Abstract
Identification of polymorphic transposable elements (TEs) is important because TE polymorphism creates genetic diversity and influences the function of genes in the host genome. However, de novo scanning of polymorphic TEs remains a challenge. Here, we report a novel computational method, called PTEMD (polymorphic TEs and their movement detection), for de novo discovery of genome-wide polymorphic TEs. PTEMD searches highly identical sequences using reads supported breakpoint evidences. Using PTEMD, we identified 14 polymorphic TE families (905 sequences) in rice blast fungus Magnaporthe oryzae, and 68 (10,618 sequences) in maize. We validated one polymorphic TE family experimentally, MoTE-1; all MoTE-1 family members are located in different genomic loci in the three tested isolates. We found that 57.1% (8 of 14) of the PTEMD-detected polymorphic TE families in M. oryzae are active. Furthermore, our data indicate that there are more polymorphic DNA transposons in maize than their counterparts of retrotransposons despite the fact that retrotransposons occupy largest fraction of genomic mass. We demonstrated that PTEMD is an effective tool for identifying polymorphic TEs in M. oryzae and maize genomes. PTEMD and the genome-wide polymorphic TEs in M. oryzae and maize are publically available at http://www.kanglab.cn/blast/PTEMD_V1.02.htm.
Keywords: polymorphic transposon, high-throughput sequencing, rice blast fungus, maize
1. Introduction
Transposable elements (TEs) are repetitive DNA sequences capable of moving in genomes. On the basis of their transposition mechanism, TEs can be divided into two major classes: RNA-based retrotransposons (Class I) and DNA transposons (Class II). Since the discovery of TEs >50 years ago, TEs have been regarded as genetic parasites.1–3 Recent studies have, however, indicated that TEs and their movements affect genome size, genome stability, gene function, gene evolution, and epigenetic regulation.4–8 Also, TEs are important factor for genome size variation in plants.9 In grasses, the diploid genome sizes vary ∼30-fold, and most of the variation is due to the amplification of LTR retrotransposons in the intergenic regions of genomes.10 It has become increasingly clear that TEs might be a major genomic source of genetic diversity that enables host genomes to respond to environmental changes.11
Despite the presence of thousands or millions of TEs in a genome, most of them are silenced or are no longer mobile. In other words, few of them are still actively transposing that create TE polymorphism in closely related individuals in a species. Nevertheless, it is the polymorphic or active TEs that play the most important role in inserting polymorphism and allelic diversity, which are essential for population dynamics.
Given the importance of polymorphic TEs, however, to the best of our knowledge, no program has been designed for de novo scanning polymorphic TEs in a genome-wide fashion. So far, programs such as RepeatMasker,12 RECON,13 Repeatscout,14 and Piler15 have been used to identify repeat sequences (including TEs). RepeatMasker, which detects repeat sequences by using both genome sequences and repeat sequence libraries,12 has been used in many genome projects.16–20 Piler is a signature-based repeat searching tool, the program searches a query sequence for particular structures or motifs that are characteristic of a known repeat sequence.15 An important limitation of RepeatMasker and Piler is that they rely on the structure and characteristics of known elements and therefore cannot detect novel elements. In contrast, Repeatscout14 uses k-mer and spaced seed approaches21 that can identify repeat families de novo based on genome sequences without the use of repeat libraries. In addition to identifying repeat sequences, some programs, like TEMP,22 RelocaTE,23 and T-lex,24 can detect TE insertion polymorphisms (TIPs) based on high-throughput pair-end short reads or next-generation sequencing (NGS) reads from a single sample or multiple samples from individuals of a population. However, all of the above-mentioned tools do not identify TEs de novo. Therefore, there is a need to develop a program for de novo identification of polymorphic TEs genome-widely.
In this study, we developed a novel method named polymorphic TEs and their movement detection (PTEMD) that identifies polymorphic TEs de novo and uncovers genome-wide TIPs using reference genome and high-throughput short reads. Our method relies on reads situated at ‘breakpoints’, i.e. reads that span inserted TEs and their flanking sequences. The new method was written in a Linux-based program. Using the PTEMD program with representative fungal and plant genomes, we have detected multiple polymorphic TE families and TIPs.
2. Materials and methods
2.1. Genome sequence data
The Magnaporthe oryzae reference genome sequence (version 8) was downloaded from the Broad Institute (http://www.broadinstitute.org). To test the PTEMD program, we re-sequenced the genomes of M. oryzae strains HM-1 and HM-2, which were isolated from rice lesions by the single-spore method (data available from the PTEMD program homepage http://www.kanglab.cn/blast/PTEMD_V1.02.htm). The strains were cultured on oatmeal medium, and the hyphae were collected. DNA was extracted from hyphae using the CTAB method,25 and the purified DNA was used for constructing Illumina sequencing libraries with about 500-bp insert size. The libraries were sequenced with the Illumina Hiseq 2000 platform to generate 101-bp pair-ends reads with about ×50 coverage of the M. oryzae genome. The re-sequence data sets of M. oryzae generations 0, 10, and 20 were downloaded from a M. oryzae re-sequencing project (GenBank accession number: one isolate of generation 0: SRX220856; three isolates of generation 10: SRX220857, SRX220858, and SRX220859; three isolates of generation 20: SRX220860, SRX220861, and SRX220862). The maize B73 reference genome sequence was downloaded from the Plant Genome Database (http://www.plantgdb.org/). Mo17 re-sequencing data set was downloaded from GenBank SRA data sets (GenBank accession number SRX245309).
2.2. Comparison of PTEMD with different programs
PTEMD-A focus is to identify polymorphic TEs genome-wide, and it is the first program that focuses on de novo identifying polymorphic TEs. The programs RepeatMasker,12 Repeatscout,14 Piler,15 and RECON13 were compared with PTEMD-A for de novo scanning of repeat sequences in M. oryzae genome. The library used in this study is the Repbase library (version 19.06; http://www.girinst.org/repbase/). The programs used in this study are RepeatMasker (version 4.05; http://www.repeatmasker.org), Repeatscout version 1.0.514, Piler (http://drive5.com/piler), and RECON version 1.08 (http://www.repeatmasker.org/RECON-1.08.tar.gz). The default or general parameters were used for the programs.
PTEMD-B focuses on identifying the TIPs, and its performance was compared with the well designed and widely used TIP scanning programs, such as RelocaTE version 1.05,23 T-lex (version 2),24 and TEMP (version 1.01).22
2.3. PCR validation of TIPs
In order to validate the predicted TIPs, 30 positions in HM-1 and 30 in HM-2 were randomly selected from the PTEMD-detected MoTE-1 insertion regions. We designed 60 specific primer pairs for the 60 selected insertion regions. PCR (Tm: 55°C, 36 cycles) was carried out to determine the TE polymorphisms between reference genome and HM-1/2. When reference genome is subjected to PCR, all of the primer pairs amplify 100- to 300-bp products. The products were resolved on 1% agarose gel through electrophoresis to distinguish the size difference due to TE insertion between HM-1/2 and the reference genome.
2.4. Runtime and memory requirement of PTEMD
The present version of PTEMD was programmed in Perl and was tested in a Linux system. We tested PTEMD using a small genome (M. oryzae), and a large and repeat-rich genome (maize; Zea mays L.). In the M. oryzae genome, the 40-Mb genome sequence and 2-Gb clean bases of high-throughput reads data (×50 coverage of the genome) were used; ∼1.5 h of CPU time was needed for detecting all of the polymorphic TE families, and 10 min of CPU time was needed for de novo detection of the new insertion sites for a single 2-kb TE sequence. When the maize genome B73 was tested (genome size ≈ 2.1 Gb), 70-Gb clean bases high-throughput reads data (accession number: SRX245309, about ×35 coverage of the genome) were used, ∼23 h of CPU time was needed for detecting all of the polymorphic TE families.
3. Results
3.1. PTEMD detection of polymorphic TEs and TIPs
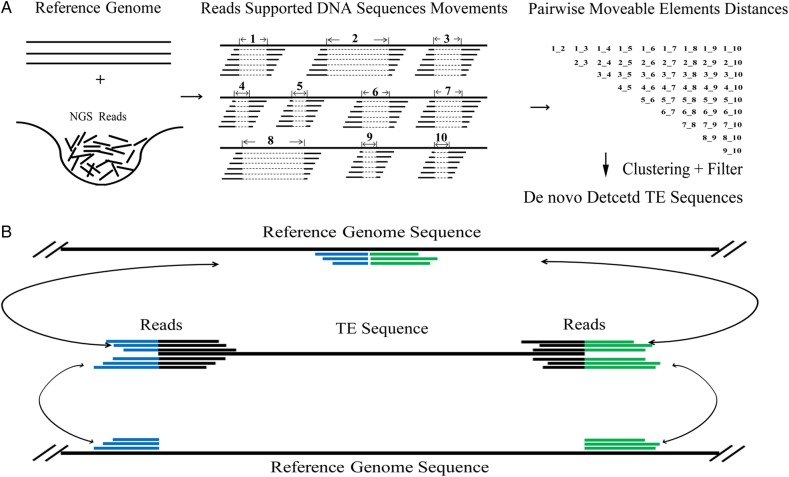
Active TEs or recently active TEs often create insertion/deletion (indel) polymorphisms in the population. PTEMD utilizes this feature to collect TEs or other movable sequences that are involved in the formation of polymorphisms. PTEMD consists of PTEMD-A (Supplementary Fig. S1A) and PTEMD-B (Supplementary Fig. S1B) pipelines. The objective of PTEMD-A is to identify the polymorphic TE families. First, high-throughput short reads will be mapped to the reference genome (Fig. 1A, left and middle part, partially assembled genomes are also been supported). For a certain locus, if there is a TE insertion in the reference genome but absent from the genome under re-sequencing, a ‘gap’ will form where reads are mapped to the flanking sequences but not the sequence in between (detail parameters refer to Pindel program).26 Those sequences in the gap represent putative mobile unit and will be retrieved from the reference genome, thereafter, PTEMD uses a pair-wise sequences alignment strategy to construct a distance matrix (Fig. 1A, right part) for all retrieved sequences. We used a time-saving method in this pair-wise sequence-alignment step. Only sequences of similar size were used for the pair-wise alignment (not exceeding 10% length), and the sequence identity was recorded as a vector in each distance-matrix calculating cycle. The sequences with low sequence identity (<75%) were not used for the alignment in the next step of the traversal cycle. This reduced the running time by 53–73% (Supplementary Fig. S2A–C) in the tested data sets. Since intact polymorphic transposons are usually highly similar, only those sequences which have at least three homologous sequences sharing ≥97% identity over 95% length will be retained and classified into different TE families, using this criterion, our result showed that only a small part of PTEMD-detected sequence families are simple repeats or truncated TEs (6.7% in M. oryzae and 16.0% in maize), which may be due to indel/homologous recombination/sequence rearrangement. In other words, most of the PTEMD-detected sequence families in M. oryzae and maize are bona fide intact transposons.
Figure 1.
The core algorithm of PTEMD. (A) PTEMD-A, de novo active TE scanning pipeline. The high-throughput short reads are first mapped to the reference genome and the genomic regions correspond to mapping gaps (the dotted line indicated the gaps) are identified and classified into different repeat families with neighbour-join clustering. (B) PTEMD-B, de novo TIPs detecting pipeline. The middle panel represents the high-throughput reads which partially mapped to the TE sequence, the un-matched part of the reads were re-located to the reference genome sequences. There are two major mapping patterns: (i) the interval distance on the reference genome between left and right mapped reads is near or equal to 0 (top panel of B), representing a TIP in the re-sequenced individual; (ii) the interval distance between left and right mapped reads is equal or almost equal to the length of the TE (bottom panel of B), representing a same TE distribution between the reference and the re-sequenced individual. This figure is available in black and white in print and in colour at DNA Research online.
To determine the exact boundary of the classified TE sequences, we first align each classified sequence cluster to a clustalw27 format file using MUSCLE28; then we use a stringent criteria (at both left and right sides of the sequence cluster, the terminal base has at least 50% consistent within the sequence cluster) to get the longest consensus sequence of the cluster (Supplementary Fig. S2D). We then annotated the sequences by aligning them with NT/NR, Repbase, and maize TE database (http://maizetedb.org/~maize/), and manually classified them based on their structure, terminal sequences, and target site duplications (TSDs). Those classified sequences are the PTEMD-A-identified polymorphic TE families.
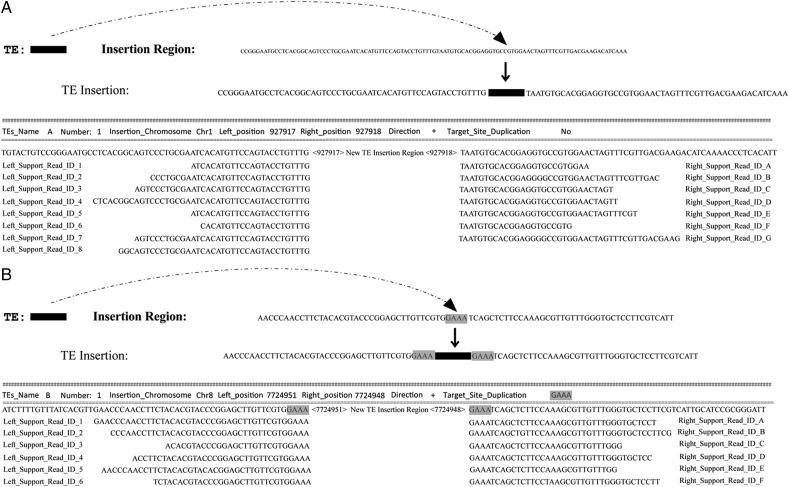
The objective of PTEMD-B is to identify the TIPs. PTEMD-B aligns the high-throughput reads to the TE sequence (Fig. 1B, middle, detail parameter refer to BWA program MEM algorithm),29 the reads which partially mapped to the terminus of the TE sequence are selected from the re-sequencing data set, the un-mapped part of those sequences (Fig. 1B, highlighted with blue and green) are isolated and re-mapped to the reference genome sequences. There are two major mapping patterns; TE insertion only in the re-sequenced genome, and TE in the same location compared with the reference genome (Fig. 1B). For the detection of TIPs, it requires both sides of the flanking sequences mapped to the same location in the genome that lacks the insertion. As a result, the detected TIPs include only polymorphism created through indels including transpositions. The TIPs do not include sequence rearrangements caused by recombination, inversion, and translocations. In addition, due to the short length of the reads, it is impossible for the program to distinguish whether the TIP is caused by a full-length TE or a TE with internal deletions. In summary, by using the two-step approach, PTEMD is able to quickly scan polymorphic TE families and their insertion landscapes from high-throughput short reads. The output format of PTEMD-B is shown in Fig. 2, the file contains the detail insertion site/s, TSD, and all of the reads situated at ‘breakpoints’.
Figure 2.
Output format of PTEMD-B. (A) TE insertion without TSD. (B) TE insertion with a TSD ‘GAAA’. All of the reads information at the breakpoint region will be included in the PTEMD-B output file.
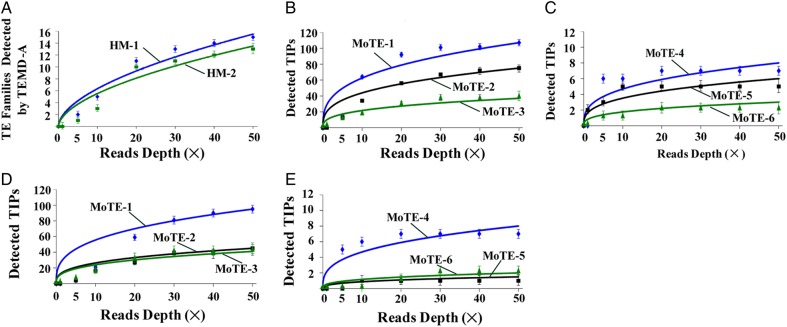
To evaluate the impact of sequencing coverage on PTEMD performance, HM-1 and HM-2 genome sequence data were randomly split into data sets with ×1, ×5, ×10, ×20, ×30, ×40, and ×50 coverage. These seven data sets were then analysed by the PTEMD, and the results were used to evaluate the performance of PTEMD on the depth of re-sequencing data sets. A polymorphic TE family was defined as the presence of at least three TIPs in the reference genome compared with the re-sequenced data sets, with ≥97% sequence identity over 95% length. As expected, the number of TE families detected was positively correlated with sequencing depth (r= 0.98 ± 0.01, P = 0.00015 ± 0.00005, Fig. 3A). Although the number of polymorphic TE families continued to increase as the coverage approached ×50, the slope decreased, and at ×50 coverage, 15 TE families were identified in the M. oryzae genome. Among the 15 TE families, 14 represent intact TEs and 1 is a TE fragment (see next section and Supplementary Table S1 for details). The total copy number of those 14 intact TE families in rice blast reference genome is 905.
Figure 3.
The PTEMD performance on polymorphic TE scanning and TIPs detection under different depths of re-sequencing data sets in M. oryzae. Y-axis represents the number of the TE families detected with the vertical line indicates standard deviation (A) and represents the number of detected TIPs with the vertical line indicates standard deviation (B–E). (A) PTEMD-A-detected TE families (HM-1 and HM-2 are two strains). X-axis: reads coverage of M. oryzae genome. (B) PTEMD-B-detected TIPs in HM-1 strain for MoTE-1–3. (C) PTEMD-B-detected TIPs in HM-1 strain for MoTE-4 to 6. (D) PTEMD-B-detected TIPs in HM-2 strain for MoTE-1–3. (E) PTEMD-B-detected TIPs in HM-2 strain for MoTE-4–6. This figure is available in black and white in print and in colour at DNA Research online.
The average sequence identity between the genomes of HM-1 and HM-2 and that of reference genome was 99.97 ± 0.01%. It is estimated that some of the PETMD-A detected polymorphic TEs may be still active or recently active. To test this, we analysed a genetically purified, single-isolated data sets.30 Using PTEMD, we detected direct breakpoint reads evidences for MoTE-1, -2, -3, -4, -5, -6, -7, and -11 within 10–20 generations, indicated that more than half of the PTEMD-detected polymorphic TE families may still be active.
To determine the false-positive rate of PTEMD-B, we used the 14 intact TE families, and identified TIPs and evaluated the false-positive of the TIPs. A TIP event was defined as a TE present in HM-1/HM-2 and absent in the rice blast reference genome.31 Using the same data sets as in Fig. 3A, we found that the number of TIPs detected was positively correlated with sequencing coverage (MoTE-1: r= 0.94 ± 0.05, P = 0.0014 ± 0.0018; MoTE-2: r= 0.95 ± 0.01, P = 0.0003 ± 0.0002; MoTE-3: r= 0.95 ± 0.007, P = 0.00045 ± 0.0002; MoTE-4: r= 0.73 ± 0.03, P = 0.04 ± 0.01; MoTE-5: r= 0.75 ± 0.03, P = 0.04 ± 0.007; MoTE-6: r= 0.90 ± 0.06, P = 0.004 ± 0.004). For example, the number of detected TIPs with MoTE-1 increased monotonically when the coverage increased but became saturated at about ×30 to ×50 in both HM-1 (Fig. 3B) and HM-2 (Fig. 3D). When the coverage increases beyond ×30, the number of PTEMD-detected TIPs still increases but at a declining rate. The number of detected TIPs for other TE families demonstrates the same tendency as MoTE-1 (Fig. 3B–E).
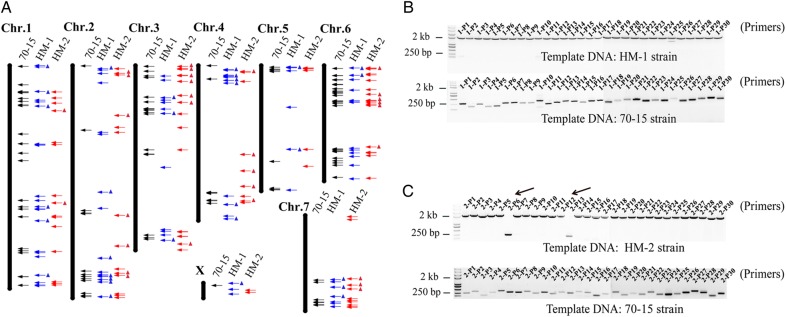
To test whether an identified TIPs was authentic, we determined the false-positive rate of the predicted TE insertions by amplifying the expected fragments in the HM-1 or HM-2 genome with PCR. For experimental validation, we selected the MoTE-1 family because the insertion polymorphic rates of MoTE-1 for all three isolates (HM-1, HM-2, and the reference genome) were 100% (Fig. 4A, all of the MoTE-1 sequences are located in different regions). Among the TIPs involving the MoTE-1 family and detected by PTEMD-B, 60 TIPs from HM-1 and HM-2 were randomly selected for validation (Fig. 4A, the blue and red triangles represent the selected TIPs). If the predicted TIP was authentic, the difference in PCR product size between a fungal isolate with the TE insertion and a isolate without the TE insertion would be the length of MoTE-1, which is ∼1.8 kb. Using M. oryzae reference genome sequences, we designed 60 primer pairs flanking the predicted insertion sites (Supplementary Table S2). The results showed that 100% of MoTE-1 TIPs in HM-1 and 93% of those in HM-2 represented true insertion polymorphism (Fig. 4B and C). To assess why two of the predicted TIPs in HM-2 (Fig. 4C, black arrows regions, 2-P6 and 2-P13) were not detected by PCR, we examined the sequence coverage for these two predicted TIPs and found that both had fewer supporting reads (2 and 3 breakpoint reads supported) than the validated insertions (≥4 breakpoint reads supported, P< 0.01). Our finding indicated that the false-positive rate of the PTEMD-B is quite low at ∼3.3% for the MoTE-1 family.
Figure 4.
TIPs of MoTE-1 among M. orzyae strains of reference genome, HM-1 and HM-2 and their validation. (A) Distributions of the MoTE-1 TIPs in the M. oryzae reference genome, HM-1 and HM-2 genomes. Black lines: M. oryzae chromosomes (X represents sequence that are not located in the seven chromosomes). Black arrows: location of the MoTE-1 TIPs in the reference genome. Gray and light gray arrows: location of MoTE-1 TIPs in HM-1 and HM-2 strains, respectively. All of the black, blue, and red arrows were located in different positions, indicated the polymorphism distribution of MoTE-1 in different strains. (B and C) PCR validating of the randomly selected 60 PTEMD-B-detected MoTE-1 TIPs [blue and red triangles in (A) represent the selected TIPs]; 30 pairs of primers complementary to flanking sequence of TIPs (B, from 1-P1 to 1-P30) for HM-1 vs. reference genome and 30 primer pairs (C, from 2-P1 to 2-P30) for HM-2 vs. reference genome were used in the testing. The length of MoTE-1 is 1.86 kb, and the length of the 60 PCR productions in reference genome varies from 100 to 300 bp (B and C, bottom panels; top panels of B and C represent the PCR production size in HM-1 and HM-2 isolates). This figure is available in black and white in print and in colour at DNA Research online.
3.2. Comparison of PTEMD with other relative programs
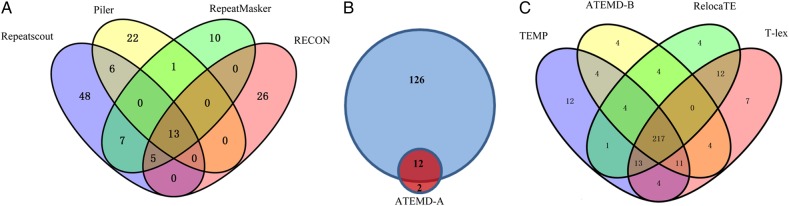
To assess the proportion of polymorphic TE families in genome, we de novo detected all of the repeat sequences families in M. oryzae genome using RepeatMasker, Repeatscout, Piler, and RECON; 36, 79, 42, and 44 repeat families were identified, respectively (Fig. 5A, Venn diagram of the four program detected repeat sequence families), about half of the families are redundant, meanwhile, total non-redundant repeat families are 138 (Fig. 5B, the blue and blue-red overlapped region). Using PTEMD-A, we detected 14 polymorphic TE families. Among the 14 TE families, 12 families have highly similar (97% identity over 95% length) homologue within the 138 non-redundant repeat families, the other two families are only detected by PTEMD-A (Fig. 5B). In other words, only ∼10% of the repeat families in M. oryzae genome are polymorphic TE families. The two polymorphic TE families that are only detected by PTEMD-A have low copy numbers (ranging from three to seven) in M. oryzae genome, and this explained why they are missed by other repeat scanning programs.
Figure 5.
Venn diagram showing the comparing of PTEMD-detected TEs and TIPs with related programs. (A) Repeat sequence families independently detected by four programs of Repeatscout, Piler, RepeatMasker, and RECON; a total number of 138 non-redundant repeat families have been identified in M. oryzae genome. (B) Comparing the 14 PTEMD-A-detected candidate active TEs with the 138 non-redundant repeat families, 12 families are overlapped. (C) Comparing the TIPs detected by PTEMD-B, TEMP, RelocaTE, and T-lex. The total non-redundant TIPs detected by all of the four tools is 301, and 217 of them are co-detected by all of the four tools. This figure is available in black and white in print and in colour at DNA Research online.
Previous result demonstrated that PTEMD-B have the best performance when the re-sequencing data are over ×30 coverage of the reference genome. Using ×50 re-sequencing data sets (HM-1 strain) and the reference genome, we detected the TIPs for the 14 intact TE families using PTEMD-B, TEMP, RelocaTE, and T-lex, and 248, 266, 255, and 268 TIPs have been detected, respectively. Most of them (217 TIPs) are co-detected by all of the four tools (Fig. 5C, Venn diagram of the four programs detected TIPs), the non-redundant TIPs are 301, and only 9% (27 of 301) of the detected non-redundant TIPs are tool specific; these indicated that all of the four methods have the high-efficiency performance under high coverage of re-sequencing data set in M. oryzae genome. As a result, PTEMD performs better for repeat identification and is comparable for scanning of TIPs compared with other programs. However, since PTEMD is the only program that assumes both functions, it is the ‘one step’ tool for identifying polymorphic TEs and their insertions without the need of any other information except genomic sequences.
3.3. Polymorphic TE families in the M. oryzae and maize genomes
Having demonstrated that PTEMD can detect polymorphic TEs that are likely to have been recently active, we next tested PTEMD with representative genomes. To assess the usability of PTEMD on genomes with widely different sizes and repeat contents, the genomes of M. oryzae (with a 40-Mb genome and 9.7% repetitive sequences)31 and maize (with a 2.1-Gb genome and over 85% repetitive sequences)32 were used.
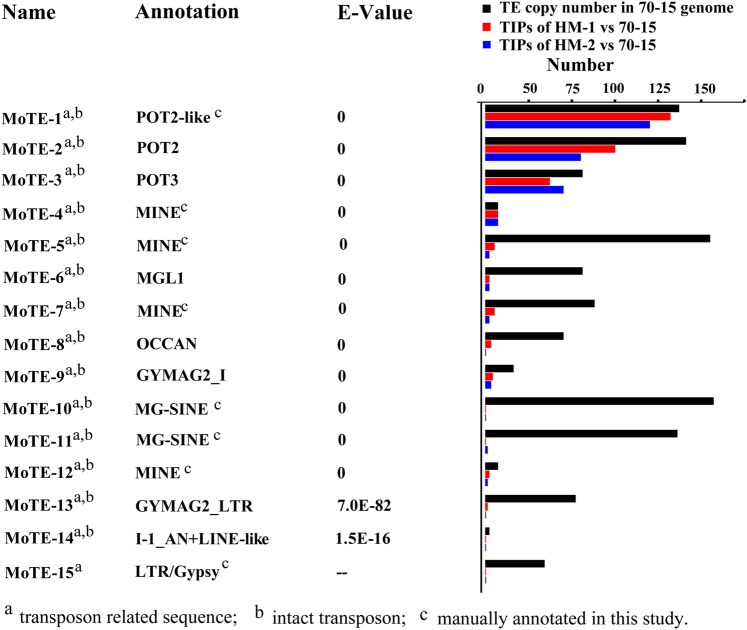
As mentioned earlier, we re-sequenced two field strains of M. oryzae (HM-1 and HM-2) at ×50 depth and compared the sequences to that of the reference genome. The average sequence identity between the genomes of HM-1 and HM-2 and that of reference genome was 99.97 ± 0.01%. Using PTEMD-A, we identified 15 TE families (Supplementary Table S1, 14 intact TE families and 1 partial TE family). To determine whether those 15 TE families were previously reported, we compared them with three databases: (i) Repbase, (ii) GenBank nucleotide sequences database (NT), and (iii) GenBank protein sequences database (NR). The results indicated that 14 families have matches in the Repbase/NT/NR databases (threshold: 1e-10) (Fig. 6). MoTE-2, -3, and -8 are previously reported transposons Pot2,33 Pot3,34 and OCCAN,35 respectively; another 11 (MoTE-1, -4, -5, -6, -7, -9, -10, -11, -12, -13, and -14) are annotated in the M. oryzae genome sequencing project;31 4 (MoTE-1, -2, -3, and -8) are DNA transposons; 2 (MoTE-6 and MoTE-14) are related to LINEs; and 2 (MoTE-9 and MoTE-13) are LTR elements. We further verified/classified all 15 elements based on their structure, TSD, and similarity to known elements at protein level (Supplementary Table S1). All the four DNA elements are Pogo-like elements that belong to Tc1/Mariner-like superfamily. Three of the elements are Gypsy-like LTR elements. MoTE-9 represents an intact LTR element, whereas MoTE-13 represents a solo LTR. MoTE-15, on the other hand, only represents part of the internal region of a LTR retrotransposon, suggesting the detection of MoTE-15 is not due to TIP, but due to structural variation like indels among individual members of an element family. In other words, the fragment represented by MoTE-15 is not present in all elements, thus creates apparent indel polymorphism and is detected in the mapping process. All the remaining elements are LINEs or their deletion derivatives. Except MoTE-14, all the other seven families are related to MoTE-6. Interestingly, the seven elements all end with simple repeats (ACT)n (n = 5–9), which is different from the poly (A) tails of most known LINEs. This result indicates that PTEMD works well with small genomes like that of M. oryzae, and all the sequences recovered are bona fide transposon sequences albeit not all of them represent intact elements.
Figure 6.
Annotation and TIPs of 15 repeat families detected by PTEMD in M. oryzae. The annotation of each TE family is based on the most similar repeats in RepBase and NCBI NT and NR databases. The third column indicates the Top-paralogous E-value of the sequence alignments between the TEs and known repeats for each family. Middle panel: PTEMD-detected TIPs in HM-1 and HM-2 strains compared with the number of TEs in the reference genome. This figure is available in black and white in print and in colour at DNA Research online.
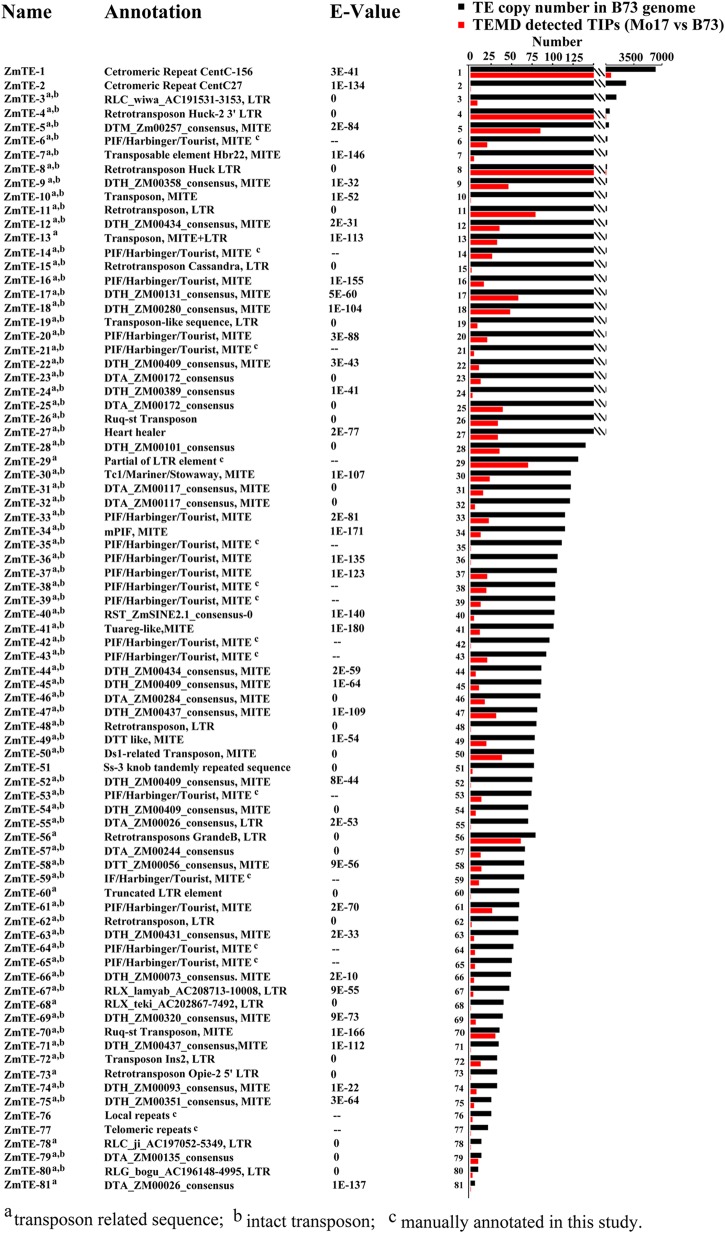
To further test PTEMD with a repeat-rich genome, we used maize as a model system. Over 85% of the maize genome, which is large in size (2.1 Gb), consists of repetitive sequences.32 In comparison with the Mo17 re-sequencing data set (GenBank accession number SRX245309) with B73 reference genome sequences, we identified 81 polymorphic repetitive sequence families (named ZmTE-1 to 81; sequences and their annotations are provided in Supplementary Table S3). The copy number in the reference genome is shown in Fig. 7. To determine the identity of those 81 sequences families, we compared them with four databases (Repbase, NT, NR, and the maize-specific TE database: http://maizetedb.org/~maize/). For those without matches in the database, we manually classify them based on their structure, terminal sequences, and TSDs. Overall, 75 (92.6%) out of the 81 families represent TEs (Fig. 7 and Supplementary Table S3) with 68 intact TEs and 7 partial TE sequences. The total copy number of those 68 TE families in B73 is 10,618. The five non-TE families are centromeric repeats, knob sequences, telomeric repeats, and local repeats. In addition, one family represents a chimeric structure from both DNA transposon and retrotransposon. Among the 75 TE families, 19 (25.3%) families are from retrotransposons and 56 (74.7%) are from DNA elements. The 19 retrotransposon families include one SINE, eight intact LTR retrotransposons, four solo LTRs, and six truncated or fragmented LTR elements (Supplementary Table S3). Two of the solo LTRs are similar to the high copy number LTR element Huck-2.36 One of the intact LTRs corresponds to Zeon-1, a moderate copy number element.37 If we consider the 6 truncated or fragmented elements likely derived from structural changes such as deletions, only 13 retrotransposons are polymorphic between Mo17 and B73 genomes. Among the 56 DNA elements, only one represents fragmented element, the remainder stand for intact elements, which are likely from recent transposition. This suggests that there are 4-fold polymorphic DNA transposons vs. retrotransposons. The 55 intact DNA elements are all small (<1 kb) non-autonomous DNA transposons including 51 miniature inverted TEs (MITEs). Notably, 41 (80%) of them are Tourist MITEs, which belong to PIF/Harbinger superfamily of DNA transposons. Several previously characterized DNA transposons are identified in this study, including Ds1, Heartbreaker, Heart healer, mPIF, Ruq-st , and the MITE that was found in maize Br2-3 allele.38–42 Finally, it is worth of mention that for either species, the output of PTEMD-A does not contain any composite elements such as nested insertions, which is likely attributed to the stringent criteria that we used to define a TE family (≥97% identity over 95% length).
Figure 7.
Annotation and TIPs of 81 sequences families detected by PTEMD in maize. The annotation of each TE/repeats family is based on the most similar repeats in RepBase, NCBI NT and NR databases, and maize TE database. Other features are shown similarly as in Fig. 6. This figure is available in black and white in print and in colour at DNA Research online.
3.4. TIPs in M. oryzae and maize
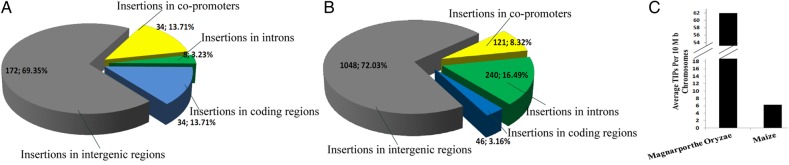
Compared with other TE sequences, polymorphic TEs create more TIPs in different individuals. To further assess the potential activity of TEs in M. oryzae and maize, we calculated the TIPs created by intact elements. By using PTEMD-B to compare the re-sequenced data sets of M. oryzae (HM-1 strain) and maize (Mo17) with their reference genomes, we detected 248 TIPs in M. oryzae (Fig. 8A, detail in Supplementary Table S4) and 1455 TIPs in maize (Fig. 8B, detail in Supplementary Table S5). The average TIPs per 10 Mb chromosome regions in M. oryzae and maize are 62.0 and 6.9 (Fig. 8C), respectively. We calculated the number of TIPs distributed in the genome (1 Mb windows was used), TIPs in both M. oryzae and maize were nonuniformly distributed (detail shown in Supplementary Figs. S3 and S4). In M. oryzae, among the detected 248 TIPs, 91.1% (226 of 248) are caused by MoTE-1, -2, -3, and -4, albeit the copy number of those four TE families in reference genome contain only 32.2% (291 of 905) of total detected polymorphic TE sequences. And MoTE-1, -2, -3 are DNA transposons and MoTE-4 is a retrotransposon. In maize, among the detected 1455 TIPs, there were a total 539 TIPs derived from retrotransposons and 916 TIPs from DNA transposons. This was translated to 44 TIPs per retrotransposon family and 16 TIPs for DNA transposons. However, a close examination indicated that the majority (351) of TIPs from retrotransposons were attributed to ZmTE-4 and ZmTE-8 (Fig. 7); both are related to Huck-2 elements. If the two families were excluded, each retrotransposon family generated 20 TIPs, which was largely comparable to that of DNA transposons. As a result, the contribution to polymorphism of individual class I and class II elements was similar with the exception of Huck-2, which generated nearly 10 times more polymorphic insertions than other elements. Despite the presence of more polymorphic DNA transposon families as well as more TIPs from DNA transposons, the average size of polymorphic DNA transposons is only 311 bp, whereas that for retrotransposon is 3.8 kb, which is a dozen-fold that of DNA transposons. This explains why DNA elements contribute to much less genome size than retrotransposons in maize.
Figure 8.
The classification of the TIPs in M. oryzae and maize genomes. (A and B) Classification of the PTEMD-B-detected TIPs in M. oryzae (A) and maize (B), respectively. Different parts represent the classification of the TIPs. (C) Average TIPs per 10 Mb chromosome regions in M. oryzae and maize. This figure is available in black and white in print and in colour at DNA Research online.
To further identify the potential effects of TE insertions on gene functions, we first grouped the TIPs into four major classes: (i) TE insertions in the gene coding regions; (ii) TE insertions in the core-promoter regions (the 500 bp upstream sequence of the gene transcription start site)43; (iii) TE insertions in the intron regions; and (iv) TE insertions in intergenic regions. Then, we downloaded the GFF files for the three genomes and developed Perl scripts (available from the author of this paper) to extract and classify the TIPs. Because TE insertions in coding regions directly interrupt the genes, we expected that few of the TIPs would be located in coding regions. Among the 248 TIPs detected in the M. oryzae genome, however, 172, 34, 8, and 34 were inserted in intergenic regions, core-promoter regions, intron regions, and coding regions, respectively, and the percentages were 69.4, 13.7, 3.2, and 13.7%, respectively (Fig. 8A and Supplementary Table S4). This indicated that 30.6% of TIPs in M. oryzae genome were located in the genic regions. Among the 1455 TIPs detected in the maize genome (Supplementary Table S5), 72.0, 8.3, 16.5, and 3.2% were inserted in the intergenic regions, core-promoter regions, intron regions, and coding regions, respectively (Fig. 8B).
4. Discussion
Two methods can be used to determine whether a DNA fragment is a potential active TE.44 In the first method, researchers detect the fragment's activity by monitoring de novo insertions in the next generation. In the second method, researchers identify TIPs among individuals. We developed PTEMD to detect both polymorphic TEs and genome-wide TIPs using reference genomes and high-throughput reads data sets. Unlike previously developed library-based or structure-based methods, PTEMD identifies polymorphic TEs (PTEMD-A) and their TIPs (PTEMD-B) by searching all of the evidences of sequence movement at the TIP sites. PTEMD-A does not depend on any TE sequence or libraries. The program detects all of the mobile sequences by comparing the re-sequencing data with the reference genome and then classifies the sequences into clusters, which are filtered and classified into TE families. Our result indicated that, although non-TE sequences are included in the PTEMD-A output file, however, the proportion of non-TE families is low (6.7% in M. oryzae and 16.0% in maize), indicating that most of the PTEMD-A-detected sequences are intact polymorphic TE families. Moreover, we analysed the genetically purified, single-isolated re-sequencing data sets, and we found the direct evidences of current mobility for more than half of the PTEMD-A-detected TE families in M. oryzae. This indicates that many of the PTEMD-A-detected polymorphic TE families could be currently active.
PTEMD-B maps the high-throughput short reads to the TE sequences and maps all of the partially mapped reads to the reference genome sequences. The position information is then analysed, and all of the TIPs including breakpoint evidence are presented in the PTEMD output file. As shown for the genome of M. oryzae and maize, only 6.7% (1 of 15) in M. oryzae and 16.0% (13 of 81) in maize of the PTEMD-detected mobile sequence families are not intact TEs; the polymorphism of those sequence families may be due to indel/homologous recombination/sequence rearrangement, in other words, most of the PTEMD-detected mobile sequence families are good candidates for active TEs.
We integrated the cores of the Pindel,26 BWA29/Bowtie2,45 Muscle,28 and BLAST46 programs into the PTEMD program as the sequence-alignment engines. BWA is a widely used high-throughput reads alignment program that can quickly map high-throughput short reads to the reference genome sequences.29 The BWT-MEM algorithm is used in PTEMD to map the high-throughput reads to both the genome and TE sequences. Muscle28 is used in PTEMD for clustering the sequence families. BLAST46 is used to align the TE sequence to the reference genomes. The PTEMD output file contains all of the detected TE families and their detailed distribution positions including all of the supporting breakpoint evidence. The PTEMD output file is therefore useful for checking the raw sequence data of the identified TE families or new TE insertions.
Previous programs have used three main methods for repeats scanning. The first method is a library-based approach, such as that used in RepeatMasker.12 The main drawback of programs that use these kinds of homology-dependent searches is that they can only detect sequences that are already known, i.e. they cannot detect completely novel elements. The second method is signature-based repeat searching, such as Piler.15 This method can search a query sequence for existing structures or motifs that are characteristic of a known repeat sequence, it can be used to find new repeats but not new classes of repeats. The third method is represented by k-mer and spaced seed approaches, such as those used in Repeatscout.14 This approach can de novo identify the repeat families based on genome sequences and has been widely used for scanning new TE families. Unlike these three methods, PTEMD is a homology-independent method that can scan active TE candidates (also including other polymorphic repeats) and their genome-wide insertion sites using high-throughput pair-end short reads and reference genomes. PTEMD therefore overcomes some important drawbacks of the previously published methods and will become an important tool for de novo polymorphic TE scanning.
M. oryzae is a model pathomycete that can quickly overcome the host resistance.47 More than 80 resistance genes have been identified in rice but most of these resistance genes remain effective for only 2–3 years.48 A previous study provided evidence that a TE insertion into the promoter region of the avirulence gene Piz-t caused an avirulent strain to become to virulent49; however, the genome-wide polymorphic TE families and their distribution landscapes among different M. oryzae strains were still largely unknown. Using PTEMD, we identified 15 polymorphic TE families in M. oryzae (14 intact TEs and 1 partial TE). In reference genome, the 14 intact TE families contain 905 copies, and the total size of those TE families is ∼1.5 Mb (3.8% of the M. oryzae genome). The top four TE families (MoTE-1, -2, -3, and -4) are widely distributed in strains HM-1 and HM-2. Further analysis of the dynamics and evolution of these top four TE families in field populations may help elucidate how these TEs affect M. oryzae virulence.
To evaluate the performance of PTEMD with large and repeat-rich genomes, we identified 68 TE families in the maize genome are polymorphic and likely active in the recent past. The total copy number of the TE sequences is 10,618 in the maize genome. Those TE sequences cover 10.1 Mb of maize genome (0.48% of genome). Our results provide novel insights about the transposon biology. First of all, TE polymorphism varies greatly between the two tested species. The highest TE polymorphism is detected in M. oryzae: its genome size is only 2% that of maize but harbours 14 families of polymorphic TE families. Second, despite that most of the maize genome consists of LTR retrotransposons, DNA elements contribute more to polymorphism among different varieties compared with LTR retrotransposons. LTR elements make up the largest component of the genome due to their large size, not high activity. Third, not a single autonomous DNA transposon has been detected to be recently mobile in maize, and all the detected DNA transposons are very small (<1 kb) non-autonomous elements. Obviously, some of the autonomous elements in maize are still functioning because they are providing transposition machinery for non-autonomous elements. It is likely their large sizes make them less competent for transposition, consistent with the notion that DNA element size is critical for the transposition activity.50–52 Finally, the polymorphic TEs discovered using PTEMD can be used as potential novel tagging tools for breeding or studying gene function. As more and more genome sequences are available in the future, PTEMD will be an important tool in identifying polymorphic TEs in different organisms.
Availability
PTEMD program, HM-1 and HM-2 strains' re-sequencing data sets (sequences in this study), and the genome-wide polymorphic TEs in M. oryzae and maize are freely and publically available at http://www.kanglab.cn/blast/PTEMD_V1.02.htm.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported in part by the National Natural Science Foundation of China to H.K. (grant numbers 31571964 and 31201476), the 973 Project (grant number 2012CB114005) of the Ministry of Science and Technology China to G.-L.W., and the US National Science Foundation to N.J. and S.-H.S. (grant number IOS-1126998) and to S.-H.S. (grant number MCB-1119778). Funding to pay the Open Access publication charges for this article was provided by the Institute of Plant Protection, Chinese Academy of Agricultural Sciences.
Supplementary Material
Acknowledgements
We thank Shuangyong Yan for reading, discussing, and editing the paper.
References
- 1.Doolittle W.F., Sapienza C.. 1980, Selfish genes, the phenotype paradigm and genome evolution, Nature, 284, 601–3. [DOI] [PubMed] [Google Scholar]
- 2.Orgel L.E., Crick F.H.. 1980, Selfish DNA: the ultimate parasite, Nature, 284, 604–7. [DOI] [PubMed] [Google Scholar]
- 3.Rebollo R., Romanish M.T., Mager D.L.. 2012, Transposable elements: an abundant and natural source of regulatory sequences for host genes, Annu. Rev. Genet., 46, 21–42. [DOI] [PubMed] [Google Scholar]
- 4.Studer A., Zhao Q., Ross-Ibarra J., Doebley J.. 2011, Identification of a functional transposon insertion in the maize domestication gene tb1, Nat. Genet., 43, 1160–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fedoroff N.V. 2012, Transposable elements, epigenetics, and genome evolution, Science, 338, 758–67. [DOI] [PubMed] [Google Scholar]
- 6.Britten R.J. 2010, Transposable element insertions have strongly affected human evolution, Proc. Natl. Acad. Sci. USA, 107, 19945–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lisch D. 2013, How important are transposons for plant evolution?, Nat. Rev. Genet., 14, 49–61. [DOI] [PubMed] [Google Scholar]
- 8.Polak P., Domany E.. 2006, Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes, BMC Genomics, 7, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piegu B., Guyot R., Picault N. et al. . 2006, Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice, Genome Res., 16, 1262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennetzen J.L., Kellogg E.A.. 1997, Do plants have a one-way ticket to genomic obesity?, Plant Cell, 9, 1509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kidwell M.G., Lisch D.R.. 2000, Transposable elements and host genome evolution, Trends Ecol. Evol., 15, 95–9. [DOI] [PubMed] [Google Scholar]
- 12.Tarailo-Graovac M., Chen N.. 2009, Using RepeatMasker to identify repetitive elements in genomic sequences, Curr. Protoc. Bioinformatics, Chapter 4, Unit 4.10. [DOI] [PubMed] [Google Scholar]
- 13.Bao Z., Eddy S.R.. 2002, Automated de novo identification of repeat sequence families in sequenced genomes, Genome Res., 12, 1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price A.L., Jones N.C., Pevzner P.A.. 2005, De novo identification of repeat families in large genomes, Bioinformatics, 21, i351–8. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R.C., Myers E.W.. 2005, PILER: identification and classification of genomic repeats, Bioinformatics, 21, i152–8. [DOI] [PubMed] [Google Scholar]
- 16.Initiative A.G. 2000, Analysis of the genome sequence of the flowering plant Arabidopsis thaliana, Nature, 408, 796. [DOI] [PubMed] [Google Scholar]
- 17.Venter J.C., Adams M.D., Myers E.W. et al. . 2001, The sequence of the human genome, Science, 291, 1304–51. [DOI] [PubMed] [Google Scholar]
- 18.Gibbs R.A., Weinstock G.M., Metzker M.L. et al. . 2004, Genome sequence of the Brown Norway rat yields insights into mammalian evolution, Nature, 428, 493–521. [DOI] [PubMed] [Google Scholar]
- 19.Goff S.A., Ricke D., Lan T.-H. et al. . 2002, A draft sequence of the rice genome (Oryza sativa L. ssp. japonica), Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 20.Yu J., Hu S., Wang J. et al. . 2002, A draft sequence of the rice genome (Oryza sativa L. ssp. indica), Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 21.Lerat E. 2010, Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs, Heredity, 104, 520–33. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang J., Wang J., Theurkauf W., Weng Z.. 2014, TEMP: a computational method for analyzing transposable element polymorphism in populations, Nucleic Acids Res., 42, 6826–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robb S.M., Lu L., Valencia E. et al. . 2013, The use of RelocaTE and unassembled short reads to produce high-resolution snapshots of transposable element generated diversity in rice, G3 (Bethesda), 3, 949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiston-Lavier A.S., Carrigan M., Petrov D.A., Gonzalez J.. 2011, T-lex: a program for fast and accurate assessment of transposable element presence using next-generation sequencing data, Nucleic Acids Res., 39, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart C., Via L.E.. 1993, A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications, Biotechniques, 14, 748–50. [PubMed] [Google Scholar]
- 26.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z.. 2009, Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads, Bioinformatics, 25, 2865–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson J.D., Gibson T.J., Higgins D.G.. 2002, Multiple sequence alignment using ClustalW and ClustalX, Curr. Protoc. Bioinformatics, Chapter 2, Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 28.Edgar R.C. 2004, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res., 32, 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows wheeler transform, Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon J., Choi J., Lee G.W., Dean R.A., Lee Y.H.. 2013, Experimental evolution reveals genome-wide spectrum and dynamics of mutations in the rice blast fungus, Magnaporthe oryzae, PLoS One, 8, e65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean R.A., Talbot N.J., Ebbole D.J. et al. . 2005, The genome sequence of the rice blast fungus Magnaporthe grisea, Nature, 434, 980–6. [DOI] [PubMed] [Google Scholar]
- 32.Schnable P.S., Ware D., Fulton R.S. et al. . 2009, The B73 maize genome: complexity, diversity, and dynamics, Science, 326, 1112–5. [DOI] [PubMed] [Google Scholar]
- 33.Kachroo P., Leong S.A., Chattoo B.B.. 1994, Pot2, an inverted repeat transposon from the rice blast fungus Magnaporthe grisea, Mol. Gen. Genet., 245, 339–48. [DOI] [PubMed] [Google Scholar]
- 34.Kang S., Lebrun M.H., Farrall L., Valent B.. 2001, Gain of virulence caused by insertion of a Pot3 transposon in a Magnaporthe grisea avirulence gene, Mol. Plant Microbe Interact., 14, 671–4. [DOI] [PubMed] [Google Scholar]
- 35.Kito H., Takahashi Y., Sato J., Fukiya S., Sone T., Tomita F.. 2003, Occan, a novel transposon in the Fot1 family, is ubiquitously found in several Magnaporthe grisea isolates, Curr. Genet., 42, 322–31. [DOI] [PubMed] [Google Scholar]
- 36.SanMiguel P., Gaut B.S., Tikhonov A., Nakajima Y., Bennetzen J.L.. 1998, The paleontology of intergene retrotransposons of maize, Nat. Genet., 20, 43–5. [DOI] [PubMed] [Google Scholar]
- 37.Hu W., Das O.P., Messing J.. 1995, Zeon-1, a member of a new maize retrotransposon family, Mol. Gen. Genet., 248, 471–80. [DOI] [PubMed] [Google Scholar]
- 38.Sutton W.D., Gerlach W.L., Peacock W.J., Schwartz D.. 1984, Molecular analysis of ds controlling element mutations at the adh1 locus of maize, Science, 223, 1265–8. [DOI] [PubMed] [Google Scholar]
- 39.Pisabarro A.G., Martin W.F., Peterson P.A., Saedler H., Gierl A.. 1991, Molecular analysis of the Ubiquitous (Uq) transposable element system of Zea mays, Mol. Gen. Genet., 230, 201–8. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q., Arbuckle J., Wessler S.R.. 2000, Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize, Proc. Natl. Acad. Sci. USA, 97, 1160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X.Y., Feschotte C., Zhang Q., Jiang N., Eggleston W.B., Wessler S.R.. 2001, P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases, Proc. Natl. Acad. Sci. USA, 98, 12572–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattramakki D., Dolan M., Hanafey M. et al. . 2002, Insertion-deletion polymorphisms in 3 ‘ regions of maize genes occur frequently and can be used as highly informative genetic markers, Plant Mol. Biol., 48, 539–47. [DOI] [PubMed] [Google Scholar]
- 43.Zou C., Sun K., Mackaluso J.D. et al. . 2011, Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana, Proc. Natl. Acad. Sci. USA, 108, 14992–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang C.R.L., Burns K.H., Boeke J.D.. 2012, Active transposition in genomes, Annu. Rev. Genet., 46, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Salzberg S.L.. 2012, Fast gapped-read alignment with Bowtie 2, Nat. Methods, 9, 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul S.F., Madden T.L., Schaffer A.A. et al. . 1997, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 25, 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dean R., Van Kan J.A., Pretorius Z.A. et al. . 2012, The Top 10 fungal pathogens in molecular plant pathology, Mol. Plant Pathol., 13, 414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W., Liu J., Triplett L., Leach J.E., Wang G.-L.. 2014, Novel insights into rice innate immunity against bacterial and fungal pathogens, Annu. Rev. Phytopathol., 52, 213–41. [DOI] [PubMed] [Google Scholar]
- 49.Li W., Wang B., Wu J. et al. . 2009, The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t, Mol. Plant Microbe Interact., 22, 411–20. [DOI] [PubMed] [Google Scholar]
- 50.Way J.C., Kleckner N.. 1985, Transposition of plasmid-borne Tn10 elements does not exhibit simple length-dependence, Genetics, 111, 705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hennig S., Ziebuhr W.. 2008, A transposase-independent mechanism gives rise to precise excision of IS256 from insertion sites in Staphylococcus epidermidis, J. Bacteriol., 190, 1488–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao D., Ferguson A., Jiang N.. 2015, Transposition of a rice Mutator-like element in the yeast Saccharomyces cerevisiae, Plant Cell, 27, 132–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.