Abstract
The contribution of repetitive elements to quantitative human traits is largely unknown. Here, we report a genome-wide survey of the contribution of Short Tandem Repeats (STRs), one of the most polymorphic and abundant repeat classes, to gene expression in humans. Our survey identified 2,060 significant expression STRs (eSTRs). These eSTRs were replicable in orthogonal populations and expression assays. We used variance partitioning to disentangle the contribution of eSTRs from linked SNPs and indels and found that eSTRs contribute 10%–15% of the cis-heritability mediated by all common variants. Further functional genomic analyses showed that eSTRs are enriched in conserved regions, co-localize with regulatory elements, and can modulate certain histone modifications. By analyzing known GWAS hits and searching for new associations in 1,685 deeply-phenotyped whole-genomes, we found that eSTRs are enriched in various clinically-relevant conditions. These results highlight the contribution of short tandem repeats to the genetic architecture of quantitative human traits.
Introduction
In recent years, there has been tremendous progress in identifying genetic variants that affect expression of nearby genes, termed cis expression quantitative trait loci (cis-eQTLs). Multiple studies have shown that disease-associated variants often overlap cis-eQTLs in the affected tissue1–3. These observations suggest that understanding the genetic architecture of the transcriptome may provide insights into the cellular-level mediators underlying complex traits4–6. So far, eQTL-mapping studies have mainly focused on SNPs and to a lesser extent on bi-allelic indels and CNVs as determinants of gene expression7–11. However, these variants do not account for all of the heritability of gene expression attributable to cis-regulatory elements as measured by twin studies, leaving on average about 20–30% unexplained8,12. It has been speculated that such heritability gaps could indicate the involvement of repetitive elements that are not well tagged by common SNPs13,14.
To augment the repertoire of eQTL classes, we focused on Short Tandem Repeats (STRs), one of the most polymorphic and abundant types of repetitive elements in the human genome15,16. These loci consist of periodic DNA motifs of 2–6bp spanning a median length of around 25bp. There are about 700,000 STR loci covering almost 1% of the human genome. Their repetitive structure induces DNA-polymerase slippage events that add or delete repeat units, creating mutation rates that are orders of magnitude higher than those of most other variant types15,17. Over 40 Mendelian disorders, such as Huntington’s Disease, are attributed to STR mutations, most of which are caused by large expansions of trinucleotide coding repeats18.
Several properties of STRs suggest they may play a regulatory role. In vitro studies have shown that STR variations can modulate the binding of transcription factors19,20, change the distance between promoter elements21,22, alter splicing efficiency23,24, and induce irregular DNA structures that may modulate transcription25. In vivo experiments have reported specific examples of STR variations that control gene expression across a wide range of taxa, including Haemophilus influenza26, Saccharomyces cerevisiae27, Arabidopsis thaliana28, and vole29. Recent studies reported that dinucleotide repeats are a hallmark of enhancers in Drosophila and are enriched in predicted enhancers in humans30. Human promoters also disproportionately harbor STRs31 and the presence of STRs in promoters or transcribed regions greatly increases the divergence of gene expression profiles across great apes32, suggesting that STRs play a key role in the evolution of expression. Several candidate-gene studies in human indeed reported that STR variations modulate gene expression19,33–37 and alternative splicing23,38,39. In one example, a recent study found that that underlying mechanism behind a GWAS signal for Ewing Sarcoma is a sequence variant in an AAGG repeat that increases the binding of the EWSR1-FLI1 oncoprotein resulting in EGF2 overexpression40. Despite these accumulating lines of evidence, there has been no systematic evaluation of the contribution of STRs to gene expression in humans.
To this end, we conducted a genome-wide analysis of STRs that affect expression of nearby genes, termed expression STRs (eSTRs), in lymphoblastoid cell lines (LCLs), a central ex-vivo model for eQTL studies. Next, we used a multitude of statistical genetic and functional genomics analyses to show that hundreds of these eSTRs are predicted to be functional. Finally, we tested the involvement of eSTRs in clinically relevant phenotypes.
Results
Initial genome-wide discovery of eSTRs
The initial genome-wide discovery of potential eSTRs relied on finding associations between STR length and expression of nearby genes. We focused on 311 European individuals whose LCL expression profiles were measured using RNA-sequencing by the gEUVADIS9 project and whose whole genomes were sequenced by the 1000 Genomes Project41. The STR genotypes were obtained in our previous study42 in which we created a catalog of STR variation as part of the 1000 Genomes Project using lobSTR, a specialized algorithm for profiling STR variations from high throughput sequencing data43. Briefly, lobSTR identifies reads with repetitive sequences that are flanked by non-repetitive segments. It then aligns the non-repetitive regions to the genome using the STR motif to narrow the search, thereby overcoming the gapped alignment problem and conferring alignment specificity. Finally, lobSTR aggregates aligned reads and employs a model of STR-specific sequencing errors to report the maximum likelihood genotype at each locus. lobSTR recovered most (r2=0.71) of the variation in STR locus lengths in the 1000 Genomes datasets based on large-scale validation using 5,000 STR genotype calls obtained by capillary electrophoresis, the gold standard for STR genotyping42. The majority of genotype errors were from dropout of one allele at heterozygote sites due to low sequencing coverage. We simulated the performance of STR associations using lobSTR calls compared to the capillary calls. This process showed that STR genotype errors reduce the power to detect eSTRs by 30–50% but importantly do not create spurious associations (Supplementary Note and Supplementary Fig. 1).
To detect eSTR associations, we regressed gene expression on STR dosage, defined as the sum of the two STR allele lengths in each individual. We opted to use this measure based on previous findings that reported a linear trend between STR length and gene expression19,34,36 or disease phenotypes44,45. As covariates, we included sex, population structure, and other technical parameters (Fig. 1a and Supplementary Methods). We employed this process on 15,000 coding genes whose expression profiles were detected in the RNA-sequencing data. For each gene, we considered all polymorphic STR variations that passed our quality criteria (Online Methods) and were within 100kb of the transcription start and end sites of the gene transcripts as annotated by Ensembl46. On average, 13 STR loci were tested for each gene (Supplementary Fig. 2), yielding a total of 190,016 STR×gene tests.
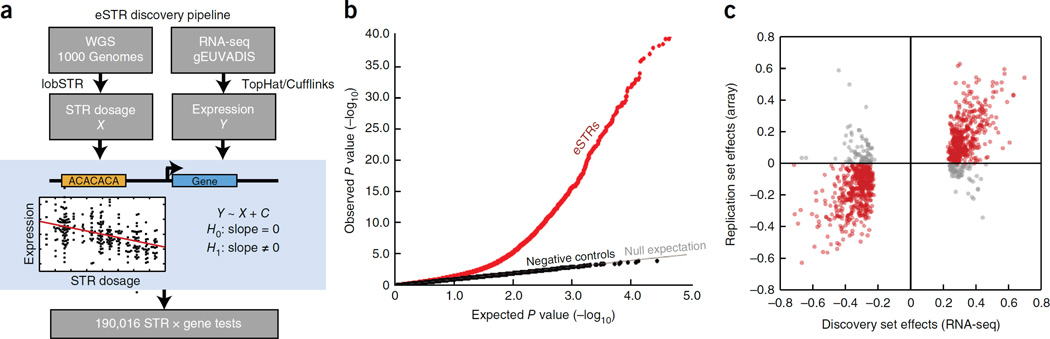
Figure 1. eSTR discovery and replication.
(a) eSTR discovery pipeline. An association test using linear regression was performed between STR dosage and expression level for every STR within 100kb of a gene (b) Quantile-quantile plot showing results of association tests. The gray line gives the expected p-value distribution under the null hypothesis of no association. Black dots give p-values for permuted controls. Red dots give the results of the observed association tests (c) Comparison of eSTR effect sizes as Pearson correlations in the discovery dataset vs. the replication dataset. Red points denote eSTRs whose directions of effect were concordant in both datasets and gray points denote eSTRs with discordant directions.
Our analysis identified 2,060 unique protein-coding genes with a significant eSTR (gene level FDR≤5%) (Fig. 1b and Supplementary Table 1). The majority of these were di- and tetra-nucleotide STRs (Supplementary Tables 2 and 3). Only 13 eSTRs fall in coding exons, but eSTRs were nonetheless strongly enriched in 5’UTRs (p=1.0×10−8), 3’UTRs (p=1.7×10−9) and regions near genes (p<10−28) compared to all STRs analyzed (Supplementary Table 4). Overall, there was no bias in direction of effect (Supplementary Table 5). We also repeated the association tests with two negative control conditions by regressing expression on (i) STR dosages permuted between samples and (ii) STR dosages from randomly chosen unlinked loci (Fig. 1b and Supplementary Fig. 3). Both negative controls produced uniform p-value distributions expected under the null hypothesis. This provides support for the absence of spurious associations due to inflation of the test statistic or the presence of uncorrected population structure. To assess the effect of low sequencing coverage on our results, we generated high coverage targeted sequencing of 2,472 promoter STRs and repeated the eSTR analysis (Online Methods). We found that association results were largely reproducible across datasets, with 80% of tested eSTRs showing the same direction of effect (p=9.9×10−12; n=126) (Supplementary Note and Supplementary Fig. 4). Three previous studies described candidate gene studies of expression STRs and involved STRs that were tested in our framework19,36,47. Our genome-wide approach was able to replicate the association between PIG3 and the pentanucleotide STR in the 5’UTR of the gene and showed the same direction of effect. However, the other two candidate genes did not meet the multiple hypothesis p-value threshold (Supplementary Table 6).
The initial discovery set of eSTRs was largely reproducible in an independent set of individuals using an orthogonal expression assay technology. We obtained an additional set of over 200 individuals whose genomes were also sequenced as part of the 1000 Genomes Project and whose LCL expression profiles were measured by Illumina expression array48. These individuals belong to cohorts with African, Asian, European, and Mexican ancestry, enabling testing of the associations in a largely distinct set of populations. The Illumina expression array allowed us to test 882 eSTRs out of the 2,060 identified above. The association signals of 734 of the 882 (83%) tested eSTRs showed the same direction of effect in both datasets (sign test p=2.7×10−94) and the effect sizes were strongly correlated (R=0.73, p=1.4×10−149) (Fig. 1c), despite only moderate reproducibility of expression profiles across platforms (Supplementary Note and Supplementary Fig. 5). For comparison, only 54% of non-eSTRs showed the same direction of effect, close to the expected value of 50% for null associations. Overall, these results show that eSTR association signals are robust and reproducible across populations and expression assay technologies.
Partitioning the contribution of eSTR and nearby variants
An important question is whether eSTR association signals stem from causal STR loci or are merely due to tagging SNPs or other variants in linkage disequilibrium (LD). Previous results reported that the average STR-SNP LD is approximately half of the traditional SNP-SNP LD42,49,50, but there are known examples of STRs tagging GWAS SNPs51.
To address this question, we partitioned the relative contributions of eSTRs versus all common (MAF≥1%) bi-allelic SNPs, indels, and structural variants (SV) in the cis region of each gene using a linear mixed model (LMM) (Fig. 2a). Multiple studies have used this approach to measure the total contribution of common variants to the heritability of quantitative traits and to partition the contribution of different classes of variants52,53. Taking a similar approach, we included two types of effects for each gene: a random effect that captures all common bi-allelic loci detected within 100kb of the gene and a fixed effect that captures the lead STR. To test whether other causal variants in the local region could inflate the estimate of the STR contribution, we simulated gene expression with one or two causal SNP eQTLs per gene while preserving the local haplotype structure. In this negative control scenario, the LMM correctly reported a median across all conditions (Supplementary Note and Supplementary Fig. 6–7), where . This suggests that other causal variants in LD do not inflate the estimator of the relative contribution of STRs. However, simulations based on capillary electrophoresis data suggest that the variance explained by STRs is downwardly biased in the presence of genotyping errors (Supplementary Note and Supplementary Fig. 8), suggesting that the reported is likely to be conservative.
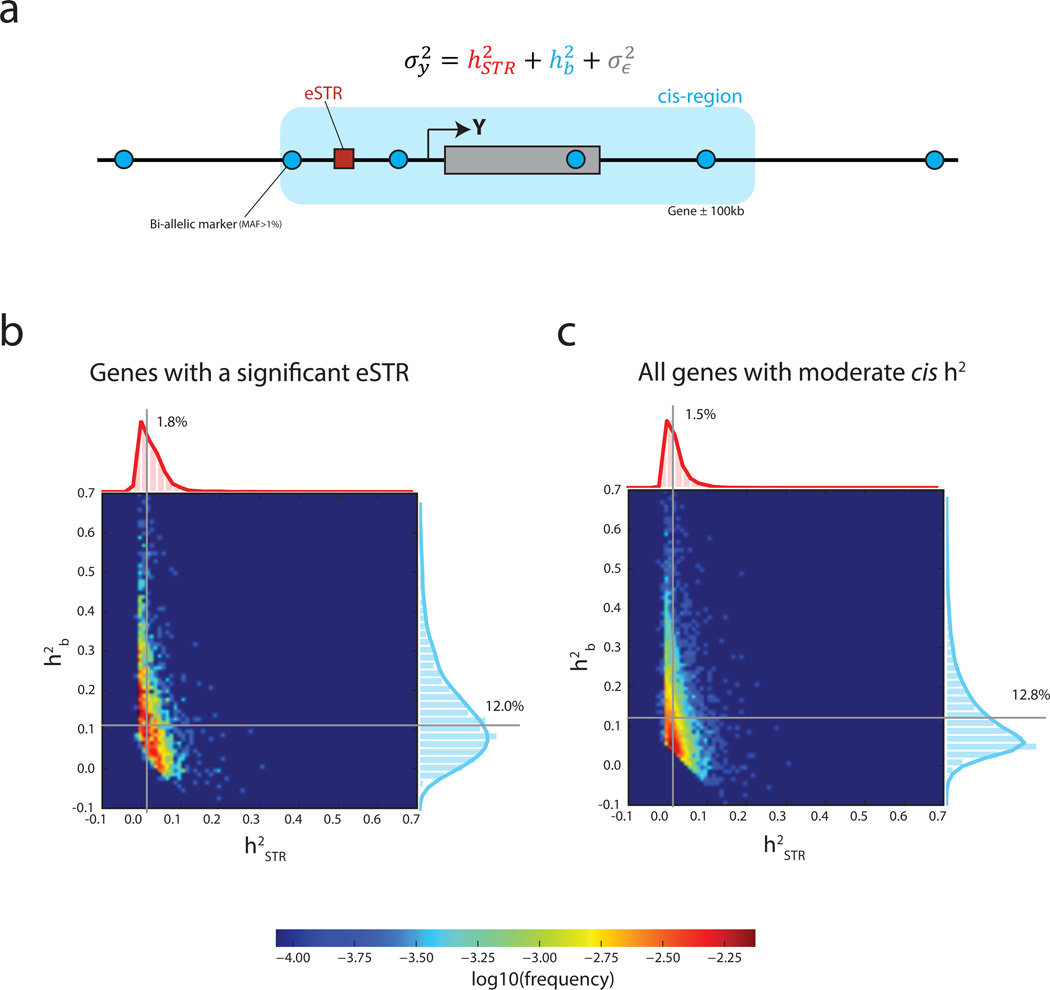
Figure 2. Variance partitioning using linear mixed models.
(a) The normalized variance of the expression of gene Y was modeled as the contribution of the best eSTR and all common bi-allelic markers in the cis region (±100kb from the gene boundaries) (b–c) Heatmaps show the joint distributions of variance explained by eSTRs and by the cis region. Gray lines denote the median variance explained (b) Variance partitioning across genes with a significant eSTR in the discovery set and (c) Variance partitioning across genes with moderate cis heritability.
The LMM results showed that eSTRs contribute about 12% of the genetic variance attributed to common cis polymorphisms. For genes with a significant eSTR, the median was 1.80%, whereas the median was 12.0% (Fig. 2b), with a median ratio of of 12.3% (CI95% 11.1%–14.2%; n=1,928) (Supplementary Table 7). We repeated the same analysis for genes with at least moderate (≥5%) cis-heritability (Online Methods) regardless of the presence of a significant eSTR in the discovery set. The motivation for this analysis was to avoid potential winner’s curse54 and to obtain a transcriptome-wide perspective on the role of STRs in gene expression (Fig. 2c). In this set of genes, eSTRs contribute about 13% (CI95% 12.2%–13.4%; n=6,272) of the genetic variance attributed to cis common polymorphisms. The median was 1.45% of the total expression variance, whereas the median was 9.10% (Table 1). Repeating the analysis while considering STRs as a random effect showed highly similar results (Supplementary Note, Supplementary Table 8, and Supplementary Fig. 9). Taken together, this analysis shows that STR variations explain a sizeable component of gene expression variation after controlling for all variants that are well tagged by common bi-allelic markers in the cis region.
The effect of eSTRs in the context of individual SNP eQTLs
To further assess the contribution of eSTRs in the context of other variants, we also inspected the relationship between eSTRs and individual cis-SNP eQTLs (eSNPs). We performed a traditional eQTL analysis using the whole genome sequencing data for 311 individuals that were part of the discovery set to identify common eSNPs [minor allele frequency (MAF) ≥5%] within 100kb of each gene. This process identified 4,290 genes with an eSNP (gene-level FDR≤5%). We then re-analyzed the eSTR association signals while conditioning on the genotype of the most significant eSNP (Fig. 3a). For each eSTR, we ascertained the subset of individuals that were homozygous for the major allele of the lead eSNP in the region. If the eSTR simply tags this eSNP, its conditioned effect should be randomly distributed compared to the unconditioned effect. Alternatively, if the eSTR is causal, the direction of the conditioned effect should match that of the original effect. We conducted this analysis for eSTR loci with at least 25 individuals homozygous for the lead eSNP and for which these individuals had at least two unique STR genotypes (1,856 loci). After conditioning on the lead eSNP, the direction of effect for 1,395 loci (75%) was identical to that in the original analysis (sign test p<4.2×10−109) and the effect sizes were significantly correlated (R=0.52; p=3.2×10−130) (Fig. 3b). This further supports the additional role of eSTRs beyond traditional cis-eQTLs.
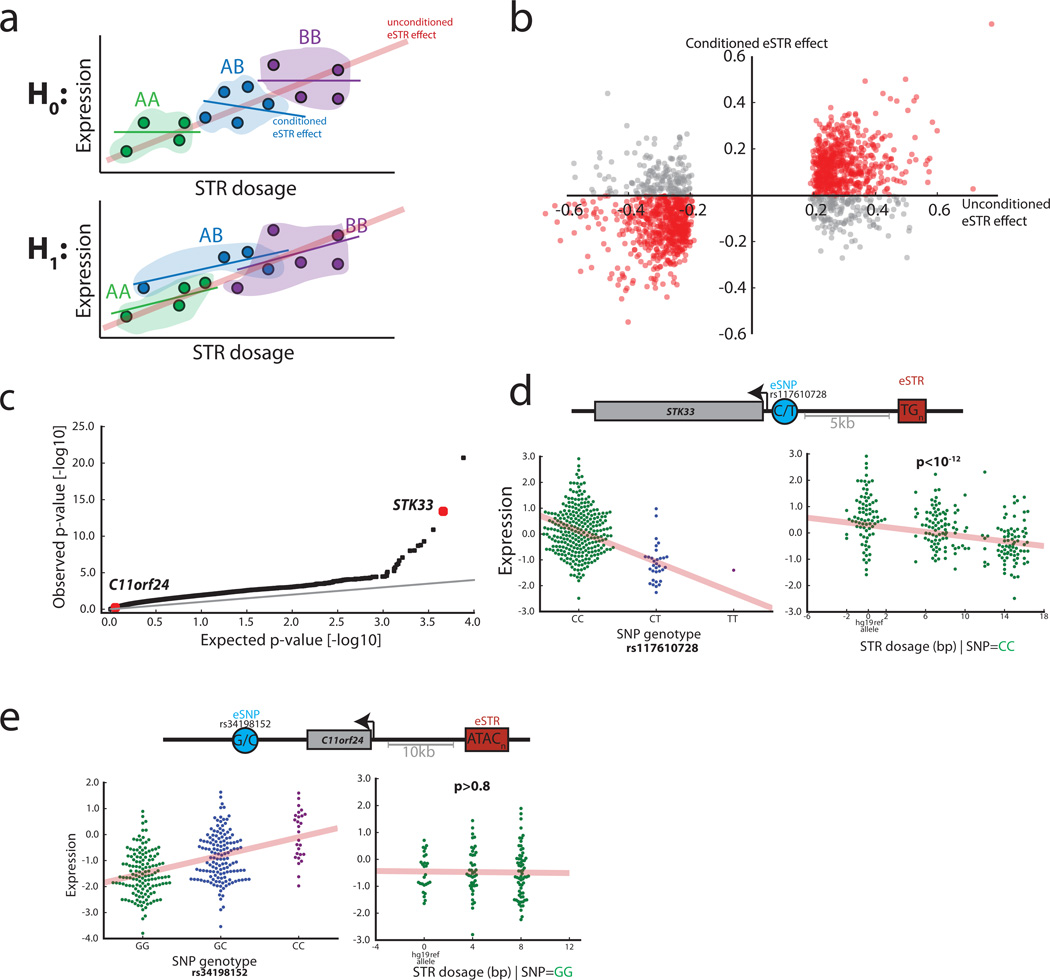
Figure 3. eSTR associations in the context of eSNPs.
(a) Schematic of the eSTR effect versus the effect conditioned on the lead eSNP genotype. Under the null expectation, the original association (red line) comes from mere tagging of eSNPs. Thus, the eSTR effect disappears when restricting to a group of individuals (dots) with the same eSNP genotype (colored patches). Under the alternative hypothesis, the effect is concordant between the original and conditioned associations (b) The original eSTR effect versus the conditioned eSTR effect. Red points denote eSTRs whose direction of effect was concordant in both datasets and gray points denote eSTRs with discordant directions (c) Quantile-quantile plot of p-values from ANOVA testing of the explanatory value of eSTRs beyond that of eSNPs (d) STK33 is an example of a gene for which the eSTR (red rectangle) has a strong explanatory value beyond the lead eSNP (blue circle) based on ANVOA. When conditioning on individuals that are homozygous for the “C” eSNP allele (bottom left, green dots), the STR dosage still shows a significant effect (bottom right) (e) C11orf24 is an example of a gene for which the eSTR was part of the discovery set but did not pass the ANOVA threshold. After conditioning on individuals that are homozygous for the “G” eSNP allele (bottom left, green dots), the STR effect is lost (bottom right).
We also found that hundreds of eSTRs in the discovery set provide additional explanatory value for gene expression beyond the lead eSNP. ANOVA model comparison showed that for 23% of the cases, a model with an eSTR significantly improved the explained variance of gene expression over considering only the lead eSNP according to an (FDR<5%) (Fig. 3c–e and Online Methods). Combined with the 183 genes with an eSTR but no significant eSNP, these results show that at least 30% of the eSTRs identified by our initial scan cannot be fully attributed to tagging of the lead eSNP. Given the reduced quality of STR compared to SNP genotypes, this analysis is likely to underestimate the true contribution of STRs. Nonetheless, our results show concrete examples for hundreds of associations in which the eSTR increases the variance explained by the lead eSNP.
Conservation and epigenetics signals support the functional role of eSTRs
To provide further evidence of their regulatory role, we analyzed eSTRs in the context of functional genomics data. First, we assessed the potential functionality of STR regions by measuring signatures of purifying selection, since previous studies reported that putatively causal eSNPs are slightly enriched in conserved regions55. We inspected the sequence conservation56 across 46 vertebrates in the sequence upstream and downstream of the eSTRs in our discovery dataset (Fig. 4a). To tune the null expectation, we matched each tested eSTR to a random STR that did not reach significance in the association analysis but had a similar distance to the nearest transcription start site (TSS). The average conservation level of a ±500bp window around eSTRs was slightly but significantly higher (p<0.03) compared to control STRs. Tightening the window size to shorter stretches of ±50bp showed a more significant contrast in the conservation scores of the eSTRs versus the control STRs (p<0.01) (Fig. 4a inset), indicating that the excess in conservation comes from the vicinity of the eSTR loci. Taken together, these results show that eSTRs discovered by our association pipeline reside in regions exposed to relatively higher purifying selection, further suggesting a functional role.
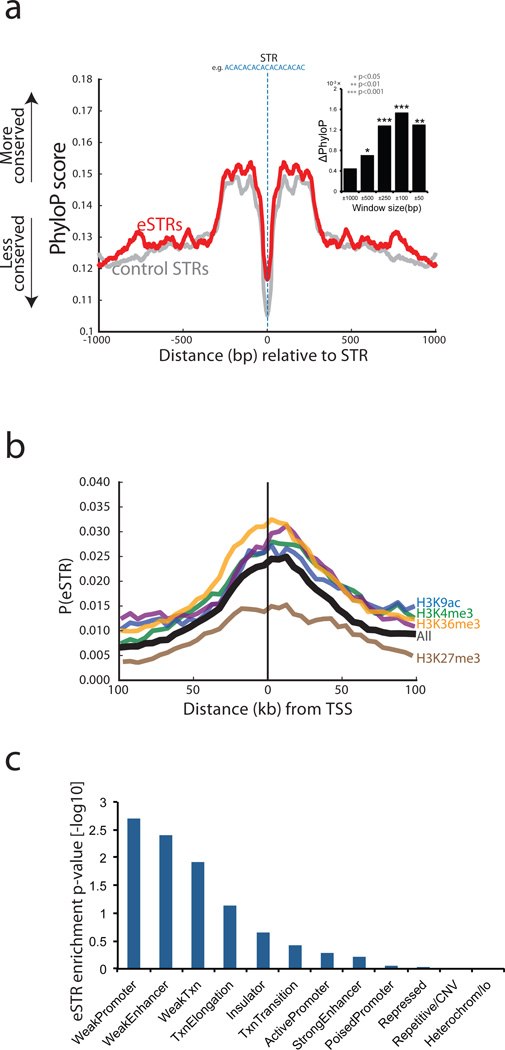
Figure 4. Conservation and epigenetic analysis of eSTR loci.
(a) Median PhyloP conservation score as a function of distance from the STR. Red: eSTR loci, gray: matched control STRs. Inset: the difference in the PhyloP conservation score between eSTRs and matched control STRs as a function of window size around the STR. (b) The probability that an STR scores as an eSTR in the discovery set as a function of distance from the transcription start site (TSS). eSTRs show clustering around the TSS (black line). Conditioning on the presence of a histone mark (colored lines) significantly modulated the probability that an STR is an eSTR (c) The enrichment of eSTRs in different chromatin states.
eSTRs substantially co-localize with functional elements. They show the strongest enrichment closest to transcription start sites (Fig. 4b) and to a lesser extent in or near predicted enhancers (Supplementary Fig. 10). We also inspected the co-localization of eSTRs with histone modifications as annotated by the Encode Consortium7 in LCLs. eSTRs were strongly enriched in peaks of histone modifications associated with regulatory regions (H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K9ac) and transcribed regions (H3K36me3) and were depleted in repressed regions (H3K27me3) (Fig. 4b). To test the significance of these signals, we constructed a null distribution for each histone modification by measuring the co-localization of eSTRs with randomly shifted histone peaks similar to the fine-mapping procedure of Trynka et al57. This null distribution controls for the co-occurrence of eSTRs and histone peaks due to their proximity to other causal variants. We found eSTR/histone co-localizations were significant (weakest p-value<0.01) after the peak shifting procedure, suggesting that these results stem from the eSTRs themselves (Supplementary Table 9). We also performed a peak-shifting analysis using ChromHMM annotations58 (Fig. 4c) which indicated that eSTRs are most strongly enriched in weak-promoters (p<0.002) and weak-enhancers (p<0.004). Again, this analysis shows overlap of eSTRs with elements that are predicted to regulate gene expression.
We also found that eSTR length variations are more likely to modulate the presence of certain histone marks (Supplementary Note and Supplementary Fig. 11). We introduced different eSTR alleles to GERV59, a machine learning approach that examines the effect of DNA sequence on histone marks. This process found that eSTRs have significantly greater effects than control STRs on predicted regulatory regions (H3K4me3 p=0.00109, DNAseI hypersensitivity p=0.00045, H3K9ac p=0.00462) and transcribed regions (H3K36me3 p=0.01336). These results are consistent with the analysis of chromatin modifications above. Importantly, since the input material for this analysis is solely STR variations that are independent of any linked variants, these results provide an orthogonal piece of evidence for the functionality of eSTRs and suggest histone mark modulation as a potential mechanism.
The potential role of eSTRs in human conditions
Encouraged by the evidence for the regulatory role of eSTRs, we wondered about their potential involvement in clinically-relevant conditions. First, we tested whether genes implicated by previous GWAS scans listed in the NHGRI GWAS catalog60 are enriched for eSTR genes. We focused on seven complex disorders: rheumatoid arthritis, Crohn’s disease, type 1 diabetes, type 2 diabetes, blood pressure, bi-polar disorder, and coronary artery disease. The first three conditions have a strong autoimmune component, rendering them more relevant to the LCL data used for eSTR discovery. To create a proper null, we compared the overlap of eSTR genes to randomly chosen sets of genes matched to the tested GWAS genes on both gene expression level in LCLs and on cis heritability.
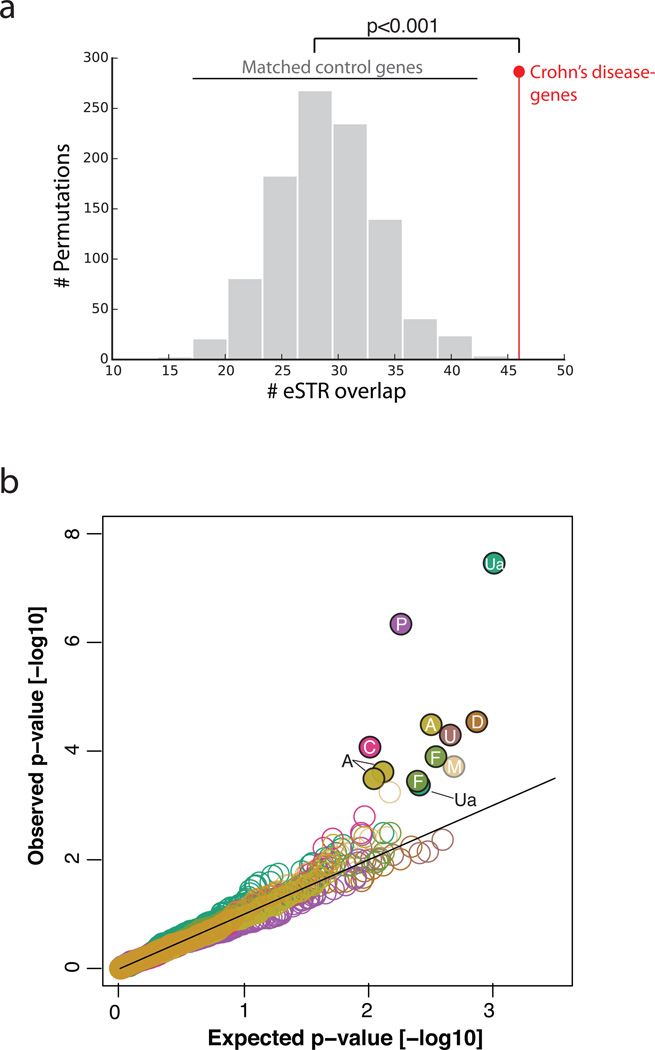
We found that GWAS genes for Crohn’s disease are significantly (p<0.001) enriched for eSTR hits (Figure 5a and Supplementary Fig. 12). Moderate enrichment for eSTRs (p=0.074) was found in GWAS genes for rheumatoid arthritis, consistent with the known role of immune function in these traits. Enrichments were 2–3 times higher for autoimmune diseases than for the other conditions (average overlap: 6%). Interestingly, for seven overlapping genes, the eSTRs explained more variance in gene expression than the lead eSNP of the gene. Furthermore, for close to thirty genes, a joint model of the lead eSTR and eSNP explained significantly more variance in gene expression than the eSNP alone, raising the possibility of an etiological role.
Figure 5. Association of eSTRs with clinical phenotypes.
(a) The overlap between eSTRs and Crohn’s disease GWAS genes (red) versus random subsets of genes (gray) matched on expression and heritability profiles in LCLs (b) quantile-quantile plots of eSTR associations in the TwinsUK data. Only traits with significant (FDR<0.1) associations are plotted. Closed circles: significant, open circles: non-significant. A: albumin; C: C-reactive protein; D: diastolic blood pressure, F: FVC, M: mean corpuscular volume, P: phosphate, U: Urea, Ua: Uric acid.
Next, we performed an association study using eSTRs to further test the hypothesis that eSTRs underlie clinically relevant phenotypes. For this, we turned to ~1,700 unrelated individuals that were sequenced to medium coverage (6×) with 100bp paired-end reads using Illumina as part of the TwinsUK cohort of the UK10K project61 and were phenotyped for a wide array of quantitative traits, primarily blood metabolites and anthropometric traits. While most of these conditions are not directly related to the immune system, we hypothesized that similar to other eQTLs3, some of the discovered eSTRs are shared across tissues and could play a role in additional tissues. After genotyping STRs with lobSTR, we tested for association between eSTRs and each of the 38 reported phenotypes, while controlling for sex, age, and population structure. To enrich for STR loci that are likely to be causal for gene expression variation, we restricted analysis to eSTRs that significantly improved the explained variance of gene expression over a model with the lead eSNP alone. In total, we obtained 499 eSTRs after applying this condition and excluding eSTRs that were genotyped in <1000 individuals.
We identified 12 significant associations (FDR per phenotype<10%) between eSTRs and the clinical phenotypes in the TwinUK data (Figure 5b and Supplementary Table 10). Only one association overlapped a known GWAS hit: an AAAC repeat on 4p16 was associated with decreased expression of SLC2A9 and increased uric acid in serum samples of the TwinsUK, which matches previous studies with SNPs62–65. The other 11 associations involved changes in blood metabolites such as albumin and C-reactive protein and physical traits such as diastolic blood pressure and FEV1 lung function and have yet to be described before in GWAS catalogs, suggesting novel loci. We caution that full validation of each of these associations will require replication in additional cohorts. Nonetheless, as we were mainly interested in the overall trend for eSTRs, we repeated the association of the 38 phenotypes in the TwinsUK cohort with a similar number of random STR loci matched on distance to transcription start sites, repeat motif, and number of genotyped samples. One hundred rounds of bootstrapping showed that eSTRs produced significantly more associations than the matched STR controls (mean for controls: 6.8 associations at FDR<10%, p<1.8×10−16). Repeating this test with a more stringent FDR of 5% revealed a similar picture: the eSTRs produced 6 associations passing this threshold (Supplementary Table 10), significantly more that the matched STR controls (mean for controls: 3.2 associations at FDR<5%, p<1.1×10−5). Taken together, our results show that eSTR signals are enriched in clinical phenotypes both in known and potentially novel GWAS hits. These results could inform future efforts for disease mapping studies.
Discussion
Repetitive elements have often been considered as neutral with no phenotypic consequences16. This coupled with the technical difficulties in analyzing these regions has led large-scale genetic studies to largely overlook the putative contribution of repeats to human phenotypes. Our study focused on short tandem repeats, one of the most polymorphic classes of loci that comprise 1% of the human genome. Despite being less abundant than SNPs, previous studies have shown that STRs are enriched in promoters and enhancers, where they frequently induce multiple base-pair variations, increasing the prior expectation of their ability to explain gene expression variation. Following these observations, we conducted a genome-wide scan for the contribution of STRs to gene expression. Our scan identified over 2,000 potential eSTRs and found that eSTRs contribute on average about 10–15% of the cis-heritability of gene expression attributed to common (MAF≥1%) polymorphisms. Functional genomics analyses provided further support for the predicted causal role of eSTRs. Finally, we found that eSTRs are enriched in clinically relevant phenotypes.
We hypothesize that there are more eSTRs to find in the genome as our analysis had several technical limitations. First, the higher genotyping error rates for STRs compared to SNPs limited our power to detect eSTRs and likely downwardly biased their estimated contribution in the LMM and ANOVA analyses. In addition, about 10% of STR loci in the genome could not be analyzed because they are too long to be spanned by current sequencing read lengths42. Second, based on previous findings in humans19,34,36, our association tests focused on a linear relationship between STR length and gene expression. However, experimental work in yeast reported that certain loci exhibit non-linear relationships between STR length and expression27, which are unlikely to be captured in our current analysis. Finally, our association pipeline takes into account only the length polymorphisms of STRs and cannot distinguish the effect of sequence variations inside STR alleles with identical lengths (dubbed homoplastic alleles66). Addressing these technical complexities would likely require phased STR haplotypes and longer sequence reads that are currently unavailable for large sample sizes. We envision that recent advancements in sequencing technologies67 will further expand the catalog of eSTRs.
Despite these technical limitations, our findings show that repetitive elements in the human genome extensively contribute to expression variation and are enriched in clinically relevant phenotypes. Our results are consistent with a recent study that reported that haplotypes of common SNPs, which capture genetic variants poorly tagged by current genotype panels, can explain substantially more heritability than common SNPs alone68. We anticipate that integrating the analysis of repetitive elements, specifically STR variations, will explain additional heritability and will lead to the discovery of new genetic variants relevant to human conditions.
Online Methods
Code availability
All code and data used for this manuscript are available on github at https://github.com/mgymrek/estrs under the GPLv3 license.
Genotype datasets
lobSTR genotypes were generated for the phase 1 individuals from the 1000 Genomes Project as described in42. Variants from the 1000 Genomes Project phase 1 release were downloaded in VCF format from the project website. HapMap genotypes were used to correct association tests for population structure. Genotypes for 1.3 million SNPs were downloaded for draft release 3 from the HapMap Consortium webpage. SNPs were converted to hg19 coordinates using the liftOver tool and filtered using Plink69 to contain only the individuals for which both expression array data and STR calls were available. Throughout this manuscript, all coordinates and genomic data are referenced according to hg19.
Targeted sequencing of promoter region STRs
We used a previously published method using capture and high-throughput sequencing70 to sequence 2,472 STRs located in gene promoters (TSS +/− 1kb) in 120 HapMap individuals of European (58 CEU individuals) and African (62 YRI individuals) ancestry. Briefly, the method uses a custom Nimblegen EZ Capture system to enrich the genomic sequence flanking, and sometimes including, the target STRs to be genotyped prior to sequencing using an Illumina Hiseq2000 instrument. We multiplexed 24 individuals per sequencing lane and utilized 100bp single-end reads. We used lobSTR version 3.0.3 to genotype STRs in these samples.
Expression datasets
RNA-sequencing datasets from 311 HapMap lymphoblastoid cell lines for which STR and SNP genotypes were also available were obtained from the gEUVADIS Consortium. Raw FASTQ files containing paired end 100bp Illumina reads were downloaded from the EBI website. The hg19 Ensembl transcriptome annotation was downloaded as a GTF file from the UCSC Genome Browser71,72 ensGene table. The RNA-sequencing reads were mapped to the Ensembl transcriptome using Tophat v2.0.773 with default parameters. Gene expression levels were quantified using Cufflinks v2.0.274 with default parameters and supplied with the GTF file for the Ensembl reference version 71. Genes with median FPKM of 0 were removed, leaving 23,803 genes. We restricted analysis to protein coding genes, giving 15,304 unique Ensembl genes. Expression values were quantile-normalized to a standard normal distribution for each gene.
The replication set consisted of Illumina Human-6 v2 Expression BeadChip data from 730 HapMap lymphoblastoid cell lines from the EBI website. These datasets contain two replicates each for 730 unrelated individuals from 8 HapMap populations (YRI, CEU, CHB, JPT, GIH, MEX, MKK, LWK) and were generated as described by Stranger et al.75. Background corrected and summarized probeset intensities (by Illumina software) contained values for 7,655 probes. Additionally, probes containing common SNPs were removed76. Only probes with a one-to-one correspondence with Ensembl gene identifiers were retained. We removed probes with low concordance across replicates (Spearman correlation ≤ 0.5). In total we obtained 5,388 probes for downstream analysis.
Each probe was quantile-normalized to a standard normal distribution across all individuals separately for each replicate and then averaged across replicates. These values were quantile-normalized to a standard normal distribution for each probe.
eQTL association testing
Expression values were adjusted for individual sex, individual membership, gene expression heterogeneity, and population structure (Supplementary Methods). Adjusted expression values were used as input to the eSTR analysis. To restrict to STR loci with high quality calls, we filtered the call set to contain only loci where at least 50 of the 311 samples had a genotype call. To avoid outlier genotypes that could skew the association analysis, we removed any genotypes seen less than three times. If only a single genotype was seen more than three times, the locus was discarded. To increase our power, we further restricted analysis to the most polymorphic loci with heterozygosity of at least 0.3. This left 80,980 STRs within 100kb of a gene expressed in our LCL dataset.
A linear model was used to test for association between normalized STR dosage and expression for each STR within 100kb of a gene (Supplementary Methods). Dosage was defined as the sum of the deviations of the STR allele lengths from the hg19 reference. For example, if the hg19 reference for an STR is 20bp and the two alleles called are 22bp and 16bp, the dosage is equal to (22−20)+(16−20) = −4bp. STR genotypes were zscore-normalized to have mean 0 and variance 1. For genes with multiple transcripts, we defined the transcribed region as the maximal region spanned by the union of all transcripts. The linear model for each gene is given by:
where y⃗g = (yg,1, …, yg,n)T with yg,i the normalized covariate-corrected expression of gene g in individual i, n is the number of individuals, αg is the mean expression level of homozygous reference individuals, βj,g is the effect of the allelic dosage of STR locus j on gene g, x⃗j = (xj,1, …, xj,n)T with xj,i the normalized allelic dosage of STR locus j in the ith individual, and ε⃗j,g is a random vector of length n whose entries are drawn from where is the unexplained variance after regressing locus j on gene g. The association was performed using the OLS function from the Python statsmodels package. For each comparison, we tested H0: βj,g = 0 vs. H1: βj,g ≠ 0 using a standard t-test. We controlled for a gene-level false discovery rate (FDR) of 5% (Supplementary Methods).
Partitioning heritability using linear mixed models
For each gene, we used a linear mixed model to partition heritability between the lead explanatory STR and other cis variants. We used a model of the form:
where:
y⃗g, αg, βj,g, x⃗j, and ε⃗j,g are as described above.
u⃗g is a length n vector of random effects and with the percent of phenotypic variance explained by cis bi-allelic variants for gene g.
Kg is a standardized n × n identity by state (IBS) relatedness matrix constructed using all common bi-allelic variants (MAF≥1%) reported by phase 1 of the 1000 Genomes Project within 100kb of gene g. This includes SNPs, indels, and several bi-allelic structural variants and is constructed as where p is the total number of variants considered, x⃗i is a length n vector of genotypes for variant i, and 1n is a length n vector of ones. Note the mean diagonal element of Kg is equal to 1.
We used the GCTA program77 to determine the restricted maximum likelihood estimates (REML) of βj,g and . To get unbiased values of , the --reml-no-constrain option was used.
We used the resulting estimates to determine the variance explained by the STR and the cis region. We can write the overall phenotypic variance-covariance matrix as:
where:
var(y⃗g) is an n × n expression variance-covariance matrix with diagonal elements equal to 1, since expression values for each gene were normalized to have mean 0 and variance 1.
In is the n × n identity matrix.
This equation shows the relationship:
where:
is the phenotypic variance, which is equal to 1.
is the variance explained by the STR. This is equal to since the STR genotypes were scaled to have mean 0 and variance 1.
is the variance explained by bi-allelic variants in the cis region. This is approximately equal to since the local IBS matrix Kg has a mean diagonal value of 1.
We estimated the percent of phenotypic variance explained by STRs, , using the unbiased estimator , where β̂j,g is the estimate of βj,g returned by GCTA, and SE is the standard error on the estimate, using the fact that β̂j,g ~ N(βj,g, SE). We estimated the percent of phenotypic variance explained by bi-allelic markers as . Note that for this analysis the STR was treated as a fixed effect. We also reran the analysis treating the STR as a random effect and found very little change in the results (Supplementary Note).
Results are reported for all eSTR-containing genes and for all genes with moderate total cis heritability, which we define as genes where . We used this approach as to our knowledge there are no published results about the cis-heritability of expression of individual genes in LCLs from twin studies. We used 10,000 bootstrap samples of each distribution to generate 95% confidence intervals for the medians.
Comparing to the lead eSNP
We identified SNP eQTLs using SNPs with MAF ≥ 1% as reported by phase 1 of the 1000 Genomes Project. We used an identical pipeline to our eSTR analysis to identify SNP eQTLs after replacing the vector x⃗j with a vector of SNP genotypes (0, 1 or 2 reference alleles) that was z-normalized to have mean 0 and variance 1. To determine whether our eSTR signal was indeed independent of the lead SNP eQTL at each gene, we repeated association tests between STR dosages and expression levels while holding the genotype of the SNP with the most significant association to that gene constant. For this, we determined all samples at each gene that were either homozygous reference or homozygous non-reference for the lead SNP. For the SNP allele with more homozygous samples, we repeated the eSTR linear regression analysis and determined the sign and magnitude of the slope. We removed any genes for which there were less than 25 samples homozygous for the SNP genotype or for which there was no STR variation after holding the SNP constant, leaving 1,856 genes for analysis. We used a sign test to determine whether the direction of effects before and after conditioning on the lead SNP are more concordant than expected by chance.
We used model comparison to determine whether eSTRs can explain additional variation in gene expression beyond that explained by the lead eSNP for each gene. For each gene with a significant eSTR and eSNP, we analyzed the ability of two models to explain gene expression:
where αg is the mean expression value for the reference haplotype, y⃗g is a vector of expression values for gene g, βeSNP,g is the effect of the eSNP on gene g, βeSTR,g is the effect of the eSTR on gene g, x⃗eSNP,g is a vector of genotypes for the lead eSNP for gene g, x⃗eSTR,g is a vector of genotypes for the best eSTR for gene g, and ε⃗j,g gives the residual term. A major caveat is that the eSNP dataset has significantly more power to detect associations than the eSTR dataset due to the lower quality of the STR genotype panel (Supplementary Note), and this analysis is therefore likely to underestimate the true contribution of STRs to gene expression. We used ANOVA to test whether the joint model performs significantly better than the SNP-only method. We obtained the ANOVA p-value for each gene and used the qvalue package to determine the FDR.
Conservation analysis
Sequence conservation around STRs was determined using the PhyloP track available from the UCSC Genome Browser. To calculate the significance of the increase in conservation at eSTRs, we compared the mean PhyloP score for each eSTR to that for 1000 random sets of STRs with matched distributions of the distance to the nearest transcription start site. For each STR, we determined the mean PhyloP score for a given window size centered on the STR. The p-value given is the percentage of random sets whose mean PhyloP score was greater than the mean of the observed eSTR set.
Enrichment of STRs and eSTRs in predicted enhancers
H3K27ac peaks produced by the ENCODE7 Project were used to determine predicted enhancers in GM12878. Peaks were downloaded from the UCSC Genome Browser and converted to hg19 coordinates using the liftOver tool. Any peak overlapping within 3kb of a transcription start site was removed to exclude promoter regions from the analysis.
Enrichment in histone modification peaks
Chromatin state and histone modification peak annotations generated by the Encode Consortium for GM12878 were downloaded from the UCSC Genome Browser. Because variants involved in regulating gene expression are more likely to fall near genes compared to randomly chosen variants, naïve enrichment tests of eSTRs vs. randomly chosen control regions may return strong enrichments simply because of their proximity to genes. To account for this, we randomly shifted the location of eSTRs by a distance drawn from the distribution of distances between the best STR and lead SNP for each gene. We repeated this process 1,000 times. For each set of permuted eSTR locations, we generated null distributions by determining the percent of STRs overlapping each annotation. We used these null distributions to calculate empirical p-values for the enrichment of eSTRs in each annotation.
Effects of eSTRs on modulating regulatory elements
One potential mechanism by which eSTRs may act is by modulating epigenetic properties. The GERV (Generative Evaluation of Regulatory Variants)59 model predicts ChIP-sequencing experiments directly from genomic sequences and optional covariates such as DNAse-seq data. We used the non-covariate version of this technique to assess the effect of STR variations on the occupancy of chromatin marks.
GERV builds on a kmer-based statistical model to predict the signal of ChIP-seq experiments from a DNA sequence context. Briefly, the model considers that each k-mer has a spatial effect on ChIP-seq read counts in a window of [−M, M−1] bp centered at the start of the k-mer. The read count at a given base is then modeled as the log-linear combination of the effects of all k-mers whose effect ranges cover that base, where k ranges from 1 to 8.
For each eSTR in our dataset, we generated sequences representing each observed allele. We filtered STRs with interruptions in the repeat motif, since the sequence for different allele lengths is ambiguous for these loci. For each mark, we used the model to predict the read count for each allele in a window of ±M bp from the STR boundaries, where M was set to 1,000 for all marks except p300, for which M was set to 200. Previous findings of GERV showed that these values of M give the best correlation between predicted and real ChIP-seq signals using cross validation. For each alternate allele, we generated a score as the sum of differences in read counts from the reference allele at each position in this window. We regressed the number of repeats for each allele on this score and took the absolute value of the slope for each locus. We repeated the analysis on a set of randomly chosen negative control loci. Control loci were chosen to match the distribution of repeat lengths and absolute signal for each mark in the reference genome. We used a Mann-Whitney rank test to compare the magnitudes of slopes between the eSTR and control sets for each mark.
Overlap of eSTR and GWAS genes
Aggregate results for seven common diseases (rheumatoid arthritis, Crohn’s disease, type I diabetes, type 2 diabetes, blood pressure, bi-polar disorder, and coronary artery disease) were downloaded from the NHGRI GWAS catalog accessed on June 12, 2015. Relevant genes were taken from the columns “Reported Gene(s)” and “Mapped_gene”. To generate a null distribution, we chose 1000 sets of randomly selected genes matched to eSTR genes on expression in LCLs (difference in RPKM < 10) and on cis heritability (difference in variance explained by cis bi-allelic variants < 5%). We compared the overlap of GWAS genes with eSTR genes vs. the 1000 control sets to determine an empirical p-value.
eSTR associations with human traits
To generate STR genotypes for each of the individuals in the UK10K TwinsUK dataset, we ran lobSTR v2.0.3 on each BAM using the options fft-windowsize=16, fft-window-step=4 and bwaq=15. The resulting BAM files were analyzed using v2.0.3 of the lobSTR allelotyper using default options, resulting in STR genotypes for 1,685 individuals.
We then performed an association test between each STR and each phenotype. To control for population structure, we adjusted STR dosages and phenotypes for the top 10 ancestry principal components based on common SNPs (MAF>=5%) after LD-pruning. Principal components were computed using EIGENSTRAT78 v5.0.1. Phenotypes were further adjusted for the age at which the phenotype was measured. Association tests were performed between the adjusted dosages and the quantile-normalized adjusted phenotypes.
We were able to analyze TwinsUK cohort for the following 38 phenotypes [in parentheses, the PMID reference given by TwinsUK to describe the phenotype measurement procedure]: Albumin (19209234), Alkaline phosphatase (19209234), Apolipoprotein A-I (15379757), Apolipoprotein B (15379757), Bicarbonate, Bilirubin (19209234), Body mass index, Creatinine (11017953), Diastolic blood pressure (16249458), Heart Rate (19587794), FEV1 (17989158), FEV1/FVC ratio (17989158), FVC (17989158), Gamma-Glutamyl Transpeptidase (19209234), Glucose (19209234), High density lipoprotein (19016618), Standing height (17559308), Hemoglobin (19862010), Hip circumference (17228025), Homocysteine (18280483), C-reactive protein (21300955), Insulin (16402267), Mean corpuscular volume (19862010), Packed Cell Volume (10607722), Phosphate (12193151), Platelet count (19221038), Red blood cell count (19820697), Sodium (18179892), Systolic blood pressure (16249458), Total cholesterol (19820914), Triglycerides (15379757), Urea (18179892), Uric acid (19209234), Waist circumference (17228025), White blood cell count (19820697), Weight (17016694), and Waist to Hip ratio.
We then examined the association in the 666 eSTR loci that contained an eSTR that significantly improved the gene expression variance when combined with the lead eSNP (nominal ANOVA p<0.05). Out of these eSTRs, 499 were genotyped in >1000 participants. For each phenotype, q values were calculated by adjusting the p-values using the Benjamini-Hochberg procedure. Only hits with a q-value < 0.1 were reported.
Supplementary Material
Acknowledgments
M.G. is supported by the National Defense Science & Engineering Graduate Fellowship. Y.E. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. This study was supported by a gift from Andria and Paul Heafy (Y.E), NIJ grant 2014-DN-BX-K089 (Y.E, T.W), and NIH grants 1U01HG007037 (H.Z), R01MH084703(J.P), R01HG006399 (A.L.P), HG006696 (A.J.S), DA033660 (A.J.S), and MH097018 (A.J.S), and a research grant 6-FY13-92 from the March of Dimes Foundation (A.J.S). We thank Tuuli Lappalainen, Alon Goren, Tatsu Hashimoto, and Dina Zielinksi for useful comments and discussions.
References and Notes
- 1.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 3.Ardlie KG, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montgomery SB, et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright FA, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–477. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Press MO, Carlson KD, Queitsch C. The overdue promise of short tandem repeat variation for heritability. Trends Genet. 2014;30:504–512. doi: 10.1016/j.tig.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellegren H. Microsatellites: simple sequences with complex evolution. Nature reviews. Genetics. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 16.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annual review of genetics. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 17.Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- 18.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 19.Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet. 2002;30:315–320. doi: 10.1038/ng836. [DOI] [PubMed] [Google Scholar]
- 20.Martin P, Makepeace K, Hill SA, Hood DW, Moxon ER. Microsatellite instability regulates transcription factor binding and gene expression. Proc Natl Acad Sci U S A. 2005;102:3800–3804. doi: 10.1073/pnas.0406805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willems R, Paul A, van der Heide HG, ter Avest AR, Mooi FR. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 1990;9:2803–2809. doi: 10.1002/j.1460-2075.1990.tb07468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yogev D, Rosengarten R, Watson-McKown R, Wise KS. Molecular basis of Mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5' regulatory sequences. EMBO J. 1991;10:4069–4079. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hefferon TW, Groman JD, Yurk CE, Cutting GR. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc Natl Acad Sci U S A. 2004;101:3504–3509. doi: 10.1073/pnas.0400182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hui J, et al. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothenburg S, Koch-Nolte F, Rich A, Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc Natl Acad Sci U S A. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiser JN, Love JM, Moxon ER. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 27.Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sureshkumar S, et al. A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science. 2009;323:1060–1063. doi: 10.1126/science.1164014. [DOI] [PubMed] [Google Scholar]
- 29.Hammock EA, Young LJ. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308:1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- 30.Yanez-Cuna JO, et al. Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res. 2014 doi: 10.1101/gr.169243.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawaya S, et al. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS One. 2013;8:e54710. doi: 10.1371/journal.pone.0054710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonay TB, et al. Tandem repeat variation in human and great ape populations and its impact on gene expression divergence. 2015 doi: 10.1101/gr.190868.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borel C, et al. Tandem repeat sequence variation as causative cis-eQTLs for protein-coding gene expression variation: the case of CSTB. Hum Mutat. 2012;33:1302–1309. doi: 10.1002/humu.22115. [DOI] [PubMed] [Google Scholar]
- 34.Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 35.Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Molecular biology and evolution. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 36.Shimajiri S, et al. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 1999;455:70–74. doi: 10.1016/s0014-5793(99)00863-7. [DOI] [PubMed] [Google Scholar]
- 37.Warpeha KM, et al. Genotyping and functional analysis of a polymorphic (CCTTT)(n) repeat of NOS2A in diabetic retinopathy. FASEB J. 1999;13:1825–1832. doi: 10.1096/fasebj.13.13.1825. [DOI] [PubMed] [Google Scholar]
- 38.Hui J, Stangl K, Lane WS, Bindereif A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 2003;10:33–37. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- 39.Sathasivam K, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grunewald TG, et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet. 2015;47:1073–1078. doi: 10.1038/ng.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willems T, et al. The landscape of human STR variation. Genome Res. 2014 doi: 10.1101/gr.177774.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gymrek M, Golan D, Rosset S, Erlich Y. lobSTR: A short tandem repeat profiler for personal genomes. Genome Res. 2012;22:1154–1162. doi: 10.1101/gr.135780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duyao M, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 45.La Spada AR, et al. Meiotic stability and genotype-phenotype correlation of the trinucleotide repeat in X-linked spinal and bulbar muscular atrophy. Nat Genet. 1992;2:301–304. doi: 10.1038/ng1292-301. [DOI] [PubMed] [Google Scholar]
- 46.Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. The Journal of biological chemistry. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 48.Stranger BE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Payseur BA, Place M, Weber JL. Linkage disequilibrium between STRPs and SNPs across the human genome. Am J Hum Genet. 2008;82:1039–1050. doi: 10.1016/j.ajhg.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawaya S, Jones M, Keller M. Linkage disequilibrium between single nucleotide polymorphisms and hypermutable loci. 2015 [Google Scholar]
- 51.Lamina C, et al. A systematic evaluation of short tandem repeats in lipid candidate genes: riding on the SNP-wave. PLoS One. 2014;9:e102113. doi: 10.1371/journal.pone.0102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gusev A, et al. Regulatory variants explain much more heritability than coding variants across 11 common diseases. bioRxiv. 2014 [Google Scholar]
- 53.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 55.Gaffney DJ, et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trynka G, et al. ARTICLE Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-coding Variants within Complex-Trait Loci. The American Journal of Human Genetics. 2015;152:97–139. doi: 10.1016/j.ajhg.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng H, Hashimoto T, Kang DD, Gifford DK. GERV: A Statistical Method for Generative Evaluation of Regulatory Variants for Transcription Factor Binding. 2015 doi: 10.1093/bioinformatics/btv565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muddyman D, Smee C, Griffin H, Kaye J. Implementing a successful data-management framework: the UK10K managed access model. Genome medicine. 2013;5:1–9. doi: 10.1186/gm504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Döring A, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nature genetics. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 63.Vitart V, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nature genetics. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 64.Wallace C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. The American Journal of Human genetics. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shin S-Y, et al. An atlas of genetic influences on human blood metabolites. Nature genetics. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber JL, Broman KW. 7 Genotyping for human whole-genome scans: Past, present, and future. Advances in genetics. 2001;42:77–96. doi: 10.1016/s0065-2660(01)42016-5. [DOI] [PubMed] [Google Scholar]
- 67.Chaisson MJ, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhatia G, et al. Haplotypes of common SNPs can explain missing heritability of complex diseases. 2015 [Google Scholar]
- 69.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guilmatre A, Highnam G, Borel C, Mittelman D, Sharp AJ. Rapid multiplexed genotyping of simple tandem repeats using capture and high-throughput sequencing. Hum Mutat. 2013;34:1304–1311. doi: 10.1002/humu.22359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barbosa-Morais NL, et al. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.