Abstract
The expression of genes involved in the pathogenesis of Staphylococcus aureus is known to be controlled by global regulatory loci, including agr, sarA, sae, arlRS, lytSR, and sarA-like genes. Here we described a novel transcriptional regulator called sarV of the SarA protein family. The transcription of sarV is low or undetectable under in vitro conditions but is significantly augmented in sarA and mgrA (norR or rat) (SA0641) mutants. The sarA and mgrA genes act as repressors of sarV expression, as confirmed by transcriptional fusion and Northern analysis data. Purified SarA and MgrA proteins bound specifically to separate regions of the sarV promoter as determined by gel shift and DNase I footprinting assays. The expression of 19 potential target genes involved in autolysis and virulence, phenotypes affected by sarA and mgrA, was evaluated in an isogenic sarV mutant pair. Our data indicated that the sarV gene product played a role regulating some virulence genes and more genes involved in autolysis. The sarV mutant was more resistant to Triton X-100 and penicillin-induced lysis compared to the wild type and the sarA mutant, whereas hyperexpression of sarV in the parental strain or the sarV mutant rendered the resultant strain highly susceptible to lysis. Zymographic analysis of murein hydrolase activity revealed that inactivation of the sarV gene results in decreased extracellular murein hydrolase activity compared to that of wild-type S. aureus. We propose that sarV may be part of the common pathway by which mgrA and sarA gene products control autolysis in S. aureus.
Staphylococcus aureus is an important human pathogen that has become a growing public health concern, primarily due to the emergence of antibiotic-resistant strains within the hospital environment (4, 5, 25). The spectrum of infections caused by this organism is extremely broad, ranging from cutaneous infections such as impetigo, folliculitis, and carbuncles to deep-seated infections such as pneumonia, endocarditis, septicemia, osteomyelitis, and other metastatic complications. Besides invasion syndromes, S. aureus can also cause toxin-mediated diseases such as food poisoning, toxic shock syndrome, and scalded skin syndrome. The primary site of infection is generally the skin or a wound from which the organism can spread to the bloodstream and subsequently disseminate into various host tissues. Once S. aureus establishes its presence in the tissue, it produces a large number of bacterial components and secreted products that include surface-associated protein adhesins, enzymes, exotoxins, and capsular polysaccharides, gene products that facilitate tissue colonization, tissue destruction, and immune evasion. The expression of many of these genes is coordinately controlled by regulatory loci (2, 5, 33, 35). These regulatory loci include agr, sarA, saeRS, sigB, lytSR, arlRS, and sarA-like genes.
The agr locus consists of two divergent transcripts, RNAII and RNAIII, which encode agrDBCA and hld, respectively (19, 20, 33). It is also a complex quorum-sensing two-component regulatory system in S. aureus. The RNAII transcriptional unit codes for four genes, of which agrBD are involved in the generation and secretion of an autoinducing octapeptide. As the autoinducing peptide reaches a threshold concentration in the growth medium during the transition from exponential to postexponential phase, activation of agrC and agrA, encoding the two-component regulatory system, would occur, leading to the transcription of RNAII and RNAIII, the agr regulatory molecule responsible for the up-regulation of exoproteins and down-regulation of cell wall-associated proteins (20, 33). The sarA locus comprises a major open reading frame (ORF), sarA, driven by three distinct promoters, resulting in three overlapping transcripts with a common terminating end (8, 29). The sarA locus up-modulates the expression of selected cell wall proteins and exoproteins (e.g., α- and β-hemolysins). DNA binding studies have revealed that SarA, the major sarA effector molecule, binds to several target gene promoters (e.g., agr, hla, and spa) to modulate gene transcription (10), thus implicating both agr-dependent and agr-independent pathways for SarA-mediated regulation.
Using affinity chromatography and genomic scan, we and others have identified several SarA homologs that are involved directly or indirectly in gene regulation. SarR, a 115-residue-long polypeptide (30), represses SarA expression during the postexponential phase by binding to the sarA promoter region. SarS (also called SarH1) is a 250-residue protein (9, 44) repressed by agr to activate spa expression and, to a lesser extent, to down-regulate hla expression. rot, also a sarA homolog, is a negative regulator for hemolysins (hla and hlb), lipase (geh), and serine proteases (sspB and splA) as well as a positive regulator for cell surface proteins (clfB/clumping factor, spa/protein A, sdrC/putative cell surface adhesin) (32, 39). SarT is a repressor of hla and RNAIII transcription (41). Contiguous to sarT, but transcribed in the opposite orientation, is sarU (sarH2), whose expression is negatively controlled by SarT. Phenotypic and transcriptional analysis revealed that sarU is a positive regulator of RNAIII and contributes to the expression of virulence genes controlled by agr (31). More recently, mgrA (also called rat or norR), a homolog of marR and also sarA, has been found to negatively regulate autolysis genes without significantly affecting sarA and agr expression (17). The mgrA gene also impacts upon the expression of type 8 capsular polysaccharide (cap8), α-hemolysin (hla), protein A (spa), norA, lipase, protease, and coagulase (27, 45).
Besides regulating virulence genes, the agr and sarA loci are also involved in the regulation of genes involved in the autolysis process, but the exact mechanism (e.g., direct versus indirect) is not known. Two-component regulatory systems such as ArlRS and LytSR have been shown to participate in the regulation of autolysis genes (6, 12, 13, 16). Downstream of the dicistronic lytSR operon is the lrgAB locus, the expression of which is dependent on activation by LytSR. Based on homology alone, it has been speculated that LrgA may function as an antiholin to inhibit murein hydrolase activity, presumably by interfering with the holin molecule that has been postulated to form a pore for the transport of murein hydrolases across the cell membrane (4, 7, 14). Although the exact mechanism of autolysis and their mode of regulation are not well defined, it is hypothesized that a diminution in transport of autolysins across the cell membrane would reduce the autolytic process.
We have identified another homolog of SarA, a 116-residue-long polypeptide called SarV (SA2062 and protein GI13702067) (21). The expression of sarV is repressed by sarA and mgrA (SA0641 and protein GI 13700577), known to be involved in the control of virulence and autolysis genes. Inactivation of sarV did not affect the expression of regulatory genes such as sarU, sarR, sarS, sarT, sarA, rot, and sae but had discernible effects on the expression of agr RNAII and RNAIII, lytSR, and arlRS. As sarA and mgrA are involved in the regulation of autolysis and virulence genes, we also evaluated the expression of 19 potential target genes involved in these phenotypes (33) (spa, coa, clfA, fnbA, fnbB, hla, atl, lytM, lytN, lrgAB, geh, scdA, abcA, pbp4, splA, scp, aur, norA, and cap8) by Northern blotting and transcriptional fusions in sarV mutants. Importantly, the sarV gene product is involved in the regulation of autolysin and protease genes (scdA, lrgB, atl, splA, and aur). Lytic assays with Triton X-100 and penicillin indicated that the sarV mutant, contrary to the effect of mgrA and sarA mutants, is more resistant to lysis than the parent, while hyperexpression of sarV renders the strain sensitive. Analysis of the sarV mutant indicated that this gene regulates extracellular and intracellular murein hydrolase activity, possibly by regulating selective genes involved for autolysis. Because sarV likely does not regulate mgrA or sarA, we propose that sarV constitutes an important “hub” for the control of autolysis by mgrA and sarA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Phage φ11 was used as a generalized transducing phage for S. aureus strains. S. aureus strain RN4220, a restriction-deficient derivative of strain 8325-4, was used as the initial recipient for the transformation of plasmid constructs. S. aureus cells were grown at 37°C with aeration in Trypticase soy broth (TSB) or 03GL agar medium supplemented with antibiotics when necessary. Luria-Bertani medium was used for growing Escherichia coli. Antibiotics were used at the following concentrations: for S. aureus, erythromycin at 5 μg/ml, kanamycin at 50 μg/ml, tetracycline at 5 μg/ml, and chloramphenicol at 10 μg/ml; for E. coli, ampicillin at 50 μg/ml, chloramphenicol at 30 μg/ml, erythromycin at 200 μg/ml, and spectinomycin at 75 μg/ml.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Reference or source | Comment |
|---|---|---|
| S. aureus strains | ||
| RN4220 | 30 | Mutant of 8325-4 that accepts foreign DNA |
| RN6390 | 30 | Laboratory strain that maintains its hemolytic pattern when propagated on sheep erythrocyte agar (parental strain) |
| RN6911 | 6 | agr mutant of RN6390 with an Δagr::tetM mutation |
| JC1413 | 43 | atl mutant of S. aureus |
| ALC1342 | Laboratory strain | A sarA mutant with deletion of ORF3 and the sarA ORF and replaced with an ermC gene, Δorf3sarA::ermC |
| ALC488 | 29 | sarA mutant of RN6390 with sarA::ermC |
| ALC1629 | Laboratory strain | ALC1342 with sarB (nt 1 to 1349 plus 180 bp of upstream sequence) integrated into the geh locus on the chromosome |
| ALC1713 | 30 | sarR mutant of RN6390 with ΔsarR::ermC |
| ALC1905 | 41 | sarT mutant of RN6390 with ΔsarT::ermC |
| ALC2272 | 31 | sarU mutant of RN6390 with ΔsarU::ermC |
| ALC1893 | 9 | sarS mutant of RN6390 with ΔsarS::ermC |
| ALC2530 | 17 | mgrA mutant of RN6390 with ΔmgrA::ermC |
| ALC2531 | 17 | ALC2530 complemented with mgrA gene into the geh locus on the chromosome |
| ALC2319 | This study | sarV mutant of RN6390 with a deletion of ORF of the sarV gene and replaced with an ermC gene |
| ALC2375 | This study | RN6390 with pALC2366 |
| ALC2376 | This study | ALC2272 with pALC2366 |
| E. coli strains | ||
| XL-1 Blue | 28 | Host strain for cloning |
| Topo InvαF′ | Invitrogen | Host strain for the TA cloning vector |
| Primers | ||
| pCL52.2 | 30 | Temperature-sensitive E. coli-S. aureus shuttle vector |
| pCR2.1 | Invitrogen | E. coli cloning vector for direct cloning of PCR products |
| pSK236 | 31 | Shuttle vector containing pUC19 cloned into the HindIII site of pC194 |
| pUC19 | 28 | E. coli cloning vector |
| pALC1484 | 18 | Modified pSK236 shuttle vector with a promoterless gfpuvr reporter gene preceded by an S. aureus ribosome binding site |
| pALC2231 | This study | pUC19 containing a 2.2-kb EcoRVIHindIII fragment containing sarV gene |
| pALC2280 | This study | pCL52.2 containing a 2.84-kb DNA fragment which has a deletion of a 565-bp BsaBl/SphI fragment that includes complete ORF of the sarV gene and an insertion of the 1.2-kb ermC gene at the BsaBI/SphI sites |
| pALC2366 | This study | pSK236 with a 1.4-kb DNA fragment containing the sarV gene with a 295-bp upstream sequence at the EcoRI site |
| pALC2489 | This study | pALC1484 with a 273-bp promoter fragment of sarV fused with the gfpuvr reporter gene at the EcoRI/XbaI sites |
Genetic manipulations in E. coli and S. aureus.
Based on homology with SarA, the sarV gene product was initially identified in the S. aureus N315 genome database (21; www.tigr.org/ and www.ncbi.nlm.nih.gov/genomics). To construct a sarV mutant, the sarV gene together with flanking sequences on both sides were amplified by PCR with the primers 5′-AACTGTCGATGGATTTAACGTTA-3′ and 5′-AGTTTAGTATTAGGTACAGCGA-3′, using chromosomal DNA from strain RN6390 as the template. The 2.2-kb PCR fragment was cloned into cloning vector pCR 2.1 (Invitrogen, San Diego, Calif.) in E. coli. The HindIII-EcoRV DNA fragment containing the 2.2-kb fragment was then cloned into the HindIII and HincII sites of pUC19. A 565-bp fragment, comprising the sarV coding region as well as 137 and 78 bp of the respective upstream and downstream regions, was deleted by restricting with BsaBI and SphI and then replacing the deleted fragment with a ∼1.2-kb ermC gene at these sites. The fragment containing the ermC replacement of the deleted sarV gene was cloned into the temperature-sensitive shuttle vector pCL52.2. Construction and selection of the putative sarV mutant (tetracycline-sensitive and erythromycin-resistant colonies) were performed as described elsewhere (30, 31). A phage φ11 lysate of the putative sarV mutant was then prepared to infect fresh strain RN6390 cells to reconstruct the sarV mutant, in an attempt to avoid any putative genomic mutations that might have occurred during the temperature shift to promote homologous recombination. The correct mutation was confirmed by PCR, Northern, and Southern hybridization with sarV and ermC probes as described previously (28, 30). One clone, designated ALC2319, was selected for further study.
To complement the sarV mutation, a 1.5-kb fragment encompassing the sarV gene and 295 bp upstream of the sarV translation start site were cloned into shuttle plasmid pSK236. The recombinant plasmid was electroporated into RN4220, selecting for chloramphenicol-resistant colonies. The correct transformant was verified by restriction analysis of the recombinant plasmid. The plasmid from RN4220 was then electroporated into parental strain RN6390 and the sarV mutant (ALC2319) to construct strains ALC2375 and ALC2376, respectively.
Isolation of RNA and Northern blot hybridization.
Total RNA from S. aureus was prepared by using a TRIzol isolation kit (Gibco BRL, Gaithersburg, Md.) and a reciprocating shaker as described elsewhere (8, 30, 31). The optical density at 650 nm OD650 of various cultures was determined with a spectrophotometer (Spectronic 20). The concentration of total RNA was determined by measuring the absorbance at 260 nm, and 10 μg of each total RNA sample was analyzed by Northern blotting as described previously (30). The genes coding for agr RNAII, agr RNAIII, sarA, sarR, sarT, sarS, sarU, rot, mgrA, sae, lytS, lytR, arlS, arlR, spa, coa, clfA, fnbA, fnbB, hla, lytM and -N, atl, lrgB, geh, scdA, abcA, pbp4, splA, scp, aur, norA, and cap8 were either amplified by PCR or excised from the plasmids containing the desired genes with restriction endonucleases. An internal fragment of the 16S rRNA gene (nucleotides [nt]777 to 1500; GenBank accession no. X68417) was used as a loading control. For detection of specific transcripts, gel-purified DNA probes were radiolabeled with [α-32P]dCTP by using the random-primed DNA labeling kit (Roche Diagnostics GmbH) and hybridized under aqueous-phase conditions at 65°C. The blots were subsequently washed and autoradiographed.
Transcriptional fusion studies of agr RNAII, agr RNAIII, hla, sarA, sarV, arlR, and lytS promoters linked to the gfpuvr reporter gene
Construction of the plasmid pALC1743, -1742, and -1740 containing agr RNAIII, agr RNAII, and hla promoter fragments linked to the gfpuvr reporter gene were described earlier (18, 31). A 268-bp sarV promoter fragment (position 21 to 288 nt upstream from the start ATG codon) was amplified by PCR using chromosomal DNA of S. aureus strain RN6390 and primers with flanking EcoRI or XbaI sites. An EcoRI and XbaI fragment containing the sarV promoter was cloned into shuttle plasmid pALC1484, generating plasmid pALC2489 containing the transcriptional fusion of the sarV promoter to the gfpuvr reporter gene. The construction of plasmid pALC1484 and modification of gfpuv (Clontech, Palo Alto, Calif.) to gfpuvr with an S65T mutation to yield a red shift (excitation maxima from 395 to 488 nm) were described earlier (18). Similarly, DNA fragments containing arlS, lytR, and mgrA P1 promoters were cloned into shuttle plasmid pALC1484, generating plasmids containing transcriptional fusions to the gfpuvr reporter gene (17). Restriction analysis and DNA sequencing confirmed the orientation and authenticity of the promoter reporter gene constructs. The recombinant plasmids, containing the respective promoter regions, were first introduced into S. aureus strain RN4220 by electroporation. Plasmids purified from RN4220 transformants were then electroporated into RN6390, its isogenic sarV::ermC (ALC2319), sarA::ermC (ALC1342), mgrA::ermC (ALC2530), mgrA::ermC/sarA::kan (ALC2535) mutants, and single-copy complemented mgrA (ALC2531) and sarA (ALC1629) strains as required.
After overnight culture, S. aureus strains harboring the recombinant plasmids were diluted 1:100 in TSB containing chloramphenicol (10 μg/ml) and grown at 37°C with shaking at 250 rpm. Aliquots (200 μl) were transferred hourly to microtiter plates to assay for OD650 and fluorescence for 10 h in an FL600 fluorescence reader (BioTek Instrument, Winooski, Vt.). Promoter activation was plotted as mean fluorescence per OD650 over time, using the average values from triplicate readings.
Purification of SarA and MgrA proteins.
The cloning and purification of the His6 tagged fusion SarA protein were described earlier (10). The 444-bp DNA fragments containing the full-length mgrA gene (SA0641of S. aureus N315) were amplified by PCR using chromosomal DNA from S. aureus RN6390 as the template and primers containing flanking restriction sites (NdeI and BamHI) to facilitate in-frame cloning into the expression vector pET14b (Novagen, Madison, Wis.). The recombinant plasmids containing the full-length mgrA coding region were confirmed by restriction digestion and DNA sequencing. The recombinant plasmids were then transformed into E. coli BL21(DE3)pLysS. The recombinant His6-MgrA protein expression and purification were done in a manner similar to that for the SarA protein as described elsewhere (10). The authenticity and purity of the purified His6-tagged MgrA fusion protein were confirmed by sodium dodecyl sulfate (SDS) gels stained with Brilliant Blue R-250 and microsequenced for the first 20 residues from the N terminus. The purified His6-tagged SarA and MgrA proteins were found to be more than 98% pure in an SDS-12% polyacrylamide gel. The concentration of the purified proteins was determined by the Bradford protein assay (Bio-Rad, Hercules, Calif.), using bovine serum albumin as the standard.
Gel shift analysis and DNase I footprinting.
To determine if the recombinant SarA or MgrA proteins bind to the sarV promoter region, a 191- or 268-bp fragment (positions 21 to 211 or 288), representing the sarV promoter region, was end labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Labeled fragment (0.1 ng or 0.5 fmol) was incubated at room temperature (RT) for 20 min with various amounts of purified SarA or MgrA protein in 25 μl of binding buffer (25 mM Tris-Cl [pH 7.5], 0.1 mM EDTA, 75 mM NaCl, 1 mM dithiothreitol, and 10% glycerol) containing 0.5 μg of calf thymus DNA (Amersham Pharmacia Biotech). The reaction mixtures were analyzed in an 8.0% nondenaturing polyacrylamide gel. The band shifts were detected by exposing dried gels to X-ray films.
Footprinting assays with template DNA fragment and DNase I were performed as previously described (3, 10, 30). To label PCR products, only one of the primers was labeled at one end. For the assay, the binding reactions were carried out in a 100-μl reaction volume containing 20 mM Tris-Cl (pH 8.0), 100 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 2 mM dithiothreitol, 10 μg of bovine serum albumin, 0.4 μg of calf thymus DNA, radiolabeled template DNA (20,000 cpm), and various amounts of the purified SarA or MgrA proteins at RT for 30 min. DNase I (0.02 U; Boehringer, Mannheim, Germany) was added and allowed to incubate for 1 min at RT. The reaction mixtures were extracted with phenol-chloroform, ethanol precipitated, washed with 70% ethanol, dried, and resuspended in loading buffer (98% deionized formamide, 10 mM EDTA [pH 8.0], 0.025% [wt/vol] xylene cyanol FF, 0.025% [wt/vol] bromophenol blue). DNA samples were denatured at 95°C for 5 min and analyzed on a 6% denaturing polyacrylamide sequence gel. The positions of the protected region were derived by comparing the footprint with the A+G sequencing ladder of the same fragment (28).
Primer extension analysis.
Mapping of the 5′ end of the sarV transcript by primer extension was performed using the primer 5′-CTTCAATTGAAATCTTATGTGTTG-3′, complementary to the sarV coding strand and located from nt positions 94 to 71 downstream from the putative start codon ATG. Reverse transcription was carried out as described elsewhere by using total RNA isolated from wild-type RN6390 or the sarA mutant as described previously (3, 30).
Lytic assays with Triton X-100 and penicillin.
These assays were performed as described elsewhere (12, 17, 27). In brief, overnight-grown bacteria were diluted to an OD650 of 0.05 in TSB containing 1 M NaCl and allowed to grow at 37°C with shaking till the OD650 reached 0.7 to 1.2. Cells were harvested, washed twice with ice-cold water, and then resuspended in 0.05 M Tris-Cl (pH 7.2) in the original volume with or without 0.05% Triton X-100. Cells were incubated at 30°C with shaking and checked for lysis by measuring the progressive decrease in absorbance (OD650) at 30-min intervals. To assess the sensitivity of the sarV mutant to penicillin, inoculations were done in TSB from overnight cultures to yield a starting OD650 of 0.05. Cultures were grown at 37°C with shaking to reach exponential phase (OD650 ∼0.5). Penicillin G was added to a concentration of 0.4 μg/ml. Cultures were incubated further, and the OD650 was measured every hour for 7 to 8 h.
Zymographic analysis.
For the detection of extracellular and intracellular murein hydrolases, SDS-polyacrylamide gel electrophoresis-based zymographic analysis was performed as described by Groicher et al. (16). In brief, various strains were grown in TSB medium for 16 h at 37°C and 250 rpm. Extracellular murein hydrolases were isolated by pelleting 16-h cultures at 6,000 × g for 20 min at 4°C. The supernatant was filter sterilized and concentrated 100-fold by using polyethylene glycol 8000, followed by dialysis in phosphate-buffered saline. To obtain intracellular and cell wall-associated murein hydrolases, the cell pellets were extracted as essentially described by Groicher et al. (16), and final fractions were dialyzed against phosphate-buffered saline buffer.
The concentrations of total proteins in each sample were determined by using the Bradford assay (Bio-Rad) according to the manufacturer's procedures. Various protein fractions were resolved on an SDS-12% polyacrylamide gel containing either Micrococcus luteus (Sigma Chemical Co.) or autoclaved and lyophilized S. aureus RN6390 cells (1 mg [dry weight] of cells per ml of gel). After electrophoresis, gels were washed with water and incubated overnight in 25 mM Tris-Cl, pH 8.0, containing 1% Triton X-100 at 37°C to allow hydrolysis of the embedded bacterial cells. After incubation, gels were stained with 1% methylene blue (Sigma Chemical Co.) and destained in water. Following destaining the gels were scanned in a scanner, and the white bands in the gels (zones of hydrolysis) indicated regions of murein hydrolase activity. Quantitative cell wall hydrolysis assays were performed as described by Rice et al. (38) as the turbidity of the samples was determined by measuring the absorbance at 580 nm with a spectrophotometer.
RESULTS
Identification of the sarV gene.
Searching for additional SarA homologs in the recently published S. aureus N315 genome, we found at least 12 proteins homologous to SarA (SA0573, protein GI 13700508), using a default setting of 30 as the cutoff for the BLAST search (21; www.ncbi.nlm.nih.gov/genomes/staphylococcus). Some of these homologs included SarT (SA2286 or SarH3, protein GI 13702448; e-value of 9e−17) (41), SarS (also called SarH1 or SA0108, protein GI 13700028; e-value of 1e−16) (9, 44), SarU (also called SarH2 or SA2287, protein GI 13702449; e-value of 2e−15) (31), SarR (SA2089, protein GI 13702095; e-value of 9e−12) (30), and Rot (SA1583, protein GI 13701558; e-value of 0.003) (32). MgrA (also called Rat, NorR, or SA0641; protein GI 13700577; e-value of 4e−04), more homologous to MarR than to SarA, has been shown to play a role in the regulation of genes involved in autolysis and virulence (17, 26, 45). The remaining homologs of unknown function were SA0668 (protein GI13700559; e-value of 1e−04 and 46% homology), SA2174 (protein GI13702335; e-value of 0.002 and 53% homology), and two more proteins (protein GI 1301144 and 13700062; e-values of 0.003 and 0.030 and homologies of 48 and 48% within a small region of proteins, respectively). SarV (SA2062 and protein GI 13702067) is the seventh protein that came up in our homology search, with SarT, -S, -U, and -R, SA0668, and MgrA being the first six hits (21). SarV is a 116-residue polypeptide (13,985 Da) that shares 52, 48, 44, 52, 51, 45, 49, and 60% homology to SarR, SarA, SarU1 (residues 1 to 124), SarU2 (residues 125 to 247), SarS1 (residues 1 to 125 of SarS), SarS2 (residues 126 to 250 of SarS), SarT, and MgrA, respectively. Based on sequence alignment, it can be observed that specific residues of SarV are conserved within the SarA protein family, including K23, L36, K48, K80, R82, D86, E87, R88, Q98, L101, and I109 of SarV (data not shown). In a BLAST comparison of SarV against other microbial genomes with the same default setting, we found a number of putative transcriptional regulators with unknown functions (e-values of 2e−06 to 9.0). Apart from S. aureus, significant homologs include Bacillus subtilis hypothetical protein ybfA (Z99105; e-value of 2 e−04 with 29% identity and 52% similarity) and a probable transcriptional regulator of Clostridium perfringens (AP003188; e-value of 0.001 with 23% identity and 50% similarity).
Expression of sarV in RN6390 and an assortment of isogenic mutants.
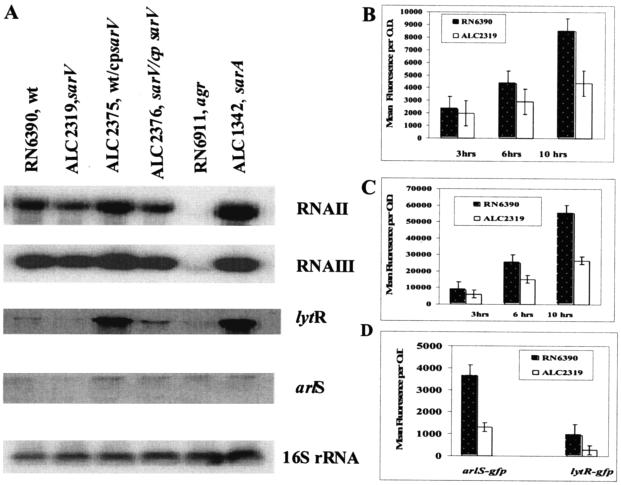
To determine the phenotypic effects of sarV, we constructed a sarV mutant in RN6390 by allelic replacement, essentially replacing the sarV gene with an ermC cassette (see Materials and Methods). A Northern blot assay with a sarV probe (350 bp) encompassing the coding region disclosed that sarV was poorly transcribed or undetectable in parental strains RN6390 and SH1000 (S. aureus 8325-4 rsbU+ strain; kindly provided by S. J. Foster). As expected, complementation of the wild type or the sarV mutant with a plasmid carrying the sarV gene (ALC2375 or ALC2376) increased the level of transcript of the sarV gene. Interestingly, the sarV transcript, sizing at ∼500 nt, was significantly enhanced in two sarA mutants (ALC488 sarA::ermC and ALC1342 Δorf3sarA::ermC) and the rat/mgrA mutant (ΔmgrA::ermC), but not in the sarR, sarS, sarT, sarU, or agr mutants (Fig. 1, top panel). The size of the transcript also hinted at the monocistronic nature of the sarV locus. Upon introducing a single copy of the mgrA gene or of the entire sarB locus into the lipase gene (geh) of the respective mgrA or sarA mutant (ALC2531 and ALC1629, respectively), the expression of sarV transcript returned to near the parental level, suggesting that the expression of sarV is truly repressed by sarA or the mgrA gene product. To determine whether the expression of mgrA was affected in these assorted mutants, we performed Northern analysis with the mgrA probe (450 bp) and found no significant changes in the expression of mgrA transcript in agr, sarT, sarU, sarV, sarS, and sarA mutants (Fig. 1, middle panel). The expression of mgrA gene transcript was slightly elevated in the sarR mutant. Notably, the mgrA transcript level was more elevated in strains (ALC2375 and ALC2376) harboring a multicopy plasmid carrying the sarV gene. Thus, besides repression by mgrA, it would seem that increased expression of sarV transcript also enhances mgrA expression, suggesting that sarV may act as an activator for the expression of mgrA transcript. However, the significance of this finding remains unclear, since we do not know the physiologic relevance of increased gene dosage in vivo (Fig. 1, top panel). There are no significant growth differences in the growth curves for the sarV mutant compared with those for the wild type and various mutants (data not shown). Growth phase variation of various mutants did not make any difference in the data presented above for the expression of sarV transcript when Northern blot hybridization was performed with total RNA isolated in the early phase of growth of various mutants (data not shown).
FIG. 1.
Northern analysis of the sarV and mgrA transcripts in the wild type, various isogenic mutants, and complemented strains at postexponential phases (OD650, ∼1.7) of growth. A total of 15 μg of cellular RNA was loaded onto each lane. The blots were probed with 360-bp sarV (upper panel), 565-bp mgrA (middle panel), and 724-bp (lower panel) fragments containing the entire ORF of the sarV and mgrA genes and internal region of 16S rDNA, respectively. cp indicates complementation with respective genes. The OD650 was determined with a spectrophotometer (Spectronic 20).
Transcriptional start sites and promoter structure of the sarV gene.
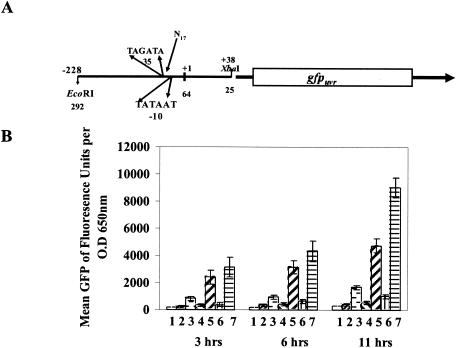
To determine the transcriptional start site and the promoter sequence, primer extension was performed with total RNA isolated from the wild-type strain RN6390 and its isogenic sarA mutant (data not shown). The transcriptional start site was mapped to an A or T which was located 62 or 63 bp upstream of the initiation codon ATG. Based upon the transcriptional start site, the predicted putative promoter boxes are TAGATA(−35)-N17-TATAAT(−10), which has close homology with the −10 and −35 consensus sequences of σA-dependent promoters (Fig. 2A; see also Fig. 4D). A strong ribosome binding site, AGGAGG, was located 9 bp upstream of the ATG translational start codon. Analysis of the downstream sequence of the sarV gene disclosed the presence of a rho-independent transcriptional terminator sequence from the position 1 to 91 bp immediately after the stop codon (TAA). Within this region, a 23-bp inverted repeat sequence from positions 1 to 23 and 68 to 91 after the stop codon of the sarV gene (AAAAATAAAAAGCATGCCAATCT-N45-AGATTGGCATGCTTTTTAAGTTTTT) could form a potential hairpin structure with a 24-nt-long heteroduplex.
FIG. 2.
Promoter activation of the sarV promoter fused to a gfpuvr reporter gene. (A) Graphical representation of the 268-bp sarV promoter fragment fused to a promoterless gfpuvr gene with an S. aureus ribosome binding site. The translational start site, labeled as +1, was identified by primer extension, and the putative promoter −10 and −35 boxed sequences are also indicated. The numbers at the line (both top and bottom) are marked according to the transcriptional start site and the translation start codon of the sarV gene, respectively. (B) The recombinant shuttle construct pALC2489 containing the 268-bp sarV promoter fragment driving the GFPuvr reporter gene was introduced into the wild type, its isogenic mutant strains, and complemented strains. Lane 1, wild-type RN6390; lane 2, sarV mutant ALC2319; lane 3, sarA mutant ALC1342; lane 4, the sarB fragment (28) integrated into the geh locus of the sarA mutant designated ALC1629; lane 5, mgrA mutant ALC2530; lane 6, the 1.5-kb DNA fragment containing the mgrA gene integrated into the geh locus of the mgrA mutant designated ALC2531; lane 7, mgrA sarA double mutant ALC2539. To minimize variations in fluorescence attributable to cell density, the data are presented as the average of reported fluorescence unit per OD650 in triplicate samples obtained at different growth points.
FIG. 4.
DNase I protection footprint analysis of SarA and MgrA binding to the sarV promoter region of S. aureus. (A) Footprint analysis of SarA binding to the sarV promoter region. The 191-bp fragment (nt positions −21 to −211 from the start codon ATG) was end labeled (top or coding strand) with [γ-32P]ATP. Lanes 1 and 6, no protein added; lanes 2 to 5, 1, 2, 3, and 4 μg of SarA, respectively. Lane M represents chemical cleavage at purine residues (A/G ladder). (B and C) Footprinting analysis of the MgrA protein binding to the 268-bp sarV promoter region (nt positions 21 to 288 from the ATG start codon). The 268-bp fragment was end labeled for top strand (B) and bottom strand (C) and incubated with DNase I in the presence or absence of proteins, as described above. No protein was added for lanes 1 and 6 in panel B or for lanes 1 and 7 in panel C. Lanes 2 to 5 (B) and 4 to 6 (C), 0.2, 0.5, 1.0, and 2.0 μg of protein was added; lane 3 of panel C, 0.3 μg of protein was added. Protected regions are marked by solid lines. (D) Nucleotide sequence of the 293-bp fragment upstream of the sarV gene was shown and marked with the putative promoter region and start site as derived from primer extension data. The putative protected regions for SarA and MgrA proteins on the promoter are marked.
sarV promoter-gfpuvr fusion studies in isogenic sarA and mgrA strains.
To confirm the repression of sarV by mgrA and sarA, we constructed a transcriptional fusion containing a 268-bp sarV promoter fragment upstream of the gfpuvr reporter gene (Fig. 2A). The construct pALC2489 was introduced into the wild-type strain RN6390, sarA mutant (ALC1342), mgrA mutant (ALC2530), sarV mutant (ALC2319), sarA/mgrA double mutant (ALC2535), complemented mgrA mutant (ALC2531), and complemented sarA mutant (ALC1629). Promoter activities, assayed as GFPuvr fluorescence (Fig. 2B), were ∼5-, 13- to 17-, and 18- to 28-fold higher at the three time points for sarA (lane 3), mgrA (lane 5), and sarA/mgrA (lane 7) mutants, respectively, compared with that of the parental strain RN6390 (lane 1). Complemented sarA and mgrA mutant strains (Fig. 2B, lanes 4 and 6) had reduced GFPuvr values compared to the parental strain. These data are consistent with the notion that the expression of sarV gene transcript is repressed by sarA and mgrA gene products. In the double sarA/mgrA mutant (Fig. 2B, lane 7), the mean GFPuvr value was higher than that for the single mutant, indicating that SarA and MgrA may repress sarV gene expression via divergent mechanisms. Collectively, these data concur with those of Northern analysis, essentially demonstrating that the expression of sarV gene transcript was undetectable in the wild type, moderately elevated in the sarA mutant, and highly expressed in the mgrA mutant. The expression of sarV is probably not dependent on its own gene product, as the mean GFPuvr value (228 to 407 ± 75 U) in the sarV mutant was comparable to that for the parental strain RN6390 (176 to 322 ± 50 U).
Binding of SarA and MgrA proteins to the sarV promoter region.
As the level of transcript of sarV was increased in sarA and mgrA mutants (Fig. 1), we speculated that SarA and MgrA may bind to the sarV promoter region to modulate sarV expression. In previous studies, we reported that members of the SarA protein family (e.g., SarA, -R, -S, and -T) have a conserved helix-turn-helix motif commonly found in DNA binding proteins (10, 30, 31). The MgrA protein, as a MarR homolog and to a lesser extent SarA, likely possesses a helix-turn-helix motif and hence is predicted to be a DNA binding protein. To verify this, we employed a 268-bp or a 191-bp sarV promoter fragment (position 21 to 288 or 21 to 211 nt upstream of the initiation codon) upstream of the ribosomal binding site for DNA binding assays. The DNA fragment (0.1 ng or 0.56 fmol per reaction mixture) was end labeled with [γ-32P]ATP and used in gel shift assays with various amounts of purified SarA and MgrA proteins (Fig. 3). Retarded protein-DNA complex could be detected with as little as 0.1 to 0.2 μg of MgrA (2.8 to 5.7 nM, assuming that MgrA is a dimeric protein like SarA or SarR) and ∼0.3 μg (10 nM) of SarA. As the concentrations of these proteins increased, the retarded protein-DNA complex became the predominant band, with complete conversion at ∼1.0 μg for both proteins. Hence, SarA and MgrA proteins can bind to the sarV promoter region, presumably acting as a repressor to sarV transcription. To verify the DNA binding specificity and to map the binding sites, we performed DNase I protection assays with the same 268- or 191-bp sarV promoter fragment labeled with γ-32P at one end in the presence of SarA or MgrA (Fig. 4). As shown in Fig. 4A, protection from DNase I digestion with SarA was mapped to a region +2 to −42 bp from the +1 transcription start in Fig. 4D. A close analysis of this sequence revealed that the protected regions (box I, −21 to −42 bp from the transcription start; box II, +2 to −19 bp from transcription start in Fig. 4D) closely resemble the SarA consensus DNA binding motif (ATTTGTATTTAATATTTATATAATTG) previously reported by our investigators (10). Box I and II have 15 of 26 and 19 of 26 matches to the SarA consensus binding sequence, respectively, thus verifying that the DNA binding site of SarA on the sarV promoter is likely specific and further confirming our group's earlier studies on the predicted SarA binding consensus sequence. We also mapped the DNA binding site of the MgrA protein on the sarV promoter. Using the same end-labeled 191-bp promoter fragment, we initially failed to identify any protected region with both strands (data not shown). However, when the assays were repeated with the larger 268-bp sarV promoter fragment that we have used for the gfpuvr fusion assay (position −21 to −288 bp from the start codon), we found two closely associated protected regions (Fig. 4B and C). The putative protected regions of MgrA are from positions −84 to −115 bp, −132 to −147 bp (Fig. 4B, top strand of DNA), and −81 to −108 bp upstream of the transcriptional start site (Fig. 4C, bottom strand of DNA, and D). DNA sequence analyses of these regions disclosed a 6-bp conserved sequence (t/aGTTGT) that was repeated four times in the top and bottom strands of the protected sequence (Fig. 4D). The sequence encompassing −84 to −115 bp upstream of the initiation codon was of interest as it contained the sequence t/aGTTGT thrice and was protected in both strands. This led us to speculate that MgrA may bind to this region to facilitate binding to other areas via conformational changes (e.g., by cooperating interaction, bending, or other means of regulation). Whether the conserved 6-bp binding region of MgrA applies to other target loci regulated by MgrA requires additional confirmation. Nevertheless, the results presented in Fig. 3 and 4 clearly demonstrated that SarA and MgrA proteins are likely to bind to different sites on the sarV promoter region with a high degree of specificity. The SarA binding sites overlap with the −10 and −35 promoter boxes, whereas the MgrA binding sites are upstream of the sarV promoter boxes.
FIG. 3.
Autoradiogram of an 8.0% nondenaturing polyacrylamide gel showing gel shifts for purified MgrA and SarA proteins with a 268-bp sarV promoter fragment. Mobility of the DNA band in the presence of increasing amounts of either MgrA or SarA protein is indicated on the top. Cold competition lanes with a 10-fold excess (molar ratio) of the unlabeled 268-bp sarV fragment are indicated and contained 1.0 μg of the purified MgrA or SarA protein.
Transcription and transcriptional fusion assays for regulatory genes in a sarV mutant and various other isogenic strains.
To determine whether inactivation of sarV affects other regulatory genes involved in virulence, we performed Northern blot analysis with 11 other regulatory loci (agr RNAII, agr RNAIII, sarA, sarR, sarS, sarT, sarU, mgrA, rot, sae, lytS, lytR, arlR, and arlS). The functions of many SarA homologs have previously been described, including SarR as a regulator of SarA and agr expression, SarS as an activator of protein A expression, SarT as a repressor of α-hemolysin expression, and SarU as an activator of RNAIII that is repressible by SarT (9, 30, 31, 41). The lytSR locus is a two-component regulatory system that negatively controls extracellular murein hydrolase activity via positive regulation of the lrgAB operon (6, 16). An arlRS mutant up-regulates the expression of autolysis genes while augmenting transcription of agr and other exoprotein genes, including those of extracellular serine protease activity and the multidrug resistance transporter gene norA (12, 13). The saeRS locus encodes another two-component regulatory system that positively regulates the expression of α- and β-hemolysins, DNase I, coagulase, and protein A (15). As shown in Fig. 5A, there was a partial reduction in RNAII and, to a lesser extent, RNAIII expression during the postexponential phase in the sarV mutant. Likewise, the levels of transcripts of lytSR and arlRS were also reduced in the sarV mutant. In contrast, the transcription levels of sarR, saeRS, sarA, sarS, sarT, rot, and sarU were not significantly altered in the sarV mutant (data not shown). However, it should be stressed that the expression of sarS, sarT, and sarU was difficult to detect even in the parental strain under routine laboratory culture conditions. Complementation of the parent and sarV mutant with a plasmid carrying the sarV gene either augmented or restored agr RNAII, agr RNAIII, lytSR, and arlRS transcripts to near or above parental levels in the recipient strain. Collectively, these data suggested that sarV might act as an activator either directly or indirectly for the expression of agr, lytSR, and arlRS.
FIG. 5.
Analysis of assorted regulatory genes in a sarV mutant. (A) Northern blots of agr RNAII and RNAIII, lytR, arlS, and 16S rRNA transcripts in the wild-type RN6390, sarV (ALC2319), trans-complemented strains (ALC2375, ALC2376), and sarA (ALC1342) and agr (RN6911) mutants. A total of 10 μg of RNA from the postexponential phase (OD650, ∼1.7) was applied to each lane, and the blots were probed with the respective gene fragment containing the coding region. (B to D) GFPuvr expression as driven by agr RNAII (B), agr RNAIII (C), and arlS-gfpuv and lytR-gfpuv (D) promoters in the wild-type RN6390 and sarV mutant ALC2319. Bar diagrams in panels B and C represent various time points, whereas in panel D the diagram represents the ∼8-h time point. The experiments were repeated at least thrice; one representative experiment is shown.
To verify reduction of agr RNAII, agr RNAIII, arlS, and lytR transcripts in the sarV mutant (ALC2319), we introduced into the isogenic sarV strains plasmids containing the agr RNAII, agr RNAIII, arlS, or lytR promoter linked to the gfpuvr gene for promoter fusion analysis. Upon monitoring bacterial growth and serial GFP expression over a 10-h period, we found that the growth rates were comparable between the two strains containing each of the constructs, with early stationary phase appearing after ∼6 h of growth. The expression of RNAII and RNAIII during the stationary phase was slightly lower in the sarV mutant than in the parental strain (Fig. 5B and C). Thus, both Northern blotting and transcriptional fusion assays were consistent with the slight reduction of levels of transcripts in agr RNAII and RNAIII expression in the sarV mutant relative to that in the parental strain. Similarly, the expression of arlS promoter-gfpuv and lytR promoter-gfpuv was found to be reduced approximately threefold in the sarV mutant compared to the wild-type strain after ∼8 h of growth (Fig. 5D). Hence, Northern and transcriptional fusion studies suggested that the sarV gene product possibly acts as an activator for the expression of agr, arlRS, and lytSR transcripts. These results were consistent in several repeated experiments. Whether this effect is direct or indirect and also physiologically relevant remains to be determined.
Northern blot analysis for the expression of target genes
The data presented above clearly indicated that the expression of sarV, normally undetectable in the parental strain, was elevated moderately in the sarA mutant and highly in the mgrA mutant. SarA and MgrA, as DNA binding proteins, bind to the sarV promoter region, presumably to repress sarV transcription. As sarA and mgrA participate in the control of autolysis as well as virulence genes (4, 8, 10, 12, 17, 26, 45), we determined the expression of target genes known to be involved in autolysis or virulence in the sarV mutant. We tested the expression of 19 potential target genes (33), with 11 genes implicated in virulence (e.g., hla, coa, geh, splA, scp, norA, aur, spa, clfA, fnbA, and fnbB) and 8 involved in autolysis (lytN, lytM, atl, lrgB, scdA, abcA, pbp2, and pbp4). The transcript levels of genes encoding many cell surface-associated proteins (fnbA, fnbB, clfA, and spa) were not markedly altered by the sarV mutation during exponential and postexponential phases of growth (data not shown). The expression levels of geh (glycerol ester hydrolase) and coa (coagulase) were also not significantly affected in the sarV mutant (data not shown). The transcript level of the hla gene was lower in the sarV mutant compared with that in the parental strain but was restored upon complementation (Fig. 6). Transcriptional fusion of hla-gfpuvr also supported the Northern blotting data (1,185 ± 250 versus 2,278 ± 500 mean fluorescence units per OD at 6 h of growth for the sarV mutant and parent strain, respectively). A significant reduction in transcripts levels of splA, encoding a putative serine protease-like gene product, aur, encoding a zinc metalloprotease, and scp, encoding staphopain (protease II) (19, 39) was observed in the sarV mutant, while transcripts levels of these genes were substantially elevated in the parental strain or returned to a near-normal level in the mutant harboring trans-complementation of the sarV gene in multicopy (Fig. 6). As predicted from earlier findings, the agr mutant also displayed a significant reduction in these proteases, while a sarA mutation led to an increase in the expression of these transcripts (19, 39).
FIG. 6.
Northern blot analysis for assorted target genes in the wild type, sarV mutant (ALC2319), trans-supplemented strains ALC2375 and ALC2376, sarA mutant ALC1342, and agr mutant RN6911. Northern blots were hybridized with hla, splA, aur, scdA, lrgB, atl, abcA, and 16S rRNA DNA fragments containing the coding regions of the respective genes. A total of 10 μg of RNA from the postexponential phase (OD650, ∼1.7) was applied to each lane, and the OD650 was determined with a spectrophotometer (Spectronic 20).
The level of transcript of scdA, a gene positively associated with decreasing peptidoglycan cross-linking and increased autolysis (7), and the autolysin gene atl, which encodes major autolysin (34, 36, 40), was found to be decreased in the sarV mutant, thus suggesting positive regulation of scdA and atl by sarV gene; complementation of the sarV mutant with a multicopy plasmid (ALC2376) increased the expression of scdA or atl in the mutant, but the effect was only partial, thus suggesting that regulation of these genes by sarV may possibly be indirect or via some unknown factor. Interestingly, a sarA mutation increased scdA and atl transcripts, while agr inactivation did not have any significant change (Fig. 6). The abcA gene, encoding an ATP binding cassette transporter, is divergently transcribed from pbp4 (encoding penicillin binding protein 4) and is associated with cell autolysis (42). Interestingly, inactivation of sarV augmented the lrgB transcript level (Fig. 6), while a sarA mutant also increased lrgB transcription, consistent with S. aureus gene array analysis (DNA chip) data (11). Northern analysis revealed little or no impact effect of sarV on abcA expression. In contrast, an agr mutant, but not a sarA mutant, displayed a significant reduction in abcA transcript level, in concordance with the data of Schrader-Fischer and Berger-Bachi (42). Under our experimental conditions, the pbp4 transcript was found to be undetectable in the isogenic sarV strains. Additionally, there was no significant difference in norA, pbp2, lytN, and lytM transcript levels attributable to the sarV mutation (data not shown). Thus, a mutation in sarV led to a reduction in transcript level of atl, the major autolysin, as well as an alteration in scdA and lrgB expression profiles consistent with an autolysis-resistant phenotype.
Phenotypic characterization of the sarV gene.
The alteration in atl, lrgB, and scdA expression levels of transcripts in the sarV mutant, coupled with the repressive effect on sarV by mgrA and sarA, suggested that sarV may play an important role in controlling autolysis mediated by mgrA and sarA in S. aureus. Accordingly, we tested the sarV mutant for susceptibility to lysis in the presence of a detergent and an antibiotic. As shown in Fig. 7A, the sarV mutant was more resistant to Triton X-100-induced lysis in 50 mM Tris-Cl buffer, pH 7.2, than the wild-type RN6390 strain. The buffer alone also induced lysis of the wild type, and this effect was amplified by 0.05% Triton X-100. In contrast, the sarV mutant strain remained less susceptible in the buffer. Interestingly, the parental strain carrying the sarV gene on multicopy plasmid (ALC2375) became slightly more lytic than the wild-type strain in the presence of Triton X-100. Similarly, the sarV mutant carrying the sarV gene on multicopy plasmid (ALC2376) was more susceptible to lysis, but slightly less than its wild-type counterpart (data not shown). We also tested the effect of sarV on penicillin-induced lysis. Penicillin G (0.4 μg/ml) was added to a growing culture at early exponential phase (OD650 = 0.5), and changes in optical density (OD650), as a percentage of initial absorbance (OD), were monitored over a 7-h period. As shown in Fig. 7B, the parental strain RN6390 exhibited sensitivity to penicillin-induced lysis while the sarV mutant was resistant, resulting in an OD higher than that of the starting culture even at 6 h after the addition of penicillin. As a positive control, the sarA and agr mutants displayed high and low sensitivities, respectively, to penicillin-induced lysis, consistent with previously published data (14, 27).
FIG. 7.
Phenotypic and trans-functional analysis of the sarV gene. (A) Effect of the sarV mutation on Triton X-100-induced autolysis. ◊ and ⧫, wild-type S. aureus strain RN6390; □ and ▪, sarV mutant ALC2319; ▵ and ▴, wild-type strain containing trans-supplemented sarV gene in multicopy plasmid pSK236, ALC2375; ○ and •, sarA mutant strain ALC488. Strains were grown as described in Materials and Methods, with 0.05% Triton X-100 (open symbols) and without Triton X-100 (filled symbols), following the decline of A650 as an indicator of lysis. (B) Effect of sarV on penicillin-induced autolysis. Penicillin G-induced autolysis of wild-type RN6390 (◊), sarV mutant ALC2319 (□), RN6390 containing trans-supplemented sarV gene, ALC2375 (Δ), agr mutant RN6911 (▪), and sarA mutant ALC1342 (•) was measured as the decline in culture turbidity versus time. Autolysis is expressed as the percent of the initial OD650 reading immediately before penicillin was added. The experiments were repeated at least thrice; one representative experiment is shown.
To determine whether the lytic resistance phenotype was attributable to variations in murein hydrolase activity, possibly due to decreased expression of atl or scdA and/or enhanced expression of lrgB, zymographic analyses of culture supernatants and intracellular proteins of various strains were performed using gels containing M. luteus or S. aureus cells to detect glucosaminidase (GL) (Fig. 8A and B) and amidase (AM) (Fig. 8C and D) activities, respectively. It is known that atl (autolysin) is expressed as a 138-kDa precursor protein that is processed by protease to yield GL and AM domains of 54 and 63 kDa, respectively (34). Comparison between various strains revealed that the culture supernatant of the sarV mutant (ALC2319) displayed decreased GL (Fig. 8A) and AM (Fig. 8C) activities compared to the wild-type RN6390 strain. Significantly, no GL and AM activities were detected in supernatant of an atl mutant, JC1413 (Fig. 8, lanes 6). The intracellular proteins of the sarV mutant showed no significant variation of GL activity in the 54-kDa-size region (Fig. 8B) or AM activity in the 63-kDa-size region (Fig. 9D), but there was an extra zone of hydrolysis for both activities in the >97-kDa-size region compared to results with RN6390. We speculated that the appearance of this band was due to the unprocessed autolysin, which is yet to be confirmed. The cell wall-associated proteins of the sarV mutant also showed similar types of variation of the GL and AM activities as were found for intracellular proteins (data not shown). The variation in autolytic activity of autolysin is controlled by multiple factors that include SspA/V8 serine protease, SspB/cysteine protease, lrgAB operon/speculated holin and antiholin, another regulatory locus, and or an undefined regulatory mechanism (37). We also speculated that various other proteases, like aur, splA, and scp, which are decreased in a sarV mutant, might be involved in the resistance of autolysis.
FIG. 8.
Zymographic analysis of various fractions of different S. aureus strains. Extracellular proteins (A and C) and intracellular proteins (B and D) were isolated, and various amounts were loaded onto an SDS-12% polyacrylamide gel containing 1.0 mg of either M. luteus (A and B) or heat-killed RN6390 cells/ml, resolved, and treated with a buffer containing 1% Triton X-100, followed by staining with 1% methylene blue. Lanes 1 and 2, 2 and 4 μg of extra- or intracellular proteins of RN6390 strain; lanes 3 to 5, 2, 4, and 6 μg of extra- or intracellular proteins of the sarV mutant (ALC2319); lane 6, 4 μg of extra- or intracellular proteins of the atl mutant (JC1413). Molecular size markers in kilodaltons are indicated to the left of each gel.
FIG. 9.
Quantitative murein hydrolase assays of the sarV mutant. Aliquots of 100 μg of the extracellular proteins of various strains were added to a suspension of M. luteus cells (1.0 mg per ml), and turbidity was monitored for 8 h. Strains used for quantitative hydrolase analysis were the wild-type RN6390 (◊), sarV mutant ALC2319 (□), and the wild-type strain containing trans-supplemented sarV in multicopy plasmid pSK236, ALC2375 (▴). ×, control in which no extracellular protein was added to the cell suspension. These data are from a single representative experiment and were reproduced several times. OD was determined at 580 nm with a spectrophotometer (Spectronic 20).
To quantify the zymographic analysis, cell wall hydrolysis assays were performed as described by Rice et al. (38). Culture supernatants (100 μg) were added to a suspension of M. luteus cells (1.0 mg per ml) in 100 mM Tris-Cl (pH 8.0) and incubated at 37°C with shaking, and the turbidity of the samples was monitored at 580 nm. As shown in Fig. 9, the extracellular murein hydrolases of ALC2319 caused no significant decrease in turbidity of M. luteus cells after 8 h of incubation, compared to about a 35% decrease in turbidity with the wild-type RN6390 extracellular murein hydrolases. In contrast, the presence of the sarV gene in multicopy plasmid in the wild-type RN6390 caused about a 40% decrease in turbidity after 8 h. These results, along with the zymographic analysis, demonstrate that inactivation of the sarV gene results in a reduction in the extracellular murein hydrolase activity produced by S. aureus.
DISCUSSION
In prior studies, we and others have characterized SarA-like proteins, including SarA (SA0573), SarR (SA2089), SarS (SA0108; SarH1), Rot (SA1583), SarT (SA2286; SarH2), and SarU (SA2287; SarH3), that are involved in the regulation of virulence genes in S. aureus. Similar to SarA, SarV has sequence similarity to other SarA homologs, such as SarA, SarR, SarT, and Rot, and also to those with two homologous halves, such as SarS and SarU (data not shown). Unlike MgrA, SarV has a lower degree of sequence similarity to MarR-like proteins (e.g., ∼20% identities with MarR-like proteins of B. subtilis and Xanthomonas axonopodis). Despite this finding, there are specific residues with the protein sequence that are conserved, similar to those found with members of the SarA protein family (see below).
We recently solved the winged-helix structure of SarR, a member of the SarA protein family (24). The dimeric wing-helix structure of SarR is a three-domain structure containing a central helical core and two winged-helix motifs. Each of the winged-helix motifs encompasses a helix-turn-helix motif (residues 52 to 73) and a β-hairpin turn (residues 80 to 95), with both predicted to be DNA binding domains, based upon sequence alignment (23). Interestingly, many of the basic residues are conserved within members of the SarA protein family, consistent with the notion that the DNA binding domains might be highly conserved within this protein family. Indeed, detailed mutational analysis of SarR indicates that many of the conserved residues (e.g., K52, K80, R82, R88, etc.) are involved in DNA binding (A. C. Manna et al., unpublished data). The structures of MarR in E. coli and SarS in S. aureus were recently solved and were found to be variants of the dimeric winged-helix structure, both containing features that are distinct from SarR (1, 23). Based on structures and sequence alignment, we propose to classify the SarA protein family into three subfamilies: (i) the single-domain proteins (e.g., SarR and SarV); (ii) the two-domain proteins (e.g., SarS and SarU); and (iii) single-domain proteins that are homologous to MarR (e.g., MgrA and its homologs). We predict that members within the same subfamily likely posses similar structures with common DNA binding domains and a divergent activation or repression domain.
The transcription of sarA-like genes is highly variable in strain RN6390. For instance, sarA, sarR, and mgrA transcripts are highly expressed in the parental strain, whereas the transcription of sarU, sarT, and sarS is not readily detectable under normal laboratory growth conditions. We also found the sarV gene to be poorly transcribed in the parental strain. Northern analyses of agr, sarR, sarT, sarU, and sarS mutants revealed transcription of the sarV gene to be low in these mutants but at a high level in the sarA mutant and more so in the mgrA mutant. The effect of a sarA mutation on sarV transcription was also supported by similarly increased mean GFPuvr values in the sarV promoter-gfpuvr fusion analysis in three different sarA mutant strains in our laboratory (data not shown). The finding that the mgrA transcript was elevated in the sarV-supplemented parent and sarV mutant strains (Fig. 1) suggested the plausibility of a feedback loop on mgrA expression by the sarV gene, possibly to fine-tune mgrA expression. This result was also supported by promoter fusion studies (mgrA P1 promoter-gfpuvr) in a pair of isogenic sarV strains, documenting a substantial reduction in mean GFPuvr fluorescence in the sarV mutant ALC2319 (7,996 ± 750 units per OD650) compared the wild-type RN6390 (11,775 ± 1,000 units per OD650). However, the applicability of this magnitude of increased sarV gene dosage to augment mgrA transcription under in vivo conditions was not defined in this study.
In gel shift studies (Fig. 3), we found that purified SarA and MgrA proteins formed complexes with the sarV promoter with fairly high affinity. As protein-DNA complexes are shifted substantially, we speculate that multiple SarA or MgrA dimers might interact with each other to bind to the sarV promoter region. This is based on our modeling data, showing that three SarA dimers are needed to bind to a single 135-bp agr promoter fragment (unpublished data). The specificity of the SarA and MgrA proteins was determined by DNase I footprinting. Two SarA binding regions (box I and II) were found within the 268-bp sarV promoter sequence, locating and overlapping with the −10 and −35 region of the sarV promoter. Interestingly, each of these two binding regions resembles the SarA consensus binding motif or the SarA box, previously reported by our group (10). In contrast, the MgrA protein has binding regions outside of the promoter boxes that are distinct from the SarA binding site. These findings are consistent with the notion that MgrA and SarA may repress sarV via divergent mechanisms.
Using DNA gene chip technology, Dunman et al. (11) have demonstrated that agr and sarA regulate the expression of over 100 genes, including known virulence factors as well as other genes involved in metabolism and/or autolysis. Many of the known virulence genes are controlled by regulators like agr, sarA, sarA homologs, sae, and arlRS. Inactivation of the sarV gene has a partial effect on the expression of agr, lytSR, arlRS, and mgrA, while the effects on sae, sarA, sarR, sarT, sarS, rot, and sarU are minimal. However, the physiological relevance and the mechanism by which the sarV gene product, normally undetectable or at a very low level in the wild type under laboratory conditions, acts as a positive regulator (e.g., agr) are not clear from these studies.
Interestingly, sarV does not have any significant effect on cell surface proteins such as protein A (spa), FnbA and -B, and clumping factor A. The regulation of secreted proteins by sarV entails up-regulation of hla, splA, and aur, concordant with the notion that sarV regulates agr. The repression of the sarV gene by mgrA and sarA suggests that the sarV gene product may play a regulatory role in cell lysis, since both mgrA and sarA negatively control genes involved in autolysis, presumably by up-regulating murein hydrolase activity in respective mutants. Remarkably, lytic assays revealed that the sarV mutant was more resistant to the lytic effect of Triton X-100 and penicillin G. Surprisingly, an examination of regulatory genes (i.e., lytSR and arlRS) known to repress murein hydrolase activity disclosed down-regulation of these genes in the sarV mutant (Fig. 5 and 6). However, other genes (i.e., scdA and atl [Fig. 6]), positively linked to increase autolysis, were down-regulated in the sarV mutant, exactly what one would expect in a strain less prone to autolysis. The expression of lrgB, a part of the lrgAB operon, which negatively controls the autolysis process, was substantially higher in the sarV mutant than in the parent. Whether lrgB may be directly involved in the lytic-resistant phenotype of the sarV mutant was not clearly defined in our study. Because atl, scdA, aur, splA, and scp expression levels are altered in the sarV mutant, we speculate that sarV might, at least in part, render S. aureus cells more susceptible to cell lysis via a mechanism distinct from mgrA or sarA. Recently, a two-component system, vraSR, has been shown to positively regulate the cell wall-associated biosynthesis genes. DNA microarray analysis of S. aureus N315 and a vraSR mutant treated with vancomycin revealed that vancomycin induces transcription of 139 genes, and SA2062 (sarV) is one of them, up-regulated about 4.2-fold in both strains (22). These data supported our finding that sarV is an important transcriptional regulator and is involved in the regulation of the virulence and autolysis genes. Collectively, these data indicate that sarV impacts negatively upon the expression of proteases (aur, splA, and scp), an autolysin (atl), and a gene involved in peptidoglycan cross-linking (scdA). These phenotypic alterations would account for the autolysis-resistant phenotype of the sarV mutant and the sensitivity of the sarV-hyperexpressing strains to autolysis. Interestingly, zymogram analysis revealed that the sarV mutant has a different profile from that of an atl mutant (Fig. 8), thus indicating that reduced atl expression alone cannot account for the autolysis-resistant phenotype of the sarV mutant. The facts that sarV is repressed by negative regulators of autolysis such as MgrA and SarA and that the sarV mutant displays a divergent autolytic phenotype from mgrA and sarA mutants indicate that SarV is likely an important regulator controlled by mgrA and sarA to modulate cell lysis.
To further define the precise function of the sarV gene product, we are in the process of constructing various knockouts of sarV with other regulatory genes, particularly with sarA and mgrA, and investigating the molecular interaction of SarV with the upstream region of its target genes. Analysis of a sarV knockout in combination with mutations in sarA, mgrA, and other regulatory genes, coupled with our studies on the structure-function relationship of these proteins, may provide a clearer understanding of the mechanism of gene regulation by SarV and other SarA homologs. By unveiling the precise knowledge of these mechanisms and how they control virulence gene expression, this might open up new perspectives for antimicrobial chemotherapy using key inhibitors of these systems.
Acknowledgments
The contribution of the S. aureus genome database at The Institute for Genomic Research, the National Institutes of Health, and at the University of Oklahoma to this work is gratefully acknowledged. We thank Brian Bateman for his technical help. We also thank other members of our lab for their support and encouragement. We thank Karen Skorupski for critical reading and insightful comments on the manuscript. We also thank Simon Foster and M. Sugai for providing strains SH1000 and JC1413, respectively.
This work was supported in part by NIH grants AI37142 and AI50678 to A.L.C.
REFERENCES
- 1.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 2.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphycoccus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Bayles, K. W. 2000. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8:274-278. [DOI] [PubMed] [Google Scholar]
- 5.Boyce, J. M. 1997. Epidemiology and prevention of nosocomial infections, p. 309-329. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human diseases, vol. 12. Churchill Livingston, New York, N.Y.
- 6.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunskill, E. W., B. L. M. de Jonge, and K. W. Bayles. 1997. The Staphylococcus aureus scdA gene: a novel locus that affects cell division and morphogenesis. Microbiology 143:2877-2882. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., K. A. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien, Y.-T., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 11.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcriptional profiling-based identification of Staphylococcus aureus genes regulated by agr and/ or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto, D. F., and K. W. Bayles. 1998. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J. Bacteriol. 180:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 16.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingavale, S. S., W. van Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 18.Kahl, B., M. Goulian, W. Van Wamel, M. Herrmann, S. Simon, G. Kaplan, G. Peters, and A. L. Cheung. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect. Immun. 68:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor SarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 403-420. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 21.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K.-I. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R.-I. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 23.Li, R., A. C. Manna, A. L. Cheung, and G. Zhang. 2003. Crystal structure of the SarS protein from Staphylococcus aureus. J. Bacteriol. 185:4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Y., A. Manna, R. Li, W. E. Martin, R. C. Murphy, A. L. Cheung, and G. Zhang. 2001. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98:6877-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowy, F. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 26.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 175:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 34.Oshida, T., M. Sugai, H. Komatsuzawa, Y.-M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human diseases. Churchill Livingston, New York, N.Y.
- 36.Ramaduri, L., K. J. Lockwood, M. J. Nadakavukaren, and R. K. Jayaswal. 1999. Characterization of a chromosomally encoded glycylglycine endopeptidase of Staphylococcus aureus. Microbiology 145:801-808. [DOI] [PubMed] [Google Scholar]
- 37.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice, K. C., B. A. Firek, J. B. Nelson, S. Yang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakuo, Y., M. Sugai, H. Komatsuzawa, S. Nakashima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrader-Fischer, G., and B. Berger-Bachi. 2001. The AbcA transporter of Staphylococcus aureus affects cell autolysis. Antimicrob. Agents Chemother. 45:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugai, M., T. Fujiwara, H. Komatsuzawa, and H. Suginaka. 1998. Identification and molecular characterization of a gene homologous to epr (endopeptidase resistance gene) in Staphylococcus aureus. Gene 224:67-75. [DOI] [PubMed] [Google Scholar]
- 44.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 45.Truong-Bolduc, Q. C., X. Zhang, and D. C. Hooper. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 185:3127-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]