Abstract
Enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC) strains are human and animal pathogens that inject effector proteins into host cells via a type III secretion system (TTSS). Cif is an effector protein which induces host cell cycle arrest and reorganization of the actin cytoskeleton. Cif is encoded by a lambdoid prophage present in most of the EPEC and EHEC strains. In this study, we analyzed the domain that targets Cif to the TTSS by using a new reporter system based on a translational fusion of the effector proteins with mature TEM-1 β-lactamase. Translocation was detected directly in living host cells by using the fluorescent β-lactamase substrate CCF2/AM. We show that the first 16 amino acids (aa) of Cif were necessary and sufficient to mediate translocation into the host cells. Similarly, the first 20 aa of the effector proteins Map, EspF, and Tir, which are encoded in the same region as the TTSS, mediated secretion and translocation in a type III-dependent but chaperone-independent manner. A truncated form of Cif lacking its first 20 aa was no longer secreted and translocated, but fusion with the first 20 aa of Tir, Map, or EspF restored both secretion and translocation. In addition, the chimeric proteins were fully able to trigger host cell cycle arrest and stress fiber formation. In conclusion, our results demonstrate that Cif is composed of a C-terminal effector domain and an exchangeable N-terminal translocation signal and that the TEM-1 reporter system is a convenient tool for the study of the translocation of toxins or effector proteins into host cells.
Enteropathogenic Escherichia coli (EPEC) is associated with diarrheal diseases in young animals and children and is an important cause of infant mortality in the developing world. This human and animal enteric pathogen is closely related to the emerging zoonotic pathogen enterohemorrhagic E. coli (EHEC), which causes acute gastroenteritis, hemorrhagic colitis, and hemolytic uremic syndrome in developed countries (32). In adhering to intestinal epithelial cells, EPEC and EHEC strains subvert the host cellular architecture to produce a histopathological feature known as attaching and effacing lesions. These are characterized by the localized destruction of brush border microvilli and intimate attachment of the bacteria to the plasma membrane of the host epithelial cells (14). The genes required for the formation of attaching and effacing lesions are clustered together in a chromosomal pathogenicity island known as the locus for enterocyte effacement (LEE), which codes for a type III secretion system (TTSS) (19, 28).
TTSSs are present in many gram-negative pathogens and symbionts. These multisubunit molecular machines are used to transfer effector proteins directly into eukaryotic cells, where the normal cellular functions are subverted for the benefit of the bacteria. The set of translocated effector molecules tends to be unique to each pathogen and reflects the needs and specific niches of each bacterial species (18). To date, seven EPEC and EHEC effector molecules have been shown to be injected into the host cell by the TTSS. Five translocated effectors are encoded by the LEE: Tir/EspE (8, 21), Map (22), EspF (29), EspG (12), and EspH (44). Two effectors are encoded outside the LEE: Cif (26) and NleA/EspI (17, 31). In addition to effectors, the LEE sequence encodes chaperones CesT and CesF, which contribute to the secretion and translocation process of Tir/Map (1, 5, 11) and EspF (13), respectively.
Cif is encoded by a lambdoid prophage present in most of the EPEC and EHEC serovars, but cif is absent or truncated in EHEC strain Sakai and in EPEC strain E2348/69 (26). In epithelial cells, Cif triggers an irreversible cytopathic effect (CPE) characterized by a progressive recruitment of focal adhesion plaques leading to the assembly of stress fibers and the inhibition of the cell cycle G2/M phase transition (10, 26, 33). The cytostatic effect can be summarized as follows. Cells progressively accumulate at 4C and 8C DNA content and do not display signs of mitosis. This cytostatic effect is not functionally related to cytoskeletal rearrangement but is linked to the maintenance of the cyclin-dependent kinase Cdk1, a key effector driving entry into mitosis, in a premitotic tyrosine-phosphorylated state (26, 33). The ability of EPEC and EHEC strains to induce both cytoskeletal alterations and to block the G2/M phase transition depends on a functional LEE type III secretion machinery but not on intimin or Tir (27, 34). The mode of action of Cif is not yet elucidated, and its functional domains remain to be defined. In addition, despite the fact that several TTSS substrates have been characterized, the mechanisms that drive Cif and other EPEC and EHEC effectors to the TTSS remain poorly understood.
In this study, we analyzed the Cif secretion and translocation domain by using TEM-1 β-lactamase as a new fluorescence-based reporter. We showed that Cif is a modular protein composed of an exchangeable N-terminal secretion and translocation signal (STS) linked to a C-terminal effector domain and that construction of fusions with the mature form of TEM-1 β-lactamase in combination with the use of fluorescent substrate CCF2/AM is a convenient tool for the analysis of TTSS effectors.
MATERIALS AND METHODS
Cell line and bacterial strains.
Human epithelial HeLa cells (ATCC CCL-2) were cultivated in Eagle minimum essential medium (MEM) supplemented with 10% fetal calf serum (FCS), l-glutamine (2 mM), and gentamicin (80 μg/ml) at 37°C in a 5% CO2 atmosphere.
E22 is a rabbit EPEC strain of serotype O103:H2 (34). E22 Δcif::FRT is a cif deletion mutant (26) obtained according to the procedure described by Datsenko and Wanner (6). E22 espB::Kan and E22 escN::Kan are E22 mutants in which the espB and escN genes are interrupted by a kanamycin resistance gene (26, 34). E22 ΔcesT::Kan is a cesT deletion mutant obtained according to the procedure described by Datsenko and Wanner (6). Bacterial strains were cultured in Luria-Bertani broth or in Dulbecco's modified Eagle medium (DMEM) buffered with 25 mM HEPES (pH 7.4). Antibiotics were used at the following final concentrations: carbenicillin, 50 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml; tetracycline, 12.5 μg/ml.
Construction of the β-lactamase TEM-1 fusion cloning plasmid and effector-TEM fusion proteins.
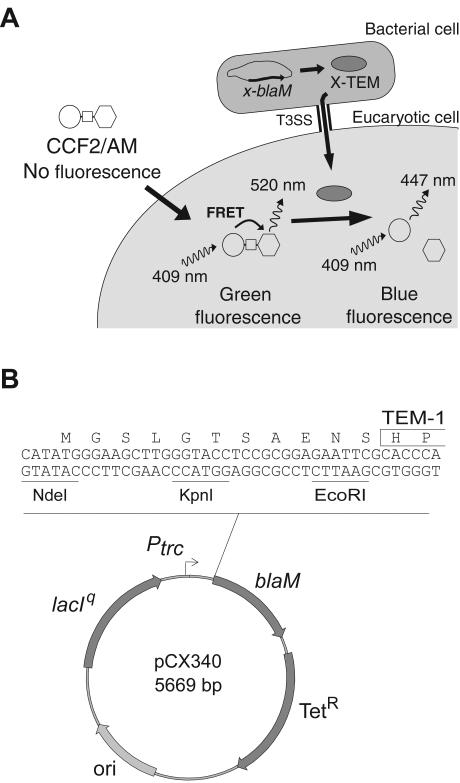
A pBR322 derivative was constructed by ligating the EcoRI-ScaI-digested plasmid pHB6 (Roche) to an EcoRI and blunt end PCR product from plasmid p46Lbla (2) obtained with primers 5′-GCG AAT TCG CAC CCA GAA ACG CTG GTG AAA GTA and 5′-GGC TCC AAT TCT TGG AGT GGT GA. This resulting plasmid carries the lacIq gene, the Ptrc promoter upstream of the blaM gene (encoding TEM-1), and the tetracycline resistance gene from pBR322. The NdeI restriction site near the ColE1 origin has been destroyed by digestion, fill-in with Klenow, and ligation. Inverse PCR with primers 5′-ATG TTA TTC CTC CTT ATT TAA TCG ATA C and 5′-ATG GGA AGC TTG GGT ACC TCC GCG G was performed to generate a unique NdeI site at the starting translational codon, thus creating a multiple cloning site (NdeI, KpnI, and EcoRI) upstream of blaM. The resulting plasmid pCX340 encodes the mature form of TEM-1 under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Ptrc promoter and allows cloning of effector protein genes with blaM to generate effector-TEM fusion proteins (Fig. 1). The cif, tir, map, and espF genes were amplified from E22 genomic DNA with primers adding an NdeI restriction site at the start codon and an EcoRI (or KpnI for tir) restriction site on the codon replacing the stop codon. PCR products were digested with NdeI and EcoRI (or KpnI for tir) and ligated to the corresponding sites in pCX340. The resulting plasmids pCX313, pCX302, pCX303, and pCX304, respectively, encode the Cif-TEM, Tir-TEM, Map-TEM, and EspF-TEM fusion proteins. The plasmid pCX329, encoding the Tir1-26-TEM fusion, was obtained by cloning an NdeI-KpnI PCR fragment amplified from plasmid pCX302 in the NdeI-KpnI-digested plasmid pCX340.
FIG. 1.
Schematic representation of the TEM-1 reporter system used to study translocation of TTSS effectors into live eukaryotic cells. (A) Upon passive entry into the eukaryotic cell, the nonfluorescent esterified CCF2/AM substrate is rapidly converted by cellular esterases in charged and fluorescent CCF2. Excitation of the coumarin moiety (represented by a circle) at 409 nm results in fluorescence energy transfer (FRET) to the fluorescein moiety (represented by a hexagon), which emits a green fluorescence signal at 520 nm. Injection of an effector fused to TEM-1 into a CCF2-loaded cell induces catalytic cleavage of the CCF2 β-lactam ring (represented by a square), disrupting FRET. This produces an easily detectable and measurable change in CCF2 fluorescence from green to blue emission. (B) Map of the effector-TEM fusion cloning vector. The blaM gene encodes the mature form of the β-lactamase TEM (the first two residues are boxed). The NdeI, KpnI, and EcoRI restriction sites are unique. The origin of replication (ori) is derived from ColE1.
Plasmids for expression of gradually truncated Cif proteins fused to TEM.
Plasmid pCX327, encoding the TEM-1 fusion with the first 16 residues of Cif (Cif1-16-TEM), was obtained by deleting the complementary domain of Cif (residues 17 to 282) by inverse PCR on the template plasmid pCX313. To facilitate screening for positive clones, an NheI restriction site was introduced at the junction between the first 16 residues of Cif and TEM-1. Other truncated forms of Cif fused to TEM-1 were obtained by the same procedure (Table 1).
TABLE 1.
List of strains and plasmids
| Strain or plasmid | Description and/or genotype | Reference or expressed protein |
|---|---|---|
| Strains | ||
| E22 | Rabbit EPEC, serotype O103:H2 | 34 |
| E22 Δcif | E22 Δcif::Kan | 26 |
| E22 espB | E22 espB::Kan | 34 |
| E22 escN | E22 escN::Kan | 26 |
| E22 ΔcesT | E22 ΔcesT::Kan | This study |
| Plasmids | ||
| pCX340 | pBR322 derivative, cloning vector used to fuse EPEC effectors to the mature form of TEM-1 β-lactamase | TEM |
| pCX311 | Negative control, fusion of MBP to TEM-1 | MBP-TEM |
| pCX312 | Negative control, fusion of GST to TEM-1 | GST-TEM |
| pCX302 | Fusion of Tir to TEM-1 | Tir-TEM |
| pCX329 | Residues 1 to 26 of Tir fused to TEM-1 | Tir1-26-TEM |
| pCX303 | Fusion of Map to TEM-1 | Map-TEM |
| pCX304 | Fusion of EspF to TEM-1 | EspF-TEM |
| pCX313 | Fusion of Cif to TEM-1 | Cif-TEM |
| pCX327 | Residues 1 to 16 of Cif fused to TEM-1 | Cif1-16-TEM |
| pCX315 | Residues 1 to 32 of Cif fused to TEM-1 | Cif1-32-TEM |
| pCX316 | Residues 1 to 62 of Cif fused to TEM-1 | Cif1-62-TEM |
| pCX317 | Residues 1 to 86 of Cif fused to TEM-1 | Cif1-86-TEM |
| pCX318 | Residues 1 to 124 of Cif fused to TEM-1 | Cif1-124-TEM |
| pCX319 | Residues 1 to 150 of Cif fused to TEM-1 | Cif1-150-TEM |
| pCX320 | Residues 1 to 206 of Cif fused to TEM-1 | Cif1-206-TEM |
| pCX321 | Residues 1 to 256 of Cif fused to TEM-1 | Cif1-256-TEM |
| pCX322 | Naturally truncated Cif of PMK5 fused to TEM-1 | CifPMK5-TEM |
| pCX323 | Naturally truncated Cif of E2348/69 fused to TEM-1 | CifE2348-TEM |
| pCX351 | Cif-TEM fusion with AMG tripeptide at position 21 | CifAMG-TEM |
| pCX352 | Tir-TEM fusion with AMG tripeptide at position 21 | TirAMG-TEM |
| pCX353 | Map-TEM fusion with AMG tripeptide at position 21 | MapAMG-TEM |
| pCX354 | EspF-TEM fusion with AMG tripeptide at position 21 | EspFAMG-TEM |
| pCX361 | Residues 1 to 20 of Cif fused to TEM-1 | Cif1-20-TEM |
| pCX362 | Residues 1 to 20 of Tir fused to TEM-1 | Tir1-20-TEM |
| pCX363 | Residues 1 to 20 of Map fused to TEM-1 | Map1-20-TEM |
| pCX364 | Residues 1 to 20 of EspF fused to TEM-1 | EspF1-20-TEM |
| pCX371 | Residues 21 to 282 of Cif fused to TEM-1 | CifΔ(1-20)-TEM |
| pCX351 | Residues 1 to 20 of Cir fused to CifΔ(1-20)-TEM | Cif1-20-CifΔ(1-20)-TEM |
| pCX372 | Residues 1 to 20 of Tir fused to CifΔ(1-20)-TEM | Tir1-20-CifΔ(1-20)-TEM |
| pCX373 | Residues 1 to 20 of Map fused to CifΔ(1-20)-TEM | Map1-20-CifΔ(1-20)-TEM |
| pCX374 | Residues 1 to 20 of EspF fused to CifΔ(1-20)-TEM | EspF1-20-CifΔ(1-20)-TEM |
Plasmids for expression of the exchangeable first 20 residues of EPEC effectors.
The Ala-Met-Gly coding sequence GCC ATG GGC, containing an NcoI restriction site (underlined), was inserted by inverse PCR at codon 21 of each effector-TEM-encoding sequence. The resulting plasmids pCX351, pCX352, pCX353, and pCX354 encode, respectively, CifAMG-TEM, TirAMG-TEM, MapAMG-TEM and EspFAMG-TEM fusions, which carry an insertion of the tripeptide Ala-Met-Gly (AMG) at position 21. Digestion of these plasmids with NcoI and EcoRI, followed by fill-in with Klenow and ligation, gave the plasmids pCX361, pCX362, pCX363, and pCX364, which encode, respectively, TEM-1 fusions with the first 20 residues of Cif, Tir, Map, and EspF (Cif1-20-TEM, Tir1-20-TEM, Map1-20-TEM, and EspF1-20-TEM). Hybrid proteins containing the first 20 residues of each effector fused to Cif with a deletion of its first 20 residues were obtained by replacing the NcoI-EcoRI fragments of pCX352, pCX353, and pCX354 with the NcoI-EcoRI fragment from plasmid pCX351. This gave plasmids pCX372, pCX373, and pCX374, encoding the Tir1-20-CifΔ(1-20)-TEM, Map1-20-CifΔ(1-20)-TEM, and EspF1-20-CifΔ(1-20)-TEM fusions. Plasmid pCX351 was digested with NdeI and blunted by treatment with mung bean nuclease, the NcoI site was filled with Klenow, and the whole molecule was ligated to itself. This gave plasmid pCX371, encoding the CifΔ(1-20)-TEM. All constructions were verified by sequencing.
Interaction between epithelial cells and bacteria for translocation analysis.
On the day before interaction, HeLa cells were trypsinized and seeded in black with clear bottom 96-well plates (Becton Dickinson) at 2 × 104 cells per well in MEM to obtain 100% confluence on the following day. The same day, bacterial strains were inoculated in Luria-Bertani broth with tetracycline. On the day of infection, overnight bacterial cultures were inoculated at a 1/100 dilution in 24-well plates in 1.5 ml of DMEM supplemented with 1% mannose, 5% FCS, and 2 mM l-glutamine. Bacterial strains were then grown as static cultures at 37°C in a 5% CO2 atmosphere for 3 h to reach an optical density at 600 nm (OD600) of 0.2 to 0.25. HeLa cells were washed twice with Hanks' balanced salt solution (HBSS) and directly infected with the DMEM bacterial culture (multiplicity of infection of about 100 bacteria per cell). After 30 min of infection, IPTG was added at a final concentration of 1 mM and the infection was allowed to proceed for an additional hour. Cell monolayers were washed twice with HBSS and covered with 100 μl of HBSS plus 20 μl of 6× CCF2/AM solution freshly prepared with the CCF2/AM loading kit (CCF2/AM final concentration, 1 μM; Invitrogen). The cells were incubated for 90 min at room temperature, fluorescence was quantified on an FL600 microplate reader (Bio-Tek) with excitation at 405 nm (10-nm band-pass), and emission was detected via 460-nm (40-nm band-pass, blue fluorescence) and 530-nm (30-nm band-pass, green fluorescence) filters. Translocation was expressed as the emission ratio at 460/530 nm to normalize the β-lactamase activity to cell loading and the number of cells present in each well. The presented data are mean values of the results from triplicate wells from two to three experiments. For statistical analyses, two-sided Student's t test was used with independent samples. P values of less than 0.05 were considered statistically significant.
Fluorescence microscopy for observation of translocation and analysis of CPE.
Cell infections were performed as described above except that 3 × 104 cells were seeded in Labtek eight-well chamber slides (Becton Dickinson) in 500 μl of MEM. For microscopic observation of translocation, cells were washed twice after infection with HBSS, loaded for 1 h with 1 μM CCF2/AM (Invitrogen), and washed twice again with HBSS. The slides were then covered with coverslips, and live cells were observed on a Leica fluorescence microscope with a 4′,6′-diamidino-2-phenylindole (DAPI) filter set (340- to 380-nm excitation and 425-nm long-pass emission). Pictures were taken under a true color camera. To analyze the Cif-related CPE, cells were infected as described above, except that after infection they were washed five times with HBSS and incubated for 3 days in MEM with 10% FCS, 2 mM l-glutamine, and 200 μg of gentamicin/ml. Cells were washed twice in phosphate-buffered saline (PBS) and fixed for 20 min with 3.7% formaldehyde. Actin stress fibers and nuclei were labeled with rhodamine-phalloidin and DAPI, respectively.
Analysis of production and secretion of TEM fusion proteins.
To detect the secretion of the different TEM-1 fusion proteins, overnight bacterial cultures were diluted to 1:100 in DMEM supplemented with 1% mannose and 2 mM l-glutamine. Bacteria were then grown as static cultures at 37°C in a 5% CO2 atmosphere for 5 h 30 min to reach an OD600 of 0.6 to 0.7 and then induced with 1 mM IPTG for an additional hour. Bacterial cultures were centrifuged for 15 min at 16,000 × g at 4°C. Culture supernatants were filtered (0.22-μm pore size, Millex GV; Millipore) and precipitated for 1 h at 4°C with 10% trichloroacetic acid. Pellets were washed twice with ice-cold acetone, resuspended in Tris-saturated sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and loaded onto a Nupage 4 to 12% gradient gel (Invitrogen). Bacterial pellets were resuspended in PBS to a final OD600 of 4. A 4-μl aliquot was boiled for 5 min in SDS-PAGE sample buffer and subjected to denaturing PAGE on a Nupage 4 to 12% gradient gel (Invitrogen). To assess the solubility of Cif-TEM fusions, bacteria from resuspended bacterial pellets were lysed by repeating freeze-thaw cycles. The lysates were sonicated to shear genomic DNA and centrifuged at 16,000 × g for 1 h at 4°C. Centrifuged lysates were subjected to PAGE as described above.
Immunoblot analysis of TEM fusion proteins.
Proteins from SDS-polyacrylamide gels were electrophoretically transferred to nitrocellulose sheets (Schleicher and Schuell) and subsequently stained with Ponceau S (Sigma) to check the loading of the lanes. Sheets were blocked with 10% nonfat dried milk in PBS with 0.1% Tween 20. Sheets were then analyzed by Western blotting with monoclonal antibody directed to the TEM-1 β-lactamase (5 μg/ml; QED Bioscience) as a primary antibody and an anti-mouse peroxidase conjugate (1/10,000; Sigma) as a secondary antibody. Nitrocellulose sheets were revealed with the Enhanced ChemiLuminescence detection system (Amersham Pharmacia Biotech) and Biomax films (Kodak). Image analyses were performed on a Macintosh computer with the public domain NIH Image program (developed at the U.S. National Institutes of Health and available at http://rsb.info.nih.gov/nih-image/).
In silico analysis of STS-encoding sequences.
EPEC effectors and their respective STS-encoding sequences (alone or fused to TEM-1) have been subjected to secondary structure prediction, producing a consensus prediction from several algorithms (3). The secondary structure predictions of the STS-encoding sequences at the mRNA level have been performed with Mfold (version 3.1) on a web server (48).
RESULTS
Translocation of EPEC effector proteins fused to TEM-1 reporter protein into live eukaryotic cells.
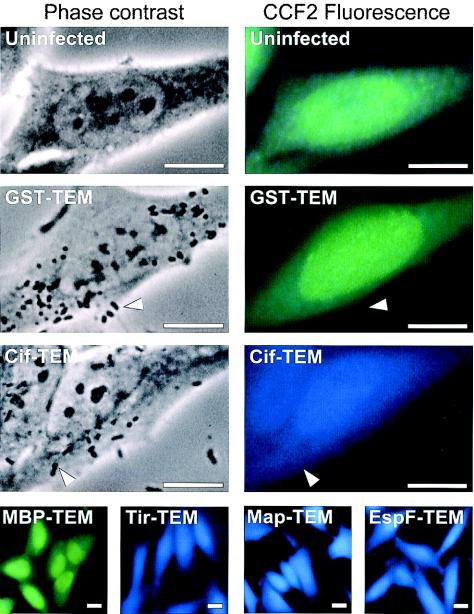
To test TEM-1 as a reporter for type III translocation, we fused TEM-1 to the C terminus of Tir, Map, EspF, and Cif, four different EPEC and EHEC effector proteins encoded or not by the LEE. As negative controls, we constructed fusions with two E. coli cytoplasmic proteins, the mature form of maltose binding protein (MBP) and glutathione S-transferase (GST). We verified the production of the fusion proteins in bacterial whole-cell lysates (data not shown) and analyzed the translocation of these fusions in infected HeLa cells (Fig. 2). EPEC strains expressing each of the fusion proteins were grown in DMEM and used to infect HeLa cells. After 90 min, the epithelial cells were washed and incubated for an additional 90 min with the β-lactamase substrate CCF2/AM. The cells were then analyzed by fluorescence microcopy with a long-pass emission filter, enabling the simultaneous observation of the green fluorescence emitted by the CCF2 substrate and the blue fluorescence emitted by the cleaved CCF2 product. Uninfected HeLa cells or cells infected with EPEC strains expressing GST-TEM or MBP-TEM fusions appeared green, indicating the absence of TEM-1 activity in these cells (Fig. 2). In contrast, cells infected with bacteria expressing the type III effector Cif fused to TEM-1 appeared blue (Fig. 2), indicating that TEM-1 was translocated into the host cells. Similar results were obtained with Tir-TEM, Map-TEM, and EspF-TEM fusions (Fig. 2).
FIG. 2.
Demonstration of the translocation of EPEC effector proteins into live HeLa cells by using TEM-1 fusions and fluorescence microscopy. HeLa cells were infected with wild-type EPEC strains expressing different TEM-1 fusion proteins. After infection, HeLa cells were washed and loaded with CCF2/AM. β-Lactamase activity in HeLa cells is revealed by the blue fluorescence emitted by the cleaved CCF2 product, whereas uncleaved CCF2 emits a green fluorescence. No detectable fluorescence arises from adherent bacteria (indicated by arrowheads). Bars, 10 μM.
Analysis of the roles of the translocator, chaperone, and STS with the TEM-1 reporter system.
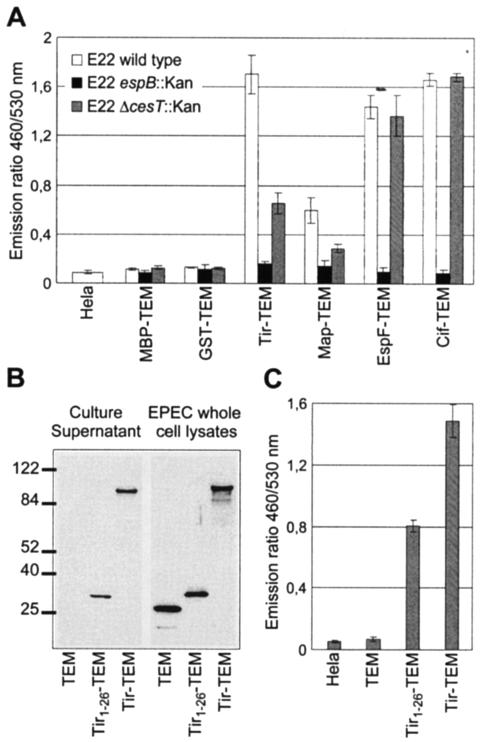
EPEC strains expressing TEM-1 fusion proteins were used to infect HeLa cells grown in 96-well tissue culture plates. After interaction with the bacteria, the HeLa cells were infected as before and then loaded with CCF2/AM, and fluorescence was measured and reported as the ratio between blue emission fluorescence (460 nm) and green emission fluorescence (520 nm) (Fig. 3A). In cells infected with EPEC strains expressing either MBP-TEM or GST-TEM fusions, the emission ratio was similar to the emission ratio of uninfected HeLa cells (Fig. 3A). In cells infected with EPEC strains expressing effector TEM-1 fusions, the emission ratio increased significantly, indicating that the different fusion proteins were translocated into the host cells (Fig. 3A). In contrast, no increase in the emission ratio was observed for all fusion proteins expressed in the espB mutant, confirming that the translocation was type III dependent and required a functional translocation pore. When the effector fusions were expressed in the cesT mutant, the emission ratios for Tir and Map fusions were significantly reduced compared to the same fusions expressed in the wild-type EPEC strain. This means that the translocation was affected but not completely abolished, in agreement with previously published results (1, 5, 11). In contrast, the emission ratio for EspF was not changed in a cesT mutant, in agreement with the fact that EspF interacts with another chaperone, namely CesF (13). Similarly, the translocation of Cif-TEM was not affected in a cesT mutant, indicating that CesT is probably not a chaperone for Cif.
FIG. 3.
Analysis of the role of the translocator, chaperone, and STS with the TEM-1 translocation reporter system. (A) Activity of the EspB translocator and the CesT chaperone on effector translocation. HeLa cells were infected with wild-type EPEC E22 strains expressing MBP-TEM or GST-TEM fusion protein or the different effector-TEM protein fusions. After infection, HeLa cells were washed and loaded with CCF2/AM. β-Lactamase activity in HeLa cells was detected by measuring cleavage of the CCF2/AM substrate with a fluorescence microplate reader and is presented as the emission ratio between blue fluorescence (460 nm) and green fluorescence (530 nm). (B) Secretion of TEM-1 fused to the secretion signal of Tir. Culture supernatants from wild-type E22 expressing TEM-1 alone, the Tir1-26-TEM fusion, and the Tir-TEM fusion were subjected to Western blot analysis with anti-TEM-1 antibody. Molecular mass markers (in kilodaltons) are indicated to the left. (C) Translocation of TEM-1 fused to the secretion signal of Tir. HeLa cells were infected with E22 strains expressing TEM-1 alone, the Tir1-26-TEM fusion, and the Tir-TEM fusion. The presented translocation data are averages of triplicate values of the results from three experiments.
In addition to the need for specific chaperone(s) and translocators, type III effector proteins have been shown to possess an STS present in the first codons. Tir was, so far, the only EPEC and EHEC effector molecule in which such a domain has been identified (4). This observation prompted us to compare the translocation and secretion of the Tir-TEM fusion and a fusion of the first 26 amino acids of Tir to TEM-1 (Tir1-26-TEM). These two fusion proteins expressed in EPEC strains were detected in culture supernatants, although Tir1-26-TEM was secreted less than Tir-TEM (Fig. 3B). When HeLa cells were infected with EPEC strains expressing Tir1-26-TEM or Tir-TEM, the ratio between blue and green emission fluorescence increased significantly, indicating that these two different fusion proteins were translocated (Fig. 3C). However, the emission ratio due to the translocation of Tir1-26-TEM was lower than the one of Tir-TEM and correlates to the lower secretion level of Tir1-26-TEM compared to that of the Tir-TEM. This is in agreement with the results of Crawford and Kaper (4) obtained with the Cya system and indicates that translocation mediated by the N-terminal domain of Tir is less efficient than the translocation mediated by the full-length effector.
Identification of the minimal N-terminal domain of Cif that can mediate translocation into eukaryotic cells.
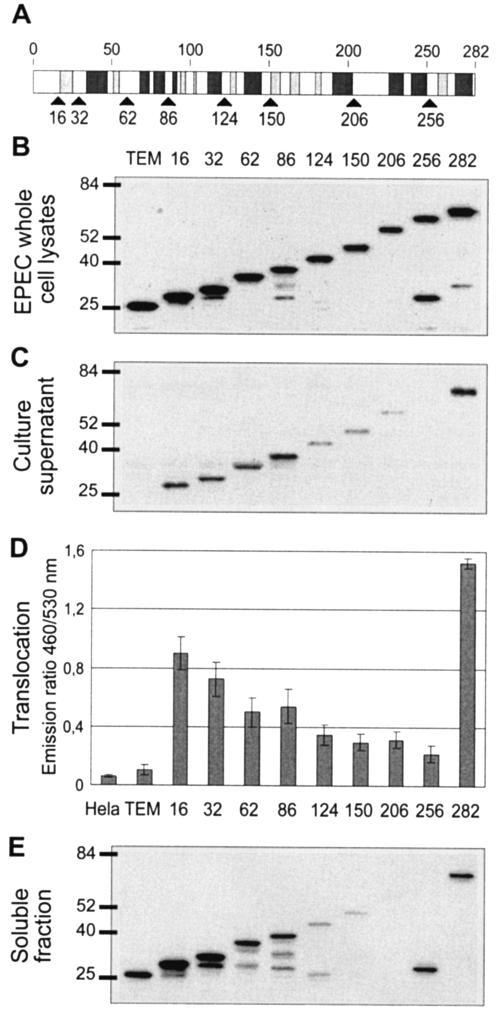
To identify the STS of Cif, we have constructed a set of truncated forms of Cif. The deletion sites were chosen to be junctions between two predicted α-helices or between the predicted α-helix and β-sheet (Fig. 4A). Plasmids expressing each fusion were transformed into an EPEC strain with a deletion of cif. Western blot analysis of the bacterial pellets revealed that the fusions were produced at a similar level (Fig. 4B). The full-length Cif-TEM was well secreted and translocated (Fig. 4C and D), whereas no secretion and no translocation were observed with TEM-1 alone (Fig. 4C and D). Analysis of the different fusions revealed that the first 16 amino acids of Cif were able to mediate the secretion and translocation of TEM-1. TEM fusions carrying the 32, 62, or 86 N-terminal Cif residues were also secreted and translocated but with a lower efficiency than full-length Cif (Fig. 4C and D). The other fusions with larger forms of Cif were not secreted and translocated or were less secreted and translocated (Fig. 4C and D). Analysis of the soluble fraction of the bacterial lysates by SDS-PAGE and immunoblotting revealed the poor solubility or absence of solubility of these fusion proteins, which were therefore less competent or not competent for secretion (Fig. 4E). In conclusion, Cif contains an N-terminal sequence which functions as an STS, but truncations in the C terminus greatly affected the solubility of the protein.
FIG. 4.
Identification of the minimal N-terminal domain of Cif that can mediate translocation into eukaryotic cells. (A) Schematic representation of the predicted secondary structure of Cif. Light and dark gray boxes represent β-sheets and α-helices, respectively. Black arrows indicate the different sites of fusion to TEM-1. (B) Expression of Cif1-X-TEM fusions in EPEC. EPEC whole-cell lysates were subjected to Western blot analysis with anti-TEM-1 antibody. Molecular mass markers (in kilodaltons) are indicated to the left. (C) Secretion of the produced Cif1-X-TEM fusions. Culture supernatants from E22 Δcif expressing TEM-1 alone and Cif1-X-TEM fusions were subjected to Western blot analysis with anti-TEM-1 antibody. (D) Translocation of Cif1-X-TEM fusions in HeLa cells. The presented data are averages of triplicate values of the results from three experiments. Numbers indicate the different sites of Cif fused to TEM-1. (E) Solubility analysis of the produced Cif1-X-TEM fusions. EPEC cells were lysed, and the soluble fraction was obtained after the removal of insoluble proteins, cell debris, and unbroken cells by centrifugation.
The first 20 codons of Cif, Tir, Map, and EspF mediate both secretion and translocation of TEM in a type III-dependent but chaperone-independent manner.
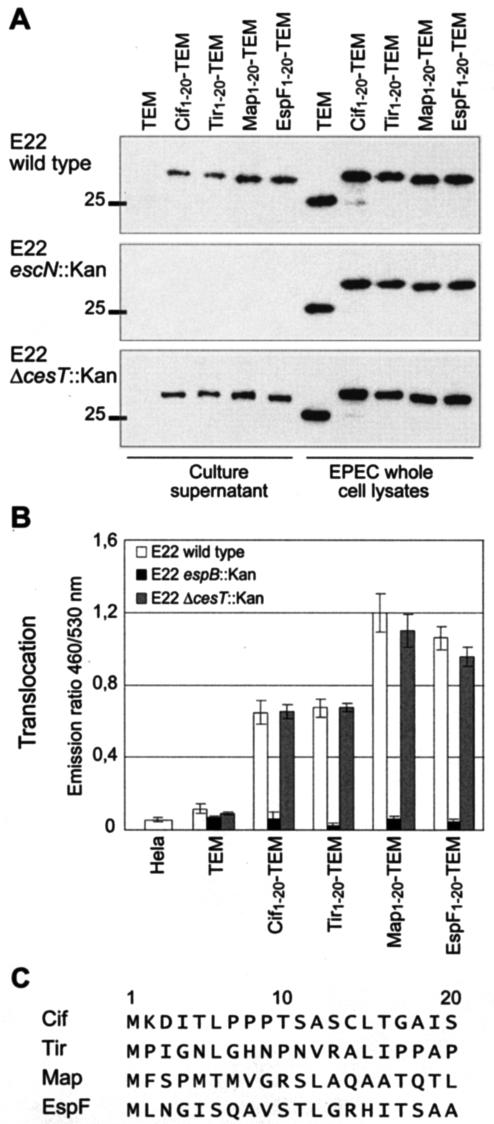
We have shown that, in addition to the LEE-encoded effector Tir, the non-LEE-encoded effector Cif also possess an N-terminal STS. In an attempt to define the structural properties of EPEC and EHEC strain STS, we tried to identify other STS in the first 20 codons of EPEC and EHEC effectors. We constructed fusions to TEM-1 of the first 20 residues of Map, EspF, Cif, and Tir. These fusions were expressed in a wild-type EPEC strain (Fig. 5A), and secretion and translocation efficiencies were measured as before (Fig. 5B). As expected, the sequence encoding the first 20 amino acids of Cif and Tir mediated both secretion and translocation. Similarly, the TEM fusions carrying the first 20 residues of Map and EspF were secreted in the culture medium. The Map1-20-TEM and EspF1-20-TEM fusions were also translocated, producing a higher emission ratio than the ones obtained for Cif1-20-TEM and Tir1-20-TEM. Thus, all analyzed effectors contain an STS encoded in their first 20 codons. To confirm that the secretion and translocation mediated by these STS were type III dependent, we transformed these constructions in an escN mutant unable to secrete the needle components of the type III apparatus and in an espB mutant unable to form the translocation pore in the eukaryotic cell membrane. As shown in Fig. 5A, the secretion of these TEM fusions was abolished in an escN mutant. Likewise, the translocation of TEM fusions was abolished in an espB mutant (Fig. 5B). However, when STS-TEM fusions were expressed in a cesT mutant, the levels of secretion and translocation were similar to those observed with the wild-type strain, although CesT is the chaperone of Tir and Map. From these results, we concluded that Cif, Tir, Map, and EspF possess an STS located in their first 20 codons and that these STS-encoding sequences direct secretion and translocation specifically via the TTSS without the need for a chaperone.
FIG. 5.
The first 20 residues of Cif, Tir, Map, and EspF mediate both secretion and translocation of TEM-1 in a type III-dependent manner. (A) Production and secretion of Cif1-20-TEM, Tir1-20-TEM, Map1-20-TEM, and EspF1-20-TEM fusions in wild-type E22, the escN mutant (TTSS defective), and the cesT mutant (defective for Tir/Map chaperone). Molecular mass markers (in kilodaltons) are indicated to the left. (B) Translocation in HeLa cells of Cif1-20-TEM, Tir1-20-TEM, Map1-20-TEM, and EspF1-20-TEM fusions in wild-type E22, the cesT mutant, and the espB mutant (translocation defective). The presented data are averages of triplicate values of the results from three experiments. (C) Alignment of the first 20 residues of Cif, Tir, Map, and EspF.
Cif is composed of a C-terminal effector domain and an exchangeable N-terminal translocation signal.
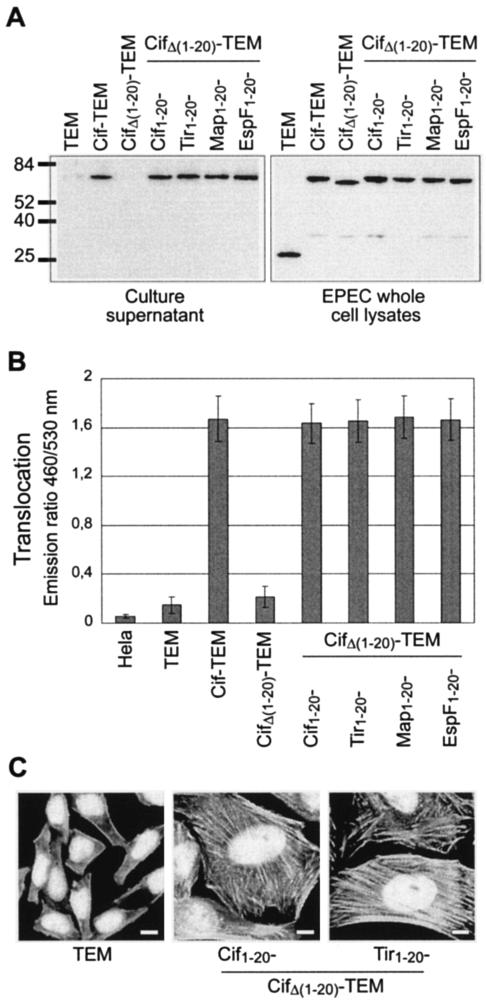
As shown above, STS from LEE-encoded or non-LEE-encoded effectors have similar behavior. To test the specificity of these STS, we exchanged the STS of the non-LEE-encoded Cif effector with the STS from the LEE-encoded Tir, Map, and EspF effectors. To do so, we constructed a Cif variant with a linker between amino acids 20 and 21, enabling the deletion or replacement of the N terminus by heterologous STS-encoding sequences from Tir, Map, and EspF or by the homologous STS-encoding sequences from Cif. The deletion of this region resulted in an N-terminally truncated form of CifΔ(1-20)-TEM fusion that was produced in a cif mutant at a level similar to that observed with the full-length Cif-TEM fusion (Fig. 6A). Solubility experiments showed that this N-terminal deletion did not alter the solubility of the protein (data not shown). However, no secretion (Fig. 6A) and no translocation could be detected (Fig. 6B). The addition of the first 20 codons of Cif restored the secretion and translocation at a level similar to that observed with the initial Cif-TEM fusion. We then examined the possibility of complementing the secretion and translocation of the CifΔ(1-20)-TEM fusion with the STS from Tir, Map, and EspF. These hybrid protein fusions were expressed in a cif mutant at a level similar to that of the original Cif-TEM fusion (Fig. 6A). The addition of the STS from Tir, Map, and EspF restored the secretion of Cif to a level similar to that observed with the Cif-TEM fusion (Fig. 6A). Moreover, cells infected with the cif mutant complemented with plasmids expressing Tir1-20-CifΔ(1-20)-TEM, Map1-20-CifΔ(1-20)-TEM, or EspF1-20-CifΔ(1-20)-TEM produced emission ratios identical to the emission ratio obtained with the cif mutant complemented with a plasmid expressing the Cif-TEM fusion, suggesting that these chimeric proteins were also injected into HeLa cells at a similar level (Fig. 6B). We then examined the ability of these protein fusions to trigger the Cif-related CPE. As expected, the nontranslocated CifΔ(1-20)-TEM fusion was not able to complement the cif mutant. In contrast, all chimeric fusions were able to fully complement the cif mutant, producing a typical CPE characterized by distended cells with large nuclei without mitotic spindles and by the formation of actin stress fibers (Fig. 6C).
FIG. 6.
Cif is composed of a C-terminal effector domain and an exchangeable N-terminal translocation signal. (A) Production and secretion of chimeric Cif-TEM fusions. Molecular mass markers (in kilodaltons) are indicated to the left. (B) Translocation in HeLa cells of chimeric Cif-TEM fusions. The presented data are averages of triplicate values of the results from two independent experiments. (C) CPE triggered by chimeric Cif-TEM fusions. HeLa cells were infected under conditions used to monitor translocation. At the end of the interaction, bacteria were killed with gentamicin and HeLa cells were incubated for a further 3 days. Actin and nuclei were stained, respectively, with rhodamine-phalloidin and DAPI. Bars, 10 μM.
Since the first 20 amino acids of Cif, Tir, Map, and EspF share no sequence homology and secondary structure similarity, these results suggest that Cif is a modular protein composed of an exchangeable N-terminal STS linked to a larger functional domain which promotes the cell cycle block and the formation of actin stress fibers.
DISCUSSION
Among LEE-encoded effectors, only the Tir effector has been studied to determine the domains involved in translocation. In this study, we analyzed the properties of Cif, the first described non-LEE-encoded effector, as a type III secretion substrate. The signals of Cif that mark the protein for secretion into the extracellular media and delivery into host cells were investigated by using the TEM-1 β-lactamase protein as a new reporter system. This approach revealed that the first 16 N-terminal residues of Cif are sufficient for secretion and delivery of the TEM-1 reporter by wild-type EPEC strains. We have also shown and confirmed that Tir, Map, and EspF contain an STS in their N termini. This result suggests that LEE-encoded and non-LEE-encoded effectors use the same molecular mechanisms to be exported from the bacterial cell via the TTSS. As shown in Fig. 5C, the amino acids of the STS of Cif, Tir, Map, and EspF share no similarity and failed to produce significant alignments. The STS from EspF and Map have been predicted to form an α-helix with an amphipathic profile, consistent with the hypothesis that amphipathicity can serve as a signal (24, 25). However, Tir and Cif have not been predicted to form α-helices, and thus, this latter hypothesis cannot be applied to these effectors. Similar analysis at the mRNA level also failed to identify conserved nucleic acid composition or particular secondary structures. The nature of the secretion signal has been the subject of many studies with Yersinia but still remains a matter of debate (for reviews and models, see references 20 and 37).
The observation that the Cif1-16-TEM fusion is less secreted and translocated than the full-length Cif-TEM is in agreement with previous studies showing that the N-terminal signal mediates secretion and translocation with lower efficiency than full-length effectors (38-40) and suggests, as for Tir, that a chaperone is required for the translocation of Cif. Preliminary studies with cosmids carrying the functional LEE from EPEC and EHEC strains suggest that the Cif chaperone (if any) is probably encoded by the LEE (data not shown). However, none of the chaperones previously shown to be involved in the translocation of LEE-encoded effectors, namely CesT for Tir and Map and CesF for EspF, could be shown to be Cif chaperones, since the translocation of Cif was not affected in a cesT mutant (Fig. 3A) and a cesF mutant (data not shown). Alternatively, it is also possible that Cif does not require a specific chaperone for its translocation into the host cell. In a recent study with the plasmid-borne minimal TTSS of Yersinia, it has been shown that translocation of YopE and YopT require their respective chaperones, whereas YopM did not require a chaperone for translocation (43). So efficient translocation could be achieved without a specific chaperone. As proposed by Gauthier and Finlay (16), the action of a chaperone, such as CesT, which directly binds EscN, would be to drive the effector to EscN to engage it into the channel. In this study, Gauthier and Finlay have also shown that Tir alone can also interact with EscN. Based on this finding, it is possible that Cif can directly bind EscN. The differential translocation efficiencies observed between Cif1-16-TEM and the full Cif-TEM fusion protein could then be explained by their differential abilities to bind EscN.
Little is known about the functional domains of Cif, but our results show that Cif lacking its STS can be targeted to the host cell by using an alternative STS from Tir, Map, or EspF. Since the first 20 amino acids of Cif, Tir, Map, and EspF share no sequence homologies and no secondary structure similarities, we believe that the translocation signal of Cif is not involved in the enzymatic or binding activity of Cif, in contrast to other effector proteins, such as YopH from Yersinia, where residues in the N-terminal domain are critical for substrate recognition (30). These results suggest that Cif is a modular protein composed of an exchangeable N-terminal STS linked to a larger functional domain which promotes the cell cycle block and the formation of actin stress fibers. The hypothesis of a C-terminal effector domain is substantiated by the fact that Cif76-282 is similar to a domain present in two putative proteins encoded by Burkholderia pseudomallei and Photorhabdus luminescens (26; data not shown). B. pseudomallei is the causative agent of melioidosis, a serious infectious disease of humans and animals that is endemic in subtropical areas. P. luminescens is a symbiont of nematodes and a broad-spectrum insect pathogen. Interestingly, these two pathogens code for one to three TTSSs, similar to secretion systems present in Xanthomonas spp. (36), Salmonella spp. (42), and Yersinia spp. (45). This observation, together with the fact that the C terminus of Cif may also have a role in protein stability and/or folding, explains the inability of wild-type strains harboring a 3′ truncated cif gene to produce a CPE on epithelial cells (26).
Several methods have been previously reported to monitor the determinants of secretion and translocation by the type III pathway. The first one was the CyaA system involving translational fusion with the calmodulin-dependent catalytic domain of the Bordetella pertussis toxin CyaA (41). This enzyme converts cellular ATP in cyclic AMP (cAMP) in the presence of the eukaryotic protein calmodulin. Also extensively used, this assay is relatively time-consuming. Another method involves a translational fusion with the phosphorylatable Elk peptide fused to the nuclear localization signal (NLS) from the large T antigen of simian virus 40 (7). The NLS sequence directs the fusion protein to the cell nucleus where the Elk tag is phosphorylated. The translocated protein can be detected by Western blotting with phosphospecific Elk peptide antibodies. Like the CyaA system, the Elk tag system uses cellular processes to detect proteins that are specifically injected into the host cell. This confers specificity but may have some limitations. For the CyaA system, some pathogenic bacteria produce toxins that are adenylate cyclases, leading to an increase of the cellular cAMP to supraphysiological levels (for example, the ExoY toxin from Pseudomonas aeruginosa) (46). This could mask the increase of cAMP converted by the CyaA fusion. In the Elk tag system, the fusion protein needs to be artificially translocated to the host cell nucleus, but many toxins are naturally targeted to other compartments such as the mitochondria (Map), the plasma membrane (Tir), or the Golgi apparatus (NleA). Thus, the system is dependent on the efficiency of the simian virus 40 NLS to alter the original intracellular targeting of the effector protein. Another method developed by Lee et al. is based on fractionation with digitonin that solubilizes the eukaryotic plasma membrane but not the prokaryotic membranes (23). As for the Cya and Elk tag systems, this requires disruption of the eukaryotic cell. In contrast, the TEM-1 is not based on a cellular process but on the differential entry of the TEM-1 substrate in bacterial and eukaryotic cells, and the use of the CCF2/AM fluorescent substrate enables translocation analysis in living cells without disruption of the host cell. In addition, this new reporter can be fused to the end of certain effectors without affecting their activity, which means that double activity tests can be carried out with efficiently translocated proteins, making the data more reliable. Because fewer than 100 molecules of TEM-1 can be readily detected within a cell (47), the system was sensitive enough to detect translocation of a weakly produced chromosomally encoded Cif-TEM fusion (data not shown).
In the context of a growing importance of TTSS in bacterial pathogenicity, TEM-1 fusion used in combination with CCF2/AM fluorescent substrate is a new powerful tool for identifying undiscovered bacterially encoded molecules that are delivered into host cells. A large number of substrates have been described for other TTSSs, such as the Salmonella sp. strain SPI-1 TTSS, which secretes at least 19 different proteins (15). The recent discovery of five other non-LEE-encoded TTSS substrates (9), in addition to Cif (26) and NleA/EspI (17, 31), raises the possibility that other type III translocated effectors may be encoded elsewhere in the EPEC and EHEC genomes. The recently published genome of EDL933 and Sakai O157:H7 EHEC strains have shown that the genome of pathogenic E. coli contains a large number of bacteriophages carrying open reading frames coding for putative proteins of unknown function (35). Thus, the number of proteins translocated by the LEE TTSS is very likely underestimated. We are currently investigating the use of TEM-1 fusion to identify other TTSS substrates. Identification of other TTSS substrates will open up new areas of investigation to increase our understanding of EHEC- and EPEC-mediated diseases.
Acknowledgments
We thank Philippe Bauchart for the construction of first-generation TEM-1 fusions and preliminary studies on translocation measurements, Jean-Philippe Nougayrède for helpful discussions and comments on the manuscript, and Ilan Rosenshine for suggesting solubility experiments. We are grateful to Neil Ledger for editorial assistance.
This work was supported by a grant from the European Union Fifth Framework Quality of Life Programme (QLK2-2000-00600).
REFERENCES
- 1.Abe, A., M. de Grado, R. A. Pfuetzner, C. Sanchez-Sanmartin, R. Devinney, J. L. Puente, N. C. Strynadka, and B. B. Finlay. 1999. Enteropathogenic Escherichia coli translocated intimin receptor, Tir, requires a specific chaperone for stable secretion. Mol. Microbiol. 33:1162-1175. [DOI] [PubMed] [Google Scholar]
- 2.Charpentier, X., C. Chalut, M. H. Remy, and J. M. Masson. 2002. Penicillin-binding proteins 1a and 1b form independent dimers in Escherichia coli. J. Bacteriol. 184:3749-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 4.Crawford, J. A., and J. B. Kaper. 2002. The N-terminus of enteropathogenic Escherichia coli (EPEC) Tir mediates transport across bacterial and eukaryotic cell membranes. Mol. Microbiol. 46:855-868. [DOI] [PubMed] [Google Scholar]
- 5.Creasey, E. A., R. M. Delahay, A. A. Bishop, R. K. Shaw, B. Kenny, S. Knutton, and G. Frankel. 2003. CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol. Microbiol. 47:209-221. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day, J. B., F. Ferracci, and G. V. Plano. 2003. Translocation of YopE and YopN into eukaryotic cells by Yersinia pestis yopN, tyeA, sycN, yscB and lcrG deletion mutants measured using a phosphorylatable peptide tag and phosphospecific antibodies. Mol. Microbiol. 47:807-823. [DOI] [PubMed] [Google Scholar]
- 8.Deibel, C., S. Kramer, T. Chakraborty, and F. Ebel. 1998. EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol. Microbiol. 28:463-474. [DOI] [PubMed] [Google Scholar]
- 9.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rycke, J., E. Comtet, C. Chalareng, M. Boury, C. Tasca, and A. Milon. 1997. Enteropathogenic Escherichia coli O103 from rabbit elicits actin stress fibers and focal adhesions in HeLa epithelial cells, cytopathic effects that are linked to an analog of the locus of enterocyte effacement. Infect. Immun. 65:2555-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, S. J., S. W. Hutcheson, M. S. Dubois, J. L. Mellies, L. A. Wainwright, M. Batchelor, G. Frankel, S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176-1189. [DOI] [PubMed] [Google Scholar]
- 12.Elliott, S. J., E. O. Krejany, J. L. Mellies, R. M. Robins-Browne, C. Sasakawa, and J. B. Kaper. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, S. J., C. B. O'Connell, A. Koutsouris, C. Brinkley, M. S. Donnenberg, G. Hecht, and J. B. Kaper. 2002. A gene from the locus of enterocyte effacement that is required for enteropathogenic Escherichia coli to increase tight-junction permeability encodes a chaperone for EspF. Infect. Immun. 70:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 15.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell. Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 16.Gauthier, A., and B. B. Finlay. 2003. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 185:6747-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruenheid, S., I. Sekirov, N. A. Thomas, W. Deng, P. O'Donnell, D. Goode, Y. Li, E. A. Frey, N. F. Brown, P. Metalnikov, T. Pawson, K. Ashman, and B. B. Finlay. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 51:1233-1249. [DOI] [PubMed] [Google Scholar]
- 18.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlinsey, J. E., J. Lonner, K. L. Brown, and K. T. Hughes. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487-497. [DOI] [PubMed] [Google Scholar]
- 21.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 22.Kenny, B., and M. Jepson. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2:579-590. [DOI] [PubMed] [Google Scholar]
- 23.Lee, V. T., D. M. Anderson, and O. Schneewind. 1998. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol. Microbiol. 28:593-601. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd, S. A., A. Forsberg, H. Wolf-Watz, and M. S. Francis. 2001. Targeting exported substrates to the Yersinia TTSS: different functions for different signals? Trends Microbiol. 9:367-371. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd, S. A., M. Sjostrom, S. Andersson, and H. Wolf-Watz. 2002. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43:51-59. [DOI] [PubMed] [Google Scholar]
- 26.Marches, O., T. N. Ledger, M. Boury, M. Ohara, X. Tu, F. Goffaux, J. Mainil, I. Rosenshine, M. Sugai, J. De Rycke, and E. Oswald. 2003. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 50:1553-1567. [DOI] [PubMed] [Google Scholar]
- 27.Marches, O., J. P. Nougayrede, S. Boullier, J. Mainil, G. Charlier, I. Raymond, P. Pohl, M. Boury, J. De Rycke, A. Milon, and E. Oswald. 2000. Role of tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 68:2171-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNamara, B. P., and M. S. Donnenberg. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71-78. [DOI] [PubMed] [Google Scholar]
- 30.Montagna, L. G., M. I. Ivanov, and J. B. Bliska. 2001. Identification of residues in the N-terminal domain of the Yersinia tyrosine phosphatase that are critical for substrate recognition. J. Biol. Chem. 276:5005-5011. [DOI] [PubMed] [Google Scholar]
- 31.Mundy, R., L. Petrovska, K. Smollett, N. Simpson, R. K. Wilson, J. Yu, X. Tu, I. Rosenshine, S. Clare, G. Dougan, and G. Frankel. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 72:2288-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nougayrede, J. P., M. Boury, C. Tasca, O. Marches, A. Milon, E. Oswald, and J. De Rycke. 2001. Type III secretion-dependent cell cycle block caused in HeLa cells by enteropathogenic Escherichia coli O103. Infect. Immun. 69:6785-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nougayrede, J. P., O. Marches, M. Boury, J. Mainil, G. Charlier, P. Pohl, J. De Rycke, A. Milon, and E. Oswald. 1999. The long-term cytoskeletal rearrangement induced by rabbit enteropathogenic Escherichia coli is Esp dependent but intimin independent. Mol. Microbiol. 31:19-30. [DOI] [PubMed] [Google Scholar]
- 35.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 36.Rainbow, L., C. A. Hart, and C. Winstanley. 2002. Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J. Med. Microbiol. 51:374-384. [DOI] [PubMed] [Google Scholar]
- 37.Ramamurthi, K. S., and O. Schneewind. 2003. Substrate recognition by the Yersinia type III protein secretion machinery. Mol. Microbiol. 50:1095-1102. [DOI] [PubMed] [Google Scholar]
- 38.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schesser, K., E. Frithz-Lindsten, and H. Wolf-Watz. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sory, M. P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 92:11998-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 42.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649-659. [DOI] [PubMed] [Google Scholar]
- 43.Trulzsch, K., A. Roggenkamp, M. Aepfelbacher, G. Wilharm, K. Ruckdeschel, and J. Heesemann. 2003. Analysis of chaperone-dependent Yop secretion/translocation and effector function using a mini-virulence plasmid of Yersinia enterocolitica. Int. J. Med. Microbiol. 293:167-177. [DOI] [PubMed] [Google Scholar]
- 44.Tu, X., I. Nisan, C. Yona, E. Hanski, and I. Rosenshine. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595-606. [DOI] [PubMed] [Google Scholar]
- 45.Waterfield, N. R., P. J. Daborn, and R. H. French-Constant. 2002. Genomic islands in Photorhabdus. Trends Microbiol. 10:541-545. [DOI] [PubMed] [Google Scholar]
- 46.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zlokarnik, G., P. A. Negulescu, T. E. Knapp, L. Mere, N. Burres, L. Feng, M. Whitney, K. Roemer, and R. Y. Tsien. 1998. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279:84-88. [DOI] [PubMed] [Google Scholar]
- 48.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]