Abstract
A combined phylogenetic and multilocus DNA sequence analysis of 26 Pseudomonas stutzeri strains distributed within the 9 genomovars of the species has been performed. Type strains of the two most closely related species (P. balearica, former genomovar 6, and P. mendocina), together with P. aeruginosa, as the type species of the genus, have been included in the study. The extremely high genetic diversity and the clonal structure of the species were confirmed by the sequence analysis. Clustering of strains in the consensus phylogeny inferred from the analysis of seven nucleotide sequences (16S ribosomal DNA, internally transcribed spacer region 1, gyrB, rpoD, nosZ, catA, and nahH) confirmed the monophyletic origin of the genomovars within the Pseudomonas branch and is in good agreement with earlier DNA-DNA similarity analysis, indicating that the selected genes are representative of the whole genome in members of the species.
The genus Pseudomonas is one of the most diverse and ecologically significant groups of bacteria on the planet (28), playing an especially important role in the carbon and nitrogen cycles. Pseudomonas stutzeri is a remarkable member of this genus, with exceptional physiological capacities, being able to metabolize a wide range of organic substrates. Members of the species mineralize organic contaminants aerobically and anaerobically as denitrifiers. P. stutzeri strains are not only able to denitrify—some strains are also able to fix dinitrogen. The species is well defined phenotypically by means of few biochemical tests that discriminate P. stutzeri from other species of Pseudomonas, but additional biochemical properties are very diverse (20). P. stutzeri is ecologically relevant, occupying many niches and being commonly isolated from environmental and clinical samples (1, 27).
Diversity within the species is not limited to physiological traits but is also reflected at the genetic level. At least nine genomovars are distinguishable within the species (6). Two members of the same genomovar share more than 70% DNA-DNA similarity, the accepted species threshold, and these values are lower and usually less than 50% when members of different genomovars are compared (22). Genomovars are also clearly separated in most cases through phylogenetic analysis of the rrn operon (8). To date, no consistent phenotypic traits have been defined in each genomovar that could justify the splitting of P. stutzeri into several species. The extremely high genetic diversity of the species, the highest so far described in any species, was demonstrated through multilocus enzyme electrophoresis (MLEE) studies (21, 27). It has also been suggested that P. stutzeri populations have a strong clonal structure (21).
Multilocus sequence typing has been proposed as a good method for population genetic analysis and to discriminate clones within a species (4). This method employs the same principles as MLEE, in that neutral genetic variation from multiple chromosomal locations is detected. This variation is identified through nucleotide sequence determination of selected loci. In an attempt to differentiate P. stutzeri populations and to clearly establish the genetic diversity and population structure of the species, a comparative analysis of gene fragments at seven loci has been carried out, following the principles of multilocus sequence analysis and also through phylogenetic analysis. Fragments of five genes metabolically relevant to the species, coding for gyrase B (gyrB), the D subunit of the sigma factor (rpoD), nitrous oxide reductase (nosZ), catechol 1,2 dioxigenase (catA), and catechol 2,3 dioxigenase (nahH), have been sequenced and analyzed, together with the 16S ribosomal DNA (rDNA) and internally transcribed spacer region 1 (ITS1) regions of the rrn operon, in 26 strains representing the 9 genomovars of the species. Molecular phylogenies inferred for each locus separately and pooled were studied to obtain an estimate of P. stutzeri genomovar phylogeny.
The isolates were obtained from clinical materials and the environment from a variety of sources and geographical regions. Three strains representative of the most closely related species (P. balearica and P. mendocina), as well as the type species of the genus (P. aeruginosa), were included as outgroups for comparative purposes.
MATERIALS AND METHODS
Microorganisms and growth conditions.
Thirty Pseudomonas isolates, corresponding to P. stutzeri (26), P. balearica (2), P. mendocina (1), and P. aeruginosa (1) species were used in this study (Table 1). Luria-Bertani broth was used routinely as growth substrate for biomass recovery. The incubation temperature was 30°C.
TABLE 1.
Bacterial strains used in this study
| Genus or species | Strain | gv. | Origin | Geographical locations | Yr isolated |
|---|---|---|---|---|---|
| P. stutzeri | CCUG 11256T | 1 | Clinical | Copenhagen | Before 1966 |
| ATCC 27951 | 1 | Yogurt | Algeria | 1960 | |
| SD55473 | 1 | Clinical | Majorca, Spain | Before 1985 | |
| A95/69 | 1 | Clinical | United Kingdom | Between 1965 and 1984 | |
| B1SMN1 | 1 | Wastewater | Menorca, Spain | 1988 | |
| S1MN1 | 1 | Wastewater | Menorca, Spain | 1988 | |
| ZoBell | 2 | Marine | Pacific | Before 1944 | |
| ATCC 17591 | 2 | Clinical | Copenhagen, | 1956 | |
| A60/72 | 2 | Clinical | United Kingdom | Between 1965 and 1984 | |
| AER 5.1 | 3 | Aircraft oil-contaminated soil | Mallorca, Spain | 1995 | |
| AN10 | 3 | Marine | Barcelona, Spain | 1982 | |
| AN11 | 3 | Marine | Barcelona, Spain | 1982 | |
| LSMN2 | 3 | Marine | Barcelona, Spain | 1988 | |
| ST27MN2 | 3 | Marine | Barcelona, Spain | 1988 | |
| 19SMN4 | 4 | Marine | Barcelona, Spain | 1988 | |
| ST27MN3 | 4 | Marine | Barcelona, Spain | 1988 | |
| DNSP21 | 5 | Wastewater | Majorca, Spain | 1988 | |
| JD4 | 5 | Garden soil | Majorca, Spain | 1995 | |
| DSM 50238 | 7 | Soil | California | Before 1966 | |
| AER 2.7 | 7 | Aircraft oil-contaminated soil | Majorca, Spain | 1995 | |
| JM300 | 8 | Soil | California | Before 1982 | |
| KC | 9 | Aquifer | California | 1990 | |
| CLN100 | 10 | Chemical production plant | Germany | 1990 | |
| Pseudomonas | PTDA | Putidoil | Siberia | Before 1990 | |
| PTDB | Putidoil | Siberia | Before 1990 | ||
| PTDE | Putidoil | Siberia | Before 1990 | ||
| P. balearica | DSM 6083T | Wastewater | Mallorca, Spain | 1988 | |
| LS401 | Marine | Barcelona, Spain | 1988 | ||
| P. mendocina | ATCC 25411T | Soil | Argentina | Before 1970 | |
| P. aeruginosa | CCM 1960T |
DNA extraction.
Bacterial genomic DNA for PCR amplifications was obtained by lysis with sodium dodecyl sulfate (SDS)-proteinase K and treatment with cetyltrimethyl ammonium bromide as described by Wilson (33).
PCR amplification and DNA sequencing.
Primers used for 16S rDNA, ITS1 and the genes rpoD, gyrB, nahH, nosZ, and catA are indicated in Table 2 and the corresponding references. Primers for the genes catA, nahH, and nosZ were designed from the conserved regions retrieved from available databases.
TABLE 2.
PCR primers used in this study
| Gene | Primer | Sequence | Source or reference |
|---|---|---|---|
| 16S rDNA | 16F27 | GAAGTCGTAACAAGG | 14 |
| 16R1488 | CAAGGCATCCACC | 14 | |
| 16fps158 | GTGGGGACAACGTTTC | 1 | |
| 16rps743 | CGATTATGACTGTGACTCCAC | 1 | |
| ITS1 | 16F945 | GGGCCCGCACAAGCGGTGG | 8 |
| 23R458 | CTTTCCCTCACGGTAC | 8 | |
| rrn16S | GAAGTCGTAACAAGG | 8 | |
| rrn23S | CAAGGCATCCACCGT | 8 | |
| gyrB | APrU | TGTAAAACGACGGCCAGTGCNGGRTCYTTYTCYTGRCA | 34 |
| M13 (−21) | TGTAAAACGACGGCCAGT | 34 | |
| UP1E | CAGGAAACAGCTATGACCAYGSNGGNGGNAARTTYRA | 34 | |
| M13R | CAGGAAACAGCTATGACC | 34 | |
| rpoD | 70F | ACGACTGACCCGGTACGCATGTAYATGMGNGARATGGGNACNGT | 34 |
| 70Fs | ACGACTGACCCGGTACGCATGTA | 34 | |
| 70R | ATAGAAATAACCAGACGTAAGTTNGCYTCNACCATYTCYTTYTT | 34 | |
| 70Rs | ATAGAAATAACCAGACGTAAGTT | 34 | |
| catA | 1C120F | CSSCGCACCATCGAAGG | 6 |
| 2C120R | SGCAAAGTCGTCCCACAG | 6 | |
| 3C120 | GGMGARTGGCCGCTGT | This study | |
| 4C120 | GGTGCAGGTGSGCGG | This study | |
| C12F | AGMTSGTCAACCGCATC | This study | |
| 8C12R | GTTGATCTGGGTGGTCAG | This study | |
| C1STF | CATGGAYGACGGYAGCG | This study | |
| C1STR | CCVGCCAGGTTGATCTG | This study | |
| nahH | 4C230 | TCCSTGGTGCTRCGYGA | This study |
| 3C230 | GATVGAKGTRTCGGTCATG | This study | |
| nosZ | U1672 | GGCCCGCTGMABCCSGARAACGANCARYTGATCGA | This study |
| L2140 | CATYTCCATGTGCAKSGCRTGGCAGAACCA | This study |
PCR amplification was performed with a personal DNA thermocycler (Eppendorf). Individual reaction mixtures contained 5 μl of a PCR buffer (Amersham Pharmacia Biotech, Inc.) and 8 μl of each of the nucleoside triphosphates (Boehringer Mannheim) at a concentration of 100 μM. A total of 2.5 μl of each of the primers was used at a concentration of 10 μM with 5 U of Taq DNA polymerase (Amersham Pharmacia Biotech, Inc.) in a total volume of 50 μl. After denaturation at 94°C for 5 min, a total of 30 cycles were performed with template DNA using the following profile. Denaturation was performed at 94°C for 1 min with primer annealing at 45 to 55°C for nosZ and nahH. For catA, temperatures varied depending on the primers used (45 to 55°C for 1C120/2C120, 55 to 59°C for C1STF/C1STR, and 50 to 55°C for 7C12F/8C12R for 1 min). Primer extension was done at 72°C for 1 min. A final elongation step was carried out at 72°C for 10 min. For sequencing reactions, the same primers and conditions were used. The amplified products were purified with MICROCON centrifugal filter devices (Amicon/Millipore) according to the manufacturer's instructions. After purification, amplified products were electrophoresed on 1.5% multipurpose agarose gels. Sequencing was carried out with an ABI PRISM Big Dye terminator cycle sequencing kit (version 3.0) and an automatic 310 Genetic analyzer DNA sequencer (Perkin-Elmer).
Sequence analysis.
The 16S rDNA gene, ITS1, nahH, catA, nosZ, gyrB, and rpoD sequence data were aligned with the closest relatives retrieved from the European Molecular Biology Laboratory nucleotide sequence database (EMBL). The alignment was done by using a hierarchical method for multiple alignments implemented in the CLUSTAL W computer program (31). Automatically aligned sequences were checked manually. Similarities and evolutionary distances were calculated with programs implemented in PHYLIP (Phylogeny Inference Package, version 3.5c) (5) Gene distance matrices were calculated from nucleotide sequences by the Jukes-Cantor method (12). Dendrograms were generated by neighbor-joining, Fitch-Margoliash, minimum-evolution, parsimony, maximum-likelihood, and bootstrap analyses (1,000 replications)—all algorithms included in programs of the PHYLIP package. Topologies of the trees were visualized with the TreeView program (19).
A hypothetical consensus multilocus tree was calculated to represent the combined molecular evolutionary relationships for five genes (rpoD, gyrB, nosZ, catA, and 16S rDNA) and ITS1 between P. aeruginosa, P. mendocina, P. balearica, and 26 strains belonging to the nine described genomovars of P. stutzeri. The evolutionary analysis was performed by calculating the corresponding distance matrices for each gene, using the algorithm of Jukes-Cantor (12). Finally, a unique matrix of distances was obtained by averaging the resulting six sets and used as an additional measure of divergence between strains (30). The consensus multilocus tree was calculated from the consensus multilocus matrix by the neighbor-joining method.
Allele diversity, nucleotide diversity, and statistical analysis.
Allele and nucleotide diversities were calculated with the DnaSP package, version 3.51 (Faculty of Biology, University of Barcelona [http://www.bio.ub.es/∼julio/DnaSP.html]) (24). For the purpose of identification, distinct allele sequences were assigned arbitrary allele numbers for each locus. For each isolate, the combination of alleles obtained at each locus defined its allelic profile or sequence type (ST).
Clustering of data obtained by nucleotide sequence analysis was performed with the START program (11). The method of split decomposition was used to assess the degree of tree-like structure present in the alleles found for each locus in the P. stutzeri isolates. The sequence alignments and the matrix of pairwise distances between the allelic profiles of all samples were converted to NEXUS files, and the split decomposition was analyzed with the SplitsTree program (Universität Bielefeld—Technische Fakultät [http://bibiserv.techfak.uni-bielefeld.de/splits][http://bibiserv.techfak.uni-bielefeld.de/splits) (10). The cophenetic correlation coefficient was calculated by using the NTSYS-PC program (F. J. Rohlf, Numerical Taxonomy and Multivariate Analysis System, version 1.80 [Exeter Software, New York, N.Y.]).
Multilocus linkage disequilibrium was estimated by measuring the index of association (IA) using the START program. A test was performed to detect selection in our population, the dN/dS ratio (described below), which was calculated as described by Nei and Gojobori (18). Relative synonymous codon usage (RSCU), codon bias index (CBI), and percent GC content analysis were calculated with the DnaSP program.
Nucleotide sequence accession number.
The nucleotide sequences determined in this study were deposited in the EMBL database under the following accession numbers: for nahH, AJ617261 to AJ617270, AJ633066 to AJ633070, AJ633092, AJ633093, and AJ539383; catA, AJ617513 to AJ617525, AJ617552 to AJ617556, AJ633071 to AJ633076, and AJ633094 to AJ633097; gyrB, AJ617557 to AJ617567, AJ617677 to AJ617680, AJ631257 to AJ631265, AJ633102 to AJ633104, AJ620493, and AJ536591; rpoD, AJ631316 to AJ631322, AJ631324 to AJ631340, and AJ518947; nosZ, AJ631971 to AJ631996 and AJ633098 to AJ633101; 16S rDNA, AJ633553 to AJ633564 and AJ544240; and ITS1, AJ635305 to AJ635313. The rest of sequences used have been deposited previously in public databases or as indicated in the respective figures.
RESULTS
Bacterial strains and genes selected.
To gain insight into the sequence diversity and structure of P. stutzeri populations, a variety of isolates of the species were selected from environmental and human habitats and from different geographical locations (Table 1). These isolates represented the nine known genomovars of the species, and most of them have been previously characterized taxonomically and through MLEE studies (8, 21). New strains included were PTDA, PTDB and PTDE, isolated as degraders of aromatics, due to differences in their colonial morphology, from a commercial product designed for bioremediation (Putidoil). P. balearica (two strains, including the type strain), P. mendocina, and P. aeruginosa type strains were included for comparative purposes.
Seven chromosomal loci were selected for study: 16S rDNA and ITS1 (representing the rrn operon), gyrB and rpoD (housekeeping genes previously included in Pseudomonas taxonomic studies) (34), catA (coding for catechol 1,2 dioxygenase, an enzyme responsible for the ortho cleavage of catechol in species of the RNA group I of Pseudomonas), nosZ (nitrous oxide reductase, a metabolically characteristic gene defining this denitrifying species), and nahH (coding for catechol 2,3 dioxygenase of the meta cleavage of catechol, a gene considered plasmid encoded in the genus Pseudomonas but chromosomally encoded in most naphthalene-degrading P. stutzeri strains studied so far) (23).
Alelle diversity.
The sequences obtained for all loci for the 26 P. stutzeri and the other reference strains were aligned. The lengths of the fragments analyzed ranged from 312 (catA) to 1,091 (16S rDNA) (Table 3). The average GC content of the different gene fragments coding for proteins ranged from 57.47 to 64.62% (data not shown). These values were slightly lower than the range of GC contents for the whole genome in strains of the species so far described (60.9 to 65.0%). With only three exceptions, the GC contents for each gene were nearly identical among strains of the same genomovar, with standard deviations less than 0.66. For instance, percent GC range in gyrB is less than 1% in members of the same genomovar, and this value for gyrB in the species is wider and ranges from 58.07 to 61.13%.
TABLE 3.
Genetic diversity of the selected loci among P. stutzeri strains analyzed in this study
| No. of strains | Locus | Fragment length (bp) | No. of alleles | Genetic diversity | No. of polymorphic sites (total)a | No. of nucleotide substitutions/nucleotide site (%) | Nucleotide diversityb | dN/dS ratio |
|---|---|---|---|---|---|---|---|---|
| 16 | nahH | 486 | 4 | 0.350 | 106 (65) | 21.8 (2.8)c | 0.03014 (0.02190) | 0.150 (0.270)c |
| 24 | catA | 312 | 18 | 0.942 | 138 (80) | 44.23 | 0.12500 (0.02100) | 0.180 |
| 26 | gyrB | 849 | 20 | 0.963 | 304 (291) | 35.8 | 0.11984 (0.00941) | 0.0236 |
| 26 | rpoD | 786 | 17 | 0.938 | 276 (204) | 35.1 | 0.11128 (0.00920) | 0.0933 |
| 26 | nosZ | 453 | 20 | 0.942 | 123 (98) | 27.1 | 0.08078 (0.00511) | 0.0813 |
| 26 | 16S rDNA | 1091 | 15 | 0.911 | 60 | 5.5 | 0.01351 (0.01368) | |
| 26 | ITS1 | 574 | 20 | 0.986 | 179 | 31.18 | NDd |
Total number of synonymous changes.
Numbers in parentheses are standard deviations.
Excluding strain ATCC 27951.
ND, not determined due to gaps and insertions in the sequences that impair the comparison of homologous positions.
All loci were highly polymorphic, and the number of polymorphic sites varied between 304 for the highest (gyrB) and 106 for the lowest (nahH) (Table 3). Excluding nahH of strain ATCC 27951, which accumulates 92 polymorphic sites, this value is only 14 for the other 15 nahH genes analyzed. The number of nucleotide substitutions per nucleotide site varied between 44.2% for catA and 21.8% for nahH (excluding ATCC 27951, with a totally different allele, this value is 2.8%). Most of the polymorphic sites were at the third position of the codon in the genes coding for proteins. The number of alleles varied in the different loci: 4 in nahH (16 strains), 18 in catA (24 strains), 20 in gyrB (26 strains), 17 in rpoD (26 strains), 18 in nosZ (26 strains), 15 in 16S rDNA (26 strains), and 20 in ITS1 (26 strains). Not considering nahH, the mean number of alleles per locus in 26 strains was extremely high, 18.7. The average number of alleles per locus and strain was 0.72.
The number of amino acid substitutions per amino acid site in the putative protein sequences was still high for CatA (37.5%), but was only 14.08% for GyrB. gyrB has a high number of nucleotide substitutions per nucleotide site, but the numbers of amino acid substitutions were the lowest of all the genes analyzed, indicating that they are evolutionarily neutral.
The extremely high nucleotide diversity of catA was manifested in the failure of PCR primers 1C12O/2C12O to amplify the target gene (450 nucleotides) in 10 of 26 strains. Five strains were amplified with primers C1STF/C1STR (320 nucleotides) designed in internal sequences of PCR primers 1C12O/2C12O. 7C12F/8C12R amplified catA from two strains. Strain ZoBell was amplified by using a combined set of primers 1C12O/8C12R (414 nucleotides). Strains CLN100 (genomovar 10 [gv. 10]) and KC (gv. 9) did not posses catA (6), as demonstrated by Southern blotting with a 622-nucleotide catA probe obtained from strain AN10 with primers 7C12F/8C12R (data not shown).
The nahH gene was the more conserved and was present in 16 of 26 P. stutzeri strains of gv. 1, 3, 4, 7, and 10. The isolates were from different origins (soils, wastewater treatment plants, and marine water) and from distant geographical locations (Siberia, California, and Majorca). Four different alleles were detected. Allele 1, with a GC content of 57.2%, was identical in 13 strains (gv. 1, 3, 4, and 7) able to degrade naphthalene. The other three strains had nahH genes that were slightly different. Strain CLN100 (gv. 10, allele 4, GC content of 57.0%) is able to degrade 2-chloronaphthalene. Its catechol 2,3 dioxygenase has two different amino acids in its composition (S189 instead of A and Y218 instead of H) (6). Strain DSM 50238 (gv. 7, allele 2, GC content of 57.2%) is not a naphthalene degrader, but is able to degrade toluate and was isolated from Californian soil. Its nahH had 18 different nucleotides, which affect the following amino acids: M23, A91, M93, D157, and L162. Strain ATCC 27951 was isolated from yogurt in Algeria (gv. 1, allele 3, GC content of 61.73%) and is not a naphthalene degrader, accumulating a high number of exclusive polymorphisms (92 of 106 polymorphic sites in the 16 strains analyzed), and 14 different amino acids (D12, V16, T19, L28, I29, R38, F53, Q63, E69, I71, N79, Q81, R84, and H102) not present in other P. sutzeri nahH genes. Q63 is unique to strain ATCC 27951 and absent in all other Pseudomonas strains compared.
dN/dS ratio.
dN indicates the number of nonsynonymous substitutions per nonsynonymous site that result in an amino acid replacement, and dS indicates the number of synonymous substitutions per synonymous site that do not change the amino acid. The dN/dS ratio was calculated in the genes coding for proteins as a measure of the degree (amount and type) of selection in P. stutzeri populations. It was less than 0.1 in three genes (gyrB, rpoD, and nosZ). The highest dN/dS ratio corresponds to catA (0.18); all of the ratios are much less than 1, indicating that these gene fragments are not under selection; that is, most of the sequence variability identified is selectively neutral. Synonymous substitutions were at least 5.5 times (1/0.18) more frequent than amino acid changes at any locus.
RSCU and CBI.
The RSCU value of a codon is the observed frequency of that codon in the gene divided by that expected under the assumption of equal usage of synonymous codons. An RSCU value of 1 indicates that the frequency of that codon is that expected for an equal codon usage; values less than 1 indicate that the codons are used less often than expected (26).
catA, gyrB, rpoD, and nosZ have the same pattern of codon usage, coinciding with those available in the databases (82,284 codons of genes of P. stutzeri strains) (17). However, the RSCU analysis demonstrated that in four amino acids, nahH utilizes codons clearly distinct from the rest of genes analyzed in this study and from those available in the databases for P. stutzeri: S shows a codon preference for TCC and AGT (it does not utilize AGC) instead of the TCG and AGC used in the rest of the genes; N uses AAT instead of AAC; I uses ATT and never ATC, the codon used preferentially in the rest of genes; Y uses only TAT; and the rest of genes preferentially use TAC. nahH exhibits a clear preference in the use of T in the third position of the codon. It does not utilize CCA or CCT for P, the codons used by the other genes. Only slight differences were observed when considering the rest of amino acids (data not shown).
CBI is a measure of the deviation from the equal use of synonymous codons. CBI values range from 0 (uniform use of synonymous codons) to 1 (maximum codon bias) (16). The CBIs for nahH are the lowest of all those for the five protein-coding genes analyzed: 0.472 (mean value considering 16 strains), 0.459 for the 13 strains of the main group, 0.482 for strain DSM 50238, 0.478 for strain CLN100, and 0.631 for strain ATCC 27951. Mean values of CBIs in the other genes are clearly higher: 0.674 for catA, 0.541 for gyrB, 0.628 for rpoD, and 0.750 for nosZ.
ST and allelic profiles.
Twenty-four STs were identified among 26 strains of P. stutzeri (Table 4). Twenty-three of them (95%) were present only once, with the most common ST (ST-3) occurring three times in members of gv. 3. The three strains of ST-3 (PTDA, PTDB, and PTDE) were isolated from the same sample due to their differences in colonial morphology. Enterobacterial repetitive intergenic consensus (ERIC)-PCR demonstrated a close genomic relationship between them (data not shown). The three most closely related strains in the allelic profile with six identical alleles, also members of gv. 3, were ST27MN2 (differing from PTD strains in 16S rDNA, ST-1), AER5.1 (differing in ITS1, ST-2), and AN10 (differing in gyrB, ST-7). Strains S1MN1 (ST-5) and B1SMN1 (ST-4) of gv. 1 also share six identical alleles, differing only in the ITS1 sequence. Strains ST27MN3 (ST-11) and 19SMN4 (ST-10) of gv. 4 have four identical alleles. The rest of the strains shared less than two alleles. No single allele was dominant in the populations studied. The highest frequencies of one allele per ST were 4 (ITS1, catA, rpoD, and nosZ) in members of gv. 3 and 3 (16S rDNA and gyrB) in members of gv. 1 and 3 (Table 4). The two members of gv. 7, AN10 (gv. 3), and DNSP21 (gv. 5), do not have any allele in common with another strain.
TABLE 4.
Sequence types at the seven loci examined in this study
| Strain | gv. | Allele at locus:
|
ST | ||||||
|---|---|---|---|---|---|---|---|---|---|
| nahH | catA | gyrB | rpoD | nosZ | 16S rDNA | ITS1 | |||
| ST27MN2 | 3 | 1 | 1 | 1 | 1 | 6 | 1 | 10 | 1 |
| AER 5.1 | 3 | 1 | 1 | 1 | 1 | 6 | 3 | 3 | 2 |
| PTD A | 1 | 1 | 1 | 1 | 6 | 3 | 10 | 3 | |
| PTD B | 1 | 1 | 1 | 1 | 6 | 3 | 10 | 3 | |
| PTD E | 1 | 1 | 1 | 1 | 6 | 3 | 10 | 3 | |
| B1SMN1 | 1 | 1 | 2 | 2 | 3 | 9 | 5 | 9 | 4 |
| S1MN1 | 1 | 1 | 2 | 2 | 3 | 9 | 5 | 7 | 5 |
| ATCC 27951 | 1 | 3 | 3 | 6 | 3 | 8 | 5 | 4 | 6 |
| AN10 | 3 | 1 | 1 | 4 | 1 | 6 | 3 | 10 | 7 |
| AN11 | 3 | 1 | 9 | 9 | 5 | 1 | 2 | 8 | 8 |
| LSMN2 | 3 | 1 | 8 | 8 | 4 | 5 | 1 | 10 | 9 |
| 19SMN4 | 4 | 1 | 5 | 3 | 2 | 3 | 4 | 6 | 10 |
| ST27MN3 | 4 | 1 | 6 | 3 | 2 | 4 | 4 | 5 | 11 |
| DSM 50238 | 7 | 2 | 7 | 7 | 7 | 2 | 6 | 1 | 12 |
| AER 2.7 | 7 | 1 | 4 | 5 | 6 | 7 | 7 | 2 | 13 |
| CCUG 11256T | 1 | 16 | 15 | 12 | 11 | 5 | 11 | 14 | |
| SD55473 | 1 | 14 | 14 | 11 | 15 | 5 | 18 | 15 | |
| A95/69 | 1 | 10 | 13 | 10 | 16 | 5 | 13 | 16 | |
| ATCC 17591 | 2 | 18 | 10 | 14 | 12 | 11 | 16 | 17 | |
| ZoBell | 2 | 15 | 12 | 15 | 12 | 12 | 17 | 18 | |
| A60/72 | 2 | 11 | 11 | 13 | 12 | 9 | 17 | 19 | |
| DNSP21 | 5 | 13 | 16 | 9 | 13 | 10 | 14 | 20 | |
| JD4 | 5 | 17 | 17 | 8 | 14 | 13 | 15 | 21 | |
| JM300 | 8 | 12 | 18 | 8 | 10 | 8 | 12 | 22 | |
| KC | 9 | 19 | 16 | 17 | 14 | 19 | 23 | ||
| CLN100 | 10 | 4 | 20 | 17 | 18 | 15 | 20 | 24 | |
Linkage disequilibrium (IA) and clonal population structure.
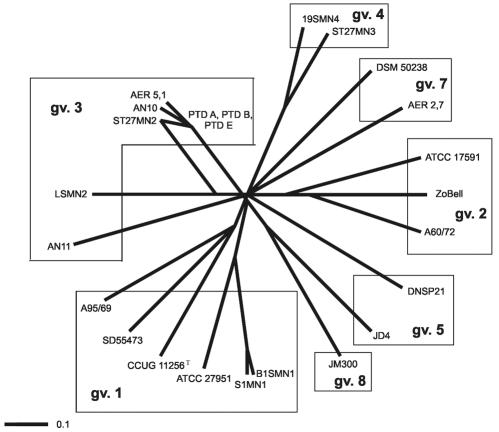
Strains KC and CLN100 were not included in the IA analysis, because their STs are unique, the strains are phylogenetically more distant from the rest of the P. stuzeri strains (6, 25), the gene catA was not present in either strain, and nahH is missing in strain KC. The IA calculated for six genes (excluding nahH, present only in 16 strains) in the other 24 strains was 2.988, indicating that there were limited recombination events and a clonal population structure. The IA value was still significantly different from 1 (2.085) when only one representative of each ST was included in the analysis, which removes bias due to taxonomic sampling. The relatedness among STs is represented in a split graph (Fig. 1), which shows a radial distribution of strains, indicating as well the clonal structure of the populations, with only possible recombination events between strains of gv. 3. If two or more strains are located in the same branch, they belong to the same genomovar of the species, with only one exception: strain JM300 (gv. 8) has an rpoD allele in common with strain JD4 (gv. 5).
FIG. 1.
Splits tree showing the distribution of the nine genomovars of P. stutzeri strains.
Gene phylogenies.
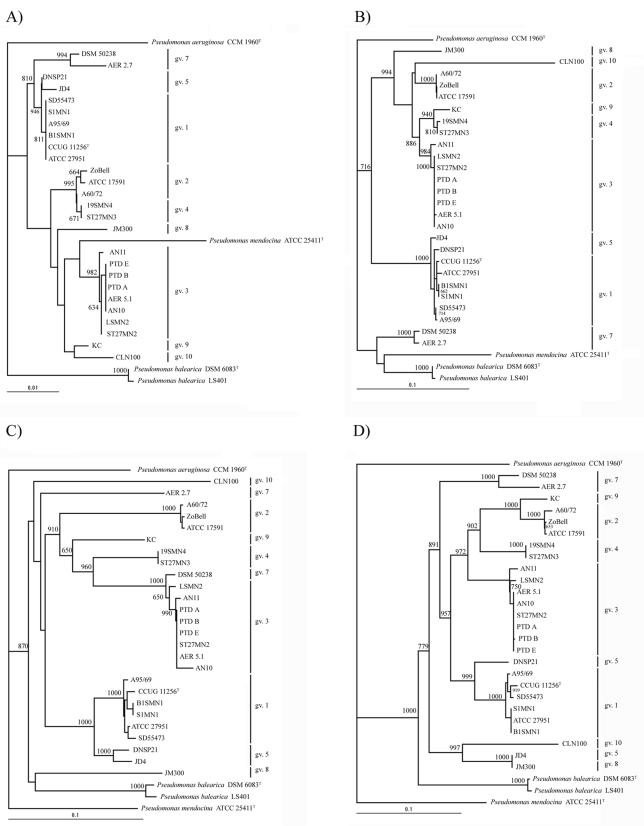
Molecular phylogenies of 26 multiple isolates belonging to P. stutzeri were obtained and referred to several closely related Pseudomonas species (P. aeruginosa, P. mendocina, and P. balearica). Seven sequences, including those of housekeeping genes (gyrB and rpoD), metabolically relevant genes (nosZ, catA, and naH), ITS1, and the 16S rRNA gene, were used to infer individual phylogenetic trees. Phylogenetic clustering of the strains based on the total number of differences among sequences was performed with either the Jukes-Cantor or Kimura algorithm and by the Fitch-Margoliash method, unweighted pair group method with arithmetic mean (UPGMA), and neighbor joining. Topologies of the trees for the same locus using different algorithms and different clustering methods were similar, and only one algorithm (Jukes-Cantor) and one clustering method (neighbor joining) are represented in Fig. 2 to avoid repetitions.
FIG. 2.
Phylogenetic tree based on the 16S rDNA (A), ITS1 (B), gyrB (C), rpoD (D), nosZ (E), catA (F), and nahH (G) genes of P. stutzeri strains. Hypothetical multilocus consensus tree showing the molecular evolutionary relationships of the genes rpoD, gyrB, nosZ, catA, 16S rDNA, and ITS1, between P. aeruginosa, P. mendocina, P. balearica, and 26 representative strains of the nine genomovars described for P. stutzeri (H). The bar indicates sequence divergence. Bootstrap values of 600 or more (from 1,000 replicates) are indicated at the nodes. Sequence accession numbers are listed in parentheses for nahH.
Most strains of the P. stutzeri complex, as defined by Yamamoto et al. (34), clustered in the same phylogenetic branch in the gene trees analyzed, and were usually separated from the three Pseudomonas species used. The only exception is nahH, which will be discussed later. P. balearica and P. mendocina were separated from P. stutzeri strains in all of the trees, with the exception of P. mendocina in the 16S rDNA tree and P. balearica and P. mendocina, which clustered with gv. 7 in the ITS1 tree. Strains belonging to the same genomovar were usually located in the same branch, with few exceptions, varying with the gene analyzed. Both strains of P. balearica (formerly gv. 6 of P. stutzeri) are grouped together in all the trees as an independent branch.
Bootstrap analysis of the 16S rDNA gave values higher than 800 only in the branching between members of gv. 1, 5, and 7. All strains of gv. 3 clustered together, and gv. 8, 9, and 10, with only one representative strain, were located in independent branches. Members of gv. 2 and 4 were grouped in the same branch.
ITS1 analysis allows a good discrimination between genomovars. Only members of gv. 1 and 5 were not clearly separated. Different ITS1 sequences were detected in strain CLN100 with microheterogeneities, and only the closest sequence was introduced in the analysis.
Phylogenetic branches in the gyrB gene were consistent with the genomovar distribution of the species, except in the two members of gv. 7 (AER2.7 and DSM 50238). Branches corresponding to genomovars were stable in the bootstrap analysis.
Bootstrap analysis of the rpoD branches was the most stable of all the genes, separating each genomovar into a different branch. Only the two members of gv. 5 were split in two different groups, clustering JD4 (gv. 5) with JM300 (gv. 8), with both strains sharing an identical allele.
In the nosZ phylogenetic tree, members of the gv. 2 grouped in one branch. One member of gv. 1 (CCUG11256) grouped in the gv. 2 branch, and the rest of the strains of gv. 1 clustered together. From gv. 3, only one member (AN11) was separated in another branch, close to strain JM300 (gv. 8). Members of gv. 4 are as distant from one another as they are from strain DSM 50238 (gv. 7). Both members of gv. 5 were distantly located in the tree, as was the case for members of gv. 7. Bootstrap values for each branch were relatively low.
Strains A95/69 (gv. 1) and A60/72 (gv. 2) are clearly separated from the rest of P. stutzeri strains in the catA analysis, being closer to the other Pseudomonas species. There is no clear distribution of strains of the same genomovar in the topology of the tree.
Thirteen of 16 P. stutzeri strains had an identical nahH gene (Fig. 2). All of them had been isolated as naphthalene degraders. The nahH alleles were slightly different from strain CLN100, isolated as a chloronaphthalene degrader, and from strain DSM 50238, a toluate degrader, which appears in the same branch as xylE of the P. putida strains. nahH allele 3 from strain ATCC 27951 (not known to be an aromatics degrader) was located more distantly in the tree. nahH sequences in the databases that were most closely related to allele 3 belonged to P. aeruginosa JI104 (88.8% identity), distantly related to the other P. stutzeri alleles. The two strains of P. balearica (naphthalene degraders) had two different nahH genes: one was included in the P. stutzeri main group (SP1402) and the other (LS401) was clearly distant.
Consensus phylogeny.
The individual distance sets obtained for six nucleotide sequences (16S rDNA, ITS1, gyrB, rpoD, nosZ, and catA) were combined to infer a composite molecular phylogeny for P. stutzeri (Fig. 2H). catA is absent in strains CLN100 and KC; therefore, the combined percentage identity was calculated ignoring catA when CLN100 and KC were compared. The three species included in the analysis are clearly separated in the resulting dendrogram. All P. stutzeri strains are located in the same phylogenetic branch, and members of each genomovar clustered together, maintaining the genomovar subdivision of the species.
DISCUSSION
16S rDNA has been used extensively as a phylogenetic marker in bacterial taxonomy for intergeneric relationships due to its extremely slow rate of evolution, and ITS1 has been selected for interspecies comparison in the genus Pseudomonas because it is not as conserved as the 16S RNA gene (8). Protein-encoding genes such as gyrB and rpoD have been reported to evolve much faster than rrn operons, thus providing higher resolution and allowing comparisons between closely related species (34). At the same time, the amino acid sequences are conservative enough to allow the comparison of taxa that are not closely related. In this study, the aforementioned genes were considered and three genes encoding catabolic enzymes (catA, nosZ, and nahH) representative of characteristic phenotypic traits of P. stutzeri were included. The analysis of this set of seven genes should provide a picture of the phylogenetic relationships between members of the species and should offer insights into the evolution of the species.
The extremely high genetic diversity of the species was manifested in the sequences analyzed. The number of nucleotide substitutions per nucleotide site was higher than in Campylobacter jejuni, Neisseria meningitidis, Streptococcus pneumoniae, Enterobacter faecium, and species of the Bacillus cereus complex, to our knowledge the highest so far described (9). The average numbers of alleles per locus and strain analyzed in the protein-coding genes were 0.72 for P. stutzeri (18.7 average alleles per locus in only 26 strains), 0.18 for C. jejuni, and 0.43 for strains of the B. cereus complex. This value is in good agreement with previous observations in MLEE studies undertaken with most of the strains analyzed in the present study, in which the genetic diversity was the highest described for a species (21). Each gene of the same genomovar has a homogeneous GC content, but is quite distinct when genes of different genomovars are compared. The species, considered as a whole, is much more diverse in GC content.
Twenty-three unique STs (95%) were observed in a total of 24 STs in 26 isolates of P. stutzeri. Only one ST was present in more than one isolate: the three strains sharing the same ST (ST-3) were identified phenotypically as members of P. stutzeri and also compared through ERIC-PCR. The resulting profiles were identical (data not shown), indicating that they might be considered as siblings. PTDA, PTDB, and PTDE were isolated as separate clones only due to differences in their colonial morphology. The multilocus sequence analysis demonstrated clearly that they are lineages of the same clone, with identical sequences, and members of gv. 3. Therefore, one different ST per P. stutzeri strain can be assumed, which is the highest possible value. Remarkably, if two strains had an allele in common they belonged to the same genomovar, with only one exception: strain JM300 (gv. 8) has an rpoD allele identical to that of strain JD4, one of the two members of gv. 5. This fact can be explained by the presence of a common ancestor for gv. 5 and 8 or a possible lateral gene transfer to JM300, a strain intensively studied due to its natural transformation (27). As revealed by the SplitsTree analysis, strains AN10, AER5.1, ST27MN2, PTDA, PTDB, and PTDE of gv. 3 present possible recombinational events.
Strains 19SMN4 and ST27MN3 of gv. 4 were very closely related in the multilocus sequence analysis with identical 16S rDNA, rpoD, and gyrB genes. Both were isolated as naphthalene degraders from samples taken in a wastewater treatment lagoon, but from different habitats (water column and sediment). Molecular typing methods demonstrated previously that both strains were genetically related, but different (1).
The SplitsTree analysis and the IA values are also in favor of an essentially clonal population structure of the species with limited recombination events, as was also deduced in the MLEE analysis.
Laterally transferred genes have often been identified on the basis of compositional features that distinguish them from ancestral genes in the genome. Recently acquired genes tend to differ in characteristics such as codon usage and GC content compared with the complete genome (3). Anomalous phylogenetic trees of these genes, when compared with other housekeeping genes, are also considered indicative of lateral gene transfer. These three characteristics indicate that catechol 2,3 dioxygenase genes in P. stutzeri and P. balearica have been acquired recently, after the differentiation of both species and after the differentiation of P. stutzeri in genomovars. It is remarkable that the GC content of nahH is the lowest of all the genes analyzed (57.47 ± 1.14) and below the range of the species (60.5 to 65.0). The preferred nucleotide in the third position of four amino acids in nahH is T (S, N, I, and Y), while C is the most frequent in the rest of proteins analyzed, indicating that the codon bias of the newly acquired gene has not been homogenized yet through evolutionary pressure. Moreover, 14 nahH genes of 16 strains analyzed were grouped very closely in the same phylogenetic branch. The topology of the tree is totally different from the rest of genes analyzed.
During the past 15 years, several groups have provided strong indications that mobile genetic elements and lateral gene transfer played an important role in the dissemination and construction of xenobiotic catabolic pathways (29). Lateral gene transfer of biodegradation genes may play a role in the adaptation of bacterial populations to organic contaminant compounds and a significant role in the acclimation of microbial communities to pollutants (15). Our results from the nahH gene in P. stutzeri, together with those presented previously (2), support the conclusion that aromatics degradation through a chromosomal meta pathway has been acquired by horizontal transfer to some strains of the species after its subdivision into genomovars, although the population structure of the species is essentially clonal.
Stability analysis using bootstrap resampling showed that the trees obtained for six genes studied were stable and well defined, clustering each species in most of them in the same phylogenetic branch (not considering nahH, a gene acquired through laterally transfer). The consensus tree of five genes and the ITS1 region is in good agreement with the discrimination of P. stutzeri from the most closely related Pseudomonas species. The present study relies on the results of the analysis of seven nucleotide sequences in the chromosome of 26 P. stutzeri strains. At least 4,551 nucleotides of each strain were sequenced and analyzed in pairwise comparisons. Due to the presence of four copies of the rrn operon in P. stutzeri strains, our data represent at least 9,546 nucleotides of the respective genomes: that is between 0.2 and 0.25% of the chromosome, depending on the genome size of the strains (between 3.75 and 4.64 Mbp) (7). Although the topologies of the individual trees do not always correspond to the genomovar subdivisions of the species, the consensus tree considering between 0.2 and 0.25% of the chromosome is in good agreement with the total DNA-DNA similarity values, on which the genomovar definition is based. The different phylogenetic distances in seven genes analyzed, together with the variable number of alleles per loci, suggest that not all loci are evolving at the same rate and that one of the genes (nahH) has been acquired through lateral gene transfer.
P. balearica is the most closely related species to P. stutzeri and was previously considered as gv. 6 of the species. Multilocus sequence analysis, together with phylogenetic analysis, clearly confirms the genomic status of P. stutzeri and P. balearica as two different species. P. balearica in all genes analyzed constitutes a clearly defined phylogenetic branch in the six trees. The only exception is the nahH gene: in strain SP1402, it is almost identical to 15 nahH genes of P. stutzeri strains, and LS401 has a different gene, clustering apart in the phylogenetic tree. It has a higher GC content (59.88) and was located near strain ATCC 27951. This result is an additional argument to reinforce the assumption of a lateral transfer of the nahH gene.
The 16S rDNA, ITS1, gyrB, and rpoD genes are relevant for the phylogenetic affiliation of one species in the bacterial domain and, in our model, into the genus Pseudomonas. The trees resulting from the analyzed sequence data, together with the consensus tree, provide an excellent data set to assess the utility of the recently proposed core genome hypothesis, which provides a genetically based approach applied to the biological species concept for bacteria (13). Following the opinion of Lan and Reeves, different types of genes might be detected in one species. (i) Genes found in most individuals of the species (the core set of genes for that species) are the genes that determine those properties characteristic of all members of the species. These genes are represented in our study of P. stutzeri and P. balearica by the aforementioned genes and nosZ and catA. They might be found in 95% or more of isolates. (ii) Additionally, each strain will have some auxiliary genes, which determine properties found in some but not all members of the species. These are, for instance, genes for new metabolic functions, without a barrier to interspecies recombination. The naphthalene catabolic genes, represented in our study by nahH, belong to this type of gene, which may be present in 1 to 95% of isolates. They indicate an adaptation to the ecological niche from which the strain was isolated. (iii) Those genes present in less than 1% of isolates are provisionally treated as foreign genes or genes being lost from the species. The proposal by Lan and Reeves has been confirmed experimentally by the analysis published by Wertz et al. (32) for seven taxa of enteric bacteria and now, in the present paper, for Pseudomonas strains.
In general, the strains of P. stutzeri identified as belonging to the same genomovar formed tight clusters in the individual gene trees, demonstrating the existence of genotypic clusters that could correspond to traditional species designations in the sense of genomic species, not considering the phenotype. Following this argument, genomovars are confirmed in this study as biological units, previously defined by total DNA-DNA similarity values, rrn sequence analysis, and MLEE.
Acknowledgments
This work was supported by project BOS2001-0303 from the CICYT (Spain). A.C. was a recipient of a predoctoral fellowship of the MEC (Spain).
We thank our colleagues M. C. Fusté, G. Lorén, and M. Farfán for many helpful discussions.
REFERENCES
- 1.Bennasar, A., C. Guasp, M. Tesar, and J. Lalucat. 1998. Genetic relationships among Pseudomonas stutzeri strains based on molecular typing methods. J. Appl. Microbiol. 85:643-656. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, R., E. García-Valdés, and E. R. B. Moore. 2000. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene 245:65-74. [DOI] [PubMed] [Google Scholar]
- 3.Daubin, V., E. Lerat, and G. Perrière. 21 August 2003, posting date. The source of laterally transferred genes in bacterial genomes. Genome Biol. 4:1-12. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.0). Cladistics 5:164-166. [Google Scholar]
- 6.García-Valdés, E., M. M. Castillo, A. Bennasar, C. Guasp, A. M. Cladera, R. Bosch, K. H. Engesser, and J. Lalucat. 2003. Polyphasic characterization of Pseudomonas stutzeri CLN100 which simultaneously degrades chloro and methylaromatics: a new genomovar within the species. Syst. Appl. Microbiol. 26:390-403. [DOI] [PubMed] [Google Scholar]
- 7.Ginard, M., J. Lalucat, B. Tümmler, and U. Römling. 1997. Genome organization of Pseudomonas stutzeri and resulting taxonomic and evolutionary considerations. Int. J. Syst. Bacteriol. 47:132-143. [DOI] [PubMed] [Google Scholar]
- 8.Guasp, C., E. R. B. Moore, J. Lalucat, and A. Bennasar. 2000. Utility of internally-transcribed 16S-23S rDNA spacer regions for the definition of Pseudomonas stutzeri genomovars and other Pseudomonas species. Int. J. Syst. Evol. Bacteriol. 50:1629-1639. [DOI] [PubMed] [Google Scholar]
- 9.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A. B. Kolsto. 2004. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 11.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 12.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N.Munro (ed.), Mammalian protein metabolism. Academic Press, Inc, New York, N.Y.
- 13.Lan, R., and P. L. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 14.Lane, D. 1991. 16S/23S sequencing, p. 115-175. In E. G. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley, Chichester, United Kingdom.
- 15.Madsen, E. L. 2002. Horizontal transfer of naphthalene catabolic genes in a toxic waste site, p. 37-44. In M. Syvanen and C. I. Kado (ed.), Horizontal gene transfer. Academic Press, London, United Kingdom.
- 16.Morton, B. R. 1993. Chloroplast DNA codon usage. Evidence for selection at the psbA locus based on tRNA availability. J. Mol. Evol. 27:273-280. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from the international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 19.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 20.Palleroni, N. J. 1984. Genus I. Pseudomonas, p. 141-199. In J. G. Holt and N.R. Krieg (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams and Wilkins Co., Baltimore, Md.
- 21.Rius, N., M. C. Fusté, C. Guasp, J. Lalucat, and J. G. Lorén. 2001. Clonal population structure of Pseudomonas stutzeri: a species with exceptional genetic diversity. J. Bacteriol. 183:736-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosselló, R., E. García-Valdés, J. Lalucat, and J. Ursing. 1991. Genotypic and phenotypic diversity of Pseudomonas stutzeri. Syst. Appl. Microbiol. 14:150-157. [Google Scholar]
- 23.Rosselló-Mora, R., J. Lalucat, and E. García-Valdés. 1994. Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains. Appl. Environ. Microbiol. 60:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 25.Sepúlveda-Torres, L., J. Zou, C. Guasp, J. Lalucat, D. Knaebel, J. Plank, and C. Criddle. 2001. Pseudomonas sp. strain KC represents a new genomovar within Pseudomonas stutzeri. Int. J. Syst. Evol. Microbiol. 51:2013-2019. [DOI] [PubMed] [Google Scholar]
- 26.Sharp, P. M., T. M. F. Tuohy, and K. R. Mosurski. 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14:5125-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sikorski, J., M. Möhle, and W. Wackernagel. 2002. Identification of complex composition, strong strain diversity and directional selection in local Pseudomonas stutzeri populations from marine sediments and soil. Environ. Microbiol. 4:465-476. [DOI] [PubMed] [Google Scholar]
- 28.Spiers, A. J., A. Buckling, and P. B. Rainey. 2000. The causes of Pseudomonas diversity. Microbiology 146:2345-2350. [DOI] [PubMed] [Google Scholar]
- 29.Springael, D., and E. M. Top. 2004. Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from eological studies. Trends Microbiol. 12:53-58. [DOI] [PubMed] [Google Scholar]
- 30.Tamames, J. 1 June 2001, posting date. Evolution of gene order conservation in prokaryotes. Genome Biol. 2:Research0020. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed]
- 31.Thompson, J., D. G. Huggins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wertz, J. E., C. Goldstone, D. M. Gordon, and M. A. Riley. 2003. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J. Evol. Biol. 16:1236-1248. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 241-242. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 34.Yamamoto, S., H. Ksai, D. L. Arnold, R. W. Jackson, A. Vivian, and S. Harayama. 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385-2394. [DOI] [PubMed] [Google Scholar]