Abstract
The WRKY genes have been identified as important transcriptional modulators predominantly during the environmental stresses, but they also play critical role at various stages of plant life cycle. We report the identification of WRKY domain (WD)-encoding genes from galegoid clade legumes chickpea (Cicer arietinum L.) and barrel medic (Medicago truncatula). In total, 78 and 98 WD-encoding genes were found in chickpea and barrel medic, respectively. Comparative analysis suggests the presence of both conserved and unique WRKYs, and expansion of WRKY family in M. truncatula primarily by tandem duplication. Exclusively found in galegoid legumes, CaWRKY16 and its orthologues encode for a novel protein having a transmembrane and partial Exo70 domains flanking a group-III WD. Genomic region of galegoids, having CaWRKY16, is more dynamic when compared with millettioids. In onion cells, fused CaWRKY16-EYFP showed punctate fluorescent signals in cytoplasm. The chickpea WRKY group-III genes were further characterized for their transcript level modulation during pathogenic stress and treatments of abscisic acid, jasmonic acid, and salicylic acid (SA) by real-time PCR. Differential regulation of genes was observed during Ascochyta rabiei infection and SA treatment. Characterization of A. rabiei and SA inducible gene CaWRKY50 showed that it localizes to plant nucleus, binds to W-box, and have a C-terminal transactivation domain. Overexpression of CaWRKY50 in tobacco plants resulted in early flowering and senescence. The in-depth comparative account presented here for two legume WRKY genes will be of great utility in hastening functional characterization of crop legume WRKYs and will also help in characterization of Exo70Js.
Keywords: Ascochyta rabiei, Exo70, WRKY domain
1. Introduction
Plants have evolved a huge-array of transcription factors (TFs) to modulate transcription of genes in response to the environmental signals. Plants have >6% of genes encoding for TFs.1 WRKY genes comprise one such TF family that has mainly expanded and diversified in higher plants. WRKY TFs are characterized by a highly conserved WRKY domain (WD) of ∼60 amino acids that preferably bind to W-box besides other DNA motifs.2 The WRKY TFs derived their name from the heptad motif sequence ‘WRKYGQK’ of WRKY DNA-binding domain. WRKY proteins are broadly divided into three groups based on the number of WDs and the type of zinc-finger motif. The group-I has two WDs with C2H2-type zinc-finger motif, group-II has one WD with C2H2-type zinc-finger motif, and group-III has one WD with C2HC-type zinc-finger motif.3 After the identification of the first WRKY gene from sweet potato4 and recognition of WRKY family in Arabidopsis,3 WRKY genes have been identified from most of the genome or transcriptome sequenced plants. Earlier, WRKY genes were considered exclusive to higher plants, nevertheless sequencing have showed their presence in many lower plants and non-plant organisms.5
The main role of WRKY TFs has been deciphered with regard to biotic stress associated processes.6 However, they are also recognized as important components of abiotic stress signalling. The WRKY proteins play a role in the antagonistic interaction of SA- and jasmonic acid (JA)-mediated signalling7 and convergence of JA and auxin signalling.8 They are also the major targets of various perturbation-activated MAPK cascades.9 Nevertheless, individual studies have shown their involvement in various stages of plant development. The AtWRKY23 gene assists in auxin distribution during root development by controlling flavonol biosynthesis,10 OsWRKY11 controls flowering time and plant height,11 and some WRKY genes are involved in secondary metabolism.12 WRKY proteins play some interesting roles also; AtWRKY40 is recruited on the host plant transformed Agrobacterium tumefaciens T-DNA for Ipt gene expression13 and AtWRKY6 restricts arsenate uptake and transposon activation.14 However, WRKY proteins function in partnership with various protein families for fine-tuned roles.9,15
Chickpea (Cicer arietinum L.) is the second largest cultivated legume crop. Debilitating fungal diseases Ascochyta blight and Fusarium wilt along with the chewing insect Helicoverpa armigera are major concerns for chickpea production.16 The WRKY TF genes, being important regulators in biotic and abiotic stress, could be utilized to improve chickpea and related legumes as reported for soybean WRKYs.17 The crop legume chickpea is closely related to model legume barrel medic (Medicago truncatula). Moreover, the availability of mutants in M. truncatula and Lotus japonicus will colossally help in functional characterization of genes. We report the identification and comparative analysis of 78 WRKYs from C. arietinum and 98 from M. truncatula. Some novel WRKY genes have been identified from legumes. Here we demonstrate phylogeny, expression, and localization study data of one such unique chimeric protein CaWRKY16. Comparative analysis of a selected region from the galegoid clade and millettioid clade legumes showed that two unique WRKY genes evolved locally due to two independent events. In addition, we have generated expression profiles of group-III WRKY genes in response to hormone treatments and Ascochyta rabiei infection. The tissue-specific expression profile of these genes is also reported. Further, we characterized a salicylic acid (SA)- and A. rabiei-induced WRKY gene with regard to W-box-binding activity, subcellular localization, transcription modulation potential, and overexpression in tobacco plants. Therefore, this study will form the basis of functional characterization of legume WRKYs and will be a foundation study for developing stress-tolerant legume crops.
2. Materials and methods
2.1. Identification of WRKY genes from C. arietinum and M. truncatula
A preliminary search for the chickpea WRKY proteins was performed on the annotated proteins of two different chickpea varieties (BioProject PRJNA175619 for ‘CDC Frontier’ and PRJNA78951 for ‘ICC4958’) available at NCBI.18,19 The C. arietinum, M. truncatula (Mt4.0v1), and A. thaliana WRKY proteins were further used as query in tblastn to find WD-encoding regions from the genome and transcriptome of ‘CDC Frontier’ and ‘ICC4958’ varieties. We also searched the predicted WRKY genes on other sources like chickpea transcriptome database (CTDB) (http://www.nipgr.res.in/ctdb.html) and http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi. Manual corrections with regard to open-reading frame (ORF) were also performed on the annotated WRKY genes of chickpea. The M. truncatula-annotated proteins were derived from http://jcvi.org/medicago/. The proteins that were not annotated as WRKY proteins in Mt4.0v1, despite the presence of WD, were manually checked and re-annotated as WRKY genes.
2.2. Exon–intron structure and phylogenetic analysis
The exon–intron organization of WRKY genes was manually checked and the ‘GT’ splice donor and ‘AG’ acceptor sites were verified. The exon–intron structure was generated using online tool Gene Structure Display Server (GSDS 2.0; http://gsds.cbi.pku.edu.cn). The largest transcript isoform of ‘CDC Frontier’ variety WRKYs was used for exon–intron display. The multiple sequence alignment of C. arietinum and M. truncatula WRKY proteins was executed using PROMALS3D (http://prodata.swmed.edu/promals3d/promals3d.php) and MUSCLE. The phylogenetic tree was constructed by maximum-likelihood method in MEGA 6.06. The branch confidence values were obtained by bootstrapping with 1000 iterations. The multiple sequence alignment for WDs of C. arietinum and A. thaliana proteins was generated using MUSCLE in MEGA 6.06.
2.3. Domain and motif identification
The SMART (http://smart.embl-heidelberg.de/) and Pfam (http://pfam.sanger.ac.uk/search) were used to predict and verify the functional domains. MEME (Multiple Expectation Maximization for Motif Elicitation) program 4.11.1 available at http://meme-suite.org/tools/meme was used to predict the potential motifs in WRKY proteins. The transmembrane (TM) helices were predicted by the use of TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
2.4. Plant materials and treatments
To grow C. arietinum variety ‘Pusa 362’ plants, four seeds were sown in each plastic pot filled with sterile agropet and vermiculite mix. They were maintained in greenhouse at 22 ± 2°C and 3-week-old healthy plants of similar phenotypes were used for spray inoculation of freshly collected A. rabiei spores, 5 mM SA, 100 µM JA, and 100 µM abscisic acid (ABA) to the individual experimental plants. The control plants were sprayed either with water or water–ethanol mix. After regular time-intervals the aerial tissue was harvested randomly. Samples were immediately frozen in liquid nitrogen and kept in −80°C for long-term storage.
2.5. RACE (random amplification of cDNA ends)
The cDNA end sequences of selected CaWRKY genes were determined by 5′- and 3′-RACE using the SMARTer™ RACE cDNA Amplification Kit (Clontech) as per the instruction manual. The gene-specific primers used for amplification are listed in Supplementary Table S1.
2.6. RNA isolation and real-time PCR analysis
Total RNA was extracted from the frozen chickpea tissue using TRIzol reagent (Life Technologies). This RNA was treated with RQ1 RNase-free DNase I (Promega, Madison, WI, USA). The first-strand cDNA synthesis was performed by oligo(dT) priming of ∼2 μg RNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies). The PCR was set in a 96 well-plate using the SYBR® Green qPCR Master Mixes (Agilent). The gene-specific primers used for RT–PCR are listed in Supplementary Table S1. The PCR was carried out on Applied Biosystems® Real-Time PCR systems using following conditions: initial denaturation at 95°C for 1 min, 40 cycles of denaturation at 95°C for 15 s, and annealing and extension at 58–61°C for 30 s. The relative expression values were calculated using β-tubulin as an internal standard.
2.7. Yeast one-hybrid analysis
The full-length ORF of CaWRKY50 was cloned in pGBKT7 vector along with three C-terminal deletion constructs and one N-terminal deletion construct. The transactivation assay was performed using yeast strain AH109 (Clontech). The β-galactosidase assay was performed as per the instructions of yeast protocol manual (Clontech). Yeast one-hybrid was performed for W-box-binding assay. The ORF of CaWRKY50 was cloned in pGADT7 vector and W-boxes were cloned in pHis2.1 using Pchn5F1/Pchn5R1 and mPchn5F1/mPchn5R1 oligonucleotide pairs. The W-box binding was checked by comparing the growth of respective clones co-transformed yeast strain Y187 cells on SD/−Leu/−Trp and SD/−Leu/−Trp/−His/+20 mM 3-AT.
2.8. Overexpression of CaWRKY50 in tobacco
The CaWRKY50 was cloned under CaMV35S promoter by replacing the uidA gene of pBI121. The obtained clone was mobilized into A. tumefaciens LBA4404. The axenically grown Nicotiana tabacum cv. Xanthi plant leaves were used for transformation. The oligonucleotide pairs of CaM35SeqF/WRKYRTR, NPTIIF/NPTIIR and WRKYRTF/pBI121R2 were used to confirm transgenic tobacco plants and expression of CaWRKY50 in tobacco.
2.9. Measurement of total chlorophyll content
The tobacco leaf senescence was assessed by measuring the total chlorophyll content of PCR confirmed transgenic 19-week-old plants along with the wild-type plants of same age. The two leaves from each plant's lowest stem axis whirl were used. They were weighed and crushed in liquid nitrogen. The chlorophyll extraction was performed using 80% acetone. The total chlorophyll amount was calculated as described by Poora.20
2.10. DNA methylation analysis
Total genomic DNA was extracted from aerial tissues of chickpea variety Pusa 362 using DNeasy® Plant mini kit (Qiagen). One microgram of this genomic DNA was treated with bisulfite using EpiTect® Bisulfite kit (Qiagen) along with an equal amount of a PCR amplified fragment as control to check the conversion efficiency. The primers, used for CaWRKY16 amplification using the bisulfite-treated DNA as template, are listed in Supplementary Table S1. The PCR products were cloned in pJET1.2 vector and sequenced. At least 10 clones, for each position of CaWRKY16's selected genomic region, were sequenced, and data were analysed online by Kismeth software (http://katahdin.mssm.edu/kismeth/revpage.pl).
3. Results
3.1. Identification of C. arietinum and M. truncatula WD-encoding genes
A simple search of proteins annotated as ‘WRKY TF’ in the C. arietinum cv. ‘CDC frontier’ protein sequences revealed 81 genes. Two of these genes were partial and four were non-WRKY genes. We further explored both Desi and Kabuli chickpea sequenced genomes to mine the WRKY genes. The presence of a WD was taken as criteria to call a gene as C. arietinum WRKY (CaWRKY). The recently released ‘ICC 4958’ CDS v2.0 was also used to check WRKY genes.21 However, of the 103 CDS annotated as WRKY, only 68 CDS had WD. It resulted in identification of 82 consensus regions among the two C. arietinum varieties. These 82 genomic regions were further checked for the expression by analysing the CTDB database (http://www.nipgr.res.in/ctdb.html), the 454 cDNA reads derived from A. rabiei inoculated aerial chickpea tissue (Kumar et al., unpublished) and chickpea RNA-seq data from NCBI. After manual inspection, four entries were removed from 82 consensus regions. Thus, in chickpea genome, 78 genes were identified to be encoding for full-length WD. The coordinates and orientations of CaWRKY genes on ‘CDC Frontier’ chromosome assembly were obtained. The CaWRKY genes were assigned numbers from CaWRKY1-70 as per their order and position on the chromosomes from top to bottom (Supplementary Fig. S1). The unplaced eight genes were named from CaWRKY71 to CaWRKY78 as per the order of scaffold contig identifiers. We checked 10 genes for full-length by RACE and 20 genes were checked for ORF amplification with 100% success rate. Further in silico analysis of CaWRKYs was performed only for ‘CDC frontier’ WRKY genes.
The closest legume to C. arietinum among the sequenced genomes is M. truncatula. Since the limited study of M. truncatula WRKY (MtWRKY) genes,22 many new genes have been annotated in Mt4.0. Hence, we decided to compare CaWRKY genes with that of M. truncatula. The M. truncatula (Mt4.0) has 111 predicted genes of WRKY gene family. Comparison with CaWRKYs revealed that few MtWRKY genes need re-annotation. The Medtr2g075680 annotated as cysteine-tRNA ligase has WD and thus included as MtWRKY gene while Medtr3g085710, Medtr6g013150, Medtr7g022360, and Medtr7g028350 are not WRKYs. Many other genes lack full-length WD while for Medtr1g077760, Medtr5g018380, and Medtr7g105000 the exon–intron junctions were corrected to get full WDs. The adjacent gene model pairs of Medtr7g104980 and Medtr7g105000 was collapsed into a single gene Medtr7g105000 with a proper WD. Thus, the MtWRKY genes encoding for WD got reduced to 98.
3.2. Phylogenetic comparison of WRKY proteins
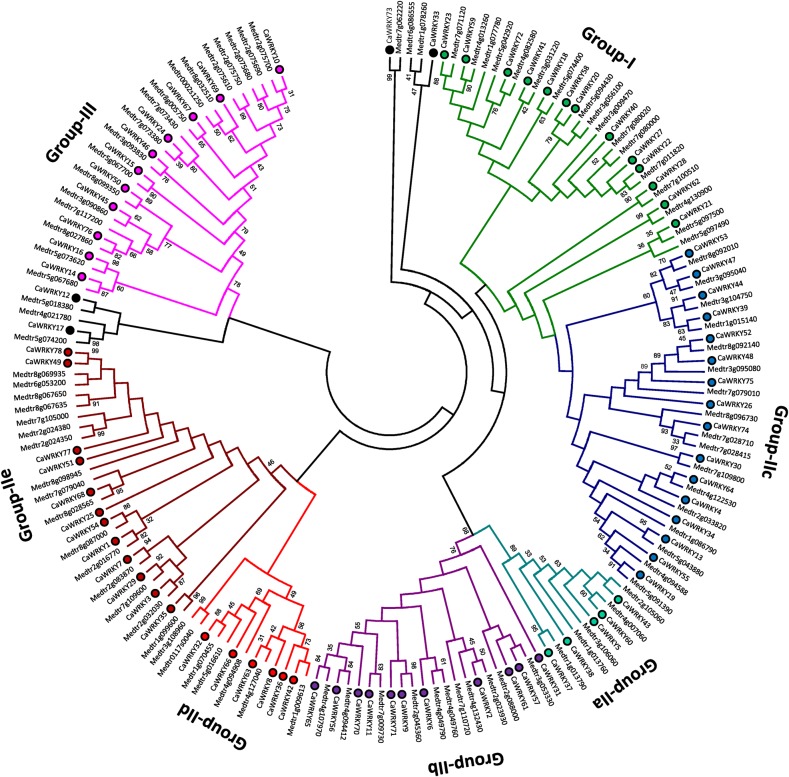
The division of WRKY proteins into three groups was earlier proposed and it was based on phylogeny and WD structure.3,23 A robust phylogeny requires a good sequence alignment. The diversity outside the WD is more prevalent; therefore, the PROMALS3D was used to align the full-length WRKY proteins. It resulted in better alignment outside the WD and reduced dependency on manual adjustments. Among the identified WRKYs of C. arietinum and M. truncatula, four proteins have chimeric sequences (CaWRKY16, CaWRKY17, Medtr5g073620, and Medtr5g074200). Therefore, only the WD sequences of these four proteins were considered for phylogenetic analysis. The phylogenetic tree clearly divided WRKY proteins into three groups: I, II, and III (Fig. 1). This phylogeny clearly showed evolutionary history of WRKY genes and re-confirmed a recent report on the evolution of WRKY genes in flowering plants.5 The group-II was complex and large; hence, it was further sub-divided into group-IIa to -IIe as done with Arabidopsis WRKYs. The group-IIc was very close to group-I while others were clearly separate (Fig. 1). This division was also supported by the number of WDs and the type of zinc-finger motif in WDs. Thus, in chickpea, group-I, -IIa, -IIb, -IIc, -IId, -IIe, and -III have 13, 5, 11, 16, 6, 12, and 11 members while barrel medic have 16, 5, 11, 18, 7, 16, and 18 members, respectively.
Figure 1.
Maximum-likelihood phylogeny of C. arietinum and M. truncatula WRKY proteins. CaWRKYs are highlighted with a filled circle against their names. The alignment was performed on PROMALS3D server, and phylogeny was constructed using MEGA6.06. Bootstrap values were derived from 1000 iterations and bootstrap values >30 are represented. The group names are indicated against deep branches. This figure is available in black and white in print and in colour at DNA Research online.
Along with clear division of most WRKY proteins, few exceptions were also present. The CaWRKY17, Medtr4g021780, Medtr5g018380, and Medtr5g074200 proteins belong to group-II based on WD structure, but these proteins clustered along with the group-III members in both neighbour-joining and maximum-likelihood methods. Another exception was CaWRKY12 with two unusual WDs, but it clubbed with group-III in phylogeny. Similar observation was for CaWRKY33, CaWRKY73, Medtr1g077780, Medtr1g078260, Medtr6g086555, and Medtr7g062220 encoded proteins. These proteins were unfit for group-I as they lack two WDs (Fig. 1; Supplementary Tables S2 and S3). These genes thus must have evolved from group-I in these legumes. The apparent expansion of MtWRKY genes when compared with CaWRKY mainly happened in group-IIe and -III. In other groups, minor differences have appeared.
3.3. WRKY gene structure and physical distribution on chromosomes
The apparent comparison of chickpea and Medicago WRKY orthologues position on the chromosomes was in agreement with the broader comparison of these two genomes.18 All assembled chickpea chromosomes except Ca08 contain WRKYs (Supplementary Fig. S1). The lack of CaWRKY on Ca08 may be attributed to partial assembly of it in ‘CDC Frontier’ when compared with Mt06. In chickpea and barrel medic, orientation of genes on chromosomes is relatively same in both directions. In chickpea, 34 CaWRKYs are in the top-to-bottom direction among the 70 genes placed on seven chromosomes. The most conserved group-I occupies four chromosomes of M. truncatula, i.e. Mt03, Mt04, Mt05, and Mt07, and group-I CaWRKYs also occupies the corresponding chromosomes of chickpea (Supplementary Fig. S1). The expansion of MtWRKY genes by tandem and segmental duplications with respect to chickpea can also be clearly visualized. We considered tandem-duplicated pairs only when genes within 200 kb genomic region belong to the same group. Chickpea CaWRKYs had four such pairs (CaWRKY14-15, CaWRKY37-38, CaWRKY47-48, and CaWRKY52-53) while M. truncatula had 11 such pairs (Supplementary Tables S2 and S3). The tandem-duplicated in barrel medic are Medtr2g075610 to Medtr2g075750 while segmental duplications are in Mt02 and Mt08 chromosomes. Thus, it explains the presence of more WRKY members in M. truncatula. In these two legumes, we established orthology for 70 pairs of genes based on homology and syntenic region around them.
The exon–intron structure of a gene family can provide additional evidences to support the phylogeny. All the WRKYs of these two legumes had intron(s) except CaWRKY17, CaWRKY49, and Medtr5g074200. The CaWRKY49 looks partial when compared with its closest CaWRKY78. The number of introns varies in WRKYs of these two legumes from one to seven. However, few genes like Medtr2g105060, Medtr8g098945, CaWRKY5, CaWRKY51, and CaWRKY61 have absence of intron in their WD. A preliminary look at the genomic structure of barrel medic and chickpea WRKYs showed high conservation at C-terminal, a region that encodes for WD (Fig. 2). The increase in size of WRKY genes has taken place towards the N-terminal of WD-encoding regions. The intron phase was two for the entire group-I C-terminal WD introns. The intron phase was also two for group-IIc, -IId, -IIe, and -III WD introns while it was zero for group-IIa and -IIb WD introns. The intron phases were same in almost all the related introns within a WRKY group (Fig. 2). The position of intron in the WD among a group is also conserved. The ‘PR’ type intron was present in group-I C-terminal WDs and group-IIc, -IId, -IIe, and -III while the WD of group-IIa and -IIb had ‘VQR’ type intron.
Figure 2.
Gene structure of M. truncatula and C. arietinum WRKY genes. For each gene, the boxes represent exons and black lines connecting them are introns. The shaded boxes in some exons of a gene represent WD-encoding regions. The N-terminal and C-terminal WD-encoding exonic regions of group-I are highlighted with different shades. The numbers 0, 1, and 2 at the start of each intron indicate its phase. The introns marked with symbol (//) were reduced in size to adjust image. This figure is available in black and white in print and in colour at DNA Research online.
3.4. Characteristics of WD and motifs
In a WD-specific phylogeny, group-I WDs separated and the remaining WDs clubbed together as per their respective group (Supplementary Fig. S2). We compared the conservation of WD in each WRKY group and found some group-specific sequence variants in both legumes (Supplementary Fig. S3). Many variants of ‘WRKYGQK’ signature-sequence were present in the WRKYs of both legumes (Supplementary Tables S2 and S3). The SMART and pfam servers revealed the presence of four domains outside the WDs. In group-IId WRKYs, a plant zinc-cluster domain (PF10533) was present before the WDs. A coiled-coil region was also predicted at the N-terminal end of CaWRKY62, CaWRKY64, CaWRKY65, and their MtWRKY orthologues. In CaWRKY16 and Medtr5g073620, a TM, an Exo70, and a WD were predicted while a TM along with a WD was present in CaWRKY17 and Medtr5g074200. A partial DUF3664 (pfam12406) was identified between SP cluster and N-terminal WD of CaWRKY22, and a partial merozoite surface domain (pfam07133) was present in CaWRKY56 at N-terminal. The motifs were also inspected in WRKYs by MEME tool and are represented as motifs of C. arietinum (MotifCa) and motifs of M. truncatula (MotifMt) followed by motif number. In chickpea, out of the 12 motifs identified, 4 (MotifCa1, 2, 3, and 5) were within the WD (conserved DNA-binding structure of ∼60 amino acids) (Supplementary Fig. S4). Interesting among chickpea motifs were MotifCa8 and MotifCa9. The MotifCa8 has leucine at regular intervals and may act like a leucine zipper, probably responsible for dimerization. It was present in 15 CaWRKYs belonging to group-IIa, group-IIb (except CaWRKY56 and CaWRKY70), and a group-III member, CaWRKY16. The motif corresponding to MotifCa8 in barrel medic was MotifMt8 (Supplementary Figs S4 and S5). The MotifCa9 and MotifMt10, present in group-I members only, were similar to the SP-cluster and D domain.9 In chickpea, MotifCa6 and MotifCa7 were present downstream to WD and were alanine-rich. Also interesting was MotifCa11 with consensus sequence of PTxTLD[LF]T after the WD of group-IIb proteins. In M. truncatula, out of the 12 motifs identified 5 (MotifMt1–5) were within WDs (Supplementary Fig. S5). The remaining motifs were of unknown nature and in future after functional characterization may be associated with specific roles.
3.5. CaWRKY16 and CaWRKY17 are unique genes
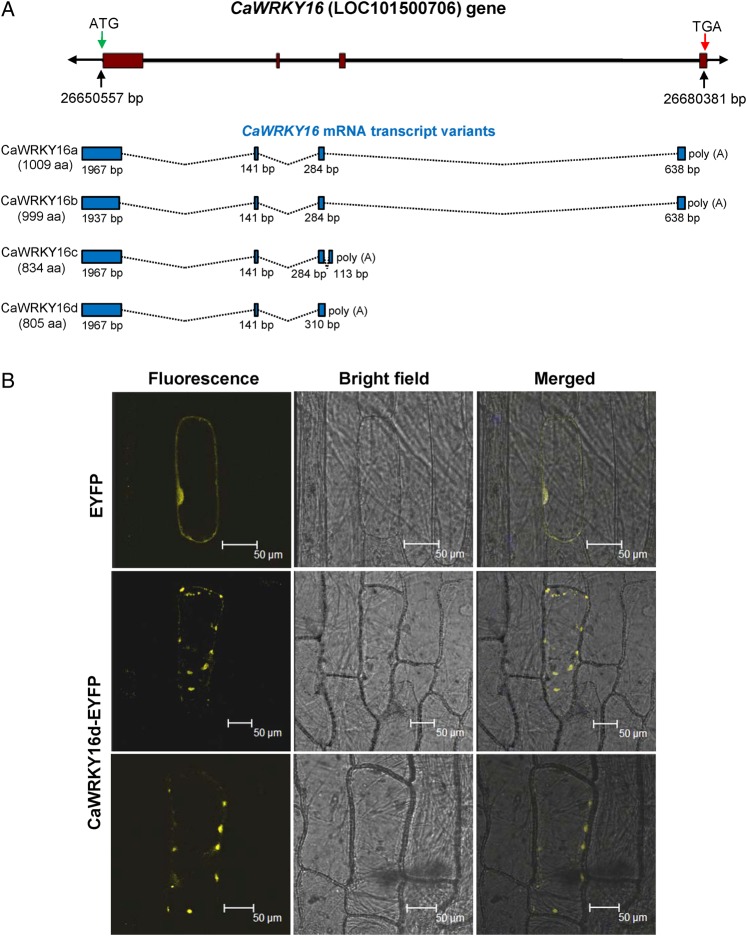
Among functionally characterized WRKY genes, AtWRKY52 (RRS1/SLH1) and AtWRKY16 (RRS1B) have unique genomic arrangement.24 Some unique Golgi apparatus localized WRKY and Exo70J proteins have been reported from legumes.25 The Golgi-targeting of these proteins is by the virtue of an N-terminal TM domain. The chickpea orthologue of GmWRP1 (Glyma14g199800) is CaWRKY17 (LOC101503578). Residing in the nearby genomic region of chickpea is another unique gene CaWRKY16 (LOC101500706) that encodes for a protein with N-terminal TM domain along with a group-III WD (Fig. 1; Supplementary Fig. S2). The WD of CaWRKY16 is flanked by partial Exo70 domains and has an N-terminal TM domain just like GmWRP1 and CaWRKY17 (Supplementary Fig. S6). The orthologues of CaWRKY16 are present in M. truncatula (Medtr5g073620), L. japonicus (Lj2g3v2314850.1), and Pisum sativum (PsCam035857_1_AA).26 However, CaWRKY16-like gene is absent in transcriptome/genome of millettioid clade legumes soybean and common bean. We were unable to amplify the annotated ORFs of CaWRKY16a and CaWRKY16b in our experiments, although in RNA-seq data the expression is known (Fig. 3A). Thus, we decided to perform 3′ RACE of CaWRKY16 using forward primers from the annotated first and second exons. Results indicated that CaWRKY16 has got an alternative polyadenylation after the predicted third exon. However, transcript isoform that we amplified was different from CaWRKY16c and therefore we named it CaWRKY16d (Fig. 3A). The shorter isoform CaWRKY16d encodes for a protein with an N-terminal TM domain followed by a partial Exo70 and a C-terminal WD. The C-terminal EYFP-decorated CaWRKY16d protein showed punctate localization in onion cells (Fig. 3B). This could be possibly the Golgi apparatus, as TM domain at N-terminal is responsible for the similar localization of GmWRP125 and TM domain in these proteins is highly conserved (Supplementary Fig. S6). Thus, our study demonstrates the presence of a unique WRKY gene in galegoid clade legumes.
Figure 3.
CaWRKY16 gene structure, transcribed mRNA isoforms, and subcellular localization. (A) CaWRKY16 genomic structure and transcribed mRNA transcript isoforms. The black line indicates introns and the filled boxes are exons. In the gene structure, position of CaWRKY16 on Ca2 chromosome of ‘CDC Frontier’ is mentioned in base pairs. Size of the protein encoded by each transcript is shown in parentheses below name of isoforms. The 5′ and 3′ UTRs are excluded in the terminal exons of mRNA transcript structures. The size of each exon in a transcript is mentioned below the exon. The transcripts CaWRKY16c and CaWRKY16d are products of alternative cleavage and polyadenylation. (B) CaWRKY16d-EYFP fusion protein showed punctate fluorescence in onion cells. Bars = 50 µm. This figure is available in black and white in print and in colour at DNA Research online.
3.6. Expression profile of group-III CaWRKYs in A. rabiei stress and hormone signalling
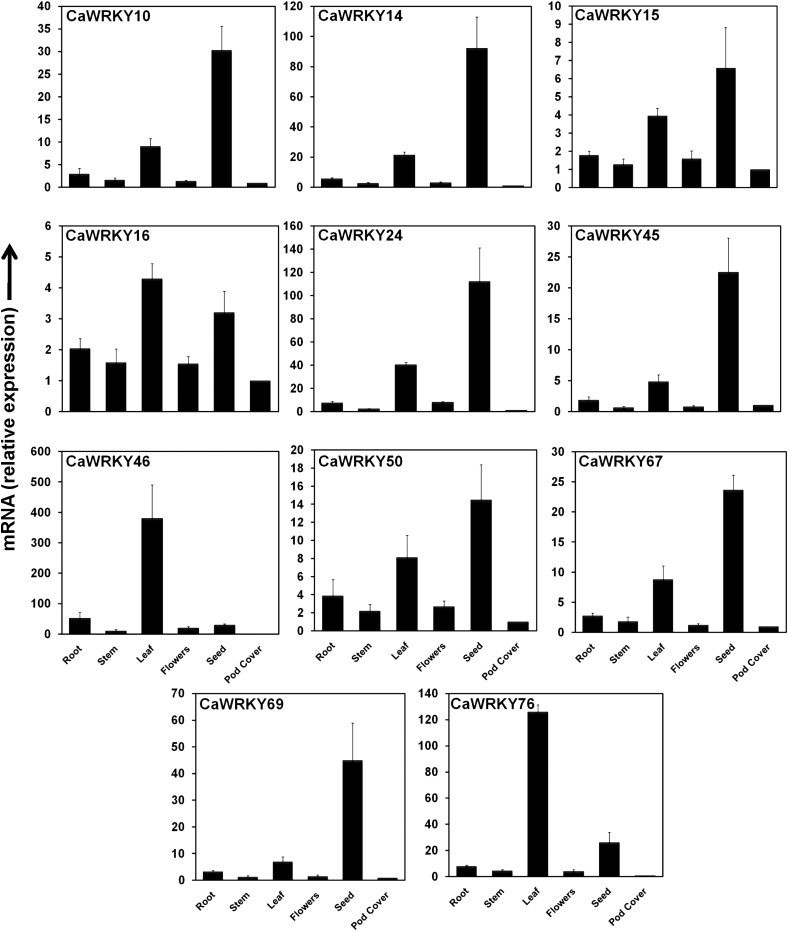
Group-III WRKY members are interesting for the aspects that they have a completely different WD zinc-finger, enormously expanded in rice and unique genes CaWRKY16 and AtWRKY52 belongs to this group. Chickpea has 11 members in this group (Fig. 1). To gain preliminary insight into the function of these chickpea WRKY genes, we investigated their transcript level using real-time PCR during the activation of stress signalling by SA, JA, and ABA treatments and A. rabiei spore inoculation (Fig. 4). Broadly they showed induction in SA and A. rabiei spore treatments. The steady high state in SA was maintained by CaWRKY16, CaWRKY46, and CaWRKY67. After 0.5 h of SA treatments, CaWRKY46 and CaWRKY50 achieved highest level. A set of group-III genes, i.e. CaWRKY14, CaWRKY45, and CaWRKY76, responded to ABA treatment also but late when compared with SA. CaWRKY45 and CaWRKY76 that responded to ABA at 24 h were the genes whose expression level steadily went down after initial induction during SA. In ABA treatment, CaWRKY14 responded very early and at higher levels. This gene along with CaWRKY76 was biphasic in ABA treatment. No significant induction was seen in the JA treatment while CaWRKY50, CaWRKY67, and CaWRKY69 got slightly down-regulated with respect to the control. This trend is consistent with the findings that in A. thaliana SA and JA signalling works antagonistically over many genes. In chickpea, infected with A. rabiei spores, the genes responded at different time intervals. High levels of transcript were maintained by CaWRKY14 and CaWRY67 at all time points with respect to control while CaWRKY45 and CaWRKY69 were biphasic. High level of initial induction (6 h after A. rabiei spore spray) of WRKYs must be due to non-specific fungal elicitors or pathogen-associated molecular patterns (PAMPs) that are released during spore germination and development.
Figure 4.
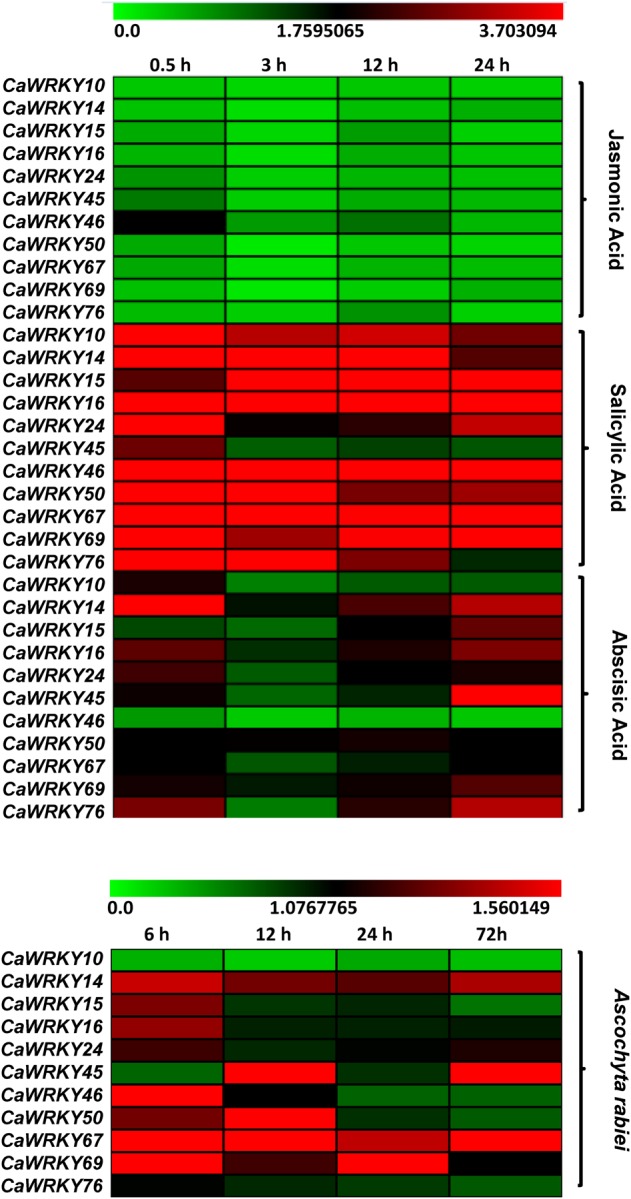
Expression profiles of C. arietinum group-III WRKY genes under hormone treatments and A. rabiei infection. Relative transcript expression values when compared with the controls were determined by qRT–PCR. The mean expression values of three biological with three technical replicates were used for representation with Mev 4.9. The expression scale is same for ABA, JA, and SA whereas for A. rabiei infection it is different. This figure is available in black and white in print and in colour at DNA Research online.
3.7. Expression pattern of group-III CaWRKYs in different tissues
The expression analysis of genes can apparently give preliminary idea about their functions. We analysed the transcript level expression of group-III CaWRKY genes under normal growth conditions in six different vegetative and reproductive tissues (root, stem, leaves, flowers, seed and pod cover) by real-time PCR (Fig. 5). The expression of group-III genes was observed in all developmental stages of plants grown under normal conditions. However, the pod cover was found to accumulate least concentration and thus was treated as control for relative expression. Interestingly, there was a high expression level for 10 genes in seed tissue, although differences in expression level existed within them. After the seed tissue, the expression level in leaves was high for the same 10 genes. Five genes (CaWRKY10, CaWRKY14, CaWRKY24, CaWRKY45, and CaWRKY67) showed preferential expression in two tissues (seed and leaf). The level of expression for CaWRKY76 was high in leaves when compared with other tissues. The genes CaWRKY15, CaWRKY16, and CaWRKY50 were expressed ubiquitously in all tissues. In root tissue, CaWRKY10, CaWRKY46, and CaWRKY50 showed considerable expression, albeit lesser than leaf or seed tissues. Among the WRKYs expressed in leaves, the expression of CaWRKY24 and CaWRKY46 was exceptionally high. Therefore, expression level quantified here suggests that some members of group-III must have tissue-specific roles.
Figure 5.
Expression analysis of group-III WRKY genes in different tissues of C. arietinum. The samples of chickpea were collected from 4-week-old plants for the root, stem, and leaf tissues. Bloomed flowers were sampled at reproductive stage while pod cover and seeds were collected at the same time points. The mRNA level expression was analysed by qRT–PCR using chickpea β-tubulin gene as internal control. The relative mRNA level was calculated with respect to the pod cover. The results presented here were obtained from three biological replicates with three technical replicates each. The error bars represent ±SD of means.
3.8. CaWRKY50 is a W-box-binding transcriptional activator
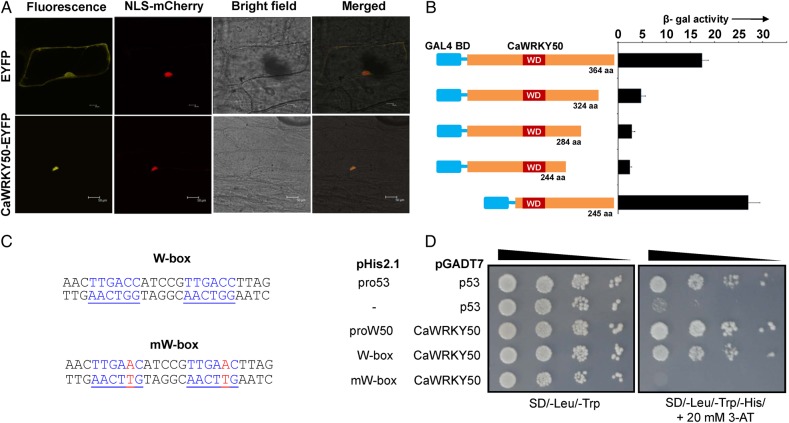
Many group-III CaWRKYs were induced at transcript level during the hormone and A. rabiei treatments, and CaWRKY50 was one of them. It was originally isolated as partial EST in suppression subtractive hybridization library screening of A. rabiei-induced chickpea genes.27 The full-length cDNA sequence was identified through RACE and submitted as the first full-length WRKY of chickpea (NCBI accession: EU049488). Thus, we further focused on characterization of SA and A. rabiei-induced CaWRKY50. The WRKY protein, being a transcription regulator, needs to localize to plant nucleus and bind regulatory elements of target genes. The CaWRKY50-EYFP fusion protein localizes in nucleus of onion cells (Fig. 6A). To further investigate whether CaWRKY50 has capability to localize to the eukaryotic nucleus by itself, we used a yeast nuclear import assay.28 In yeast strain L40, a fusion protein of mutated bacterial LexA (mLexA) TF and CaWRKY50 localizes into yeast nucleus and activates the reporter gene LacZ (Supplementary Fig. S7). Thus, these two results suggest that CaWRKY50 can enter eukaryotic nucleus and resides in plant cell nucleus.
Figure 6.
CaWRKY50 subcellular localization in plant cell and yeast one-hybrid assays. (A) CaWRKY50-EYFP fusion protein localizes to onion cell nucleus. The CaWRKY50-EYFP fusion and NLS-mCherry nuclear marker genes were transiently expressed in onion cells and visualized after 2 days. Bar is 20 µm for EYFP panel and 50 µm for CaWRKY50-EYFP panel. (B) CaWRKY50 transactivates reporter gene by its C-terminal region in yeast one-hybrid system. Full-length and four truncated forms of CaWRKY50 were fused with GAL4 DNA-binding domain in pGBKT7 vector. Yeast transformation and β-gal activity (in miller units) were quantified to check the strength of transcativation. (C) Sequences of pChn5 oligonucleotides cloned in pHis2.1 vector, underlined are two W-boxes and their mutated mW-box forms. (D) Yeast one-hybrid analysis of CaWRKY50 against its own 1.2 kb promoter (proW50) and pChn5 W-boxes. Positive (pHis2.1-pro53 + pGADT7-p53) and negative (pHis2.1 + pGADT7-p53; pHis2.1-mW-box + pGADT7-CaWRKY50) controls are also included. The growth of yeast cells co-transformed with positive controls, proW50, and W-box clones suggests interaction of CaWRKY50 with own promoter and W-box. This figure is available in black and white in print and in colour at DNA Research online.
Sequences outside WD(s) vary among members of WRKY group thus providing functional diversity and unique characters. We used yeast one-hybrid system to check the transcription activation and W-box-binding activity of CaWRKY50. The Gal4BD-CaWRKY50 fusion protein can activate LacZ reporter in the yeast cells. The β-gal activity comparison among the yeast cells transformed with various C-terminal truncations of CaWRKY50 showed that the transcription activation activity mainly resides in 40 amino acids of C-terminal end. However, it was interesting to note that a single N-terminal deletion results in more β-gal activity than the full-length CaWRKY50 for unknown reasons (Fig. 6B). To check whether CaWRKY50 can bind to W-box in yeast, we cloned a fragment of a chitinase promoter having two W-boxes, along with mutated W-boxes as negative control (Fig. 6C). The promoter region of CaWRKY50 was also used as a separate clone in this experiment. The growth of yeast on auxotrophic selection (SD/−Leu/−Trp/+20 mM 3-AT) clearly showed that CaWRKY50 binds to W-box in a sequence-specific manner and its own promoter (Fig. 6D). Thus, from these experiments we inferred that CaWRKY50 is a plant nuclear localized transcriptional activator and it can bind to W-box.
3.9. Overexpression of CaWRKY50 gene in tobacco
Transformation of some legume species for functional genomics is a challenging task. Therefore, to investigate the role of CaWRKY50 in plant growth and development, we decided to transform it in tobacco. However, during transformation process, the number of shoots regenerated from calli was very less using CaWRKY50 overexpression construct compared with vector control (pBI121). Shoot regeneration in CaWRKY50 construct became comparable to vector control after increased supplementation of cytokinin in media. It suggests that ectopic overexpression of CaWRKY50 in tobacco plant interferes with hormone signalling.
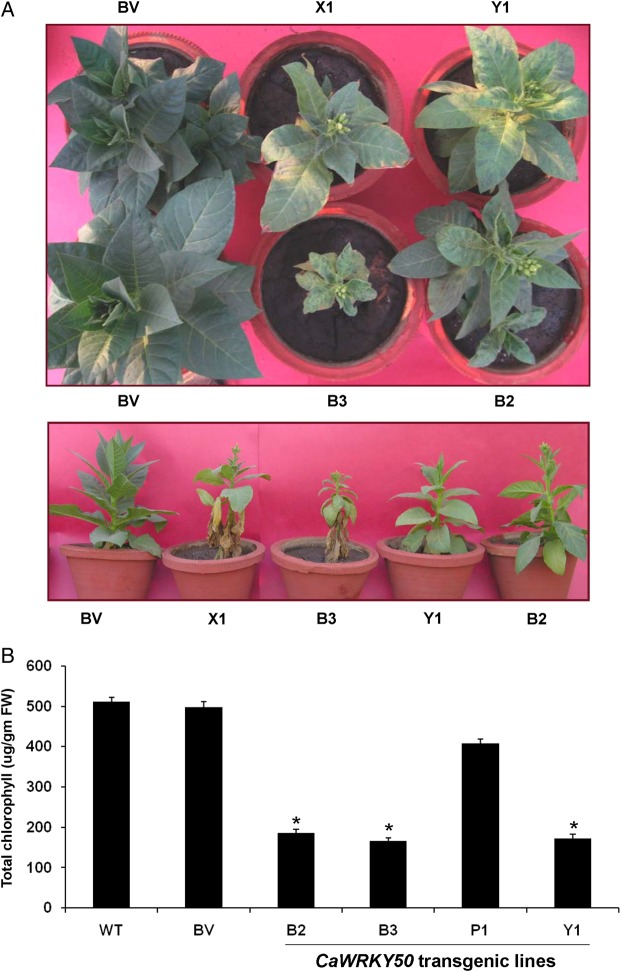
Most of the PCR-positive CaWRKY50 transgenic tobacco plants showed phenotypic differences when compared with the vector control plants, during hardening and seed setting under greenhouse conditions. The tobacco plants overexpressing CaWRKY50 were dwarf with chlorotic lesions on leaves and matured early, i.e. early flowering (Fig. 7A). The extent of these phenotypes correlated with the expression level of CaWRKY50 transcript, i.e. more in overexpression lines B2 and B3 (Supplementary Fig. S8A). The appearance of similar phenotypes in early generations of AtWRKY53 overexpressing Arabidopsis plants that disappeared in subsequent generations has been described in earlier report.29 Such lesions on plant leaves are often associated with oxidative stress. Thus, the leaves of tobacco (T0 transgenic) lines with chlorotic lesions were analysed for the presence of H2O2. The 3,3′-Diaminobenzidine staining clearly showed H2O2 accumulation in chlorotic areas of high CaWRKY50 expressing B3 and Y1 lines when compared with the low CaWRKY50 expressing P1 line, vector control (BV), and wild-type leaves (Supplementary Fig. S8B). For subsequent analysis, five transgenic lines B2, B3, P1, and Y1 with varying level of CaWRKY50 transcripts along with BV line were selected. However, in the subsequent generations (T1 and T2), dwarfism and chlorotic lesions gradually disappeared (Supplementary Fig. S9) despite the expression of CaWRKY50 but the early flowering and senescence phenotypes were regularly observed. We utilized the T2 transgenic plants to check the developmental effects of CaWRKY50 ectopic expression in tobacco. We observed that flower buds in B2, B3, and Y1 transgenic lines (T2 generation) appeared 21 ± 5 days prior to BV line and wild type. However, in the P1 line, flower buds appeared 10 ± 3 days prior to BV line and wild-type plants. We measured early senescence phenotype on leaves of T2 transgenic plants by estimating total chlorophyll content from the lower stem axis leaves of tobacco as previously reported in Arabidopsis.29 The early senescence phenotype showed a positive correlation with the expression of CaWRKY50 in tobacco. The lower leaves of CaWRKY50 overexpressing B2, B3, and Y1 line tobacco plant had 2.5–3 times less chlorophyll when compared with the wild-type and vector control plants of the same age (Fig. 7B). Thus, our results clearly demonstrated that CaWRKY50 overexpression in tobacco results in early senescence and early maturation (appearance of flower buds). This corroborates the studies on its Arabidopsis orthologue AtWRKY53, a master regulator of age-induced leaf senescence.
Figure 7.
Overexpression of CaWRKY50 in tobacco leads to early flowering and senescence. (A) The top and side view of early flowering and senescence phenotype in representative CaWRKY50 overexpressing T0 lines (B2, B3, X1 and Y1) with respect to pBI121 vector transformed controls (BV). The dwarf phenotype with chlorotic lesions on leaves seen in T0 plants were not retained in T2 generation (B). The senescence phenotype was quantified at 19th week after germination by measuring the total chlorophyll content of T2 generation line (B2, B3, P1, and Y1), BV, and wild-type tobacco leaves. The total chlorophyll content correlates with the level of CaWRKY50 expression in transgenic lines. The data presented here were derived from the means of three biological replicates with each replicate having a minimum of 10 plants for each line. Error bars represent ±SD of means. *Indicates significant difference (P < 0.001) when compared with wild type or blank vector control. This figure is available in black and white in print and in colour at DNA Research online.
4. Discussion
4.1. WRKY genes in C. arietinum and M. truncatula
Identification and functional characterization of WRKY genes in plants gained pace with the advent of next-generation sequencing technology. We identified 78 and 98 WRKYs in C. arietinum and M. truncatula, respectively. Number of WRKY genes is more in chickpea when compared with L. japonica (61) while they are very less with regard to palaeopolyploid soybean (182). However, M. truncatula has evolved more WRKYs than C. arietinum mainly by tandem duplication in contrast to soybean where the expansion was due to segmental duplication.30 Number of WRKYs in chickpea is close to that of A. thaliana (72). Some novel WRKY genes in both legumes were difficult to be grouped, with confidence, in three known WRKY groups and thus remained unclassified (Supplementary Tables S2 and S3). An identified feature of WDs is the divergence from conserved sequence ‘WRKYGQK’. But, these differences in the heptad sequence became relevant after reports that an amino acid in WD heptad sequence is acetylated by bacterial effector for virulence.31 We identified many variants of this WD conserved motif in chickpea and barrel medic (Supplementary Tables S2 and S3). The importance of these variants is also highlighted by the facts that NtWRKY12 having a divergent heptad ‘WRKYGKK’ binds to a different motif (TTTTCCAC)32 and variants of soybean GmWRKY6 and GmWRKY21 do not bind normally.17 Tools are now available to decipher the DNA motifs occupied by these variants with reduced cost of ChiP-Seq. Therefore, future studies may find new motifs to which these proteins bind. In general, the conserved group-I N-terminal WD has zinc-finger consensus of Cx4Cx22HxH, but in CaWRKY41, it is Cx4Cx23HxH due to a triplet repeat-induced change. Thus, CaWRKY41's N-terminal WD may also bind to DNA. It is expected that the local interaction with pathogens and environment may have influenced expression, sequence divergence, and co-evolving interacting partners. Thus, functional studies will be of worth use in these evolved WRKY genes.
4.2. WRKY genes and retrotransposon elements
Transposons are known to modulate arrangement of genes in genomes, and sometimes species-specific amplification of transposons gives unique features to particular species. While analysing orthologues of CaWRKYs in M. truncatula, we also matched the surrounding genomic regions of up to three genes to analyse the synteny around WRKY genes between these two legumes. We were surprised to find transposon remnants, tandem duplication events, and GDSL-motif encoding genes wherever synteny breaks. The remnants of transposons were found around the 31 CaWRKY genes mostly belonging to group-II and -III. Thus, evolution of WRKY and their surrounding regions is influenced by transposons.
Based on our comparative analysis, we speculated that evolution of CaWRKY16-like genes was by the action of transposons. Presence of transposon can alter the expression level of nearby genes and can modulate transcript isoforms, if present in introns. Such cases exist for wheat,33 A. thaliana RPP7 gene,34 and oil palm.35 The CaWRKY16 has three predicted transcript isoforms and we cloned the fourth CaWRKY16d isoform (Fig. 3A). The CaWRKY16c and CaWRKY16d isoforms are the products of alternative cleavage and polyadenylation possibly due to the presence of an inactive Ty1-copia retrotransposon sequences. Sequence with homology to UBN2 superfamily protein is present ∼100 bp downstream to CaWRKY16d transcript cleavage site. The presence of transposon sequences can modulate the methylation status of genomic DNA, may add new cis-elements to be recognized by cleavage factors, and can influence nucleosome positioning. All these changes can contribute to alternative cleavage and polyadenylation.36 We analysed DNA methylation status of ∼1.6 kb region around the CaWRKY16c and CaWRKY16d isoforms cleavage and polyadenylation region. The common exonic region of CaWRKY16c and CaWRKY16d isoforms has higher percentage of GC content, but it showed only ∼50% methylation of all three types (Supplementary Fig. S10). The region downstream to the cleavage site has more fraction of methylated DNA although percentage GC is less. Thus, there is differential methylation at DNA sequences of exons, introns, and region after cleavage site, which also harbour transposon sequences (Supplementary Fig. S10).
4.3. Group-III WRKY gene expression
The WRKY genes are mainly characterized with respect to plant stresses. In accordance with this, the transcript level of many WRKY genes gets modulated during stresses.6 We checked barrel medic's group-III WRKYs expression in online available data.37 However, probe set IDs of nine WRKY members were not found including a set of five genes (Medtr2g075610–Medtr2g075750) that show signatures of tandem duplication within a genomic region of ∼50 kb. More data are available from the experiments related to root symbiosis, an area highly explored in this model legume. In barrel medic, Medtr7g073380's expression was high in most of the tissues and Medtr3g093830 maintained higher levels in leaf and root. Interesting was the higher expression of Medtr8g005750 in root hairs in response to nod factors and Sinorhizobium meliloti 1021 while Medtr3g093830 also responded to these signals. Only Medtr5g073620 and Medtr7g117200 were induced 3 weeks after mycorrhizal treatment while remaining six genes got down-regulated. The down-regulation of these genes could be related to the suppression of defence responses. Such type of down-regulation is seen during symbiosis and establishment of biotrophic lifestyle on host plants by pathogens. An Exo70I protein has recently been shown to be required for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis.38 Hence, the analysis of Medtr5g073620 (encoding for a novel Exo70) mutants could reveal its novel function during symbiosis. Six genes except Medtr5g073620 and Medtr8g005750 responded to salt stress. Analysis of group-III WRKY genes in A. thaliana has earlier revealed that their expression is independent of phylogeny. In chickpea and barrel medic also, this seems to be true as group-III orthologues express differently. The CaWRKY14-15 tandem-duplicated genes responded to A. rabiei and SA treatments but at different level. In a set of duplicated WRKY gene, Medtr7g073430 probably evolved its role during nodulation as it gets expressed higher during nodulation. Analysis of 1.5 kb promoter region's cis-acting elements in chickpea and barrel medic failed to reveal reasons for the tissue-related expression. However, the genes of chickpea responding to SA and A. rabiei were having at least four W-boxes and two TGA TF-binding sites similar to the barrel medic four elicitor responsive genes (Medtr3g090860, Medtr3g093830, Medtr7g117200, and Medtr8g099350). A resemblance was found in chickpea and barrel medic with respect to expression of WRKYs upon JA treatment. They were either down-regulated or did not respond significantly to JA treatment. This could be due to the TGA factors that regulate antagonistic nature of SA and JA signalling through NPR1.39 The early response of some WRKY genes upon A. rabiei spore inoculation is most likely due to the PAMP-activated signalling. The WRKY genes are in fact part of PAMP-activated signalling by being the downstream targets of MAPKs.9 A systemic analysis of the promoter will reveal the regulatory aspect of group-III WRKY genes during stress signalling or may help in identification of some novel elicitor responsive elements.
4.4. CaWRKY16 gene evolved in galegoid clade
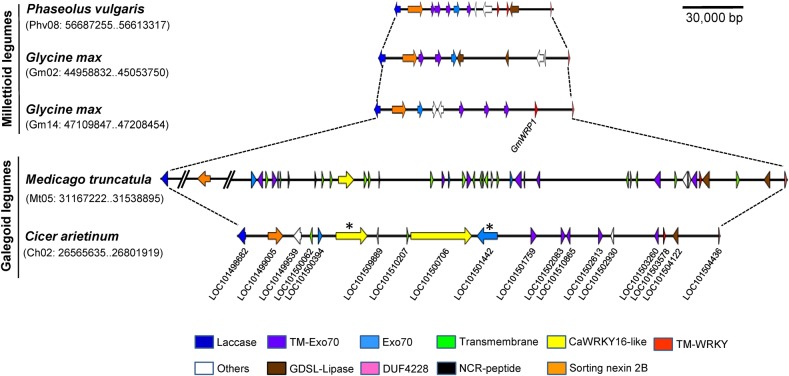
Unique proteins with a Golgi-targeting TM domain were reported recently from soybean.25 The other domains in these soybean proteins with Golgi-targeting TM domain are Exo70 (in seven GmExo70Js) and WRKY (in GmWRP1). We have reported here another unique gene CaWRKY16 that is located in same genomic region that havens legume-specific TM domain-encoding genes and chickpea orthologue of GmWRP1 i.e. CaWRKY17 (Fig. 8). Presence of two unique WRKY genes with different WDs within ∼127 kb genomic region of chickpea Ca02 suggests towards their independent origin. CaWRKY17 gene evolved in a progenitor of legumes prior to the diversification of millettioid and galegoid clade legumes as its orthologues are present in both clade members (Fig. 8) and also in Arachis duranensis (http://peanutbase.org). Based on the fact that CaWRKY16-like gene is absent in soybean and common bean transcriptome/genomic sequences while it is present in four analysed galegoid legumes, we propose that CaWRKY16-like genes must have evolved in galegoids after separation of these legumes from millettioid legumes.
Figure 8.
Annotated genes of Phaseolus vulgaris (Ch08), Glycine max (Ch02 and Ch14), M. truncatula (Ch05), and C. arietinum (Ch02) are represented as arrow or triangle. Base pair coordinates of the genomic regions on assembled chromosomes are indicated in parenthesis to the left of each genomic segment. Orthologue genes at ends of genomic segment are connected by dotted lines. Genes marked with asterisk have stop codons in C. arietinum. In M. truncatula, two unique segments flanking sorting nexin 2B gene were removed to adjust the whole region. The size of genomic region and gene are up to the scale mentioned. This figure is available in black and white in print and in colour at DNA Research online.
We have discussed here the localized evolution of CaWRKY16 and CaWRKY17-like genes in a specific genomic region of legumes. The unique WRKY gene evolution hypothesis presented here is backed by comparing phylogenies of WDs (Supplementary Fig. S2) and Exo70 proteins (Supplementary Fig. S11). We also compared genomic region among barrel medic, chickpea, common bean, and soybean to support our hypothesis on CaWRKY16 evolution. An apparent look at these genomic regions shows that this region in barrel medic region is highly dynamic with many events of tandem duplication and presence of NCR-peptide encoding genes (Fig. 8). Phylogeny revealed that WD of CaWRKY16 is close to CaWRKY14's WD while WD of CaWRKY17 is close to the N-terminal WD of CaWRKY18 (Supplementary Fig. S2). As these four genes are placed in a nearby genomic region, CaWRKY16 and CaWRKY17-like genes must have evolved by the local events in this variable region of legumes. CaWRKY17 possibly evolved from a TM domain-encoding gene that gained WD by partial duplication of CaWRKY18 N-terminal WD or by an event of unequal crossing-over between them. Such events of unequal crossing-over within the tandem-duplicated genes can generate new genes.40 The evolution of CaWRKY16 seems to be complex and possibly involves the action of transposing elements as two large introns in CaWRKY16 showed homology with inactive copia-type retrotransposons. The association of WRKY genes with transposons is already known in regard to zinc-finger motifs of WD.41 The introns of CaWRKY16-orthologue from L. japonicus are small, which suggest a possible transposon excision event in Lj2g3v2314850.1. Thus, the two WD-encoding exons may have appeared between an Exo70J gene by the action of transposons, which carried WD exons along with them. Later, these transposons got inactivated in chickpea while in barrel medic and L. japonicus they got excised. Soybean Exo70J gene (LOC100816208) is closest to CaWRKY16 (Supplementary Fig. S11) and it must be a progenitor of CaWRKY16-like gene of the galegoid clade legumes.
4.5. Molecular functions of CaWRKY16
Our expression analysis suggested that CaWRKY16 gene is ubiquitously expressed in all chickpea tissues, and it responds to SA and A. rabiei (Figs 4 and 5). Thus, it should have a role in SA-induced plant responses against pathogens. The possible molecular role of these unique multi-domain proteins can be understood from their motifs and domains. A TM domain of CaWRKY16 may localize it to Golgi apparatus while group-III WDs are known for DNA binding. The Exo70 domains of both CaWRKY16 and CaWRKY17 are close to well-characterized AtExo70B members. In Arabidopsis, Exo70B subclade genes AtExo70B1 and AtExo70B2 control the immune signalling against bacterial, oomycetes, and fungal pathogens.42,43 Recent reports also indicate towards the importance of Exocyst and its component Exo70 in plant–pathogen interaction. The OsExo70F members interact with AVR-Pii to regulate plant immunity44 and Phytophthora infestans effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity.45 The TIR-NBS2 or associated molecules monitor the integrity of AtExo70B1, making it an important component of plant defense43 and AtExo70B1 also interacts with RIN4.46 A model in which unique chimeric genes function as decoy or help in pathogen effector recognition has emerged in cases like rice Pikp-1 interacts with AVR-PikD and two linked pairs with atypical WRKY genes AtWRKY16 and AtWRKY52 recognize effector activity.24,31,47 Thus, being so close to Exo70B group CaWRKY16 in association with CaWRKY17 may behave as decoy to detect pathogen effectors. The regulation of plant genes by CaWRKY16 is another aspect but for that it needs to move from Golgi apparatus to nucleus after specific signal perception. The group-III member CaWRKY16 despite its ‘WKKYGKK’ variant WD may bind to regulatory regions of target genes.
4.6. CaWRKY50 is a functional orthologue of AtWRKY53
In Arabidopsis, group-III WRKY genes are involved in plant senescence and defence. Among them, role of AtWRKY30, -53, -54, and -70 is documented for plant senescence. AtWRKY53 is a positive regulator whereas AtWRKY54 and AtWRKY70 are the negative regulators of plant senescence. Our overexpression analysis in tobacco also demonstrates that CaWRKY50 can induce plant senescence making it a functional orthologue of AtWRKY53 from chickpea (Fig. 7). This functional closeness is also supported by phylogeny. Both AtWRKY53 and CaWRKY50 are induced by SA and pathogens, and they also act as transcriptional activators in yeast. Another gene AtWRKY4148 is also close to CaWRKY50 and it plays role in PAMP-induced signalling. However, AtWRKY41 responds at a lower level to SA signalling compared with AtWRKY53.49 To confirm whether AtWRKY53 and CaWRKY50 are similar at molecular level, CaWRKY50's interaction was checked with a CaMEKK as in Arabidopsis AtMEKK1 physically associates with AtWRKY53.50 However, in yeast two-hybrid analysis chickpea CaMEKK (LOC101498916), closest to AtMEKK1, failed to interact with CaWRKY50 and CaWRKY73 (Supplementary Fig. S12). This could be attributed to the divergence of MEKKs outside kinase domain or variation in Arabidopsis and chickpea WRKYs or evolution of a unique case in Arabidopsis. Thus, sequence comparison and CaWRKY50 overexpression in tobacco leading to early senescence and flowering showed that CaWRKY50 is a functional orthologue of AtWRKY53 despite differences at molecular level.
Taken together, our research has helped in the identification of WRKY genes in agriculturally important legumes and seems to be the first report on a comparative account of WRKY genes from two legumes. Functional characterization of legume WRKY genes will also hasten in model plants L. japonicus and M. truncatula, where transposon mutant lines are available for many genes. Identification of legume-specific WRKY genes will also help promoting the research to identify and functionally characterize chimeric WRKY genes that have independently evolved in other plant systems. The CaWRKY group-III genes expression study presented here will serve as reference in the expression of WRKY genes. Our study also demonstrates that despite sequence divergence in WRKY genes many functional similarities do exist. We will also further focus our study to elucidate role of CaWRKY16 and CaWRKY50 in plant–pathogen interaction using model legumes.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was funded by Department of Biotechnology, Government of India through research grant for the Challenge Program on Chickpea Functional Genomics project (Sanction No. BT/AGR/CG-Phase II/01/2014) and core grant from National Institute of Plant Genome Research, New Delhi, India. Funding to pay the Open Access publication charges for this article was provided by the National Institute of Plant Genome Research, New Delhi, India.
Supplementary Material
Acknowledgements
The authors acknowledge help extended by Prof. Vitaly Citovsky of Stony Brook University, USA in providing the yeast nuclear import system; Dr Tsuyoshi Nakagawa of Shimane University, Japan for donating pGWB441 vector; and Prof. Inhwan Hwang of POSTECH, Republic of South Korea for NLS-mRFP. We are also thankful to Central Instrument Facility of National Institute of Plant Genome Research for their technical help. S.P. acknowledges UGC, India for fellowship.
References
- 1.Franco-Zorrilla J.M., Lopez-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R.. 2014, DNA-binding specificities of plant transcription factors and their potential to define target genes, Proc. Natl. Acad. Sci. USA, 111, 2367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machens F., Becker M., Umrath F., Hehl R.. 2014, Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana, Plant Mol. Biol., 84, 371–85. [DOI] [PubMed] [Google Scholar]
- 3.Eulgem T., Rushton P.J., Robatzek S.. 2000, The WRKY superfamily of plant transcription factors, Trends Plant Sci., 5, 199–206. [DOI] [PubMed] [Google Scholar]
- 4.Ishiguro S., Nakamura K.. 1994, Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato, Mol. Gen. Genet., 244, 563–71. [DOI] [PubMed] [Google Scholar]
- 5.Rinerson C.I., Rabara R.C., Tripathi P., Shen Q.J., Rushton P.J.. 2015, The evolution of WRKY transcription factors, BMC Plant Biol., 15, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D., Amornsiripanitch N., Dong X.. 2006, A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants, PloS Pathog., 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shim J.S., Jung C., Lee S. et al. . 2013, AtWRKY44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling, Plant J., 73, 483–95. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y., Liang G., Yang S., Yu D.. 2014, Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence, Plant Cell, 26, 230–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishihama N., Yoshioka H.. 2012, Post-translational regulation of WRKY transcription factors in plant immunity, Curr. Opin. Plant Biol., 15, 431–7. [DOI] [PubMed] [Google Scholar]
- 10.Grunewald W., Smet I.D., Lewis D.R. et al. . 2012, Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis, Proc. Natl. Acad. Sci. USA, 109, 1554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Y., Chen X., Xie K. et al. . 2014, Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice, PloS One, 9, e102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schluttenhofer C., Yuan L.. 2015, Regulation of specialized metabolism by WRKY transcription factors, Plant Physiol., 167, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Lee C.W., Wehner N. et al. . 2015, Regulation of oncogene expression in T-DNA-transformed host plant cells, PloS Pathog., 11, e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castrillo G., Sanchez-Bermejo E., de Lorenzo L. et al. . 2013, WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis, Plant Cell, 25, 2944–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi Y., Yang Y., Zhou Y. et al. . 2013, Protein-protein interactions in the regulation of WRKY transcription factors, Mol. Plant, 6, 287–300. [DOI] [PubMed] [Google Scholar]
- 16.Singh A., Singh I.K., Verma P.K.. 2008, Differential transcript accumulation in Cicer arietinum L. in response to a chewing insect Helicoverpa armigera and defence regulators correlate with reduced insect performance, J. Exp. Bot., 59, 2379–92. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Q.Y., Tian A.G., Zou H.F. et al. . 2008, Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants, Plant Biotechnol. J., 6, 486–503. [DOI] [PubMed] [Google Scholar]
- 18.Varshney R.K., Song C., Saxena R.K. et al. . 2013, Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement, Nat. Biotechnol., 31, 240–6. [DOI] [PubMed] [Google Scholar]
- 19.Jain M., Misra G., Patel R.K. et al. . 2013, A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.), Plant J., 74, 715–29. [DOI] [PubMed] [Google Scholar]
- 20.Poora R.J. 2002, The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b, Photosynth. Res., 73, 149–56. [DOI] [PubMed] [Google Scholar]
- 21.Parween S., Nawaz K., Roy R. et al. . 2015, An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.), Sci. Rep., 5, 12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song H., Nan Z.. 2014, Genome-wide identification and analysis of WRKY transcription factors in Medicago truncatula, Hereditas (Beijing), 36, 152–68. [DOI] [PubMed] [Google Scholar]
- 23.Rushton P.J., Somssich I.E., Ringler P., Shen Q.J.. 2010, WRKY transcription factors, Trends Plant Sci., 15, 247–58. [DOI] [PubMed] [Google Scholar]
- 24.Saucet S.B., Ma Y., Sarris P.F., Furzer O.J., Sohn K.H., Jones J.D.G.. 2015, Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4, Nat. Commun., 6, 6338. [DOI] [PubMed] [Google Scholar]
- 25.Chi Y., Yang Y., Li G., Wang F., Fan B., Chen Z.. 2015, Identification and characterization of a novel group of legume-specific, Golgi apparatus-localized WRKY and Exo70 proteins from soybean, J. Exp. Bot., 66, 3055–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alves-Carvalho S., Aubert G., Carrere S. et al. . 2015, Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species, Plant J., 84, 1–19. [DOI] [PubMed] [Google Scholar]
- 27.Jaiswal P., Cheruku J.R., Kumar K. et al. . 2012, Differential transcript accumulation in chickpea during early phases of compatible interaction with a necrotrophic fungus Ascochyta rabiei, Mol. Biol. Rep., 39, 4635–46. [DOI] [PubMed] [Google Scholar]
- 28.Rhee Y., Gurel F., Gafni Y., Dingwall C., Citovsky V.. 2000, A genetic system for detection of protein nuclear import and export, Nat. Biotechnol., 18, 433–7. [DOI] [PubMed] [Google Scholar]
- 29.Miao Y., Laun T., Zimmermann P., Zentgraf U.. 2004, Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis, Plant Mol. Biol., 55, 853–67. [DOI] [PubMed] [Google Scholar]
- 30.Yin G., Xu H., Xiao S. et al. . 2013, The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups, BMC Plant Biol., 13, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roux C.L., Huet G., Jauneau A. et al. . 2015, A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity, Cell, 161, 1074–88. [DOI] [PubMed] [Google Scholar]
- 32.van Verk M.C., Pappaioannou D., Neeleman L., Bol J.F., Linthorst H.J.M.. 2008, A novel WRKY transcription factor is required for induction of PR1-a gene expression by salicylic acid and bacterial elicitors, Plant Physiol., 146, 1983–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kashkush K., Feldman M., Levy A.A.. 2003, Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat, Nat. Genet., 33, 102–6. [DOI] [PubMed] [Google Scholar]
- 34.Tsuchiya T., Eulgem T.. 2013, An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication, Proc. Natl. Acad. Sci. USA, 110, E3535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong-Abdullah M., Ordway J.M., Jiang N. et al. . 2015, Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm, Nature, 525, 533–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cowley M., Oakey R.J.. 2013, Transposable elements re-wire and fine-tune the transcriptome, PloS Genet., 9, e1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He J., Benedito V.A., Wang M. et al. . 2009, The Medicago truncatula gene expression atlas web server, BMC Bioinformatics, 10, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Pumplin N., Ivanov S., Harrison M.J.. 2015, EXO70I is required for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis, Curr. Biol., 25, 2189–95. [DOI] [PubMed] [Google Scholar]
- 39.Spoel S.H., Koornneef A., Claessens S.M.C. et al. . 2003, NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol, Plant Cell, 15, 760–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelesko J.G., Harper R., Furuya M., Gruissem W.. 1999, Rare germinal unequal crossing-over leading to recombinant gene formation and gene duplication in Arabidopsis thaliana, Proc. Natl. Acad. Sci. USA, 96, 10302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babu M.M., Iyer L.M., Balaji S., Aravind L.. 2006, The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons, Nucleic Acids Res., 34, 6505–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pecenkova T., Hala M., Kulich I. et al. . 2011, The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction, J. Exp. Bot., 62, 2107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao T., Rui L., Li J. et al. . 2015, A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant, PloS Genet., 11, e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujisaki K., Abe Y., Ito A. et al. . 2015, Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity, Plant J., 83, 875–87. [DOI] [PubMed] [Google Scholar]
- 45.Du Y., Mpina M.H., Birch P.R.J., Bouwmeester K., Govers F.. 2015, Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity, Plant Physiol., 169, 1975–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afzal A.J., Kim J.H., Mackey D.. 2013, The role of NOI-domain containing proteins in plant immune signaling, BMC Genomics, 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kroj T., Chanclud E., Michel-Romiti C., Grand X., Morel J.B.. 2016, Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread, New Phytol., doi:10.1111/nph.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Higashi K., Ishiga Y., Inagaki Y., Toyoda K., Shiraishi T., Ichinose Y.. 2008, Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana, Mol. Genet. Genomics, 279, 303–12. [DOI] [PubMed] [Google Scholar]
- 49.Besseau S., Li J., Palva E.T.. 2012, WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana, J. Exp. Bot., 63, 2667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miao Y., Laun T.M., Smykowski A., Zentgraf U.. 2007, Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence related WRKY53 transcription factor on the protein level and can bind to its promoter, Plant Mol. Biol., 65, 63–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.