Abstract
The turbot is a flatfish (Pleuronectiformes) with increasing commercial value, which has prompted active genomic research aimed at more efficient selection. Here we present the sequence and annotation of the turbot genome, which represents a milestone for both boosting breeding programmes and ascertaining the origin and diversification of flatfish. We compare the turbot genome with model fish genomes to investigate teleost chromosome evolution. We observe a conserved macrosyntenic pattern within Percomorpha and identify large syntenic blocks within the turbot genome related to the teleost genome duplication. We identify gene family expansions and positive selection of genes associated with vision and metabolism of membrane lipids, which suggests adaptation to demersal lifestyle and to cold temperatures, respectively. Our data indicate a quick evolution and diversification of flatfish to adapt to benthic life and provide clues for understanding their controversial origin. Moreover, we investigate the genomic architecture of growth, sex determination and disease resistance, key traits for understanding local adaptation and boosting turbot production, by mapping candidate genes and previously reported quantitative trait loci. The genomic architecture of these productive traits has allowed the identification of candidate genes and enriched pathways that may represent useful information for future marker-assisted selection in turbot.
Keywords: turbot, genome sequencing, genetic map, genome evolution, productive traits
1. Introduction
The turbot (Scophthalmus maximus) is a flatfish naturally distributed around the European coast from the East and Northeast Atlantic Ocean, including the Baltic Sea, along the Mediterranean up to the Black Sea. As a flatfish, the turbot is adapted to a benthic lifestyle, and thus lives in shallow waters mainly during larvae and juvenile stages, moving to deep waters upon reaching adulthood. It displays a cryptic body colour, is covered by a mucous scaleless skin, follows a carnivorous narrow prey-spectrum diet, and undergoes a complete metamorphosis to adapt to benthic life (https://aquatrace.eu/leaflets/turbot). Genetic evidence of flatfish adaptation to sea bottom has been reported from the whole genome sequencing (WGS) of the tongue sole (Cynoglossus semilaevis).1 However, adaptations to the low oxygen concentration, low light levels, microbiota, and prey range characteristic of the sea bottom may have involved different strategies. This diversity may be related either to a rapid adaptive radiation of flatfish or to a polyphyletic origin.2
The great commercial value of turbot has promoted its intensive farming during the last decade (∼75,000 t) (https://aquatrace.eu/leaflets/turbot). The main goals of breeding programmes in this species are increased growth rate, control of sex ratio, and improved disease resistance. Study of the variation in these traits is also essential for understanding local adaptation of turbot populations.3 Although quantitative trait variability has traditionally been attributed to many loci with small effects, genome association screening has demonstrated the existence of specific genomic regions that explain an important proportion of the phenotypic variance of these traits.4 The detection of quantitative trait loci (QTL) in turbot has recently been initiated for growth,5,6 sex determination (SD),7 and resistance to the main industrial diseases.8–10 While increasing amounts of turbot genomic data have begun to enable a deeper knowledge of the genomic basis of these traits, a reference genome sequence is essential to achieve more efficient selection and sustainable management of wild resources.
In this study, we report a high-quality genome of the turbot by integrating high-throughput genome sequencing and previous genomic resources.11,12 Our assembly of the turbot genome spans 544 Mb (∼96% of estimated genome content), is highly contiguous with a scaffold N50 of 4.3 Mb, and has a transposable element (TE) content representing ∼5% of the genome. We identified 22,751 protein-coding genes transcribed into 28,189 mRNAs by using gene prediction and empirical transcriptomic data. We anchored 80% of the genome assembly to the turbot genetic map,13,14 thus enabling comparative mapping with other fish genomes and facilitating the consolidation of the turbot map in accordance with its karyotype. We found a nearly 1:1 relationship between turbot linkage groups (LGs) and those of other Percomorpha, although we also observed important micro-reorganizations within each LG. Paralogous relationships showed the conservation of important collinear blocks within the turbot genome, most likely the result of the ancestral teleost genome duplication. Some of these paralogous genes may represent genetic expansions related to adaptations to benthic lifestyle and to low temperatures. The genomic architecture of growth, SD, and resistance to pathologies was investigated using previously reported QTL as a reference. We mapped candidate genes using the established relationship between physical and genetic maps, and also identified new candidates and enriched functions by gene mining around the most strongly associated markers. Our results provide new insights into flatfish evolution and enable the development of more refined genomic breeding tools.
2. Materials and methods
The statistical and bioinformatics approaches as well as public repositories used in this study are outlined in this section. Most details on statistical packages, software and the parameters applied for each analysis are included in Supplementary Methods, File S1.
2.1. Genome sequencing and assembly
WGS was performed using the Illumina Genome Analyzer IIx and HiSeq2000 sequencing instruments. The standard Illumina protocol with slight modifications was followed for the construction of short-insert paired-end (PE) and mate pair (MP) libraries. A fosmid library was constructed in the pNGS vector (Lucigen Corp.) and the DNA was processed for end sequencing following the Lucigen protocol. Genome size was estimated by analysing genomic k-mer content.
PE reads were assembled into unitigs, then into contigs and finally into scaffolds. At each assembly step, sequences were filtered from contaminating sequences, their ends trimmed and finally subjected to a misassembly detection routine. Scaffolds were reassembled using MP and fosmid-end sequences and gap-filled at each step.
2.2. Comparative mapping
To anchor scaffolds to the genetic map, we performed a similarity search of the genomic sequences corresponding to linkage mapping markers against the turbot genome. The correspondence between LGs of the turbot genetic map and chromosomes/scaffolds of the model species was drawn with MAPCHART 2.2.15 Sequences of orthologous genes from anchored scaffolds were compared with the updated versions from model fish genomes downloaded from ftp://ftp.ensembl.org: Tetraodon nigroviridis v.8.61, Oryzias latipes v.1.61 and C. semilaevis, in order to identify syntenies. Orthologous relationships among species and paralogous relationships within the turbot genome were represented with circle diagrams using CIRCOS.16 Non-anchored scaffolds were predictively mapped on turbot LGs by comparing nucleotide sequences of orthologous genes against model fish genomes. Those scaffolds matching in the same homologous position with at least three reference species were predictively mapped at specific turbot LGs.
2.3. Repetitive elements
The repetitive component of the turbot genome was analysed using a combination of ab initio and sequence homology-based methods. The assembled genome was analysed to determine low-complexity sequences, short repetitive motifs, and interspersed repeats. The genome sequence was further analysed to extract the nucleotide sequences of the putatively full-length insertions (>80% length of the closest canonical query sequence), as described elsewhere,17 using RepBase database sequences as a query. All TE-derived sequences were named according to the RepBase element sequence that produced the highest match score and classified.
2.4. Protein-coding gene annotation
Turbot RNA-seq, ESTs, cDNAs, and mRNAs deposited in public databases and Percomorpha proteins from Uniprot were combined with ab initio gene predictions to produce a set of consensus coding sequence models, which were then updated to include untranslated (UTR sequences) and annotated alternative splice forms.
2.5. RNA genes annotation
Non-coding RNA (ncRNA) genes were annotated by running the program cmsearch v1.1 within Infernal (http://www.ncbi.nlm.nih.gov/pubmed/24008419) against the RFAM database of RNA families (v12.0) (http://www.ncbi.nlm.nih.gov/pubmed/23125362). Also, transfer RNAs (tRNA)-scan-SE (v1.23) (http://nar.oxfordjournals.org/content/25/5/0955.full) was run in order to detect the transfer RNA genes present in the genome assembly of S. maximus.
2.6. Proteome functional annotation
For the functional annotation of the identified protein-coding genes, we used an in-house developed automatic pipeline. An electronic inference of function was performed based on the sequence similarity between our proteins and other sequences annotated in different public repositories. Different strategies and software were employed to obtain gene ontology (GO) terms and KEGG orthology (KO) groups. KO identifiers were then used to retrieve KEGG relevant functional annotations. GO terms distribution was determined according to functional categories.
2.7. Orthologues and paralogues identification: similarity approach
The turbot proteome was compared using BLASTP (E-value: 1e−05) against those of medaka (O. latipes), Tetraodon (T. nigroviridis), and tongue sole (C. semilaevis) to identify orthology relationships, and against the turbot genome itself to look for paralogy. Orthology relationships were established following a reciprocal best-match strategy: we first looked for orthology between turbot and two model fish (medaka and Tetraodon) and then, from the obtained list, we investigated orthology with tongue sole. Paralogy relationships between turbot proteins were deduced using a reciprocal second best-match strategy. Pairs of paralogues were annotated using BlastKOALA (http://www.kegg.jp/blastkoala/) for KO codes. Only those pairs with the same KEGG annotation were considered to be consistent paralogues for further analysis.
2.8. Phylogenomics
The turbot phylome was reconstructed using the phylomeDB pipeline.18 For each turbot gene, a search was performed against the proteome database of the 17 selected species (Supplementary File 1: Table S1). Multiple alignments of homologous sequences were built and then used for phylogenetic tree reconstruction. Paralogy and orthology predictions for the species considered were analysed based on the turbot phylome. Gene duplications in the turbot lineage were identified and used to estimate gene enrichment using the turbot proteome as a reference. A total of 389 genes with one-to-one orthologues in each studied species were selected and their alignments concatenated to derive the species tree. We also reconstructed a super-tree from all single gene trees in the turbot phylome using a parsimony strategy.
2.9. Adaptation to benthic life-style
Paralog relationships identified from the similarity and phylogenetic approaches were combined to obtain a consistent set of paralogues resultant from specific duplications in the turbot or flatfish (incorporating C. semilaevis) genome lineages. A filtering was performed on that list to avoid artefacts from genome assembling after detecting an excess of paralogues in small scaffolds with 100% homology with the major ones. So, all paralogues included in scaffolds <20 kb with >98% homology to the biggest ones were discarded for this analysis. This set of paralogues was evaluated to identify functional enriched pathways using the turbot transcriptome as a background (false discovery rate (FDR) <5%) and gene family expansions that could be related to particular adaptations of flatfish to benthic lifestyle. Phylogenetic analysis of selected gene sequences was performed by the neighbour joining method, using the JTT amino acid substitution model. Selective pressures on particular genes or gene families that could explain adaptation to the benthic life-style were investigated both through evaluation of gene family expansions from the set of paralogues, as well as by analysing the ratio of non-synonymous (Ka) vs. synonymous (Ks) substitution rates (Ka/Ks) on candidate genes with the closely related and well-annotated species medaka (O. latipes), Tetraodon (T. nigroviridis), and tongue sole (C. semilaevis).
2.10. Genetic architecture of growth, resistance to diseases, and SD
Based on previous reports and our own data, a list of candidate genes for sex-, growth-, and immune-related traits in fish and vertebrates was obtained. These genes were located in the turbot genome and then in the genetic map using the established relationship between them both. Analyses were focused on major QTL associated with growth, SD, or disease resistance, and on those overlapping for different traits. Mining analysis around selected QTL for these traits was performed to identify additional candidates and enriched pathways or functions using the turbot transcriptome (22,751 genes) as a background (FDR <5%) within the extracted gene lists. Selected candidate genes identified across QTL regions and traits were represented in the turbot genetic map13 using Mapchart 2.2.15
3. Results
3.1. Genome sequencing and assembly
We sequenced the turbot genome using a whole genome shotgun approach. To facilitate hierarchical assembly, we constructed two PE libraries, two MP libraries and a fosmid-end library and sequenced them on the Illumina platform (Supplementary File 1: Table S2). The final assembly (‘sm5’) has a total length of 544 Mb, including 14 Mb of gaps. The contig N50 is 31.2 kb and the scaffold N50 is 4.3 Mb. The largest scaffold is 19 Mb long and 95% of the assembly is found in 287 scaffolds ≥166 kb. The GC content is 43.3%. The percentage of complete and partial core eukaryotic genes (248 genes in total) according to the CEGMA pipeline is 98 and 99%, respectively. Using the depth of the main 17-k-mer peak (114 from a total of 129,589,789,624 k-mers), we estimated the turbot genome size to be 568 Mb (Supplementary File 1: Fig. S1), so our assembly represents 95.8% of this estimation. Sequencing data are available in the European Nucleotide Archive (ENA) with the following study accession: PRJEB11743.
Repetitive sequences make up to 8.5% of the turbot genome (Supplementary File 1: Table S3). These are grouped into three main categories: simple repeats (3%), low-complexity motifs (0.5%), and TEs (5%). The TE-derived fraction is very similar to that found in the other known flatfish genome (C. semilaevis; 5.8% TEs).1 The turbot displays a higher proportion of TEs than T. nigroviridis and Fugu rubripes (<3%), but much lower than that observed in other fish such as Danio rerio (>40%).19,20
The genome assembly was compared with the turbot genetic map (486 mapped markers out of 513 genotyped; Supplementary Table S4, File 1: Fig. S2).13,14 All genetic markers with available sequence (492 of 513; 96%) match unique positions in 162 scaffolds. Using genetic markers, 150 of these scaffolds were anchored to single LGs. Six scaffolds were anchored to two LGs; the break points in which were confirmed by comparative mapping with model species (see below), suggesting assembly errors. In total, 156 scaffolds were anchored, representing 79.7% of the assembly. The remaining six scaffolds with markers contain only matches to unlinked markers.
3.2. Gene and functional annotation
Combining empirical transcriptomic data and ab initio predictions, we identified a preliminary set of 22,751 genes, whose 28,189 transcripts encode 26,823 unique proteins (Supplementary Table S5). Of the 28,189 transcripts, 24,239 are supported by protein alignments, 22,597 by GMAP transcript alignments, and 25,737 (91%) by either protein or GMAP transcripts. The rest are supported by ab initio gene predictions. We also identified a total of 5,808 ncRNA genes in the turbot genome (Supplementary Table S6), including 176 ribosomal RNAs genes, 766 tRNA genes, 285 microRNAs (miRNA), 779 small nuclear RNAs, one small cytoplasmic RNA gene, and 12 long non-coding RNAs genes.
A total of 23,460 (87.5%) proteins are assigned some type of annotation feature. We identified 18,571 (69.2%) proteins using Blast2GO and 11,307 (42.2%) using KEGG, and 19,445 (72.5%) have both Blast2GO and KEGG annotations. In total, 23,245 (86.6%) proteins exhibited some type of protein signature using InterProScan (Supplementary File 1: Table S7). The correspondence between the protein length and the number of annotated and non-annotated proteins makes it evident that non-annotated sequences tend to be smaller (Supplementary File 1: Fig. S3), which is likely due to incomplete open reading frames erroneously predicted by the automatic annotation pipeline. Using the three available resources (InterPro, KEGG and Blast2GO), we associated at least one GO term to 20,927 (78.0%) proteins: 16,590 to Biological Process, 14,489 to Cellular Component, and 18,522 to Molecular Function (Supplementary File 1: Fig. S4).
3.3. Orthologues and paralogues: comparative genomics
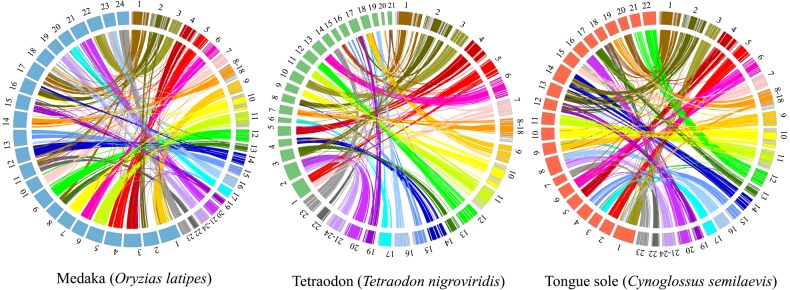
A list of 10,050 orthologous genes was obtained by identifying the best common reciprocal BLAST match of the 22,751 turbot predicted genes against the proteomes of two closely related and well-annotated species (O. latipes and T. nigroviridis). This list was additionally used to look for orthology with C. semilaevis, so that 7,090 orthologous genes were identified (Supplementary Table S8). The relationship between the turbot genetic map and the reference genomes of these species was evaluated by comparing the position of ∼8,000 orthologues included in the 156 anchored scaffolds to the turbot genetic map. We observe large syntenic blocks, indicative of the high evolutionary conservation of chromosome organization within Percomorpha (Fig. 1). Most turbot LGs show a 1:1 relationship with specific chromosomes of the three reference genomes, indicating a conserved macrosyntenic pattern. Only a few major reorganizations resulting from chromosome fusions or fissions, and some minor translocations, were detected. According to this analysis, LG8 and LG18 on the one hand, and LG21 and LG24 on the other, match to single orthologous chromosomes in the three reference genomes, which confirms their merging into a single chromosome in accordance with the turbot karyotype (2n = 22).21 However, when the sequences of the 10 major turbot scaffolds and the genome of the two closest species, the flatfish tongue sole (C. semilaevis) and stickleback (Gasterosteus aculeatus) were compared using BLAST (E-value <e−100) (see Phylogenomics), a notable amount of chromosome micro-reorganizations were detected (Supplementary File 1: Fig. S5). Interestingly, many more reorganizations were observed with regard to tongue sole than to stickleback, which suggests a high chromosome evolutionary rate within Pleuronectiformes in accordance with previous molecular information.22
Figure 1.
Macrosyntenic relationship pattern between turbot (S. maximus) and three related Percomorpha species (medaka, Tetraodon, and tongue sole). LG 8 and 18 (LG8 − 18) and 21 and 24 (LG21 − 24) are grouped indicating their merging in single chromosomes as suggested by the syntenic patterns observed with the three species.
Comparative mapping with the aforementioned fish species allowed us to: (i) detect the misassembly of four major scaffolds (s00001, s00003, s00008, and s00016), which were in turn disassembled and then anchored to specific LGs (Supplementary File 1: Fig. S2, Table S4); (ii) anchor 61 additional scaffolds, thus increasing the mapping coverage up to 489 Mb (89.9% of turbot genome) (Supplementary Table S9); and (iii) assign 20 of 27 unlinked markers to specific LGs (Supplementary Table S4).
Paralogous relationships between the 22,751 turbot genes were initially hypothesized using a second best hit reciprocal BLAST strategy, leading to 3,035 putative paralogous pairs. An additional filtering was applied and only those pairs with the same assigned KO code were retained, constituting a consistent set of 1,858 pairs of paralogues (Supplementary Table S10). A circle diagram (Supplementary File 1: Fig. S6) and an oxford plot (Supplementary File 1: Fig. S7) were constructed with paralogous pairs using the 156 anchored scaffolds, and important syntenic blocks were identified between pairs of chromosomes. While in some cases, a 1:1 relationship was highly supported (LG3–LG23, LG4–LG6, LG10–LG11, LG12–LG14), in other cases, three or more LGs were involved, suggesting genomic reorganizations of homeologous chromosomes arisen from the teleost genome duplication.1
3.4. Phylogenomics
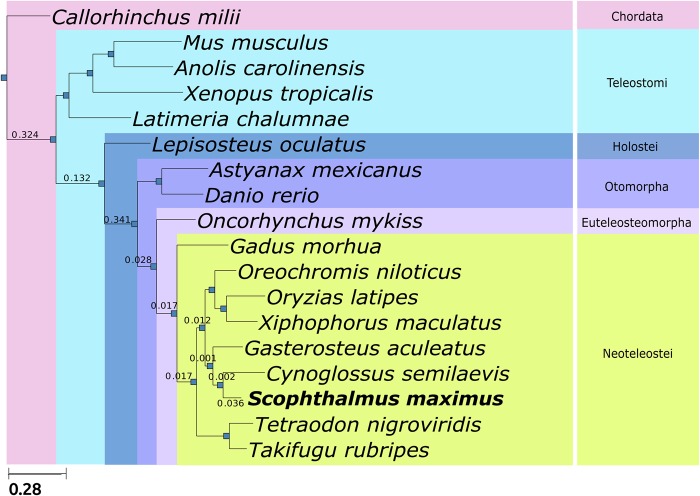
To ascertain the evolution of the turbot genome, we reconstructed the evolutionary histories of all identified turbot genes along with those of another 17 sequenced teleosts (Supplementary File 1: Table S1) with the phylomeDB pipeline.18 Gene family trees were analysed to predict orthologous and paralogous relationships, thus complementing the previous analysis. This approach enabled us to detect and date duplication events and to transfer functional annotations from one-to-one orthologues. All trees and alignments are available through PhylomeDB (www.phylomeDB.org, PhylomeID=18). We reconstructed the evolutionary relationships of the species under study by concatenating the alignments of 389 genes that were present in single copy in all analysed species and by building a super-tree combining all individual gene trees from the phylome. Both approaches resulted in the same, highly supported topology (Fig. 2), which is largely consistent with the reported relationships of the studied species.23 Our phylogeny shows that tongue sole is the closest species to turbot and that these two species are grouped with G. aculeatus, as previously suggested.24 This group is closer to the clade containing Oryzias, Oreochromis, and Xiphophorus than to the one containing Tetraodon and Takifugu, a relationship not recovered when a smaller set of genes was used.23 Then we mapped to this species tree the duplications detected in all gene trees and computed duplication densities per branch in all lineages leading to turbot (Fig. 2). Our results show the largest peaks of duplication densities at the base of vertebrates and at the base of teleosts (only represented by Otomorpha species in this study), which correspond to the known rounds of ancestral genome duplications. Interestingly, a notable duplication density was detected within Pleuronectiformes when comparing turbot against tongue sole (0.036). We found that genes families duplicated at the teleost whole genome duplication are enriched in ion transporters (calcium, potassium) related to osmoregulation as previously reported.25
Figure 2.
Phylogenetic tree obtained from the concatenation of 389 single copy, widespread genes in 17 selected fish species. All nodes are maximally supported (100% bootstrap). Numbers on the branches indicate gene duplication densities (average number of duplications per gene and per lineage) in the turbot (S. maximus) lineage.
3.5. Adaptation to benthic life style
As the genetic basis of adaptation to the particular benthic life conditions of this species was unknown, we examined genomic expansions and signals of positive selection from turbot paralogues and candidate genes, respectively. We identified gene families related to vision and to cell membrane composition that duplicated in the turbot genome when compared with other fish species. Also, 25 significantly enriched KEGG pathways (FDR <5%) were identified in the list of paralogues associated with immune system, membrane functions, lipid and amino acid metabolism, olfactory system and detoxification processes (Supplementary Table S11).
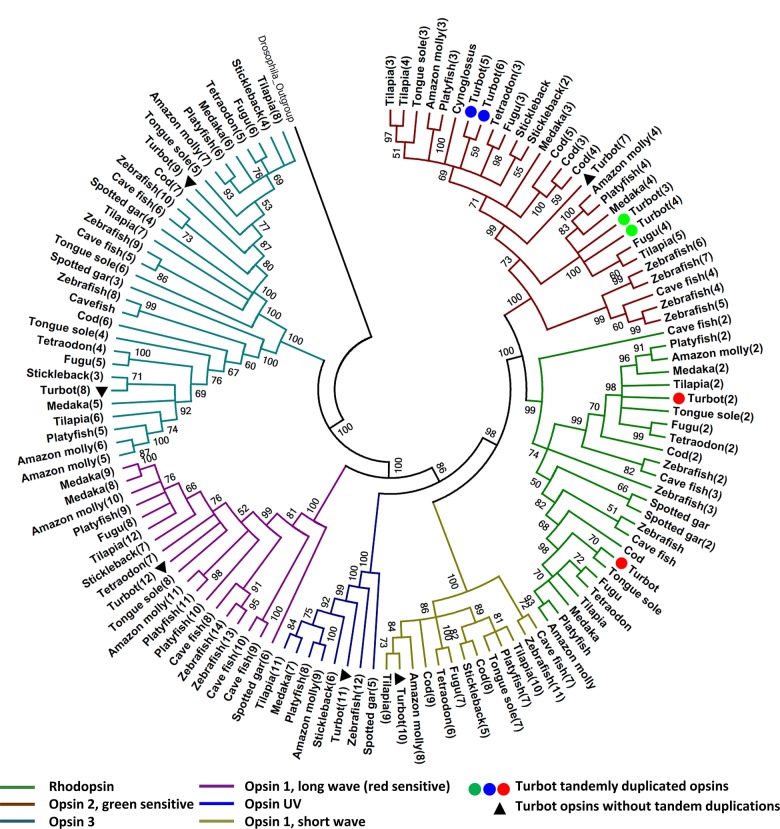
Rhodopsin (RH), and specifically the green-sensitive opsin, is the primary pigment of rod photoreceptors enabling vision in low light conditions, such as those existing in the sea bottom.26 We found duplications of two green-sensitive RH genes in the turbot genome (rh2a, rh2b) when compared with the genomes of 10 different fish species, including the flatfish tongue sole1 (Fig. 3). An additional fifth RH rh2 gene and another one sensitive to ultraviolet light were also detected in the turbot genome but they were missing in the tongue sole. Altogether, turbot shows the highest number of green-sensitive opsins (5) among the fish species studied. Additionally, turbot showed six copies of β-crystalline b (crybb) genes, a number similar to model fish, while the tongue sole has only one. Interestingly, we detected in turbot 15 copies of gamma-crystalline m2 (crygm2) and 10 copies of gamma-crystalline m3 (crygm3), genes that usually appear in only two copies in model teleosts, excluding zebrafish. No signals of positive selection were detected between duplicated opsin 2 genes of turbot with regard to other fish species from the estimated ratio of non-synonymous (Ka) vs synonymous (Ks) rates (Ka/Ks = 0.071–0.258; Supplementary Table S12; Fig. 3), nor with the remaining opsin genes (opsins UV: 0.140–0.390; RHs: 0.053–0.183).
Figure 3.
Phylogenetic relationships of the RH genes of turbot (S. maximus) with other fish with available genome. Branches with support lower than 50 were collapsed.
The most important biochemical response of poikilothermic organisms to environmental cooling is to increase unsaturated fatty acids of both membrane and depot lipids.27 Accordingly, tissues of marine fish contain higher amounts of membrane polyunsaturated fatty acids (PUFA) than other vertebrate taxa.28 PUFA content within fish is also variable according to their optimal living temperature. Since PUFA are particularly susceptible to peroxidation, phospholipid hydrolysis by phospholipase A2 (PLA2) and subsequent reduction by glutathione, dependent-glutathione peroxidase (GPx) are required to prevent oxidative damage.29 In turbot, we identified five copies of pla2 and two copies of gpx1 genes (Fig. 4). Furthermore, we could detect signals of positive selection in three pla2 genes (turbot2, turbot3, and turbot4; Fig. 4) which showed consistent positive Ka/Ks values between them (Supplementary Table S12). The only fish species with more pla2 copies than turbot is the cod (Gadus morhua) (6) which lives in very cold waters, while all other fish show two copies at most. Two copies of gpx1 are also found in other fish species, but not in the tongue sole (1 copy). We also identified two copies of the glutathione synthetase gene (gss) in turbot (Fig. 4), an enzyme involved in glutathione synthesis, thus being the only fish species with two gss copies.
Figure 4.
Phylogenetic analysis of GPx1, phospholipase A2 (A), and glutathione synthetase (B). (C) Scheme showing the gene number for phospholipase A2, GPx, and glutathione synthetase in cod, turbot, and tongue sole. Red dots: turbot genes. Green triangles: tandem repeated genes. DE, duplication event. The green arrow indicates the dependence on temperature for optimal growth.
3.6. Genetic architecture of SD, growth, and resistance to diseases
QTL related to SD, resistance to diseases and growth were previously reported in turbot.6–10 We used the turbot genome and the previously reported QTL to investigate the genomic architecture of these traits (Supplementary Tables S13–S17; Fig. 5).
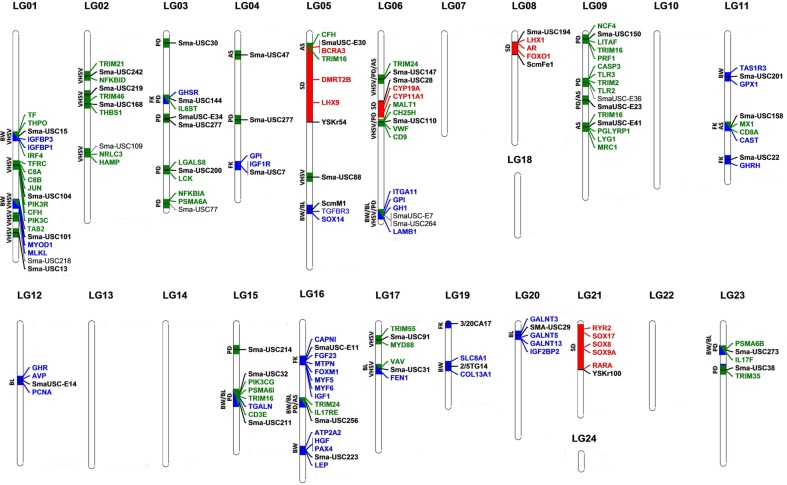
Figure 5.
Genetic architecture (QTL and candidate genes) of sex determination (red; SD), growth (blue; BW: body weight; BL: length; FK: Fulton's Factor) and resistance to diseases (green; VHSV, virus of the hemorrhagic septicaemia; AS, A. salmonicida; PD, P. dicentrarchi) in turbot (S. maximus).
Sex determination behaves like a complex trait and shows a high evolutionary turnover in fish.30 A major SD region at LG5 and another three minor ones at LG6, LG8, and LG21 were previously reported in turbot7 and data suggest that the turbot SD region is of recent origin.24 Understanding SD is relevant for turbot industry because females largely outgrow males.
We selected 186 candidate genes involved in sexual differentiation, and, with a broader perspective, in reproduction, to be mapped in the turbot genome and predictively located in the genetic map (Supplementary Table S14A; Fig. 5). Only one, stra8 (stimulated by retinoic acid gene 8), was not found in the turbot genome. In higher vertebrates, meiosis is triggered by the induction of stra8 through the retinoic acid pathway.31 Within fish, stra8 has been identified in catfish (Silus meridionalis), but not in model fish genomes. A stra8-independent signalling pathway, also based on retinoic acid, has been proposed to regulate meiotic initiation in those teleosts,31 and it could also be functioning in turbot.
The highest concentration of candidate genes was detected at LGs where the SD-QTL had been reported. Some of the genes mined around these QTL (Supplementary Table S15A), like lhx9, ar, and members of the sox family (sox8a, sox9a, and sox17), would deserve special attention because they have been specifically related to SD or early gonad development in other fish species.32–34 In particular, sox9, which plays a key role at the initial steps of testis differentiation, has been suggested as SD in brill (Scophthalmus rombus), a close relative to turbot.35
Growth is under complex genetic control and influenced by environmental, metabolic, and physiological factors.36,37 Growth traits constitute the main target of breeding programmes in fish38 and have been related to adaptive variation in wild populations of turbot.3,39 Growth-associated QTL markers6,10 (Supplementary Table S13B) and 208 selected candidate genes from fish and vertebrates36,37 were mapped in the turbot genome, and most of them predictively located in the turbot genetic map (85%) (Supplementary Table S14B; Fig. 5). A main cluster was detected at LG16, where several growth-related QTL had been reported.6 Some genes located within a major Fulton's factor (FK)-QTL at LG16 (myf5, myf6, and igf1) were previously used for growth-assisted selection in livestock and other aquaculture species.36,40 This region has also been associated with body length (BL) in turbot,5 body weight (BW) in brill (S. rhombus),41 and FK and BW in salmonids.42,43 Other genes (Supplementary Tables S14B and S15B) and functions related to regulation of muscle development and growth were found at specific LGs (Supplementary File 1: Results).
Disease control represents one of the main challenges for turbot aquaculture, and variation at immune genes has also been invoked to explain genetic differentiation of wild populations of turbot.44 Most genes involved in the major vertebrate immune response were identified in the turbot genome. The crucial role of Toll-like receptors (TLRs) in the innate immune response is well known.45 Teleost TLR families share common properties with their mammalian counterparts, but they vary largely among fish species.46 A complete repertoire of bacterial and viral TLR receptors were identified in the turbot genome (Supplementary File 1: Fig. S8). Nevertheless, the mammalian orthologues of tlr4, tlr6, and tlr10 were not identified, as occurs in the other teleost species. In spite of tlr4 and its accessory molecules (cd14 and md2) being missing in the turbot genome, the acute-phase LPS Binding Protein (lbp) was successfully identified. In zebrafish, belonging to the only known fish group containing tlr4a and tlr4b (cyprinids), these genes do not seem to be the functional receptors of LPS,47 unlike in mammals, and therefore the mechanism underlying LPS recognition in fish remains unclear and is an interesting question for future investigations.
Previous QTL screening for resistance and survival to a bacterium, Aeromonas salmonicida (AS), a parasite, Philasterides dicentrarchi (PD), and a virus, haemorrhagic septicaemia virus revealed specific genomic regions explaining a significant proportion of trait variance,8–10 thus representing a reference to tackle their genetic basis. Relevant immune genes were detected at QTL related to the studied diseases (Supplementary File 1: Results, Table S14C; Fig. 5), and some of them identified at QTL associated with different pathogens (Supplementary Table S15C), which suggests a general role in immune response. Trim16 (at LG5 and LG9) and trim24 (at LG6 and LG16) were detected in AS- and PD-resistance QTL. Recent studies have associated the tripartite motif family with resistance to the infectious salmon anaemia virus in Atlantic salmon (Salmo salar).48 Genes related to the phosphoinositide 3-kinase pathway were identified in VHS- (pik3r, pik3c, adcy1; LG1) and PD- (pik3cg, adcy3; LG23) QTL. Recently, five genes belonging to this pathway were found to be associated with resistance to Flavobacterium columnare in catfish.49 Another interesting point is the high representation of proteasome and ubiquitin-related proteins in QTL related to resistance to the three pathogens [e.g. three copies of the proteasome subunit α type-6 (psma6) at several QTL], suggesting a potential role of the ubiquitin-proteasome system in the resistance, likely through the antigen presentation process.
4. Discussion
The turbot (S. maximus, Scophthalmidae) belongs to the order Pleuronectiformes, a group of fish with notable adaptations to demersal life and great commercial value. The turbot genome presented here (×219 sequence coverage; 95% of estimated genome size in scaffolds) represents one of the highest quality draft genomes among aquaculture species, very similar to that reported for tongue sole (C. semilaevis, Cynoglossidae; ×212).1 The estimated genome size of turbot (568 Mb) is very close to our previous estimation (600 Mb)50 and also very similar to that of tongue sole (male: 495 Mb; female: 545 Mb). Flatfish genomes are among the most compact fish genomes, and accordingly, the percentage of TEs (∼5%) was slightly higher than pufferfish (∼3%), the smallest reported vertebrate genome (∼400 Mb).20
Important genomic resources have also been reported in other farmed flatfish species of different families: halibut (Hipoglossus hipoglossus, Pleuronectidae), Japanese flounder (Paralichthys olivaceus; Paralichthyidae), and Senegalese and common soles (Solea senegalensis and S. solea; Soleidae), including genetic maps of medium–high-density applied for QTL screening, comprehensive transcriptomic and miRNA databases applied for gene expression evaluation, and ongoing whole genome projects.51 Pleuronectiformes represents along with Salmoniformes the fish order with the most genomic resources among aquaculture fish. The integration of all these data involving five of the principal flatfish families will help to improve production as well as facilitate the investigation of flatfish origin and adaptation to demersal life.
Flatfish suffer a drastic environmental change from the pelagic larval stage up to juvenile and adult stages, living in the sea bottom at progressively higher depth. In addition to the particular metamorphosis changes from the bilateral symmetry to flat morphology common to all flatfish, other adaptations related to a new prey-spectrum, low light conditions, and a different microbiota environment need to be faced, and little information exists on the particular strategy developed by each flatfish species or family.
Teleosts show a wide range of visual adaptations related to the wide environmental diversity they face. The whole genome duplication, which occurred early in the evolution of ray-finned fish, provided raw genetic material to refine gene functions. Hence, many teleost families have specialized their duplicated vision-related genes to adapt to their particular environment.52 Functional specialization of paralogues has occurred especially within the green-sensitive opsins related to low light environments.53 After the metamorphosis of Pleuronectiform fish, environmental light substantially changes from shorter to longer wavelengths. Although further research should be done, our data suggest an improved vision of turbot to adapt to benthic life supported by tandem gene duplications of some vision-related genes. The detected green-sensitive opsin duplications may be adjusted to different wavelengths as reported in tilapia53 or zebrafish54 to refine turbot vision in the low light conditions of the sea bottom. The presence of spectrally distinct visual pigments that provide diverse visual sensitivities, and the differential ontogenetic expression of the opsin genes in several fish species53,55,56 suggest the adaptation to different habitats during the fish life cycle. Additionally, the presence of multiple crystalline genes could improve the eye structure itself enhancing vision accuracy. These observations contrast with the decayed visual system reported in tongue sole suggested by the absence or pseudogenization of several crystalline genes and the absence of the green-sensitive opsin duplications observed in turbot.1 In tongue sole, adaptation to benthic life seems to have occurred by enhancing other sensorial alternatives, like the increased lateral-line sensitivity and species-specific sensory papillae.1 Genomic expansions have been reported in other genome projects relating to adaptation to particular lifestyles, as the expansion of stress-related genes in the oyster genome57 or osmo-regulatory genes in the sea bass genome.25
Another important difference of turbot with tongue sole is related to the metabolic machinery involved in preventing oxidation of PUFA membrane lipids, which shows signals of gene expansion and positive selection in turbot. As increasing PUFA is the main adaptive response of poikilotherms to environmental cooling, these differences may be related to the colder and wider temperature range environment of turbot. While the optimal growth temperature of tongue sole is 22°C,58 it is 18°C for the turbot. Furthermore, turbot exhibits a high growth rate between 13 and 20°C,59 thus showing better adaptation not only to lower temperatures but also to temperature fluctuations. The same happens with cod, which, like turbot, harbours several copies of pla2 and gpx1, and lives at temperatures ranging from −1 to 20°C.60 Important changes necessary for adaptation to benthic lifestyle should be shared by Pleuronectiformes, especially if a monophyletic origin is true but, at the same time, a fast adaptive radiation occurring ∼40 MYA may have contributed to the specific evolutionary history of each family or species.
We also investigated the genomic architecture of three main productive traits (SD, growth, and resistance to pathogens) that are essential for understanding adaptation of turbot populations to environmental variation.3,39 The three can be considered complex traits but with notable architectural differences. While major genetic effects (SD master genes) and high heritabilities are involved in SD,30 moderate heritabilities and more widespread, although sometimes important, genetic effects have been documented for BW and length, and usually low heritabilities and widespread minor genetic factors for disease resistance traits, although with notable exceptions.38
Nearly, all selected candidate genes for the three investigated traits (>1,000) were found in the turbot genome and most of them predictively mapped. Only two genes, one related to meiosis triggering (stra8) and another one to a key immune pathway (tlr4) were missing. This observation is consistent with similar information in other teleosts which indicates alternative pathways to cover these functions and highlights the functional diversification of teleosts.
An important dispersion of candidate genes and QTL was detected in the turbot genome for the three traits in accordance with their complex condition. However, relevant gene clusters were identified for the three traits associated to specific QTL. These regions were also detected in other fish and vertebrate species, supporting their relevance and trans-specific conservation.4 Also, some genes or gene families were associated with resistance QTL for different pathogens, evidencing their generalist component, and making them essential in order to get more robust broodstock through marker-assisted selection programmes. Finally, processes related to growth, immunity, and gonadal differentiation overlapped at the same genomic regions, denoting genetic correlations between different traits, either positive or negative, an issue of major productive importance.
5. Conclusion
The genome sequence of turbot (S. maximus) we report here represents an important contribution to understand the organization and evolution of fish genomes. This genome is among those with highest coverage within fish, similar to that of the tongue sole, and among the most compact ones within vertebrates. The integration of the turbot genetic and physical maps and their comparison with model fish genomes provide powerful tools for future studies in turbot, fish and vertebrates. Our in silico analysis of the turbot genome has permitted to advance interesting hypotheses on flatfish adaptation and diversification to cope with demersal life challenges. Finally, this genome has shed light on the genetic basis of relevant productive traits, and represents a milestone for boosting turbot production through more efficient marker-assisted selection programmes.
Data availability
Sequencing data are available in the ENA with the following study accession: PRJEB11743.
Authors’ contributions
A.F. and P.M. organized the work and built up the consortium. T.A. established the strategy of genome sequencing assembly and annotation and wrote the corresponding section. The bioinformatic analysis of that section was done by T.A., A.C., J.G.G., M.G., and I.G.G., B.G., and J.L.G. generated the fosmid libraries. X.M. designed and wrote the work on repetitive elements with the collaboration of X.B. and J.L.A.F. M.H., C.B., and P.M. designed the comparative mapping analysis, which was developed by M.H., C.F., and B.G.P. M.H. and P.M. wrote the corresponding section of the manuscript. D.R., A.G.T., J.A.A.D., and P.M. designed and developed the work on gene orthology and paralogy following the similarity approach. D.R. wrote the corresponding part of the manuscript. R.G., A.H.P., and A.V. performed the work on functional annotation and wrote the corresponding section. T.G. designed and wrote the phylogenomics section, according to the work developed by L.C. and M.M.H. J.A.R. carried out the work and wrote the section on adaptation to demersal life in collaboration with D.R. and P.M. C.B., A.V., A.F., and B.N. designed the work related to the genetic architecture of productive traits. The growth section was developed by C.B., D.R., and C.F., and written by C.B. Sex determination was developed and written by A.V., X.T., and D.R. Resistance to diseases was developed and written by P.P., G.F.C., A.F., and B.N. P.M. wrote common sections in collaboration with C.B. and A.F., and integrated all the sections of the manuscript. All the authors approved the final version of the manuscript.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was funded by the Spanish Government: projects Consolider Ingenio: Aquagenomics (CSD2007-00002) and Metagenoma de la Península Ibérica (CSD2007-00005), Ministerio de Economía y Competitividad and European Regional Development Funds (AGL2012-35904), and Ministerio de Economía y Competitividad (AGL2014-51773 and AGL2014-57065-R); and Local Government Xunta de Galicia (GRC2014/010). P.P. and D.R. gratefully acknowledge the Spanish Ministerio de Educación for their FPU fellowships (AP2010-2408, AP2012-0254). Funding to pay the Open Access publication charges for this article was provided by the Ministerio de Economía y Competitividad (AGL2014-51773) and Xunta de Galicia (GRC2014/010).
Supplementary Material
References
- 1.Chen S., Zhang G., Shao C. et al. 2013, Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle, Nat. Genet., 46, 253–60. [DOI] [PubMed] [Google Scholar]
- 2.Campbell M.A., Chen W.J., López J.A.. 2014, Molecular data do not provide unambiguous support for the monophyly of flatfishes (Pleuronectiformes): a reply to Betancur-R and Orti, Mol. Phyl. Evol., 75, 149–53. [DOI] [PubMed] [Google Scholar]
- 3.Vandamme S.G., Maes G.E., Raeymaekers J.A. et al. 2014, Regional environmental pressure influences population differentiation in turbot (Scophthalmus maximus), Mol. Ecol., 23, 618–36. [DOI] [PubMed] [Google Scholar]
- 4.Mackay T.F.C. 2001, The genetic architecture of quantitative traits, Annu. Rev. Genet., 35, 303–39. [DOI] [PubMed] [Google Scholar]
- 5.Ruan X., Wang W., Kong J., Yu F., Huang X.. 2010, Genetic linkage mapping of turbot (Scophthalmus maximus L.) using microsatellite markers and its application in QTL analysis, Aquaculture, 308, 89–100. [Google Scholar]
- 6.Sánchez-Molano E., Cerna A., Toro M.A. et al. 2011, Detection of growth-related QTL in turbot (Scophthalmus maximus), BMC Genomics, 12, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez P., Bouza C., Hermida M. et al. 2009, Identification of the major sex-determining region of turbot (Scophthalmus maximus), Genetics, 183, 1443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodríguez-Ramilo S.T., Toro M.A., Bouza C. et al. 2011, QTL detection for Aeromonas salmonicida resistance related traits in turbot (Scophthalmus maximus), BMC Genomics, 12, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodríguez-Ramilo S.T., Fernández J., Toro M.A. et al. 2013, Uncovering QTL for resistance and survival time to Philasterides dicentrarchi in turbot (Scophthalmus maximus), Anim. Genet., 44, 149–57. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez-Ramilo S.T., de la Herrán R., Ruiz-Rejón C. et al. 2014, Identification of quantitative trait loci associated with resistance to viral haemorrhagic septicaemia (VHS) in turbot (Scophthalmus maximus): a comparison between bacterium, parasite and virus diseases, Mar. Biotechnol., 16, 265–76. [DOI] [PubMed] [Google Scholar]
- 11.Pereiro P., Balseiro P., Romero A. et al. 2012, High-throughput sequence analysis of turbot (Scophthalmus maximus) transcriptome using 454-pyrosequencing for the discovery of antiviral immune genes, PLoS ONE, 7, e35369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribas L., Pardo B.G., Fernández C. et al. 2013, A combined strategy involving Sanger and 454 pyrosequencing increases genomic resources to aid in the management of reproduction, disease control and genetic selection in the turbot (Scophthalmus maximus), BMC Genomics, 14, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermida M., Bouza C., Fernández C. et al. 2013, Compilation of mapping resources in turbot (Scophthalmus maximus): a new integrated consensus genetic map, Aquaculture, 414-415, 19–23. [Google Scholar]
- 14.Martínez P., Hermida M., Pardo B.G. et al. 2008, Centromere-linkage in the turbot (Scophthalmus maximus) through half-tetrad analysis in diploid meiogynogenetics, Aquaculture, 280, 81–8. [Google Scholar]
- 15.Voorrips R.E. 2002, MapChart: software for the graphical presentation of linkage maps and QTLs, J. Hered., 93, 77–8. [DOI] [PubMed] [Google Scholar]
- 16.Krzywinski M., Schein J., Birol I. et al. 2009, Circos: an information aesthetic for comparative genomics, Genome Res., 19, 1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartolomé C., Bello X., Maside X.. 2009, Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes, Genome Biol., 10, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huerta-Cepas J., Capella-Gutierrez S., Pryszcz L.P. et al. 2011, PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions, Nucleic Acids Res., 39, D556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe K., Clark M.D., Torroja C.F. et al. 2013, The zebrafish reference genome sequence and its relationship to the human genome, Nature, 496, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aparicio S., Chapman J., Stupka E. et al. 2002, Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes, Science, 297, 1301–10. [DOI] [PubMed] [Google Scholar]
- 21.Taboada X., Pansonato-Alves J.C., Foresti F. et al. 2014, Consolidation of the genetic and cytogenetic maps of turbot (Scophthalmus maximus) using FISH with BAC clones, Chromosoma, 123, 281–91. [DOI] [PubMed] [Google Scholar]
- 22.Castro J., Pino A., Hermida M. et al. 2008, A microsatellite marker tool for parentage assessment in Senegal sole (Solea senegalensis): genotyping errors, null alleles and conformance to theoretical assumptions, Aquaculture, 261, 1194–203. [Google Scholar]
- 23.Broughton R.E., Betancur-R R., Li C., Arratia G., Ortí G.. 2013, Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution, PLoS Curr., doi:10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouza C., Hermida M., Pardo B.G. et al. 2012, An Expressed Sequence Tag (EST)-enriched genetic map of turbot (Scophthalmus maximus): a useful framework for comparative genomics across model and farmed teleosts, BMC Genet., 13, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tine M., Kuhl H., Gagnaire P.A. et al. 2014, European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation, Nat. Commun., 5, 5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama S. 2008, Evolution of dim-light and color vision pigments, Annu. Rev. Genomics Hum. Genet., 9, 259–82. [DOI] [PubMed] [Google Scholar]
- 27.Hazel J.R., Williams E.E.. 1990, The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment, Prog. Lipid Res., 29, 167–227. [DOI] [PubMed] [Google Scholar]
- 28.Sargent J.R., McEvoy L.A., Estevez A., Bell G.J., Bell M.V., Henderson R.J.. 1999, Lipid nutrition of marine fish during early development: current status and future directions, Aquaculture, 179, 217–9. [Google Scholar]
- 29.van Kuijk F.J., Handelman G.J., Dratz E.A.. 1985, Consecutive action of phospholipase A2 and glutathione peroxidase is required for reduction of phospholipid hydroperoxides and provides a convenient method to determine peroxide values in membranes, J. Free Radic. Biol. Med., 1, 421–7. [DOI] [PubMed] [Google Scholar]
- 30.Martínez P., Viñas A.M., Sánchez L., Díaz N., Ribas L., Piferrer F.. 2014, Genetic architecture of sex determination in fish: applications to sex ratio control in aquaculture, Front. Genet., 5, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng R., Fang L., Cheng Y. et al. 2015, Retinoic acid homeostasis through aldh1a2 and cyp26a1 mediates meiotic entry in Nile tilapia (Oreochromis niloticus), Sci. Rep., 5, 10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eshel O., Shirak A., Weller J.I., Hulata G., Ron M.. 2012, Linkage and physical mapping of sex region on LG23 of Nile tilapia (Oreochromis niloticus), G3: Genes Genom. Genet., 2, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takasawa K., Kashimada K., Pelosi E. et al. 2014, FOXL2 transcriptionally represses Sf1 expression by antagonizing WT1 during ovarian development in mice, FASEB J., 28, 2020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujii J., Kodama M., Oike A. et al. 2014, Involvement of androgen receptor in sex determination in an amphibian species, PLoS ONE, 9, e93655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taboada X., Hermida M., Pardo B.G., Viñas A., Bouza C., Martínez P.. 2014, Fine mapping and evolution of the sex determination region in turbot (Scophthalmus maximus), G3: Genes Genom. Genet., 4, 1871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Santis C., Jerry D.R.. 2007, Candidate growth genes in finfish—where should we be looking?, Aquaculture, 272, 22–38. [Google Scholar]
- 37.Johnston I.A., Bower N.I., Macqueen D.J.. 2011, Growth and the regulation of myotomal muscle mass in teleost fish, J. Exp. Biol., 214, 1617–28. [DOI] [PubMed] [Google Scholar]
- 38.Gjedrem T. 2005, Selection and Breeding Programs in Aquaculture, Springer, Dordrecht, The Netherlands. [Google Scholar]
- 39.Vilas R., Vandamme S.G., Vera M. et al. 2015, A genome scan for candidate genes involved in the adaptation of turbot (Scophthalmus maximus), Mar. Genomics, 23, 77–86. [DOI] [PubMed] [Google Scholar]
- 40.Tsai H.Y., Hamilton A., Guy D.R., Houston R.D.. 2014, Single nucleotide polymorphisms in the insulin-like growth factor 1 (IGF1) gene are associated with growth-related traits in farmed Atlantic salmon, Anim. Genet., 45, 709–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermida M., Rodríguez-Ramilo S.T., Hachero-Cruzado I. et al. 2014, First genetic linkage map for comparative mapping and QTL screening of brill (Scophthalmus rhombus), Aquaculture, 42,0-421, S111–20. [Google Scholar]
- 42.Moghadam H.K., Ferguson M.M., Rexroad C.E. III, Coulibaly I., Danzmann R.G.. 2007, Genomic organization of the IGF1, IGF2, MYF5, MYF6, and GRF/PACAP genes across Salmoninae genera, Anim. Genet., 38, 527–32. [DOI] [PubMed] [Google Scholar]
- 43.Wringe B.F., Devlin R.H., Ferguson M.M., Moghadam H.K., Sakhrani D., Danzmann R.G.. 2010, Growth-related quantitative trait loci in domestic and wild rainbow trout (Oncorhynchus mykiss), BMC Genet, 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vilas R., Bouza C., Vera M., Millán A., Martínez P.. 2010, Variation in anonymous and EST-microsatellites suggests adaptive population divergence in turbot, Mar. Ecol. Prog. Ser., 420, 231–9. [Google Scholar]
- 45.Kawai T., Akira S.. 2010, The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors, Nat. Immunol., 11, 373–84. [DOI] [PubMed] [Google Scholar]
- 46.Star B., Nederbragt A.J., Jentoft S. et al. 2012, The genome sequence of Atlantic cod reveals a unique immune system, Nature, 477, 207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan C., Charette J., Catchen J. et al. 2009, The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions, J. Immunol., 183, 5896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J., Boroevich K.A., Koop B.F., Davidson W.S.. 2011, Comparative genomics identifies candidate genes for infectious salmon anemia (ISA) resistance in Atlantic salmon (Salmo salar), Mar. Biotechnol., 13, 232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geng X., Sha J., Liu S. et al. 2015, A genome-wide association study in catfish reveals the presence of functional hubs of related genes within QTLs for columnaris disease resistance, BMC Genomics, 16, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuñado N., Terrones J., Sánchez L., Martínez P., Santos J.L.. 2002, Synaptonemal complex analysis in spermatocytes and oocytes of turbot, Scophthalmus maximus (Pisces, Scophthalmidae), Genome, 44, 1143–7. [DOI] [PubMed] [Google Scholar]
- 51.Cerdà J., Manchado M.. 2013, Advances in genomics for flatfish aquaculture, Genes Nutr., 8, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furutani-Seiki M., Wittbrodt J.. 2004, Medaka and zebrafish, an evolutionary twin study, Mech. Dev., 121, 629–37. [DOI] [PubMed] [Google Scholar]
- 53.Spady T.C., Parry J.W., Robinson P.R., Hunt D.M., Bowmaker J.K., Carleton K.L.. 2006, Evolution of the cichlid visual palette through ontogenetic subfunctionalization of the opsin gene arrays, Mol. Biol. Evol., 23, 1538–47. [DOI] [PubMed] [Google Scholar]
- 54.Chinen A., Hamaoka T., Yamada Y., Kawamura S.. 2003, Gene duplication and spectral diversification of cone visual pigments of zebrafish, Genetics, 163, 663–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortesi F., Musilová Z., Stieb S.M. et al. 2015, Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes, Proc. Natl Acad. Sci. USA, 112, 1493–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valen R., Edvardsen R.B., Søviknes A.M., Drivenes Ø., Helvik J.V.. 2014, Molecular evidence that only two opsin subfamilies, the blue light- (SWS2) and green light-sensitive (RH2), drive color vision in Atlantic cod (Gadus morhua), PLoS ONE, 9, e115436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang G., Fang X., Guo X. et al. 2012, The oyster genome reveals stress adaptation and complexity of shell formation, Nature, 490, 49–54. [DOI] [PubMed] [Google Scholar]
- 58.Fang J., Tian X., Dong S.. 2010, The influence of water temperature and ration on the growth, body composition and energy budget of tongue sole (Cynoglossus semilaevis), Aquaculture, 299, 106–14. [Google Scholar]
- 59.Imsland A., Foss A., Gunnarsson S. et al. 2001, The interaction of temperature and salinity on growth and food conversion in juvenile turbot (Scophthalmus maximus), Aquaculture, 198, 353–67. [Google Scholar]
- 60.Rose G.A., Atkinson B.A., Baird J., Bishop C.A., Kulka D.W.. 1994, Changes in the distribution of Atlantic cod and thermal variations in Newfoundland waters, 1980–1992, ICES Mar. Sci. Symp., 198, 542–52. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available in the ENA with the following study accession: PRJEB11743.