Abstract
The duplicated hasR and hasL genes of Streptomyces ambofaciens encode alternative sigma factors (named σBR and σBL) belonging to the σB general stress response family in Bacillus subtilis. The duplication appears to be the result of a recent event that occurred specifically in S. ambofaciens. The two genes are 98% identical, and their deduced protein products exhibit 97% identity at the amino acid level. In contrast with the coding sequences, their genetic environments and their transcriptional control are strongly divergent. While hasL is monocistronic, hasR is arranged in a polycistronic unit with two upstream open reading frames, arsR and prsR, that encode putative anti-anti-σ and anti-σ factors, respectively. Transcription of each has gene is initiated from two promoters. In each case, one promoter was shown to be developmentally controlled and to be similar to those recognized by the B. subtilis general stress response sigma factor σB. Expression from this type of promoter for each of the has genes dramatically increases during the course of growth in liquid or on solid media and following oxidative and osmotic stresses. Reverse transcription-PCR measurements indicate that hasR is 100 times more strongly expressed than hasL from the σB-like promoter. Transcription from the second promoter of each gene (located upstream of arsR in the case of the hasR locus) appears to be constitutive and weak. Quantitative transcriptional analysis in single and double has mutant strains revealed that σBR and σBL direct their own transcription as well as that of their duplicates. Only a slight sensitivity in response to oxidative conditions could be assigned to either single or double mutants, revealing the probable redundancy of the σ factors implied in stress response in Streptomyces.
Gene duplication is a key mechanism for genome evolution. In contrast to eukaryotes, duplication of genes in prokaryotes is considerably less well documented. However, according to the work of Ikeda et al. (20), 35% of the 7,574 Streptomyces avermitilis open reading frames (ORFs) cluster into 721 paralogous families, with membership ranging from 2 to 91 genes per family. The membrane-spanning components of the ATP binding cassette (ABC) transporters, the two-component transcriptional regulator systems, and the σ factors are the most prominent examples of such gene families (20). The Streptomyces coelicolor A3(2) (4) and S. avermitilis (20) genomes each contain 65 and 60 σ-factor-encoding genes, respectively, many more than the 7 reported in Escherichia coli (5), 16 in Bacillus subtilis (23), and 13 in Mycobacterium tuberculosis (9), the genus most closely related to Streptomyces. According to the classic duplication-degeneration-complementation model (14), gene duplication can lead to the overproduction of a gene product. The conservation of duplicated genes on the evolutionary scale might also involve subfunctionalization of both members of the pair. Each duplicate can undergo complementary degenerative mutations involving reduction or specialization of its expression. The combined action of both loci would be necessary to ensure the ancestral function. In Streptomyces, the large-scale duplication phenomenon might correlate with a complex life cycle associated with a high level of differentiation at both the morphological (formation of aerial mycelium and sporulation) and biochemical (production of numerous secondary metabolites) levels.
Among the 65 sigma factor genes in S. coelicolor, nine, homologous to the B. subtilis stress response factor σB, were found to group into a subfamily with the deduced proteins exhibiting 38 to 73% identity (4) (sigF/SCO4035, sigG/SCO7341, sigH/SCO5243, sigI/SCO3068, sigJ/SCO0600, sigK/SCO6520, sigL/SCO7278, sigM/SCO7314, and sigN/SCO4034; sig gene names are according to the Kelemen nomenclature; G. Kelemen, personal communication). A similar situation was revealed in S. avermitilis (20), showing that the σB subfamily resulted from duplication events that probably arose in the S. coelicolor-S. avermitilis common ancestor.
One member of this family, called sigB (8) or sigJ (40) in S. coelicolor and SAV741 in S. avermitilis, was found in two copies in Streptomyces ambofaciens (13). The two copies were named has for homologous to alternative sigma factors and are 98% identical at the nucleotide level (13). Such a high level of identity between two duplicated genes is rare in a Streptomyces species. For example, the glgBI and glgBII genes of S. coelicolor exhibit only 73% nucleotidic identity (glgBI is involved in glycogen metabolism during vegetative growth, while glgBII participates in the accumulation of glycogen in spores and is expressed only in late phases of differentiation) (6). Clustering of the S. coelicolor ORFs by the Basic Local Alignment Search Tool protein clustering program (excluding mobile genetic element-derived products; BLASTCLUST [2]) indeed revealed only 10 gene pairs encoding products which share the same high level of amino acid identity as the products of the S. ambofaciens has genes (data not shown). None of the 10 pairs are predicted to encode σ factors.
Taking into account the phylogenetic relationships among S. avermitilis, S. coelicolor, and S. ambofaciens, the last two being most closely related, the duplication of the has genes appears to be a recent event and therefore constitutes a useful model to study the fate of duplicated genes. With this in mind, we investigated the role of the has genes in S. ambofaciens using transcriptional and mutational analyses.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Strains and plasmids used are described in Table 1. All Streptomyces culture conditions, media, and selective conditions are as previously described (22). Growth curves were carried out in Hickey-Tresuer (HT) liquid medium. Exponential, transition, and stationary phases were defined as being after 9, 16, and 24 h of growth, respectively. Stress conditions were applied as follows: cells were grown to mid-exponential phase in liquid HT medium and exposed to the stressing agent for 30 min (10 mM H2O2, 500 mM NaCl, 34% sucrose, 0.7 M ethanol, or 30% cumene hydroperoxide). High-temperature shocks consisted of a shift from 30 to 42°C for 20 min. Oxidative stress experiments were also performed on solid medium by the disk paper method as described previously (29), with H2O2 concentrations ranging from 0.1 to 10 mM. Luria-Bertani and SOB liquid media were used for growing E. coli, and Luria-Bertani medium was used for growing B. subtilis (34). UV treatments were carried out on spores, spread on HT medium, by irradiation (0 to 400 J/m2) from GTE Sylvania UV lamps (254 nm, 64 mW/cm2). After irradiation, plates were incubated at 30°C in the dark to avoid photoreactivation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Property(-ies)a | Reference |
|---|---|---|
| Strains | ||
| S. ambofaciens | ||
| ATCC 23877 | wt | 31 |
| ETH11317 | wt | 19 |
| DSM40697 | wt | 19 |
| NSAR1, NSAR2, NSAR3 | hasR replaced by aac(3)IV/oriT cassette | This study |
| NSAL1, NSAL2, NSAL3 | hasL replaced by aac(3)IV/oriT cassette | This study |
| NSARL1, NSARL3 | hasR and hasL replaced by aac(3)IV/oriT and aadA/oriT cassettes, respectively | This study |
| NSALR2, NSALR7 | hasL and hasR replaced by aac(3)IV/oriT and aadA/oriT cassettes, respectively | This study |
| NSAR2-s1, NSAR2-s2 | NSAR2 with pSETΩ integrated into the chromosomal attB site of φC31 | This study |
| NSAR2-sR1, NSAR2-sR2 | NSAR2 with pSETΩsR integrated into the chromosomal attB site of φC31 | This study |
| E. coli | ||
| DH5α | General cloning strain | 34 |
| ET12567 | Used for intergeneric conjugation | 27 |
| BW25113 | Used for PCR-targeting mutagenesis | 10 |
| Plasmids and cosmids | ||
| Supercos1 | bla neo | 11 |
| 14C4 | Recombinant Supercos1 containing hasL locus | 13 |
| 25E1 | Recombinant Supercos1 containing hasR locus | 13 |
| pIJ790 | gam bet exo | 15 |
| pIJ773 | oriT aac(3)IV | 15 |
| pIJ778 | oriT aadA | 15 |
| pSETΩ | oriT attP int aadA | 28 |
| pSETΩsR | pSETΩ + hasR cloned into the EcoRI site | This study |
gam, Gam (host's exonuclease V inhibitor) gene; bet, Bet (single-stranded-DNA binding protein) gene; exo, exonuclease gene; aadA, spectinomycin and streptomycin resistance gene; aac(3)IV, apramycin resistance gene; neo, kanamycin resistance gene; bla, ampicillin resistance gene; int, integrase gene of φC31; oriT, origin of transfer.
Nucleic acid manipulations.
Standard or pulsed-field gel electrophoresis DNA preparations from S. ambofaciens were made as described earlier (24, 25). Cosmid and plasmid DNAs were extracted from E. coli by the alkaline method (34). All restriction enzymes, PCR reagents, and other molecular biology reagents were purchased from New England Biolabs or Roche Diagnostics.
Digoxigenin-DNA labeling (digoxigenin dUTP), hybridization, washings, and detection were performed according to the recommendations of the manufacturer (Roche). Light emission was acquired with a Fluor’S MultiImager (Bio-Rad Laboratories). DNA sequencing was performed using the dye terminator cycle sequencing kit on an ABI Prism 310 system and analyzed with the Sequencing Analysis software (Applied Biosystems). All sequence similarity searches were performed in the nonredundant (NR) database by using the BLAST programs (2) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Total RNA was isolated from HT liquid or solid (growth on cellophane) medium-grown cultures of S. ambofaciens with the TRI reagent (Sigma), followed by a chloroform extraction. High-resolution S1 nuclease mapping experiments were performed according to the method of Kieser et al. (22) with probes consisting of PCR products (primer sequences and localization are as described in Table 2 and Fig. 1). Reverse transcription-PCR (RT-PCR) experiments were carried out according to the method of Pang et al. (30).
TABLE 2.
Primers used in this work
| Usage | Primer | Nucleotide sequencea (5′-3′) | Use |
|---|---|---|---|
| Construction of mutant strains | HasR1 | GCCGTGAGCGGGGCCGATCGACTGGGAGGGAACATC ATGTGTAGCCTGGACCTCCTTC | Replacement of hasR |
| HasR2 | ACCGCCGCGGGAAGGACAGCCGCCGCCTGAGCGGCG CTCAATTCCGGGGATCCGTCGACC | Replacement of hasR | |
| HasL1 | GTGACGAGCAGGCAGTTCGAACAGGGAGGGCCCATC ATGTGTAGGCTGCAGCTGCTTC | Replacement of hasL | |
| HasL2 | GCCGCCCCGGTGTGCCCACGGTGAGGGGCCCTCGCC TCAATTCCGGGGATCCGTCGACC | Replacement of hasL | |
| Complementation | CoR1 | TATGAATTCTGTGACGGTGTGCGGATG | Complementation of hasR mutant |
| CoR2 | AATGAATTCCCGGGCGGTCGAGGATGA | Complementation of hasR mutant | |
| Real-time RT-PCR | Rqu1 | CCGATGAGGCCGGCCGT | Gene expression of hasRp1 and hasRp2 |
| Rqu2 | GATGTTCCCTCCCAGTCGATCGG | Gene expression of hasRp1 and hasRp2 | |
| Rqu3 | CCGACGAGACTGCGCTTAC | Gene expression of hasRp2 | |
| Rqu4 | GGCGAGACAGACCGAGAAAA | Gene expression of hasRp2 | |
| Lqu1 | CTGCTGACGAGCAGGCAG | Gene expression of hasLp1 and hasLp2 | |
| Lqu2 | GATGGGCCCTCCCTGTT | Gene expression of hasLp1 and hasLp2 | |
| Lqu3 | CCCGAGGCCGGGGGTATTG | Gene expression of hasLp2 | |
| HrdB-R | CGCGAGCCCATCTCGCTGC | Gene expression of hrdB | |
| HrdB-F | CGTCGAGGGTCTTCGGCTG | Gene expression of hrdB | |
| Probes for S1 mapping | R5 | GTCATCCACGGTGGTGGCAGCGGCA | Probes for hasR (PR1, PR3) |
| G | GTCCTTGGGCGCCACCTTGGACGGG | Probe for hasR (PR3) | |
| E | GGCATGCCCGTGGTGGTGTGCTCGG | Probes for hasR and hasL (PL, PR1) | |
| L5 | CACGCACAGCGTGTACGTGGTGTCC | Probe for hasL (PL) | |
| arsR1 | CGGTGTCAACGTAGACGGA | Probe for hasR (PR2) | |
| arsR2 | TCTCCACTGGCCCGGATAC | Probe for hasR (PR2) |
The sequence in boldface corresponds to hasR or hasL locus sequences (including the start and stop codons, which are underlined), while the sequence in lightface matches the end of the disruption cassette.
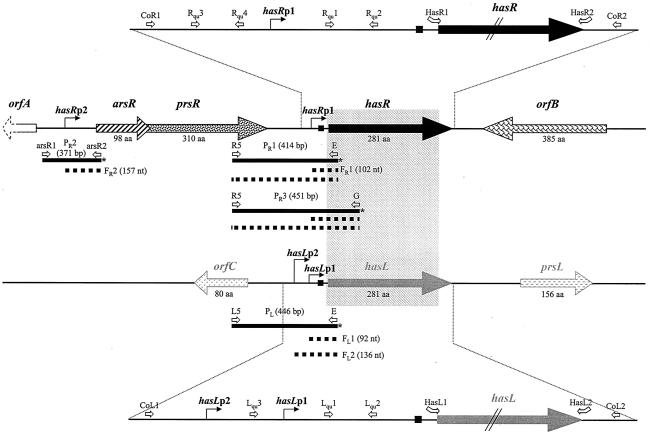
FIG. 1.
Genetic organization of the S. ambofaciens DSM49697 hasR and hasL loci. The gray rectangle represents the homologous area between the two has loci. The ribosome binding sites are symbolized by the small black squares. orfA is incompletely sequenced (dashed arrow). S1 probes (PR1, PR2, PR3, and PL) are presented as black bars with an asterisk symbolizing the radiolabeled end. The dotted thick lines represent the protected fragments obtained with each S1 probe. The different primers used in real time RT-PCR experiments, for construction of S1 probes, or for the PCR-targeted mutagenesis are symbolized by the open arrows (primer sequences are in Table 2). aa, amino acids.
Real-time PCR.
Real-time PCRs were carried out on an iCycler iQ real-time PCR detection system (Bio-Rad Laboratories), and the data were analyzed using the software provided by the supplier. Sequences of primers and localizations are indicated in Table 2 and Fig. 1. Assays were performed using 5 μl of cDNA, 12.5 μl of Platinum Quantitative PCR SuperMix-UDG (Invitrogen), 3.3 μl of SYBR Green I (10,000× dilution; Sigma), and 10 pmol of each primer in a final volume of 25 μl. Thermal cycle conditions were as follows: 2 min at 50°C and 10 min at 95°C followed by 40 cycles of a maximum of 30 s at 95°C and 1 min at 60°C (annealing and extension) and then 80 steps of 10 s with temperature increasing by 0.5°C for each step from 55 to 94°C (determination of the melting curve). For each run, a standard dilution of the cDNA was used to check the relative efficiency of primers. A negative control (distilled water) was included in each real-time PCR assay. Each experiment was performed in duplicate. The hrdB gene was used as an internal control to quantify the relative expression of the target genes. The expression levels of the downstream promoters (hasRp1 and hasLp1) corresponded to the total gene expression (quantified with Rqu1-Rqu2 and Lqu1-Lqu2, Fig. 1 and Table 2) deduced from those of the upstream promoters (hasRp2 quantified with Rqu3-Rqu4 and hasLp2 quantified with Lqu3-Lqu2 [Fig. 1 and Table 2]).
PCR-targeted mutagenesis.
Gene disruptions were achieved using the PCR-targeting system developed by Gust et al. (15). In order to replace an 840-bp sequence corresponding to the hasR coding region (included in the cosmid 25E1), two primers, HasR1 and HasR2 (Table 2 and Fig. 1), were used to amplify the cassettes aac3(IV)/oriT and aadA/oriT from plasmids pIJ773 and pIJ778, respectively (15). Similarly, the HasL1 and HasL2 primers (Table 2 and Fig. 1) were used to replace the 840-bp hasL sequence in cosmid 14C4. Allelic exchanges were confirmed by Southern blotting and PCR analysis.
Complementation.
The hasR gene (promoter and coding sequence) was amplified using the CoR1-CoR2 primer set (Table 2 and Fig. 1) and cloned into the conjugative and integrative vector pSETΩ (28) to give pSETΩsR (Table 1). The integrity of the insert was checked by sequencing. The recombinant plasmid was then introduced into the single mutant strains by conjugal transfer. Conjugation and screening of S. ambofaciens exconjugants were carried out according to the method of Kieser et al. (22), except that HT containing MgCl2 (10 mM) was used instead of soy flour mannitol (SFM) MgCl2.
BLASTCLUST.
Analysis with use of the Basic Local Alignment Search Tool protein clustering program (BLASTCLUST) (2) was performed on the complete set of S. coelicolor ORFs excluding transposases (http://www.sanger.ac.uk) using the following parameters: minimum 95% identity and 100% length coverage.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the hasR (4,429-nucleotide [nt]) and hasL (3,659-nt) loci were deposited in the database under accession numbers AF050150 and AF0150151, respectively.
RESULTS AND DISCUSSION
Characterization of the S. ambofaciens DSM40697 hasR and hasL duplicated genes.
Each of the duplicated has genes is located on a different arm of the linear chromosome, hasR at about 850 kb from the right chromosomal end and hasL at 450 kb from the left end. The has genes constitute a substrate for homologous recombination and were discovered as a result of their implication in DNA rearrangements (13). The homology between the two has loci starts 4 nt upstream of the start codon and stops 8 nt before the stop codon (Fig. 1). The nucleotide sequences of the duplicated has ORFs are 98% identical over 846 bp.
The putative proteins, σBR and σBL, differ by only 8 amino acids over 281 residues (sequencing of the chromosomal loci resulted in the identification of four additional divergent amino acid positions in addition to those in the initial report [13]) and share 100% identity in regions 2-4 and 4-2, which are involved in the recognition of the −10 and −35 regions of promoters (26). Homologues of σBR and σBL are listed in Table 3. The σBR and σBL factors exhibit the highest similarity with σB of S. coelicolor (8), also named SigJ by Viollier et al. (40). The genes controlled by σB are reported to play a role in osmoprotection and in the erection of aerial mycelium (8).
TABLE 3.
Homologues of S. ambofaciens σBR and σBL
| Protein (reference) | % Similarity (% identity of regions 2-4 and 4-2, respectively)
|
|
|---|---|---|
| S. ambofaciens DSM40697 σBR | S. ambofaciens DSM40697 σBL | |
| S. ambofaciens DSM40697 σBL | 97 (100, 100) | |
| S. coelicolor A3(2) σB (SigJ) (8) | 94 (100, 100) | 93 (100, 100) |
| S. avermitilis SAV741 (20) | 80 (100, 78) | 80 (100, 78) |
| S. coelicolor A3(2) SigL (SCO7278a) (4) | 73 (95.5, 74) | 71 (95.5, 74) |
| Streptomyces setonii CrtS (21) | 64 (100, 62.5) | 64 (100, 62.5) |
| S. coelicolor A3(2) SigF (32) | 46 (73, 62.5) | 48 (73, 62.5) |
| B. subtilis σB (39) | 34 (73, 50) | 33 (73, 50) |
SCO7278 is named SigL according to the nomenclature of G. Kelemen.
The orthologues of the has genes are named sigB (sigJ) in S. coelicolor (94 and 93% amino acid identity with σBR and σBL, respectively) and SAV741 for S. avermitilis (80% amino acid identity with σBR and σBL). In each organism, they belong to a gene family consisting of nine members: sigF, sigG, sigH, sigI, sigJ (sigB), sigK, sigL, sigM, and sigN in S. coelicolor and SAV4185, SAV7400, SAV3013, SAV3492, SAV741, SAV1087, SAV1873, SAV1151, and SAV4186 in S. avermitilis.
In both S. coelicolor and S. avermitilis, the has gene orthologue is not duplicated. Indeed, the closest homologues to σB (SigJ) in S. coelicolor and SAV741 in S. avermitilis within their respective genomes are the sigma factors SigL and SAV115, respectively. The two pairs share only 73 and 70% identity, while σBR and σBL of S. ambofaciens DSM40697 are 97% identical to each other at the amino acid level.
Furthermore, the has genes were also found to be duplicated in two other isolates of S. ambofaciens (ATCC 23877 and ETH11317). For each strain, the nucleotidic identity between the two has genes was 97% (data not shown).
Since S. avermitilis is a species more phylogenetically distant from S. coelicolor than is S. ambofaciens (http://avermitilis.ls.kitasato-u.ac.jp/), these data suggest that the duplication of the has genes is a recent event which has presumably occurred following divergence from the last common ancestor of the S. coelicolor and S. ambofaciens species.
As shown in Fig. 1, the sequenced region of the hasR locus (4,429 nt) contains five ORFs named orfA, arsR, prsR, hasR, and orfB. Notably, the two ORFs upstream of hasR, arsR and prsR, are predicted to encode homologues of anti-anti-σ and anti-σ factor proteins, respectively. Thus, arsR is 91% similar to the anti-anti-σB encoded by rsbB of S. coelicolor (8), and prsR exhibits 89% similarity to anti-σB encoded by rsbA of S. coelicolor (8).
The genetic organization of the hasR locus is identical to that of sigB (sigJ) in S. coelicolor (Fig. 2A). The initiation codon of prsR overlaps the termination codon of arsR (as in S. coelicolor), suggestive of coregulation, and this was subsequently demonstrated (see below). Interestingly, the conserved sigB (sigJ) locus is found on the left chromosomal arm of S. coelicolor 640 kb from the end (4), while the hasR locus is located 850 kb from the end of the right chromosomal arm in S. ambofaciens. Thus, the divergence of the two species was probably accompanied by a large terminal rearrangement.
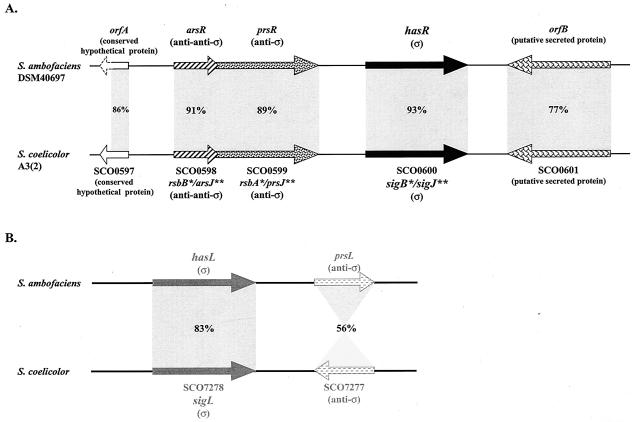
FIG. 2.
Comparison of the organization of the has loci in S. ambofaciens DSM40697 and S. coelicolor A3(2). The percentages correspond to the similarities between the deduced products of the genes. The 86% similarity observed between the products of orfA and ScF55-21c applies to the first 104 amino acids of the proteins only, since it was incompletely sequenced (dashed arrow) in S. ambofaciens. *, according to the work of Cho et al. (8); **, according to the work of Viollier et al. (40) or G. Kelemen (personal communication).
The genetic organization of the hasL locus is significantly different from that of hasR and of sigB (sigJ) in S. coelicolor (Fig. 2B). The product of hasL shares 71% amino acid identity with SigL (SCO7278) of S. coelicolor (one of the nine σ factors of the family previously described), but no homology could be found between the two loci when the upstream regions were compared. Immediately downstream of hasL, prsL encodes a putative anti-σ protein showing only 56% similarity with the product of SCO7277, located downstream from sigL. Furthermore, the two ORFs lie in the opposite orientation compared to their upstream σ factor-encoding genes.
Mapping of the TSPs.
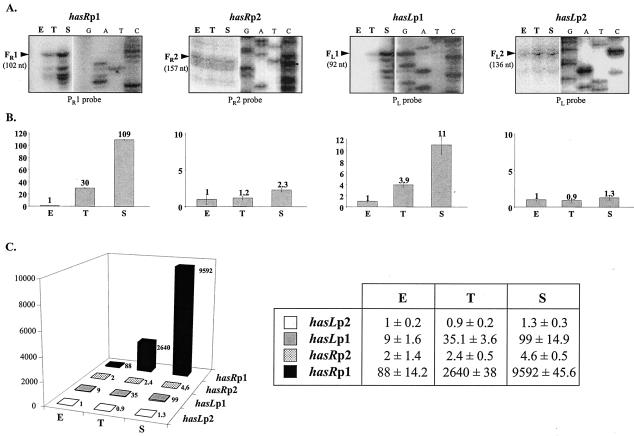
For hasR, S1 nuclease mapping experiments using the PR1 probe (Fig. 1), consisting of 414 bp overlapping the intergenic region between hasR and prsR, revealed two protected fragments. The shortest one, FR1 (102 nt), revealed a transcriptional start point (TSP) located 53 bp upstream of the hasR start codon (Fig. 3A), while the longer one (414 bp) could correspond either to probe-probe reannealing or to a readthrough transcript originating from the arsR promoter region (i.e., from hasRp2 [data not shown]). RT-PCR experiments with the primers used for the construction of the PR1 probe confirmed the existence of a second promoter (data not shown). A second TSP (hasRp2) was indeed found 114 nt upstream of the arsR start codon with the use of the probe PR2 (fragment FR2 of 157 nt; Fig. 1 and 3A). These data are consistent with the chromosomal organization of the hasR locus, which suggests that hasR is part of a three-gene operon, arsR-prsR-hasR, although transcription from a promoter located between hasRp1 and hasRp2 (Fig. 2A) cannot be excluded. However, in S. coelicolor, a single operon including the homologous system consisting of the rsbB, rsbA, and sigB genes was also reported (8).
FIG. 3.
Identification and quantification of the four has transcripts. (A) Identification of the transcription start sites of hasR (left) and hasL (right) genes by S1 nuclease mapping and determination of the level of transcription of the has genes during growth in liquid culture. Total RNAs were prepared from the wt strain at the exponential (E), transition (T), and stationary (S) phases of growth in liquid HT medium and analyzed using the S1 probes PR1, PR2, and PL. Only the part of the autoradiographic film corresponding to each protected fragment (FR1, FR2, FL1, and FL2) is shown, alongside a panel showing the DNA sequence ladder. The experiments were performed using three independent RNA samples for each growth phase, and representative results are shown. (B) Quantification of transcription from the two hasR (left) and hasL (right) genes during growth in liquid HT medium with the use of real-time RT-PCR (see Materials and Methods). hrdB was used as an internal reference, as it is expressed at a constant level throughout growth. The level of expression of each transcript in the exponential phase was arbitrarily fixed at 1. These experiments were performed using the same three independent RNA samples as detailed for panel A above. (C) Comparison of the levels of expression of each has transcript during growth in liquid HT medium with the use of real-time RT-PCR. The level of expression of the least expressed transcript—corresponding to the hasLp2-initiated transcript in the exponential-phase sample—was arbitrarily fixed at 1.
For hasL, two protected fragments of 92 (FL1) and 136 (FL2) nt were detected using the PL probe (446 bp; Fig. 1 and 3A). They correspond to transcripts initiated from two TSPs, hasLp1 and hasLp2, located 43 and 87 bp, respectively, upstream of the start codon.
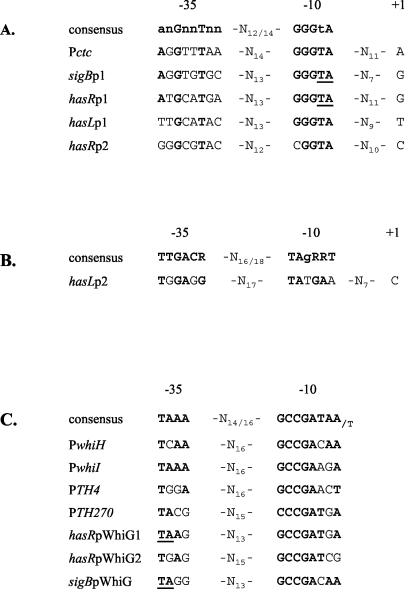
Interestingly, two boxes matching the consensus sequence of promoters recognized by the B. subtilis general stress response σB factor (39) and by σB of S. coelicolor (8) were identified upstream of both hasRp1 and hasLp1 (Fig. 4A). A putative promoter sequence for the principal σ factor, σHrdB of S. coelicolor (36), was identified upstream from the TSP hasLp2 (Fig. 4B). Only a degenerate σB promoter consensus was found upstream from hasRp2 (Fig. 4A). Interestingly, two sequences that are almost identical to the promoters recognized by the sigma factor WhiG were identified upstream of hasR (Fig. 4C). One of these putative promoters (hasRpWhiG1) overlaps the hasRp1 promoter. However, no transcription start site could be associated with these promoters with the use of either the PR1 or the PR3 probe under the experimental conditions used (Fig. 1). However, WhiG is known to be involved in regulating the early stages of sporulation in S. coelicolor (7), and the samples analyzed were from liquid-grown cells that do not undergo morphological differentiation. However, analysis of RNA samples prepared from surface-differentiated mycelium also did not reveal the expected transcript. The same sequence is also present upstream of sigB and overlapping the sigBp1 promoter in S. coelicolor (Fig. 4C). Further experiments might confirm the activation of these potential promoters under some specific conditions.
FIG. 4.
Promoter sequences. (A) Alignment of the hasRp1, hasLp1, and hasRp2 promoters with the previously reported σB-type promoters Pctc of B. subtilis (33) and sigBp1 of S. coelicolor (8). The consensus is according to the work of Cho et al. (8). (B) Alignment of the hasLp2 promoter sequence with the consensus recognized by the principal sigma factor σHrdB in S. coelicolor (36). (C) Alignment showing putative WhiG-type promoters identified upstream of the hasR coding region and upstream of sigB (sigJ) in S. coelicolor. PwhiH and PwhiI are the promoter sequences of the whiH and whiI genes known to be regulated by WhiG in S. coelicolor (1). PTH4 and PTH270 are two sequences identified as targets of WhiG in S. coelicolor (38). The consensus is according to the work of Tan and Chater (38). The nucleotides in boldface are those in common with the promoter consensus sequence. The two nucleotides underlined (TA) in the −10 region of the hasRp1 (or sigBp1) promoter and in the −35 region of the hasRpWhiG1 (or sigBpWhiG) promoter are shared by the two putative promoter sequences.
Developmental control of the has gene transcription.
S1 nuclease mapping showed that, while the levels of hasRp2 and hasLp2 transcripts were rather constant, transcription from hasRp1 and hasLp1 increased significantly in transition and stationary phase (Fig. 3A). This induction was confirmed and quantified by real-time RT-PCR experiments (Fig. 3B). The most remarkable increase was observed in stationary phase, where abundance of the hasRp1 and hasLp1 transcripts was 109- and 11-fold higher than in the exponential-phase samples, respectively. When the levels of expression of the has genes are compared, it appears that hasR is expressed more highly than is hasL, with a maximum difference of 100-fold occurring between the hasRp1 and hasLp1 promoters in stationary phase (Fig. 3C). Analogous data were obtained using RNAs prepared from surface-grown mycelium (data not shown). The net levels of transcription of the has genes during growth of the wild-type (wt) strain are reported in Fig. 5 and are incorporated in a global hypothesis presented in Fig. 6. For clarity, the hasRp2 and hasLp2 promoters responsible for basal expression of the has genes are not considered in this model.
FIG. 5.
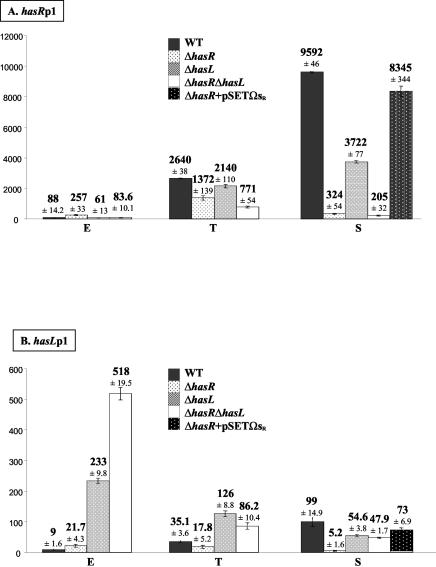
Quantification of the transcription levels from hasRp1 (A) and hasLp1 (B) during growth and the effect of mutations in hasR and hasL genes. The genetic contexts are named ΔhasR, ΔhasL, or ΔhasR ΔhasL and result from the analysis of three independent strains of each type (Table 1). The complementation experiments correspond to the ΔhasR+pSETΩsR data and result from the analysis of two independent strains (Table 1). For each strain, three independent RNA samples have been analyzed by real-time RT-PCR. The values are relative to the level of expression from hasLp2 in exponential phase, arbitrarily fixed at 1.
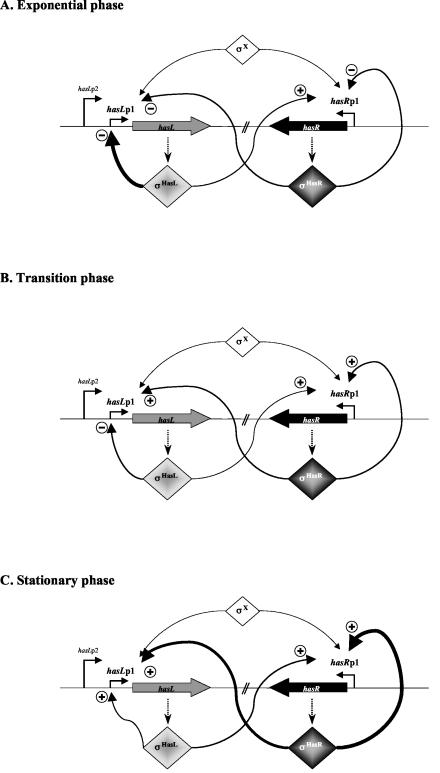
FIG. 6.
Model for the control of expression of the has genes during growth in liquid culture. The three growth phases are depicted: exponential (A), transition (B), and stationary (C). The thickness of the lines symbolizes the level of control as deduced from real-time RT-PCR quantification (Fig. 5). + and − indicate positive or negative effects on promoter transcription, respectively, of the factors σBR/L as deduced from expression analysis in the mutant strains (Fig. 5).
Positive autoregulation and antagonist effects.
The has genes encode σB-like sigma factors, and the hasRp1 and hasLp1 promoters are homologous to those recognized by B. subtilis σB. We therefore tested the ability of both the σBR and σBL factors to regulate expression from their own, and each other's, promoters. For this purpose, single knockout mutant strains were first constructed by replacing either hasR or hasL coding sequence with a cassette containing an apramycin resistance gene, as described in Materials and Methods (Table 1). A double mutant strain was derived from the single mutants by replacing the remaining intact has gene with a cassette harboring the spectinomycin resistance gene. All the gene replacements were confirmed by Southern hybridization and PCR (see Materials and Methods), and the absence of large chromosomal rearrangements was assessed using pulsed-field gel electrophoresis (data not shown). All further experiments were carried out using three representative mutants of each type (Table 1). Transcripts initiated from hasRp1 and hasLp1 in the single and double mutant strains during exponential-, transition-, and stationary-phase growth in liquid HT medium were quantified (see Materials and Methods), and the results are presented in Fig. 5.
In the ΔhasR mutant, hasRp1 transcript abundance in the stationary-phase sample was 30-fold lower than that of the parent strain at the same time point and 1.9-fold lower in transition phase (Fig. 5A). These data indicate that σBR positively activates the transcription of its own gene from hasRp1 at these growth stages, as illustrated in Fig. 6. This situation is common for stress response sigma factors. For example, in S. coelicolor, sigB (promoter P1) and sigH (promoter P2) expression show an autoregulatory activity in response to an osmotic stress (8, 35). The same phenomenon was reported for σB of B. subtilis and of Listeria monocytogenes (3, 16). Surprisingly, in the exponential-phase samples, the level of transcription measured from hasRp1 was 2.9-fold greater in the mutant than in the wt strain (Fig. 5A), suggesting that σBR might exert an antagonistic effect on the transcription of its own gene during vegetative growth (Fig. 6A). This antagonistic effect might result from competition of two (or more) σ factors for the same promoter sequence or for the RNA polymerase core enzyme as described for E. coli between σS and σ70 (12, 17) and for σX and σW in B. subtilis (18).
Transcription initiated from hasLp1 in the ΔhasL mutant exhibited a slightly different pattern of transcription from that observed for hasRp1 in ΔhasR (Fig. 5). Transcription from hasLp1 was only 1.8-fold lower in the mutant during stationary phase, while a significant antagonistic effect on transcription was observed in both transition- and exponential-phase samples, which showed a 3.6- and 20-fold increase, respectively.
To confirm that the observed altered transcriptional levels are specifically the result of a deficiency in σBR/L, a wt copy of hasR under the control of its own promoter was introduced into the NSAR or NSAL [hasR or hasL replaced by aac(3)IV/oriT cassette, respectively, in S. ambofaciens DSM40697] strain, respectively, with the integrative vector pSETΩ (28) (see Materials and Methods). Transcription was nearly fully restored in both the complemented strains: e.g., the transcription levels from hasLp1 and hasRp1 were 74 and 87% restored, respectively, in a complemented ΔhasR strain in stationary phase (Fig. 5).
Cross-regulation of has gene transcription.
Similar net positive or negative influences on the levels of transcription were noticed when cross-regulation between the two has systems was surveyed (Fig. 5). In the ΔhasL mutant, a slight but significant decrease in hasRp1 transcription was observed in all three growth phases: transcription was reduced by 1.4-fold in exponential phase and by 1.2- and 2.6-fold in the transition- and stationary-phase samples, respectively. Reciprocally, in the ΔhasR mutant strain, the transcription level from hasLp1 decreased by 19-fold in the stationary phase of growth (Fig. 5B) and by twofold in transition phase. These data revealed a positive activation of hasRp1 transcription by σBL and reciprocally, and perhaps more strongly, of σBR on hasLp1 transcription as illustrated in Fig. 6. Surprisingly however, a 2.4-fold increase in the level of hasLp1 transcription was observed in exponential growth phase in the ΔhasR mutant.
In the ΔhasR ΔhasL double mutant strains, a variable effect on the level of transcription from hasRp1 and hasLp1 was observed (Fig. 5), strongly suggesting that additional σ factors recognize the hasRp1 and hasLp1 promoter sequences (noted as σX in Fig. 6). It is particularly interesting to note the additional effects of the absence of σBR and σBL in the double mutant ΔhasR ΔhasL compared to the single mutant strains (Fig. 5). For example, when a net transcriptional induction was observed in both single mutant strains compared to the wt, this induction was even higher in the double mutant strain (e.g., hasLp1 transcription in the exponential-phase samples). Furthermore, when antagonistic effects were observed in the single mutant strains, the net level of transcription in the double mutant was averaged (e.g., hasRp1 and hasLp1 transcription in exponential- and transition-phase samples, respectively). Finally, when the deficiency of each σ factor was accompanied by a reduction in transcription, this decrease was greater in the double mutant (e.g., hasRp1 transcription in transition and stationary phase and hasLp1 in stationary phase).
Note that the regulation model can reach a higher degree of complexity if the posttranscriptional regulation is overlaid (anti-anti-σ and anti-σ factors) as reported for many stress response σ factors (41).
The has genes belong to a network involved in stress response.
The single and double mutant strains were compared to the wt when grown on both rich (HT, nutrient agar [NA], SFM, R2, and R2-yeast extract) and minimal (minimal medium mannitol [MMM] and minimal medium glucose [MMG]) media. No difference in growth rate or differentiation was detected, however, nor was any difference observed when strains were grown either at high temperature (37°C instead of the 30°C classically used) on solid medium (HT, SFM, MMM, or MMG) or in acid or alkaline conditions (pH 5.0 [acid] or pH 9.0 [alkaline]). The mutants showed no significant sensitivity to UV radiation exposure (0 to 400 J/m2), and no impairment of growth could be detected on exposure to osmotic stress conditions in liquid or solid medium. These results are significantly different from those reported by Cho et al. (8), where the sigB mutant in S. coelicolor was shown to be deficient both in aerial mycelium formation (on R2-yeast extract and NA) and in its response to osmotic stress. In addition, mutation of another member of the σB family, SigH, also led to slightly different conclusions. While Viollier et al. (40) could not distinguish the phenotypes of a ΔsigH mutant from those of the wt strain in response to osmotic stress, Sevcikova et al. (35) reported that sigH is required for correct morphological differentiation and growth under conditions of high osmolarity.
Finally, a slight but reproducible sensitivity to oxidative stress conditions distinguished the has mutant strains (single or double) from the wt strain. The most significant difference was observed using the paper disk method with a concentration of H2O2 of 10 mM, where the diameter of the zone of growth inhibition around the disks was 11.2% (±5.6%) higher for the double mutant strain than for the wt strain (the wt level varying by 2.8%). Furthermore, the deletion of the prsR gene (which encodes an anti-σ factor) seemed to confer a weak resistance to oxidative stress (data not shown). These data are consistent with the involvement of σBR/L in the stress response, since the anti-σ factor may titrate the σBR/L factors and prevent the induction of target genes involved in the stress response. This hypothesis is supported by the report of the probable posttranslational control of three related σ factors (σΗ, σL, and σF) by the same anti-σ factor, RshA, in Streptomyces griseus (37).
The transcription levels of the has genes under stress conditions were measured by real-time PCR and expressed as induction factors (IF; transcription level following stress/transcription level without stress; see Materials and Methods) (Table 4). In the wt strain, transcription from hasRp1 and to a lesser extent from hasLp1 was increased following exposure to oxidative stress with 10 mM H2O2 (IF, 17 and 7.5, respectively) as well as to osmotic stress with 500 mM NaCl (IF, 25 and 21, respectively) or with 34% sucrose (IF, 22.5 and 23, respectively). In the same experimental conditions, no significant induction was observed from hasRp2 and hasLp2 TSPs (IF ranging between 1 and 2.5). No increase in transcription was observed after ethanol (4%), cumene hydroperoxide (30%), or heat shock (42°C) treatments for any of the four promoters.
TABLE 4.
Induction of the has gene transcription levels under stress conditions
| Promoter and strain | Transcription level (IFa) under stress condition:
|
||
|---|---|---|---|
| H2O2 | NaCl | Sucroseb | |
| hasRp1 | |||
| wt | 17 | 25 | 22.5 |
| ΔhasR | 14 | 18 | ND |
| ΔhasL | 13.2 | 28.4 | ND |
| ΔhasR ΔhasL | 16.5 | 19.1 | 23.9 |
| hasLp1 | |||
| wt | 7.5 | 21.1 | 23 |
| ΔhasR | 9 | 19.3 | ND |
| ΔhasL | 7.1 | 18 | ND |
| ΔhasR ΔhasL | 6.9 | 18.4 | 18.2 |
IFs correspond to the ratio of the transcription level following stress to the transcription level without stress.
ND, not determined.
It appears that the IFs from hasRp1 and hasLp1 are not significantly altered in the single and double mutants compared to the wt (Table 4). Indeed, the IFs following oxidative stress (H2O2, 10 mM) ranged between 13.2 and 16.5 for hasRp1 and between 6.9 and 9 for hasLp1 in the double (or single) mutant strains (versus 17 and 7.5, respectively, in the wt strain). The induction response following osmotic stress (data not shown) was also retained (500 mM NaCl and 34% sucrose).
These data suggest that the induction of transcription from the hasRp1 and hasLp1 promoters under stress conditions results at least in part from the action of alternate σ factors (noted as σX in Fig. 6). This also provides an explanation for the absence of a marked phenotype in response to stress in the wt and mutant strains while the has transcription is induced under the same conditions. This is consistent with the residual transcriptional activity observed from hasRp1 and hasLp1 in the double mutant strain ΔhasR ΔhasL (Fig. 5). These data are also consistent with those reported by Viollier et al. (40) suggesting the existence of shared promoter recognition specificity within the stress response σ factors and notably among SigH, SigI, and SigJ (orthologue of σBR of S. ambofaciens) implied in the osmotic stress response in S. coelicolor. The stress responses might therefore rely on the activity of several specific σ factors, particularly for the response to osmotic stress. Hence, a sensory system that can coordinate the activity of multiple paralogous σ factors would be implied in the expression of genes belonging to the same stress regulon (40).
In conclusion, the conservation of the duplicated has genes in S. ambofaciens might illustrate the subfunctionalization phenomenon described in the duplication-degeneration-complementation model (14). The two σBR/L factors are 97% identical at the amino acid level (and 100% in the domains involved in the −35 and −10 box recognition) and probably recognize the same regulatory signals and consequently coregulate the same genes. However, the transcriptional control revealed during this study clearly shows that the two has copies are differently expressed, suggesting the specialization of the two has genes. The redundancy of the σB-like σ factors in Streptomyces might increase the complexity of the response to stress. Further, the duplication of the sigB (sigJ) gene in S. ambofaciens would correspond to a recent event of this evolutionary process and might allow the fine-tuning of the response.
Acknowledgments
V.R. and T.W. were fellows of the Ministère de l'Education Nationale, de la Recherche et de la Technologie (MENRT). T.W. and B.A. were also recipients of a grant from the Association pour la Recherche sur le Cancer (ARC) and EMBO, respectively.
We thank B. Gust, K. Chater, and T. Kieser (JIC) for providing the PCR-targeting system and A. Hesketh for his help in preparing the manuscript. We are grateful to G. Kelemen for critical reading of the manuscript and for permission to quote unpublished data as a personal communication.
REFERENCES
- 1.Ainsa, J. A., H. D. Parry, and K. F. Chater. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 34:607-619. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bruton, C. J., K. A. Plaskitt, and K. F. Chater. 1995. Tissue-specific glycogen branching isoenzymes in a multicellular prokaryote, Streptomyces coelicolor A3(2). Mol. Microbiol. 18:89-99. [DOI] [PubMed] [Google Scholar]
- 7.Chater, K. F., C. J. Bruton, K. A. Plaskitt, M. J. Buttner, C. Méndez, and J. D. Helmann. 1989. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility σ factor of B. subtilis. Cell 59:133-143. [DOI] [PubMed] [Google Scholar]
- 8.Cho, Y. H., E. J. Lee, B. E. Ahn, and J. H. Roe. 2001. SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol. Microbiol. 42:205-214. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, G. A., K. Lewis, and B. E. Rothenberg. 1989. High efficiency vectors for cosmid microcloning and genomic analysis. Gene 79:9-20. [DOI] [PubMed] [Google Scholar]
- 12.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, G., T. Wenner, B. Decaris, and P. Leblond. 1998. Chromosomal arm replacement generates a high level of intraspecific polymorphism in the terminal inverted repeats of the linear chromosomal DNA of Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 95:14296-14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan, and J. Postlethwait. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecker, M., W. Schumann, and U. Volker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 17.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 18.Huang, X., K. L. Fredrick, and J. D. Helmann. 1998. Promoter recognition by Bacillus subtilis σW: autoregulation and partial overlap with the σX regulon. J. Bacteriol. 180:3765-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hütter, R. 1967. Classification of the streptomycetes with special regard to the antibiotics formed from them. Bibl. Microbiol. 6:1-382. [PubMed] [Google Scholar]
- 20.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 21.Kato, F., T. Hino, A. Nakaji, M. Tanaka, and Y. Koyama. 1995. Carotenoid synthesis in Streptomyces setonii ISP5395 is induced by the gene crtS, whose product is similar to a sigma factor. Mol. Gen. Genet. 247:387-390. [DOI] [PubMed] [Google Scholar]
- 22.Kieser, T., M. J. Bibb, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Centre, Norwich, United Kingdom.
- 23.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 24.Leblond, P., P. Demuyter, J. M. Simonet, and B. Decaris. 1991. Genetic instability and associated genome plasticity in Streptomyces ambofaciens: pulsed-field gel electrophoresis evidence for large DNA alterations in a limited genomic region. J. Bacteriol. 173:4229-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leblond, P., G. Fischer, F. X. Francou, F. Berger, M. Guérineau, and B. Decaris. 1996. The unstable region of Streptomyces ambofaciens includes 210 kb terminal inverted repeats flanking the extremities of the linear chromosomal DNA. Mol. Microbiol. 19:261-271. [DOI] [PubMed] [Google Scholar]
- 26.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacNeil, D. J. 1988. Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J. Bacteriol. 170:5607-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connor, T. J., P. Kanellis, and J. R. Nodwell. 2002. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 45:45-57. [DOI] [PubMed] [Google Scholar]
- 29.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Mol. Microbiol. 42:1007-1020. [DOI] [PubMed] [Google Scholar]
- 30.Pang, X., B. Aigle, J. M. Girardet, S. Mangenot, J. L. Pernodet, B. Decaris, and P. Leblond. 2004. Functional angucycline-like antibiotic gene cluster in the terminal inverted repeats of the Streptomyces ambofaciens linear chromosome. Antimicrob. Agents Chemother. 48:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinnert-Sindico, S., L. Ninet, J. Preud'Homme, and C. Cosar. 1955. A new antibiotic: spiramycin. Antibiot. Annu. 1954-1955:724-757. [Google Scholar]
- 32.Potuckova, L., G. H. Kelemen, K. C. Findlay, M. A. Lonetto, M. J. Buttner, and J. Kormanec. 1995. A new RNA polymerase sigma factor, sigma F, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 17:37-48. [DOI] [PubMed] [Google Scholar]
- 33.Ray, C., R. E. Hay, H. L. Carter, and C. P. Moran, Jr. 1985. Mutations that affect utilization of a promoter in stationary-phase Bacillus subtilis. J. Bacteriol. 163:610-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sevcikova, B., O. Benada, O. Kofronova, and J. Kormanec. 2001. Stress-response sigma factor sigma(H) is essential for morphological differentiation of Streptomyces coelicolor A3(2). Arch. Microbiol. 177:98-106. [DOI] [PubMed] [Google Scholar]
- 36.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takano, H., K. Hosono, T. Beppu, and K. Ueda. 2003. Involvement of sigma(H) and related sigma factors in glucose-dependent initiation of morphological and physiological development of Streptomyces griseus. Gene 320:127-135. [DOI] [PubMed] [Google Scholar]
- 38.Tan, H., and K. F. Chater. 1993. Two developmentally controlled promoters of Streptomyces coelicolor A3(2) that resemble the major class of motility-related promoters in other bacteria. J. Bacteriol. 175:933-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatti, K. M., and C. P. Moran, Jr. 1984. Promoter recognition by sigma-37 RNA polymerase from Bacillus subtilis. J. Mol. Biol. 175:285-297. [DOI] [PubMed] [Google Scholar]
- 40.Viollier, P. H., G. H. Kelemen, G. E. Dale, K. T. Nguyen, M. J. Buttner, and C. J. Thompson. 2003. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol. Microbiol. 47:699-714. [DOI] [PubMed] [Google Scholar]
- 41.Viollier, P. H., A. Weihofen, M. Folcher, and C. J. Thompson. 2003. Post-transcriptional regulation of the Streptomyces coelicolor stress responsive sigma factor, SigH, involves translational control, proteolytic processing, and an anti-sigma factor homolog. J. Mol. Biol. 325:637-649. [DOI] [PubMed] [Google Scholar]