase that is homologous to members of the P4 family of integrases (13). Integrases mediate PAI integration into the chromosome (19, 26) and are also essential for PAI excision (11, 20, 27). However, little is known about how the excision of PAIs is regulated.
The regulation of integrase-mediated excision of mobile genetic elements, in the cases where it is understood, follows two distinct mechanisms. In the majority of cases, excision is stimulated by a class of proteins collectively known as excisionases or recombination directionality factors (RDFs) (12). RDFs have been identified mostly in phage systems but have also been identified in transposons (22) and plasmids (14). Typical members of this family include the bacteriophage lambda excisionase (Xis) and bacteriophage P2 Cox proteins. RDFs are usually small DNA-binding proteins encoded by genes in close proximity to the integrase genes carried on mobile elements. In the best-studied case, that of lambda Xis, the protein regulates excision by binding directly to DNA in the vicinity of the recombination site and to integrase to form a complex that stimulates excision and inhibits integration (23). The excision of CP4-57, a genomic island of E. coli K-12, is stimulated by an alternative mechanism that relies instead on the transcriptional activation of the CP4-57 integrase gene by a protein designated AlpA (25). The resulting increase in integrase expression is responsible for the stimulation of excision. In the present study, we tested whether excision of the SRL PAI was also regulated and investigated the mechanism by which this regulation occurs.
Sequence analysis of AlpA homologues.
Analysis of the SRL PAI sequence revealed two open reading frames (ORFs) with corresponding sequence similarity to proteins that regulate the excision of other mobile elements, namely, ORF3, which was subsequently termed Rox (regulator of excision), and ORF41 (13). Rox shows 66% sequence similarity to AlpA from CP4-57, while ORF41 is more closely related (53% similarity) to Vis, another AlpA homologue from the satellite phage P4. Vis is a multifunctional protein that represses transcription from the PLL late promoter of P4 and activates transcription of Psid (16). In addition, Vis represses transcription of the P4 integrase gene and is also an excisionase that promotes integrase-dependent P4 excision (D. Ghisotti, personal communication).
The recent sequencing of the enterohemorrhagic E. coli (EHEC) strain O157:H7 EDL933 has revealed AlpA homologues, Z1124 and Z1563, on duplicate O islands 43 and 48, which have an integrase nearly identical with that encoded by the SRL PAI. Unlike alpA, the EHEC homologues show significant nucleotide identity with rox. EHEC EDL933 O islands 43 and 48 encode the proteins required for tellurite resistance and urease biosynthesis and share no similarity with the SRL PAI aside from the presence of a number of putative prophage ORFs (13). Database probing also revealed Rox homologues on other previously described elements, including the she PAI in S. flexneri and the high pathogenicity island (HPI) of Yersinia pestis. An alignment of these seven homologues shows two areas of conservation (Fig. 1). The first region corresponds to a predicted DNA-binding, helix-turn-helix motif in all homologues except Roxshe and ORF41, with Dodd and Egan scores (5) for RoxSRL, AlpA, Vis, EHEC Z1124 or Z1563, and YP01904 of 6.35, 3.98, 5.25, 4.94, and 3.45, respectively, where a value of ≥2.5 is indicative of a likely helix-turn-helix motif. Database and motif probing has provided no clues as to the potential role of the second conserved region. Mutational analysis may provide clues as to the potential function of this region.
FIG. 1.
Multiple alignment of Rox homologues. Amino acids that are identical or similar in at least two-thirds of the sequences are rendered as white characters on a black or grey background, respectively. Z1124 and Z1563 are encoded by EHEC EDL933 O islands 43 and 48, respectively. YPO1904, Roxshe, Vis, and AlpA are proteins encoded by the HPIs of Y. pestis, the S. flexneri she PAI, bacteriophage P4, and cryptic prophage CP4-57, respectively. The proposed helix-turn-helix (HTH) motif is indicated by a bar. Alignment was performed by using Clustal W (24).
Recently, an xis gene was identified on the SXT element of V. cholerae and found to have a role in SXT excision (4). SXT Xis did not belong to any of the RDF subgroups previously described by Lewis and Hatfull (12). We compared a member of each previously described subgroup of RDFs, including SXT Xis, with Rox, ORF41, and the homologues described above. Clustal W analysis suggested that these homologues are most closely related to the SLP1 subgroup, which is typified by an RDF encoded by the SLP1 plasmid from Streptomyces coelicolor (12), but that they form a distinct and thus a new subgroup (Fig. 2). Of the new subgroup, Roxshe, YPO1904, and Vis have been shown experimentally to have a role in excision (10, 20; D. Ghisotti, personal communication).
FIG. 2.
Phylogenetic tree representing subgroups of RDFs. Members of each subgroup were chosen based on the Lewis and Hatfull definition of RDF subgroups (12). The proposed new subgroup is boxed. Mapping was performed by using Clustal W (24). B.melitensis, Brucella melitensis; M.loti, Mesorhizobium loti; S.melioti, Sinorhizobium meliloti; M.leprae, Mycobacterium leprae.
Effect of Rox and ORF41 on SRL PAI excision.
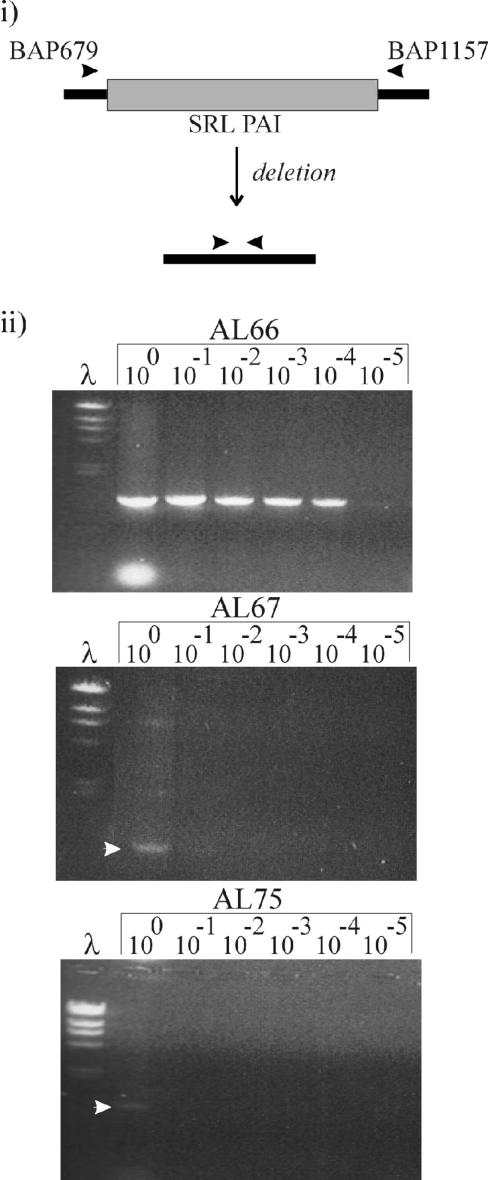
In order to determine if Rox or ORF41 had a role in the excision of the SRL PAI, assays were performed to test SRL PAI excision when these genes were overexpressed. rox and orf41 were amplified by PCR from S. flexneri YSH6000 genomic DNA and cloned directionally into the EcoRI and BamHI sites of pPBA1100 (pUC18 carrying kan from pMK3), giving rise to plasmids pAL34 and pAL31, respectively. Accurate amplification of PCR products was confirmed by sequencing the inserts of the plasmids obtained. These plasmids, along with pPBA1100, were transformed separately into S. flexneri 2a strain YSH6000, resulting in strains AL66(pAL34), AL67(pAL31), and AL75(pPBA1100). Each strain was grown overnight with aeration in 2.5 ml of 2YT medium supplemented with 50 μg of kanamycin/ml and 10 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Extraction of genomic DNA was performed as described previously (2). Excision of the SRL PAI was assayed by semiquantitative PCR with inward-facing primers flanking the SRL PAI (BAP679 [5′-GTGCTGCTTTCGGTGTGC-3′] and BAP1157 [5′-GCCAGCATTTCAACAGGAGG]) (Fig. 3). The PCR amplification of recA (BAP1643 [5′-CTACGCACGTAAACTGGGCG-3′] and BAP1644 [5′-ACCGGTAGTGGTTTCCGGG-3′]) served as controls for template concentration. Although recA controls indicated that the concentration of chromosomal DNA was 10-fold greater in the AL66 template than in the wild-type and AL67 templates (data not shown), the level of SRL PAI excision was 104-fold greater in the AL66 template, where Rox was overexpressed, than in the wild-type template (Fig. 3). In contrast, overexpression of orf41 had no effect on SRL PAI excision. The PCR product obtained from strain AL66 was sequenced and shown to be the reconstitution of serX with a single copy of the 14-bp direct repeat that flanks the SRL PAI, as was previously observed upon deletion of the SRL PAI (27). Therefore, Rox, but not ORF41, stimulates SRL PAI excision.
FIG. 3.
The effect of RoxSRL and ORF41 on SRL PAI excision. (i) The shaded box denotes the SRL PAI, and the lines denote YSH6000-flanking chromosomal DNA. The primers used to determine if the PAI has excised are shown by arrowheads. (ii) Lane λ, lambda HindIII markers. Other lanes contain 10-fold serial dilutions of genomic DNA. AL66, AL67, and AL75 are S. flexneri 2a strains overexpressing rox, overexpressing orf41, and carrying an empty plasmid, respectively. The SRL PAI excisions in AL67 and the plasmid control can be seen as PCR products and are indicated by the arrowheads.
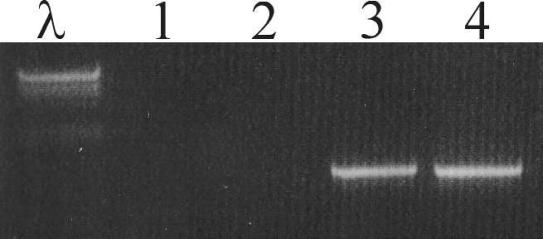
In order to determine if the effect of Rox on PAI excision was dependent on the SRL PAI integrase, a PCR excision assay was performed on an int mutant, AL11 (27). AL11 was transformed with pUC19-Tp and pAL85 (pUC19-Tp carrying rox), giving rise to strains AL296 and AL295, respectively. The excision assay was performed as described previously, with strains AL325 (wild-type int with pAL85), YSH6000 (parent), and SBA1363 (SRL PAI−) as controls. The results showed that both Rox and Int have critical roles in SRL PAI excision, as excision was not observed in AL295, the int mutant strain that overexpressed Rox. In contrast, SRL PAI excision was clearly visible when rox was overexpressed in the presence of an intact int gene (AL325) (Fig. 4).
FIG. 4.
The effect of Int and RoxSRL on SRL PAI excision. Lane λ, lambda HindIII markers. The various strains used as templates for PCR were as follows: AL296 (int mutant, pUC19-Tp) (lane 1), AL295 (int mutant, pUC19-Tp/rox) (lane 2), SBA1363 (SRL PAI−) (lane 3), AL325 (wild-type int, pUC19-Tp/rox) (lane 4).
Does Rox affect int transcription?
A potential binding site (YYRTTCGRNRY) for the bacteriophage P4 Vis protein was proposed by Polo et al. (16). This binding site is present upstream of slpA, the gene encoding the CP4-57 integrase, and the SRL PAI int gene. The similarity to AlpA and the presence of a potential binding site upstream of int suggested that Rox may be required for the regulation of int transcription. In order to test this hypothesis, the int gene of S. flexneri 2a strain SBA1366, an antibiotic-sensitive derivative of YSH6000, was disrupted by a Campbell insertion with pJP5603 as previously described (27), giving rise to strain AL467. It was necessary to inactivate int to avoid potential complications associated with SRL PAI excision, leading to the loss of the PAI and therefore the int gene. AL467 was transformed with pBAD30 (6) and pAL216 (pBAD carrying rox), giving rise to strains AL468 and AL469, respectively. The use of pBAD allowed the tight regulation of Rox expression. RNA was extracted as described previously (21) with the following modifications: cells were grown for 1 h in a solution containing Luria broth, 50 μg of kanamycin/ml, and 100 μg of ampicillin/ml and then induced with 1 mM arabinose for an additional 2 h before RNA extraction. Real-time reverse transcriptase PCR was performed as described by Boyce et al. (3). Assays of gyrB transcription served as controls for RNA concentration. Levels of transcription of rox were also compared between strains, and as expected, the level of rox transcription was significantly higher (27-fold) in AL469 than in AL468. However, there was no statistically significant change (P value = 0.385; two-tailed t test) in int transcription when rox was overexpressed following arabinose induction. Therefore, although Rox stimulates integrase-dependent SRL PAI excision, it is not an activator of int transcription. Multiple attempts to purify Rox for further characterization and DNA-binding experiments were prevented by the exceptional instability of the protein.
Little is known about the mechanisms of PAI mobility. However, Turner et al. (27) showed that the SRL PAI int gene is required for excision of the SRL PAI, and it has been demonstrated that HPI int and SRL PAI int modules are capable of site-specific integration (19, 26). Together, these results support the significance of int genes in PAI mobility. This study investigated the role of Rox in the regulation of SRL PAI excision. Our findings suggest that although Rox stimulates the excision of the SRL PAI, regulation of excision does not follow the model proposed for CP4-57 (10). Since Rox does not act like AlpA, the transcriptional activator of CP4-57, we propose based on its homology to Vis and its presently proven ability to potently stimulate excision of the SRL PAI that it probably has direct excisionase activity. On the basis of sequence comparisons, RoxSRL appears to be a member of a new subgroup of RDFs that includes Roxshe, Vis, YP01904, and Z1124 or Z1563.
REFERENCES
- 1.Al-Hasani, K., K. Rajakumar, B. Dieter, R. Robins-Browne, B. Adler, and H. Sakellaris. 2001. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb. Pathog. 30:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrus, V., and M. K. Waldor. 2003. Control of SXT integration and excision. J. Bacteriol. 185:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guzman, L.-M., D. Belin, M. J. Garson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Häcker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 8.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaper, J. B., and J. Hacker (ed.). 1999. Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 10.Kirby, J. E., J. E. Trempy, and S. Gottesman. 1994. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J. Bacteriol. 176:2068-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesic, B., S. Bach, J.-M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 52:1337-1348. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, J. A., and G. F. Hatfull. 2001. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 29:2205-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin, C., P. Mazodier, M. V. Mediola, B. Gicquel, T. Smokvina, C. J. Thompson, and J. Davies. 1991. Site-specific integration of the Streptomyces plasmid pSAM2 in Mycobacterium smegmatis. Mol. Microbiol. 5:2499-2502. [DOI] [PubMed] [Google Scholar]
- 15.Moss, J. E., T. J. Cardozo, A. Zychlinsky, and E. A. Groisman. 1999. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol. 33:74-83. [DOI] [PubMed] [Google Scholar]
- 16.Polo, S., T. Sturniolo, G. Deho, and D. Ghisotti. 1996. Identification of a phage-coded DNA-binding protein that regulates transcription from late promoters in bacteriophage P4. J. Mol. Biol. 257:745-755. [DOI] [PubMed] [Google Scholar]
- 17.Purdy, G. E., and S. M. Payne. 2001. The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J. Bacteriol. 183:4176-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajakumar, K., C. Sasakawa, and B. Adler. 1997. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect. Immun. 65:4606-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakin, A., C. Noeiting, P. Schropp, and J. Heesemann. 2001. Integrative module of the high-pathogenicity island of Yersinia. Mol. Microbiol. 39:407-415. [DOI] [PubMed] [Google Scholar]
- 20.Sakellaris, H., S. N. Luck, K. Al-Hasani, K. Rajakumar, S. A. Turner, and B. Adler. 2004. Regulated site-specific recombination of the she pathogenicity island of Shigella flexneri. Mol. Microbiol. 52:1329-1336. [DOI] [PubMed] [Google Scholar]
- 21.Simms, D., P. E. Cizdziel, and P. Chomczynski. 1993. TRIzol: a new reagent for optimal single-step isolation of RNA. Focus (Gaithersburg) 4:99-102. [Google Scholar]
- 22.Su, Y. A., and D. B. Clewell. 1993. Characterization of the left 4 kb of conjugative transposon Tn916: determinants involved in excision. Plasmid 30:250. [DOI] [PubMed] [Google Scholar]
- 23.Swalla, B. M., E. H. Cho, R. I. Gumport, and J. F. Gardner. 2003. The molecular basis of co-operative DNA binding between lambda integrase and excisionase. Mol. Microbiol. 50:89-99. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trempy, J. E., J. E. Kirby, and S. Gottesman. 1994. Alp suppression of Lon: dependence on the slpA gene. J. Bacteriol. 176:2061-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner, S. A., S. N. Luck, H. Sakellaris, and B. Adler. 2004. Role of attP in integrase-mediated integration of the Shigella resistance locus pathogenicity island of Shigella flexneri. Antimicrob. Agents Chemother. 48:1028-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner, S. A., S. N. Luck, H. Sakellaris, K. Rajakumar, and B. Adler. 2001. Nested deletion events of the SRL pathogenicity island of Shigella flexneri 2a. J. Bacteriol. 183:5535-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesan, M. M., M. B. Goldberg, D. J. Rose, E. J. Grotbeck, V. Burland, and F. R. Blattner. 2001. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69:3271-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vokes, S. A., S. A. Reeves, A. G. Torres, and S. M. Payne. 1999. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol. Microbiol. 33:63-73. [DOI] [PubMed] [Google Scholar]