Abstract
Clostridium perfringens is the cause of several human diseases, including gas gangrene (clostridial myonecrosis), enteritis necroticans, antibiotic-associated diarrhea, and acute food poisoning. The symptoms of antibiotic-associated diarrhea and acute food poisoning are due to sporulation-dependent production of C. perfringens enterotoxin encoded by the cpe gene. Glucose is a catabolite repressor of sporulation by C. perfringens. In order to identify the mechanism of catabolite repression by glucose, a mutation was introduced into the ccpA gene of C. perfringens by conjugational transfer of a nonreplicating plasmid into C. perfringens, which led to inactivation of the ccpA gene by homologous recombination. CcpA is a transcriptional regulator known to mediate catabolite repression in a number of low-G+C-content gram-positive bacteria, of which C. perfringens is a member. The ccpA mutant strain sporulated at a 60-fold lower efficiency than the wild-type strain in the absence of glucose. In the presence of 5 mM glucose, sporulation was repressed about 2,000-fold in the wild-type strain and 800-fold in the ccpA mutant strain compared to sporulation levels for the same strains grown in the absence of glucose. Therefore, while CcpA is necessary for efficient sporulation in C. perfringens, glucose-mediated catabolite repression of sporulation is not due to the activity of CcpA. Transcription of the cpe gene was measured in the wild-type and ccpA mutant strains grown in sporulation medium by using a cpe-gusA fusion (gusA is an Escherichia coli gene encoding the enzyme β-glucuronidase). In the exponential growth phase, cpe transcription was two times higher in the ccpA mutant strain than in the wild-type strain. Transcription of cpe was highly induced during the entry into stationary phase in wild-type cells but was not induced in the ccpA mutant strain. Glucose repressed cpe transcription in both the wild-type and ccpA mutant strain. Therefore, CcpA appears to act as a repressor of cpe transcription in exponential growth but is required for efficient sporulation and cpe transcription upon entry into stationary phase. CcpA was also required for maximum synthesis of collagenase (kappa toxin) and acted as a repressor of polysaccharide capsule synthesis in the presence of glucose, but it did not regulate synthesis of the phospholipase PLC (alpha toxin).
Clostridium perfringens is a gram-positive anaerobic bacterium readily found in soil, sediments, and the intestinal contents of humans and animals (19). It is the cause of several human diseases, including gas gangrene (clostridial myonecrosis) and enteritis necroticans (41). C. perfringens is also the third most common source of bacterial food poisoning in the United States (34, 40). After ingestion of contaminated food containing vegetative cells, food poisoning symptoms are caused by production of a potent enterotoxin protein (C. perfringens enterotoxin [CPE]) made by sporulating cells in the gastrointestinal tract. The enterotoxin interacts with epithelial cell tight junction proteins in a series of steps, leading to cell death and the symptoms of diarrhea and intestinal cramping characteristic of the disease (33). Enterotoxin-positive strains of C. perfringens have increasingly been identified as a significant cause of non-food-borne and antibiotic-associated diarrhea (1, 21, 23, 48). A strong correlation between the location of the cpe gene and disease has been observed: acute food poisoning isolates carry the cpe gene on the chromosome, while isolates obtained from cases of non-food-borne or antibiotic-associated diarrhea have a plasmid-borne copy of the cpe gene (13, 14, 48). Whether located on the chromosome or on a plasmid, CPE production is always correlated with sporulation by C. perfringens (35, 43).
Because sporulation and enterotoxin production are inextricably linked, one approach to dealing with the disease is to identify agents that block sporulation and, therefore, CPE production and disease. Glucose has been shown to act as a catabolite repressor of sporulation in C. perfringens (29, 45). The mechanism of catabolite repression of sporulation by glucose in C. perfringens has not been determined, but in Bacillus subtilis, another gram-positive spore-forming bacterium, many catabolite repressor (CR) effects from glucose, including sporulation, have been shown to be mediated by the transcriptional regulator CcpA, a member of the LacI/GalR family of repressor proteins (11, 20). Homologues of CcpA have been found across a broad spectrum of low-G+C-content gram-positive bacteria, including the clostridia (10, 15). CcpA binds to cis elements termed cre (catabolite responsive elements) and functions either as a transcriptional repressor or activator (51). However, CcpA often has weak, nonspecific affinity for DNA when added alone in in vitro experiments (17). A corepressor of CcpA is HPr-ser-phosphate (17). The HPr protein is phosphorylated at Ser 46 by HPr serine kinase/phosphatase, and the kinase function is activated by fructose 1,6-bis-phosphate (FBP). The HPr-ser-P-CcpA complex then binds to cre elements and regulates transcription of genes in the CcpA regulon. By regulating HPr ser kinase/phosphatase activity, the intracellular concentration of FBP provides a link between the metabolic state of the cell and CcpA transcriptional activity.
A ccpA mutant strain of B. subtilis exhibits partial relief of CR effects of glucose on sporulation, but mutants lacking active CcpA still show some glucose-mediated repression of sporulation (11, 37), suggesting that other mechanisms of CR are involved. Using a whole-genome transcriptional analysis approach, Moreno et al. identified many genes that were subject to CR by glucose in a CcpA-independent manner and found that most of these were involved in sporulation (38). Because sporulation is necessary for enterotoxin synthesis (i.e., food poisoning) by C. perfringens (35), we investigated the role that CcpA plays in CR of sporulation and CPE synthesis. Unlike the situation in B. subtilis, where CcpA partially mediates CR of sporulation by glucose, CcpA is needed for efficient sporulation and CPE synthesis in C. perfringens whether glucose is present or not, establishing a novel role for CcpA in sporulating bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. C. perfringens was grown in a Coy anaerobic chamber (Coy Laboratory Products) in PGY medium (30 g of proteose peptone per liter, 20 g of glucose per liter, 10 g of yeast extract per liter, and 1 g of sodium thioglycolate per liter) as described previously (36). E. coli was grown in Luria-Bertani broth (10 g of tryptone per liter, 5 g of NaCl per liter, and 5 g of yeast extract per liter) on plates or in liquid.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| DH10B | F−mcrA D(mrr-hsdRMS-mcrBC) F80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara, leu)7697 galU galK λ−rpsL endA1 nupG | Gibco/BRL Corp. |
| HB101 | supE44 ara14 galK2 lacY1 Δ(gpt-proA)62 rpsL20 (Strr)xyl-5 mtl-1 recA13 Δ(mcrC-mrr) HsdS-(r− m−) | D. Ohman |
| C. perfringens | ||
| NCTC 8798 | CPE+ | C. Duncan |
| SM101 | High-frequency electroporation derivative of NCTC 8798 | 51 |
| SM120 | ccpA mutant derivative of SM101 | This study |
| Plasmids | ||
| pBluescript SK− | Stratagene Corp. | |
| pBluescript SK+ | Stratagene Corp. | |
| pSF3 | Source of mob region | D. Ohman |
| pJIR750 | C. perfringens-E. coli shuttle vector | 7 |
| pJIR751 | C. perfringens-E. coli shuttle vector | 7 |
| pYZ67 | pBluescript SK-/catP | This study |
| pSM300 | pBluescript SK-/ermBP | This study |
| pSM300M | pSM300/mobilization (mob) region from pSF3 | This study |
| pSM310 | pBluescript SK-/ccpA | This study |
| pSM310Cm | pSM310/catP | This study |
| pSM225 | pSM300M/ccpA-catP | This study |
| pSM237 | pJIR751/cpe-gusA fusion | This study |
| pSM257 | pJIR751/ccpA | This study |
| pDOB13 | pJIR751/ccpA | This study |
Sporulation assays were done based on previously described methods (2, 28). Briefly, overnight cultures grown at 37°C in fluid thioglycolate medium (FTG) were added to prewarmed serum bottles containing 50 ml of Duncan-Strong sporulation medium (DSSM) with raffinose, which contained, per liter, 4 g of yeast extract, 15 g of proteose peptone, 5.4 g of Na2HPO4, 1 g of sodium thioglycolate, and 4 g of raffinose (28). After 24 h, the cultures grown in DSSM were serially diluted and plated on PGY medium to determine the total number of CFU. To determine the number of spores in the culture, samples were also heated at 75°C for 15 min prior to plating on PGY medium. In order to ascertain the effect of sugars on sporulation, stock solutions of each sugar were added to give a final concentration of 5 mM.
Cloning the ccpA gene of C. perfringens.
The published sequences of ccpA genes from several Bacillus species and the sequence encoding the CcpA homolog from Clostridium saccharobutylicum (formerly designated a strain of C. acetobutylicum [24]), named RegA by Davison et al. (15), were used to design two degenerate primers to amplify, by PCR, a ∼250-bp region near the N terminus of the ccpA gene of C. perfringens strain NCTC 8798 (S. B. Melville, unpublished data). DNA sequencing confirmed the fragment was part of the ccpA gene, because the sequence showed a high level of homology to other ccpA genes. The fragment was then used as a probe for Southern blot analyses of chromosomal DNA isolated from strain NCTC 8798. For Southern blot analyses, 10 μg of chromosomal DNA was digested to completion and agarose gel electrophoresis and transfer to a nylon membrane were done as previously described (42). To detect hybridization to the probes, the Phototope Star detection system was used according to the manufacturer's instructions (New England Biolabs). A single 3.8-kb HindIII fragment was found to hybridize to the probe. A plasmid library was constructed in E. coli containing C. perfringens HindIII-digested chromosomal DNA fragments 3 to 5 kb in size and was cloned into the vector pBluescript SK+. E. coli clones containing the library were then screened by colony hybridization with the 32P-labeled ccpA gene fragment as probe. One clone was identified that contained a 3.8-kb HindIII insert and was named pSM310. Sequencing of the insert showed it contained the entire ccpA gene and flanking sequences (Fig. 1A).
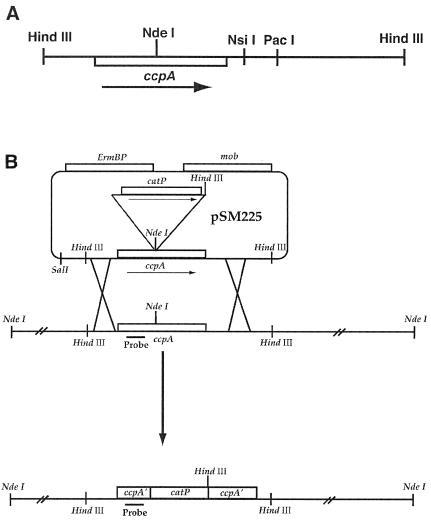
FIG. 1.
(A) Schematic diagram showing restriction sites and ccpA gene orientation in the 3.8-kb HindIII insert in pSM310. (B) Diagram showing the strategy used for allele replacement of the wild-type copy of the ccpA gene of C. perfringens with an insertionally inactivated copy. The location of the ccpA-specific probe is shown beneath the N-terminal coding region of the ccpA gene.
Plasmid constructs.
The 3.8-kb HindIII fragment from pSM310 was subcloned into the E. coli-C. perfringens shuttle vector pJIR751 (6) to make plasmid pDOB13. Plasmid pDOB13 was digested with PacI and KpnI, the overhanging ends were removed by T4 DNA polymerase, and the plasmid was self ligated. This resulted in plasmid pSM257, which contains only the ccpA gene and its promoter region (Fig. 1A). Plasmid pYZ67 was made by PCR amplification of the catP gene from plasmid pJIR750 (6), using primers with an EcoRI site at the upstream end and HindIII at the downstream end, digesting the PCR product with EcoRI and HindIII, and ligating the catP gene to pBluescript SK− at the EcoRI-HindIII sites of the polylinker. Plasmid pSM310Cm was made by cloning the EcoRI-HindIII catP gene fragment from pYZ67, blunt ending with the Klenow fragment of DNA polymerase, and ligating to pSM310 digested with NdeI that also had the ends filled in by the Klenow fragment. The unique NdeI site in pSM310 lies near the center of the ccpA gene (Fig. 1A). The entire 5.1-kb ccpA-catP construct was cloned into the mobilizable suicide vector pSM300M by digesting pSM310Cm with BamHI and SalI, isolating the ccpA-catP fragment, and ligating it to BamHI-SalI-digested pSM300M to make pSM225. pSM300M was made in two steps. First, the bla gene from pBluescript SK− was replaced with the ermBP gene from pJIR751 by ligating the blunt-ended ermBP gene fragment into DraI (at position 1912)- and SspI (at position 442)-digested pBluescript SK− to create pSM300. Next, the mob locus from plasmid pSF3 (44), cut with EcoRI and BglII, was blunt ended and ligated to pSM300 (which had been partially digested with PvuII) at the PvuII site at position 997 to make pSM300M.
The cpe-gusA fusion vector, pSM237, was made by digesting pSM104 (36), which encodes chloramphenicol resistance, with PstI and EcoRI and isolating the fragment containing the cpe-gusA gene fusion. This fragment was then ligated to the E. coli-C. perfringens shuttle vector, pJIR751 (6), which encodes erythromycin resistance, that had been digested with PstI and EcoRI. Plasmid pSM237 was used to quantify cpe promoter activity by measuring β-glucuronidase enzyme activity, the product of the gusA gene (22).
Construction of a ccpA mutation in C. perfringens.
A ccpA mutant strain was constructed by using allele replacement techniques by conjugational transfer of plasmid pSM225 from E. coli into C. perfringens strain SM101 (52). pSM225 lacks an origin of replication that functions in C. perfringens. The recombination strategy used is shown in Fig. 1B. First, plasmid pSM225 was transformed into E. coli strain HB101 carrying plasmid pRK2013 (16). pRK2013 provided all of the tra gene functions in trans to mobilize pSM225 into C. perfringens. Plasmid pSM225 was conjugationally transferred by biparental mating of E. coli strain HB101 carrying pRK2103 and pSM225 with C. perfringens strain SM101. Conjugation was carried out on filters at a ratio of donor to recipient of 100:1. Filters were then placed on PGY plates and were incubated in an anaerobic chamber overnight. The following day the filters were washed with PGY liquid medium to detach the bacteria and the cells were pelleted by centrifugation and plated on the C. perfringens-selective media, tryptose sulfite cycloserine agar, containing erythromycin (30 mg/liter) and chloramphenicol (20 mg/liter). Several transformants were isolated from the mating. Two of these transformants were then grown in PGY liquid cultures with chloramphenicol only and were spread on PGY chloramphenicol plates. Replica plating techniques were used to screen >3,000 colonies for the loss of erythromycin resistance and maintenance of chloramphenicol resistance, the pattern expected if the cell had undergone the double recombination event shown in Fig. 1B. Two erythromycin-sensitive chloramphenicol-resistant isolates were obtained, and results are described for one of them, SM120.
Enzyme and polysaccharide capsule assays.
Cell cultures grown for extracellular toxin assays were incubated in either PGY or PY (PGY without added glucose) medium. All cultures were inoculated with a 1% aliquot of a culture grown overnight in PGY and grown to late log phase before samples were removed for toxin assays. Alpha toxin activity was measured through a slightly modified version of a previously published method (49). Ten-microliter samples of the appropriate filtered cell culture supernatants were added to 100 μl of a reaction mixture containing 1.0 mM CaCl2, 0.1 mM ZnCl2, and 10 mM p-nitrophenylphosphorylcholine (NPPC) in phosphate-buffered saline (pH 7.2). Reaction mixtures were incubated at 37°C for 1 h and then were quenched with 1 ml of cold 0.02 N NaOH, and the A410 was then determined.
Kappa toxin activity assays were based on previously described methods (5, 32). Overnight cultures of C. perfringens were transferred into 50 ml of either PGY or PY at a 1% inoculum and were incubated anaerobically for 4 h at 37°C. The cultures were centrifuged, and the supernatants were collected into 250-ml centrifuge bottles. Ammonium sulfate (Fisher Scientific) was added to the solution to saturation to precipitate proteins. After 1 h of incubation at room temperature, the samples were centrifuged at 10,000 × g and the supernatants were poured off. The pellets were resuspended in 25 ml of sodium borate buffer (0.2 M boric acid, 0.15 M NaCl) and were transferred to 50-ml centrifuge bottles. Ammonium sulfate was added again until saturation was achieved, at which time samples were incubated for 1 h at room temperature. Upon centrifugation, the supernatant was removed and the pellet was resuspended in 500 μl of sodium borate buffer supplemented with 30 μM ZnCl2. These concentrated protein solutions were dialyzed four times against 1-liter volumes of zinc-supplemented sodium borate buffer. After dialysis, collagenase activity was determined by measuring the release of azo-dye from Azocoll (Sigma). For each sample, 6 mg of Azocoll was placed in a microcentrifuge tube and was washed with sodium borate buffer. Four-hundred microliters of concentrated culture supernatant was added to each tube, along with 800 μl of sodium borate buffer. The reaction mixtures were incubated for 3 h at 37°C with horizontal shaking. After incubation, samples were centrifuged at 13,000 × g for 15 min and the supernatants were transferred to new tubes. The A595 for each sample was determined in a Thermospectronics Genesys spectrophotometer. Specific activity for collagenase is shown as A595 units per milligram of protein. Protein concentrations were determined by using the Bio-Rad protein assay kit (Hercules, Calif.). β-Glucuronidase enzyme assays to detect cpe-gusA transcriptional activity in C. perfringens were done as previously described (36). Glucose was added to the growth medium at a final concentration of 5 mM as described for some experiments.
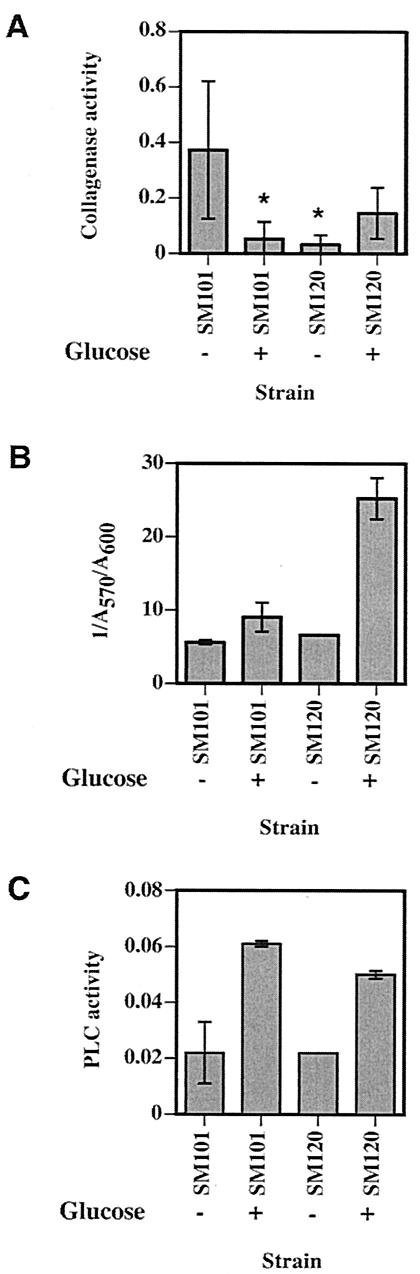
The levels of polysaccharide capsule produced by SM101 and the ccpA mutant were measured by the ability of cellular polysaccharides to bind trypan blue by the method previously published by Black and Yang (9), with slight modifications. Cells were grown overnight in T-soy broth with and without 14 mM glucose (Difco, Detroit, Mich.) and were transferred into fresh medium at a 5% inoculum size. After 4 to 6 h of growth, cultures were centrifuged, the pellets were washed once with modified morpholinepropanesulfonic acid (MOPS) buffer (10 mM MOPS [pH 7.6], 2 mM MgSO4, 1 mM CaCl2) and resuspended in 3.5 ml of modified MOPS buffer, and the optical density at 600 nm (OD600) was obtained for each sample. Duplicate assays were performed by adding 1.35 ml of cell suspension to a 2-ml microcentrifuge tube with 150 μl of a 50-μg/ml trypan blue solution (Biowhittaker, Walkersville, Md.). The tubes were incubated with horizontal shaking for 30 min at room temperature and were centrifuged at 13,000 × g. For each sample, 200 μl of the supernatant was loaded in triplicate in a 96-well plate and the A570 was obtained by using a SPECTRAfluor plus plate reader (Tecan, Salzburg, Austria). The amount of dye bound (the more dye that bound to the capsule, the lower the A570 observed) was divided by the OD600 values for each sample to give dye-binding units as A570/OD600. In order to provide a positive correlation to the amount of capsule material, the results were presented as the ratio 1/A570/OD600 in Fig. 4.
FIG. 4.
Levels of synthesis of collagenase (A), polysaccharide capsule (B), and PLC (C) in the wild-type (SM101) and ccpA mutant (SM120) strains under the conditions shown in the figure. For panels A and C the medium was PGY or PY (PGY without glucose), while in panel B the medium was T-soy broth with 14 mM glucose added as indicated. Shown are the means and standard deviations of triplicate (A) and duplicate (B and C) sample values. The asterisks denote a statistically significant difference (P < 0.05) between strain SM101 without glucose and the indicated values, determined with the t test. There was no statistically significant difference in the values compared with those of any of the other possible combinations shown in panel A.
Measurement of raffinose utilization.
Raffinose concentrations were determined as described in protocols published by Boehringer Mannheim for the UV spectroscopic determination of raffinose in foodstuffs (catalog number 428167). Samples were taken from the cell cultures at the designated times. Supernatants containing soluble carbohydrates were separated from cells by centrifugation at 10,000 × g for 5 min, followed by filtration through a 0.2-μm-pore-size syringe filter. Filtered supernatants were stored at −20°C until analyzed.
Nucleotide sequence accession number.
The C. perfringens ccpA gene sequence from strain SM101 has been deposited in GenBank under the accession number AF309566. Since the time of this submission to GenBank, the entire sequence of the genome of strain SM101 has been determined by The Institute of Genomic Research (http://www.tigr.org/tdb/mdb/mdbinprogress.html). The ccpA gene sequence in AF309566 is identical to the sequence obtained by TIGR for the ccpA gene.
RESULTS
C. perfringens ccpA gene.
The C. perfringens ccpA gene from strain SM101 was predicted to encode a protein of 332 amino acids with a molecular mass of 37,200 Da. The CcpA protein of C. perfringens exhibited the highest level of identity (∼70%) to CcpA orthologs from C. saccharobutylicum (termed RegA) (15) and C. tetani (10) but exhibited significantly less identity (≤44%) to other CcpA orthologs from gram-positive bacteria in the sequence databases. Kraus et al. (25) noted the significant sequence difference between the RegA protein of C. saccharobutylicum and other members of the CcpA subfamily of the LacI/GalR family of transcriptional regulators.
The coding region for the ccpA gene in strain SM101 lies 717 bp downstream from a divergently transcribed conserved hypothetical protein and 250 bp upstream of a hypothetical gene transcribed in the same direction as ccpA (http://www.tigr.org/tdb/mdb/mdbinprogress.html). Thirty base pairs downstream of the coding region for ccpA there is a potential 28-bp stem-loop structure followed by a string of seven thymidines, possibly indicating the presence of a rho-independent terminator that can function in C. perfringens (52).
Allele replacement of the wild-type ccpA gene with an inactivated copy of ccpA.
Strain SM101 was used to study the effects of a ccpA mutation because it sporulates well, is CPE positive, and is relatively easily transformed by electroporation. A conjugational system (30) was used to transfer the suicide plasmid, pSM225, from E. coli into C. perfringens. The allele replacement strategy, following integration of pSM225 into the chromosome, is shown in Fig. 1B. To confirm that the recombination events had taken place as shown in Fig. 1B, chromosomal DNA was isolated from the ccpA mutant strain SM120, digested with restriction enzymes, and subjected to a Southern blot analysis, which confirmed that the recombination events shown in Fig. 1B had occurred (data not shown). To our knowledge, this is the first report of a mutation made in a gene in C. perfringens by using conjugational transfer of nonreplicating plasmids from E. coli to C. perfringens. This technique may be useful for constructing mutations in C. perfringens strains that are not efficiently transformed by electroporation.
The CcpA protein is needed for efficient sporulation in C. perfringens but does not mediate glucose CR of sporulation.
To determine the role CcpA plays in regulating glucose-mediated CR of sporulation in C. perfringens, the wild-type (SM101) and ccpA mutant (SM120) strains were grown in DSSM-raffinose sporulation medium with or without added glucose. In the absence of glucose, the wild-type strain sporulated at 68% while the ccpA mutant strain produced spores at 1.1%, a 60-fold decrease compared to spore production levels of the wild type (Table 2). The presence of a plasmid (pSM257) carrying a wild-type copy of the ccpA gene in the ccpA mutant strain restored sporulation only to 11%, still sixfold less than the wild-type level (Table 2). However, the presence of plasmid pSM257 in the wild-type strain lowered sporulation to a level (6.5%) similar to that seen with the complemented ccpA mutant strain, suggesting multiple copies of the ccpA gene are inhibitory to normal sporulation functions (Table 2). The origin and replication functions of pSM257, which are derived from plasmid pIP404, probably result in a plasmid copy number of 20 to 25 per cell in C. perfringens (41). The addition of 5 mM glucose to the DSSM-raffinose medium lowered the sporulation of the wild-type strain to 0.033%, a 2,000-fold reduction, while the ccpA mutant strain sporulated at 0.0014%, an 800-fold reduction compared to spore production levels of the ccpA mutant strain in the absence of glucose (Table 2). Because the ccpA mutant strain shows a level of CR due to glucose similar to that of the wild-type strain, it appears that CcpA does not mediate CR effects of glucose on sporulation. The presence of pSM257 in the wild-type strain caused a 26-fold reduction in sporulation when glucose was present, which is in the same range as the 10-fold decrease seen in the absence of glucose (Table 2). Compared to the ccpA mutant strain, however, the ccpA mutant strain with pSM257 exhibited a 10-fold increase in sporulation in the absence of glucose while a 12-fold decrease was seen in the absence of glucose (Table 2). We do not have an explanation for these contrasting results, but it is probably due to aberrant CcpA regulation due to multicopy effects, because it was seen only at very low levels of sporulation with strains containing plasmid pSM257.
TABLE 2.
Spore production by C. perfringens wild-type and ccpA mutant strains in sporulation medium in the presence or absence of 5 mM glucosea
| Strain | Without glucose
|
With glucose
|
||||
|---|---|---|---|---|---|---|
| Total no. of CFU | No. of spores | Sporula- tion (%) | Total no. of CFU | No. of spores | Sporula- tion (%) | |
| SM101 | 3.2 × 107 (6.4 × 107-6.3 × 106) | 2.2 × 107 (4 × 107-1.9 × 106) | 68 | 2.9 × 107 (4.3 × 107-2.5 × 106) | 9,500 (15,000-5,200) | 0.033 |
| SM101(pSM257) | 1.1 × 107 (1.3 × 107-3.0 × 106) | 7.2 × 105 (1.4 × 106-1.0 × 103) | 6.5 | 1.8 × 107 (3.6 × 107-4 × 106) | 227 (640-100) | 0.0013 |
| SM120 | 5.2 × 106 (1.5 × 107-1.2 × 106) | 5.9 × 104 (2.8 × 105-7.5 × 102) | 1.1 | 5.6 × 106 (1 × 107-2 × 106) | 78.3 (160-10) | 0.0014 |
| SM120(pSM257) | 9.1 × 106 (2.5 × 107-9.0 × 105) | 1.0 × 106 (8.4 × 106-5.0 × 102) | 11 | 2.7 × 107 (8 × 107-7 × 106) | 28.0 (50-10) | 0.0001 |
Values shown are the means, with the range in parentheses.
A ccpA mutant strain is able to utilize raffinose during growth in DSSM-raffinose sporulation medium.
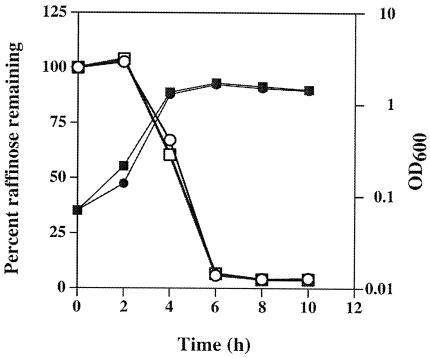
The medium used to induce sporulation of strain SM101 contains 7.9 mM raffinose, which was shown previously to be necessary for efficient sporulation and CPE production by the parent strain of SM101, NCTC 8798 (36). One possibility as to why the ccpA mutant strain failed to sporulate efficiently in DSSM-raffinose medium was that it could not utilize the raffinose as a carbohydrate source to sporulate. This hypothesis was tested by measuring raffinose consumption by SM101 and the ccpA mutant strain during growth in DSSM-raffinose. These strains used raffinose at the same rate and to the same extent, beginning in the late exponential phase of growth (Fig. 2). Therefore, an inability to metabolize raffinose was not the reason the ccpA mutant strain failed to sporulate efficiently in DSSM-raffinose medium.
FIG. 2.
Growth (closed symbols) and utilization of raffinose (open symbols) by strains SM101 (wild type) (squares) and SM120 (ccpA mutant) (circles). Representative data are shown from one of two identical experiments.
Other sugars show different effects on sporulation by wild-type and ccpA mutant strains of C. perfringens.
Shih and Labbe (45) demonstrated that other sugars besides glucose could have a CR on sporulation in C. perfringens. Therefore, we examined the effects of the additions of 5 mM (each) mannose, lactose, and galactose on sporulation by the wild-type and ccpA mutant strains of C. perfringens (Table 3). For the wild-type strain, mannose and lactose repressed sporulation 280- and 29-fold, respectively, compared to sporulation seen in the absence of sugars, while galactose stimulated sporulation 2.4-fold (compare the top row in Table 3 to the top row in Table 2). The extremely high sporulation efficiency seen with galactose, 170%, probably represents incomplete germination of spores in the experiment that measured the total CFU in which the cells were not subjected to germination-inducing heat treatment (see Materials and Methods). We saw with the ccpA mutant strain that mannose repressed sporulation 5,600-fold in comparison to the same strain in the absence of sugars, while ≤10 spores/ml could be detected in the medium with added lactose and galactose (compare the bottom row in Table 3 to the third row in Table 2). The results seen with galactose were surprising, because galactose stimulated sporulation in the wild-type strain as described above.
TABLE 3.
Sporulation by C. perfringens wild-type and ccpA mutant strains in the presence of sugars other than glucosea
| Strain | Mannose
|
Lactose
|
Galactose
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total no. of CFU | No. of spores | Sporulation (%) | Total no. of CFU | No. of spores | Sporulation (%) | Total no. of CFU | No. of spores | Sporulation (%) | |
| SM101 | 7.5 × 107 (1.9 × 108- 1.4 × 107) | 1.8 × 105 (2.8 × 105- 1.5 × 10) | 0.24 | 2.00 × 107 (2.3 × 107- 1.8 × 107) | 4.8 × 105 (1.4 × 106- 1.4 × 104) | 2.4 | 2.2 × 107 (4.5 × 107- 7 × 106) | 3.0 × 107 (4.3 × 107- 2.5 × 107) | 170 |
| SM120 | 4.7 × 107 (7.7 × 107- 3 × 107) | 72 (110-95) | 0.0002 | 9.7 × 106 (1.3 × 107- 3 × 106) | ≤10 | ∼0 | 5.7 × 106 (1.0 × 107- 2.0 × 106) | ≤10 | ∼0 |
Values shown are the means, with the range in parentheses.
Transcription of the cpe gene is repressed in exponential phase but induced in stationary phase by CcpA in C. perfringens grown in sporulation medium.
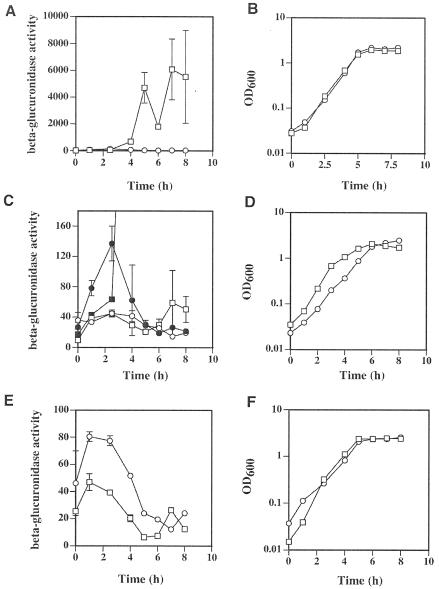
We used a cpe-gusA protein fusion on a plasmid (pSM237) to measure cpe expression in wild-type and ccpA mutant strains of C. perfringens. Transcription and translation from the cpe promoter was measured as units of β-glucuronidase activity, which is the gene product of the E. coli gusA gene (see Materials and Methods). In early and mid-exponential phase the wild-type and ccpA mutant strains showed relatively low levels of β-glucuronidase activity (but not the same levels; see below) (Fig. 3A and B). Beginning in late log phase and extending throughout stationary phase, there was a dramatic induction in β-glucuronidase activity in the wild-type strain but not in the ccpA mutant strain; the activity in the wild-type strain was as much as 230 times as high as that seen in the ccpA mutant strain. In contrast, in early and mid-exponential phases the levels of β-glucuronidase activity in the ccpA mutant strain were actually about twice that seen in the wild-type strain (Fig. 3C and D), suggesting that CcpA may act as a repressor of cpe transcription in exponential phase. The addition of 5 mM glucose to the sporulation medium prevented the strong induction seen in stationary phase in the wild-type strain and abolished the twofold difference seen in exponential phase between the ccpA mutant and wild-type strains (Fig. 3C and D). The presence of glucose resulted in a slower growth rate in the ccpA mutant strain compared to that of the wild-type strain, but similar levels of growth were achieved by the time the cells reached stationary phase (Fig. 3D). Because we observed that the ccpA mutant strain had higher levels of expression of the cpe-gusA fusion in exponential phase than did the wild-type strain in sporulation medium, we measured the levels of expression in PGY medium, which does not induce sporulation in C. perfringens (Fig. 3E and F). Throughout the exponential phase the ccpA mutant strain exhibited a twofold higher level of expression than did the wild-type strain, suggesting that CcpA can act as a repressor of cpe expression when the cells are growing in the vegetative state.
FIG. 3.
Expression from the cpe-gusA fusion in wild-type and ccpA mutant strains of C. perfringens. β-Glucuronidase activity derived from the gusA gene was used as an indicator of cpe transcription. Expression (A) and growth (B) of the wild-type (squares) and ccpA mutant strain (circles) in DSSM sporulation medium are shown. Also shown are expression (C) and growth (D) of the wild-type (open squares) and ccpA mutant strains (open circles) in DSSM sporulation medium with 5 mM glucose added to the medium. Superimposed in panel C are the values shown in panel A (closed symbols, with wild-type [squares] and ccpA mutant [circles]) so that direct comparisons can be made at these low levels of expression. Expression (E) and growth (F) of the wild-type (squares) and ccpA mutant strains (circles) in PGY medium are also shown. Note the difference in scale between panel A and panels C and E. The means and standard deviations of triplicate samples are shown in panels A, C, and E. Representative growth curves from one of three experiments are shown in panels B, D, and F.
Regulation of synthesis of collagenase (kappa toxin) and polysaccharide capsule are dependent on CcpA, but PLC (alpha toxin) is not.
We examined whether other virulence factors besides CPE are subjected to CR by glucose and whether CcpA mediates these effects. In the absence of glucose the wild-type strain produced, on average, 12 times as much collagenase as the ccpA mutant strain (Fig. 4A). Production of collagenase by C. perfringens was repressed by the addition of glucose to the medium, but this effect was not relieved in the ccpA mutant strain (Fig. 4A). Strain SM101 produces a polysaccharide capsule (data not shown), as does its parent strain, NCTC 8798 (Hobbs serotype 9) (12). The polysaccharide capsule of strain NCTC 8798 has been determined to be composed of glucose, galactose, and galactosamine in a molar ratio of 1:1.6:1.1 (12). Without glucose, the ccpA mutant strain produced about 1.2-fold more capsule than the wild-type strain (Fig. 4B). With the addition of glucose, the ccpA mutant strain produced 2.8 times as much capsule material as the wild-type, suggesting that CcpA exhibits glucose-mediated CR on capsule synthesis. In the wild-type strain, PLC synthesis increased about threefold with the addition of glucose, but the ccpA mutant strain exhibited the same pattern and levels of activity, suggesting that CcpA was not the mediator of the activation effect shown by glucose (Fig. 4C).
DISCUSSION
The goal of this work was to investigate the role of the transcriptional regulator CcpA in CR of sporulation and enterotoxin synthesis in C. perfringens. The ccpA gene of C. perfringens was cloned and sequenced. An allele replacement strategy was used to introduce a mutation into the ccpA gene of C. perfringens, and our analysis of this mutant suggests that CcpA regulates sporulation in a manner different from that seen in B. subtilis: CcpA was necessary for efficient sporulation in C. perfringens (Table 2), whereas in B. subtilis it only mediates CR by glucose and is not directly involved in sporulation (11, 37).
The ccpA mutant strain sporulated at a frequency 60-fold less than that of the wild-type strain in the absence of glucose (Table 2). The sporulation medium we used in these experiments, DSSM, contains ∼8 mM raffinose. Typically, C. perfringens sporulation media, including DSSM, contain moderate amounts of nutrient-rich ingredients (e.g., proteose peptone and yeast extract) in combination with a slowly utilizable carbohydrate source (e.g., starch or raffinose) (26-29, 45). Therefore, carbohydrate metabolism appears to be an important part of the initiation and/or completion of sporulation by C. perfringens. However, the utilization rate of raffinose did not differ between the wild-type and ccpA mutant strains (Fig. 2), indicating that there must be another CcpA-mediated effect that regulates sporulation. The question of why CcpA is necessary for sporulation remains to be answered, but one approach to solving the problem will be to look for second-site mutations that restore sporulation to wild-type levels in the ccpA mutant strain.
The addition of glucose to the sporulation medium resulted in a ∼2,000-fold reduction and ∼800-fold reduction in sporulation by the wild-type and ccpA mutant strains, respectively (Table 2). Because the amount of repression was similar in the two strains, we interpret these results to mean that CcpA did not mediate the glucose-mediated CR effect seen with sporulation. This parallels the CcpA-mediated CR effect by glucose seen in B. subtilis, where a ccpA mutant strain was only partially derepressed for sporulation in the presence of glucose (11, 37). As mentioned in the Introduction, Moreno et al. identified many genes in B. subtilis that were subject to CR by glucose in a CcpA-independent manner and found that most of these were involved in sporulation (38). A similar situation seems to exist in C. perfringens.
As seen with glucose, the addition of mannose, lactose, and galactose showed decreasing levels of CR on sporulation in the C. perfringens wild-type strain in the following order: glucose > mannose > lactose > galactose (Tables 2 and 3). In fact, galactose provided a 2.4-fold increase in sporulation efficiency compared to that of the wild-type strain. In the ccpA mutant strain, the results were very different, with sporulation being much lower under all conditions; the level of CR was in the following order: lactose or galactose > mannose > glucose (Tables 2 and 3). One model to partially explain these results is shown in Fig. 5A. In this model, the presence of sugars signals to an unknown regulator (X in the figure) to repress sporulation. CcpA has dual roles in this model, where it can negatively affect the activity of X and can act directly on the sporulation cycle as an activator. Therefore, in a ccpA mutant strain, X would be derepressed and CcpA would not activate sporulation genes, leading to a powerful repressive effect on sporulation.
FIG. 5.
Model illustrating the involvement of CcpA in regulation of sporulation (A) and cpe transcription (B). Plus symbols indicate an activating effect, and minus symbols indicate a repressive effect. (B) The arrows in front of the cpe gene represent three independent promoters identified upstream of the cpe gene (52). See the text for detailed descriptions of the models.
During sporulation, the level of transcription of the cpe gene generally was in agreement with the amount of sporulation that occurred under each condition (Table 2 and Fig. 3). By far the highest amount of cpe transcription occurred during sporulation by the wild-type cells in the absence of glucose. This is consistent with previous results where the highest level of cpe induction always occurred in concert with sporulation (35, 36, 52). Transcription of the cpe gene was greatly reduced in the ccpA mutant strain in the absence of glucose and in both strains in the presence of glucose (Fig. 3A, C, and E). During the vegetative stage of growth in sporulation medium or in growth in PGY, the amount of cpe promoter activity, although low, was twice as high in the ccpA mutant strain as it was in the wild-type strain (Fig. 3C and E). This is the first time to our knowledge that a regulator of cpe transcription during vegetative growth has been identified. A model summarizing the effects of CcpA on cpe transcription is shown in Fig. 5B. During vegetative growth CcpA acts directly or indirectly to repress cpe transcription. However, under sporulating conditions CcpA is an activator of sporulation functions which leads to synthesis of active forms of the mother cell-specific sigma factors, SigE and SigK. We have identified three promoters upstream of cpe that are responsible for the majority of sporulation-dependent transcription, and the −10 and −35 promoter recognition sequences of these promoters have a high level of homology to consensus −10 and −35 recognition sequences from σE- and σK-dependent promoters identified in B. subtilis (18, 52). Therefore, our hypothesis is that CcpA activates cpe transcription during sporulation indirectly by activating or derepressing genes that lead to the synthesis of SigE and SigK (Fig. 5B).
We also compared the ccpA mutant and wild-type strains for differences in expression of the toxin collagenase. Collagenase activity was repressed by glucose in the wild-type strain but not in the ccpA mutant strain, but CcpA was needed for expression even in the absence of glucose (Fig. 4A). This suggests that other factors are involved in regulating collagenase activity. Transcription of the gene encoding the collagenase of C. perfringens, colA, has been shown to be regulated by the global two-component regulatory factors VirR/VirS as well as a regulatory RNA, VR-RNA (7, 8, 39, 46). It may prove valuable to determine if these alternative regulators are subjected to CR effects mediated by CcpA.
The role of the capsule as a virulence factor in C. perfringens pathogenicity has been controversial (47). Polysaccharide capsule synthesis was induced only 1.2-fold by the addition of glucose to the medium in which the wild-type strain was grown, but it was induced 3.8-fold in the ccpA mutant strain when glucose was added (Fig. 4B). This suggests that capsule synthesis is more strongly induced by glucose in the absence of CcpA. Because glucose is one of the components of the capsule of strain SM101 (12), it is not surprising that it induced the synthesis of more capsular material, but the regulatory mechanism appears to involve more functions than CcpA.
Synthesis of PLC, the phospholipase/sphingomyelinase of C. perfringens, was induced about threefold by the addition of glucose to the medium, but this effect was independent of CcpA. As with colA, transcription of the plc gene has been shown to be regulated by VirR/VirS and VR-RNA (7, 8, 31, 39, 46). Because PLC has been shown to be the most important virulence factor in gas gangrene infections caused by C. perfringens (3, 4, 50), it would be of interest to determine the mechanism of glucose-mediated induction of this virulence factor.
In summary, we have identified CcpA as an important regulator that is necessary for sporulation and CPE production in C. perfringens. This could provide a valuable insight into potential therapeutic strategies to block sporulation and CPE production and to relieve the symptoms of patients infected with non-food-borne enteritis caused by C. perfringens.
Acknowledgments
We thank Wesley Black for assistance with the polysaccharide capsule assays.
This work was supported by grants #98-02844 and #2000-02621 from NRICGP/USDA awarded to S.B.M.
REFERENCES
- 1.Abrahao, C., R. J. Carman, H. Hahn, and O. Liesenfeld. 2001. Similar frequency of detection of Clostridium perfringens enterotoxin and Clostridium difficile toxins in patients with antibiotic-associated diarrhea. Eur. J. Clin. Microbiol. Infect. Dis. 20:676-677. [DOI] [PubMed] [Google Scholar]
- 2.Ando, Y., T. Tsuzuki, H. Sunagawa, and S. Oka. 1985. Heat resistance, spore germination, and enterotoxigenicity of Clostridium perfringens. Microbiol. Immunol. 29:317-326. [DOI] [PubMed] [Google Scholar]
- 3.Awad, M. M., A. E. Bryant, D. L. Stevens, and J. I. Rood. 1995. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15:191-202. [DOI] [PubMed] [Google Scholar]
- 4.Awad, M. M., D. M. Ellemor, R. L. Boyd, J. J. Emmins, and J. I. Rood. 2001. Synergistic effects of alpha-toxin and perfringolysin O in Clostridium perfringens-mediated gas gangrene. Infect. Immun. 69:7904-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Awad, M. M., and J. I. Rood. 1997. Isolation of alpha-toxin, theta-toxin and kappa-toxin mutants of Clostridium perfringens by Tn916 mutagenesis. Microb. Pathog. 22:275-284. [DOI] [PubMed] [Google Scholar]
- 6.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:233-235. [DOI] [PubMed] [Google Scholar]
- 7.Banu, S., K. Ohtani, H. Yaguchi, T. Swe, S. T. Cole, H. Hayashi, and T. Shimizu. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 35:854-864. [DOI] [PubMed] [Google Scholar]
- 8.Ba-Thein, W., M. Lyristis, K. Ohtani, I. T. Nisbet, H. Hayashi, J. I. Rood, and T. Shimizu. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 178:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black, W. P., and Z. Yang. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J. Bacteriol. 186:1001-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruggemann, H., S. Baumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. Martinez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. USA 100:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambliss, G. H. 1993. Carbon source-mediated catabolite repression, p. 213-220. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 12.Cherniak, R., and H. M. Frederick. 1977. Capsular polysaccharide of Clostridium perfringens Hobbs 9. Infect. Immun. 15:765-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Canard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 15.Davison, S. P., J. D. Santangelo, S. J. Reid, and D. R. Woods. 1995. A Clostridium acetobutylicum regulator gene (regA) affecting amylase production in Bacillus subtilis. Microbiology 141:989-996. [DOI] [PubMed] [Google Scholar]
- 16.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita, Y., Y. Miwa, A. Galinier, and J. Deutscher. 1995. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol. Microbiol. 17:953-960. [DOI] [PubMed] [Google Scholar]
- 18.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatheway, C. L. 1990. Toxigenic clostridia. Clin. Microbiol. Rev. 3:66-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol. Microbiol. 5:575-584. [DOI] [PubMed] [Google Scholar]
- 21.Hogenauer, C., et al. 1998. Mechanisms and management of antibiotic-associated diarrhea. Clin. Infect. Dis. 27:702-710. [DOI] [PubMed] [Google Scholar]
- 22.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, S., and D. N. Gerding. 1997. Enterotoxemic infections, p. 117-140. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, Inc., San Diego, Calif.
- 24.Keis, S., R. Shaheen, and D. T. Jones. 2001. Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. Int. J. Syst. Evol. Microbiol. 51:2095-2103. [DOI] [PubMed] [Google Scholar]
- 25.Kraus, A., E. Kuster, A. Wagner, K. Hoffmann, and W. Hillen. 1998. Identification of a co-repressor binding site in catabolite control protein CcpA. Mol. Microbiol. 30:955-963. [DOI] [PubMed] [Google Scholar]
- 26.Labbe, R., E. Somers, and C. Duncan. 1976. Influence of starch source on sporulation and enterotoxin production by Clostridium perfringens type A. Appl. Environ. Microbiol. 31:455-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labbe, R. G., and C. L. Duncan. 1975. Influence of carbohydrates on growth and sporulation of Clostridium perfringens type A. Appl. Microbiol. 29:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labbe, R. G., and D. K. Rey. 1979. Raffinose increases sporulation and enterotoxin production by Clostridium perfringens type A. Appl. Environ. Microbiol. 37:1196-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labbe, R. G., and N.-J. R. Shih. 1997. Physiology of sporulation of clostridia. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, Inc., San Diego, Calif.
- 30.Lyras, D., S. B. Melville, and J. I. Rood. 1997. Conjugative transfer of shuttle and suicide vectors from Escherichia coli to Clostridium perfringens, p. 73. In S. Cole, B. McClane, J. Rood, and R. Titball (ed.), Proceedings of the 2nd International Meeting on the Molecular Genetics and Pathogenesis of the Clostridia, Chateau Domaine de Seillac, France. Société Française de Microbiologie, Paris, France.
- 31.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 32.Matsushita, O., K. Yoshihara, S. Katayama, J. Minami, and A. Okabe. 1994. Purification and characterization of Clostridium perfringens 120-kilodalton collagenase and nucleotide sequence of the corresponding gene. J. Bacteriol. 176:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClane, B. A. 2001. The complex interactions between Clostridium perfringens enterotoxin and epithelial tight junctions. Toxicon 39:1781-1791. [DOI] [PubMed] [Google Scholar]
- 34.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melville, S. B., R. E. Collie, and B. A. McClane. 1997. Regulation of enterotoxin production in Clostridium perfringens, p. 471-490. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, Inc., San Diego, Calif.
- 36.Melville, S. B., R. Labbe, and A. L. Sonenshein. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect. Immun. 62:5550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miwa, Y., M. Saikawa, and Y. Fujita. 1994. Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology 140:2567-2575. [DOI] [PubMed] [Google Scholar]
- 38.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 39.Ohtani, K., S. K. Bhowmik, H. Hayashi, and T. Shimizu. 2002. Identification of a novel locus that regulates expression of toxin genes in Clostridium perfringens. FEMS Microbiol. Lett. 209:109-114. [DOI] [PubMed] [Google Scholar]
- 40.Olsen, S. J., L. C. MacKinnon, J. S. Goulding, N. H. Bean, and L. Slutsker. 2000. Surveillance for foodborne-disease outbreaks-United States, 1993-1997. Morb. Mortal. Wkly. Rep. 49:1-62. [PubMed] [Google Scholar]
- 41.Rood, J. I., and S. T. Cole. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55:621-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 44.Selvaraj, G., T. C. Fong, and V. N. Iyer. 1984. A portable DNA sequence carrying the cohesive site (cos) of bacteriophage λ and the mob (mobilization) region of the broad-host-range plasmid RK2: a module for the construction of new cosmids. Gene 32:235-241. [DOI] [PubMed] [Google Scholar]
- 45.Shih, N.-J., and R. G. Labbe. 1994. Effect of glucose on sporulation and extracellular amylase production by Clostridium perfringens type A in a defined medium. Curr. Micobiol. 29:163-169. [Google Scholar]
- 46.Shimizu, T., K. Shima, K. Yoshino, K. Yonezawa, and H. Hayashi. 2002. Proteome and transcriptome analysis of the virulence genes regulated by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 184:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, L. D. S. 1975. Clostridium perfringens, p. 115-176. In L. D. S. Smith (ed.), The pathogenic anaerobic bacteria. C. C. Thomas, Springfield, Ill.
- 48.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens, D. L., B. E. Troyer, D. T. Merrick, J. E. Mitten, and R. D. Olson. 1988. Lethal effects and cardiovascular effects of purified alpha- and theta-toxins from Clostridium perfringens. J. Infect. Dis. 157:272-279. [DOI] [PubMed] [Google Scholar]
- 50.Stevens, D. L., R. K. Tweten, M. M. Awad, J. I. Rood, and A. E. Bryant. 1997. Clostridial gas gangrene: evidence that alpha and theta toxins differentially modulate the immune response and induce acute tissue necrosis. J. Infect. Dis. 176:189-195. [DOI] [PubMed] [Google Scholar]
- 51.Tobisch, S., D. Zuhlke, J. Bernhardt, J. Stulke, and M. Hecker. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J. Bacteriol. 181:6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]