Abstract
To further understand Mycobacterium tuberculosis pathogenesis, the regulation of potential virulence genes needs to be investigated. The eis gene of M. tuberculosis H37Rv enhances the intracellular survival of Mycobacterium smegmatis, which does not contain eis, within macrophages (J. Wei, J. L. Dahl, J. W. Moulder, E. A. Roberts, P. O'Gaora, D. B. Young, and R. L. Friedman, J. Bacteriol. 182:377-384, 2000). Experiments were done to characterize the eis promoter in M. smegmatis and M. tuberculosis H37Ra. The putative −10 and −35 regions matched the Escherichia coli σ70 consensus 67 and 83%, respectively, making it a group A/SigA-like mycobacterial promoter. Expression of site-directed variants of the core promoter region, determined by flow cytometry using gfp as a reporter, showed that the putative −10 region is essential for eis expression. In addition, site-directed alteration of the eis promoter to the consensus E. coli σ70 promoter elements increased gfp transcription to levels similar to that driven by the heat shock promoter, phsp60, of Mycobacterium bovis BCG. Upstream promoter deletion analysis showed that a 200- and 412-bp region of the promoter was necessary for maximum expression of gfp in M. smegmatis and M. tuberculosis H37Ra, respectively. Random mutagenesis of the 412-bp eis promoter, using a catechol 2,3-dioxygenase screen and activity assay, defined nucleotides upstream of the core promoter region that are essential to eis expression in both M. smegmatis and M. tuberculosis H37Ra, including a region homologous to a DinR cis element.
Tuberculosis (TB) continues to be the world's most destructive human bacterial infectious disease. Current estimates show that more than two million people die from TB each year, and TB remains a major cause of premature death (18). Mortality due to TB is a major global health crisis due to AIDS and the increasing prevalence of multidrug-resistant strains of Mycobacterium tuberculosis, although effective treatments are available. Despite the elucidation of the genome sequence of several M. tuberculosis strains (5, 14) and available genetic tools to identify genes involved in TB pathogenesis (13, 16, 23, 26), the molecular basis of its ability to survive within host cells and evade host immune responses is unknown. Understanding the molecular mechanisms of pathogenesis is essential for the development of better methods of diagnosis, treatment, and prevention. One way to heighten our understanding of M. tuberculosis pathogenesis is to examine the regulation of potential virulence genes, in particular the promoters and other elements that govern their expression.
Approximately 130 mycobacterial promoters have been characterized to date, but only 76 have been categorized into the four groups (A to D) of mycobacterial promoters (15). Identified promoters comprise less than 3% of the potential promoters in the genome of M. tuberculosis. The majority of the categorized promoters are from Mycobacterium smegmatis and Mycobacterium paratuberculosis, with less than 25% derived from M. tuberculosis itself. This indicates that we know little about promoter function in mycobacteria and even less about promoters from M. tuberculosis. Therefore, there is a need for detailed studies on M. tuberculosis promoter characterization and function, especially for genes that may play a role in the survival of M. tuberculosis within macrophages.
The eis gene of M. tuberculosis H37Rv was found to enhance the survival of saprophytic M. smegmatis during repeated passage through the human macrophage-like cell line U-937 (32). How eis confers this survival phenotype on M. smegmatis, which does not contain eis, is unknown because it is not homologous to any gene of known function. Fractionation studies and immunoblot analyses performed on culture-grown M. tuberculosis found that the Eis protein was distributed throughout the bacterium, including the cell wall and cytoplasm (7). More recent studies have shown that eis is differentially expressed in a clinical strain of M. tuberculosis upon infection of activated human macrophages (4). These results do not suggest a function for the Eis protein or indicate how eis may be regulated. Our goal here is to understand the expression of eis as a foundation for eventually understanding its proposed role in M. tuberculosis virulence.
MATERIALS AND METHODS
Bacterial strains and growth media.
M. smegmatis 1-2c, a derivative of strain mc26 selected for improved transformation efficiency (35), was grown in Middlebrook 7H9 broth (Difco) supplemented with 2% glucose and 0.05% Tween 80. M. tuberculosis H37Ra, an avirulent derivative of H37Rv, was cultured in Middlebrook 7H9 broth (Difco) supplemented with 10% oleic acid albumin dextrose catalase and 0.05% Tween 80. M. smegmatis and M. tuberculosis H37Ra were also plated on Middlebrook 7H10 agar (Difco) supplemented with either 2% glucose or 10% oleic acid albumin dextrose catalase, respectively. Kanamycin at a concentration of 25 μg/ml (Boehringer Mannheim) was used in both liquid and solid mycobacterial media to maintain vector constructs. Luria-Bertani broth or agar with 50 μg of kanamycin/ml was used for selection and growth of E. coli DH10B (Invitrogen) transformants. Hygromycin B was added at 50 μg/ml to maintain mycobacteria containing the eis clone p69 (32).
RNA isolation and primer extension.
Twenty-five-milliliter culture pellets from log-phase grown M. smegmatis(p69), M. tuberculosis H37Ra(p69), and M. tuberculosis H37Ra (wild type) were resuspended in 1 ml of RLT lysis buffer (QIAGEN). Cells were disrupted three times in a FastPrep FP120 angular reciprocating shaker (Bio 101) at maximum settings using 0.1-mm zirconium beads. Total RNA was isolated using an RNeasy mini kit (QIAGEN), and RNA yield and purity were calculated using spectrophotometry after on-column DNaseI digestion, according to the manufacturer's instructions. The primer Pxt was end labeled with 30 μCi of [32P-γ]ATP using T4 polynucleotide kinase. Unincorporated radionucleotides were removed using a G-25 Sephadex spin column (Boehringer Manheim). Primer extension was performed using 5 μg of total RNA and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Primer extension reactions were incubated for 1 h at 44°C. Primer extension products were run alongside a Redivue 33P-labeled DNA sequencing reaction performed with Pxt, using p69 as a template on a 6% polyacrylamide, 8 M urea gel. The gel was exposed to BioMax autoradiograph film (Kodak) for 48 h at −80°C prior to development.
Site-directed mutagenesis of putative core eis promoter.
Site-directed mutagenesis was performed using the Quikchange site-directed mutagenesis kit (Stratagene) with pEP412 as a template, according to the manufacturer's protocol. All site-directed mutations in pSKM constructs were verified by DNA sequencing using the pF1 primer.
Construction of pEP vector series.
The promoterless gfp vector pFPV27 (see Fig. 3A) was used as the primary cloning vector for these studies. Variable regions of the eis promoter upstream from the start codon of the gene, and not including the putative Shine-Dalgarno sequence, were amplified using PCR. The reverse primer, ApaIr, which contains an ApaI restriction sequence at the 5′ end, was used for all amplifications. Forward primers containing a BamHI restriction sequence were paired with ApaIr for amplification of eis promoter variants, and amplicons were cloned into the BamHI/ApaI site of pFPV27 (1) to create the pEP vector series (see Fig. 3A). The numbers associated with each construct indicate the size of the amplicon. Primers were obtained from Invitrogen. PCRs contained: 1× PWO polymerase buffer with 2 mM MgSO4 (Roche), 0.6 mM deoxynucleoside triphosphates, 4 μM (each) primer specific for the desired product, 2.5 U of PWO proofreading polymerase (Roche), and 5% dimethyl sulfoxide in a final volume of 50 μl. Thermocycling reactions were performed in a Bio-Rad iCycler thermocycler with the following parameters: an initial denaturation at 95°C for 5 min, 30 cycles of 95°C for 1.5 min, 56°C for 1.5 min, and 72°C for 1.5 min, followed by a final extension at 72°C for 5 min.
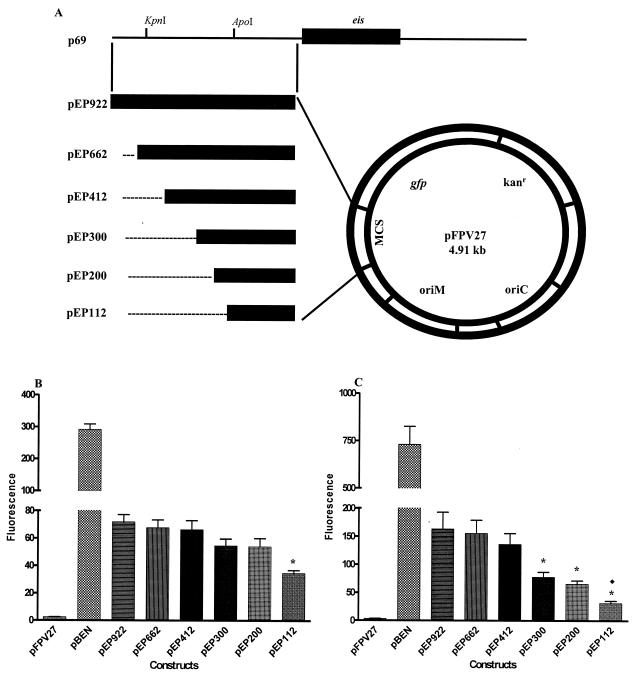
FIG. 3.
(A) Construction of the pEP vector series. A 922-bp fragment upstream of eis in p69 was PCR amplified and cloned into the promoterless gfp shuttle vector pFPV27 to create the full-length eis promoter construct pEP922. Subsequent 5′ deletions of the eis promoter (pEP662, pEP412, pEP300, pEP200, and pEP112) were amplified and cloned into pFPV27. Flow-cytometric analysis of gfp expression by the pEP vector series in M. smegmatis (B) and M. tuberculosis H37Ra (C), including the negative control pFPV27 and the positive control pBEN, is shown. Data represent results from at least three independent experiments performed in triplicate. In panel B, the asterisk denotes a significant difference in the fluorescence between pEP112 and pEP922 at a P value of <0.0001 using a paired t test, and in panel C, it indicates significant differences between pEP112, pEP200, and pEP300 versus pEP922 at a P value of <0.0001 using one-way ANOVA. In panel C, the diamond indicates a significant difference in fluorescence at a P value of <0.05, using a paired t test, between pEP112 and pEP200. Note that different fluorescence scales are used for results presented in panel B versus panel C.
Electroporation of E. coli, M. smegmatis, and M. tuberculosis H37Ra.
Electroporations were performed using a Gene Pulser electroporator (Bio-Rad). Forty microliters of a 1:5 dilution of Max Efficiency E. coli DH10B cells in cold 10% sterile glycerol were mixed with a 1/10 volume of the appropriate ligation reaction and placed on ice for 10 min. Electroporation into E. coli was performed at settings of 1.8 kV, 25 μF, and 100 Ω in a 0.1-cm cuvette (Bio-Rad) that had been prechilled on ice for 20 min. Electrocompetent M. smegmatis 1-2c (0.4 ml) was mixed with approximately 1 μg of DNA from each construct, pipetted into a 0.4-cm cuvette (Bio-Rad), and placed on ice for 30 min. Electroporations were performed at settings of 2.5 kV, 25 μF, and 1,000 Ω. Conditions identical to those used for M. smegmatis were used for M. tuberculosis H37Ra except that all steps were performed at room temperature, as previously described (31).
Flow cytometry of mycobacteria.
M. smegmatis and M. tuberculosis H37Ra cultures containing the pEP constructs were grown to log phase, harvested, vortexed for 10 s using 3-mm glass beads to reduce clumping, and diluted to an optical density at 650 nm (OD650) of 0.05 to 0.250. The appropriate dilution to obtain 108 cells in 1 ml for each sample was calculated by using the equivalence that an OD650 of 0.100 equals 108 cells (32). Dilutions were prepared in 12- by 75-mm polystyrene round-bottom tubes (Becton Dickinson) containing 10 to 12 3-mm sterilized glass beads, and samples were vortexed for 10 s just prior to flow cytometry to reduce clumping. A Becton Dickinson FACScan 8383 with a 488-nm argon laser was gated to detect the presence of individual mycobacteria producing green fluorescent protein (GFP). A total of 10,000 events from each sample was measured, with at least 2,000 gated events recorded to ensure statistical significance. All parameters for data acquisition were identical for both M. smegmatis and M. tuberculosis H37Ra. Mycobacteria containing pFPV27 were used as negative controls for these experiments. Positive controls were mycobacteria containing the vector pBEN, which contains gfp driven by phsp60, a strong heat shock promoter from Mycobacterium bovis BCG (2, 27). Levels of fluorescence were plotted as the geometric mean of the histograms.
Random PCR mutagenesis and PCR of mutated inserts for sequencing.
To create the pTKep mutants, the primers Epxyl-F and Epxyl-R were used to amplify the 412-bp eis promoter. Mutagenic PCR pools of the eis promoter were cloned into the promoterless xylE-containing vector pTKmx, as described previously (19). Mutagenic PCR was performed using conditions previously described (20), with the following modifications. Briefly, 0.4 mg of each primer/ml was added to a reaction mixture containing a 0.25:1 ratio of dATP to dGTP, dCTP, and dTTP to attain approximately one nucleotide change per 400 bp. The primers Kep-F and PCR2 were used to amplify eis promoter mutants from pTKep clones. Crude extracts of mycobacteria to be used as PCR templates were prepared as described elsewhere (16). Two and a half microliters of crude extract was added to a final volume of 20 μl containing 2 μl of 10× reaction buffer (Roche), 4 μl of 1.25 mM deoxynucleoside triphosphates, 0.27 U of Taq polymerase (Roche), 20 pmol of each primer, and 5% dimethyl sulfoxide. Mutations were verified by DNA sequencing of both strands from each mutant using the primers Kep-seq and Epxyl-R.
Qualitative and quantitative catechol 2,3-dioxygenase activity assays.
For qualitative assays, M. smegmatis(pTKep) and M. tuberculosis H37Ra(pTKep) clones, grown on their respective agar, were sprayed with 100 mM catechol in 50 mM potassium phosphate buffer (pH 7.5). Catechol 2,3-dioxygenase (CDO) converts catechol into 2-hydroxymuconic semialdehyde, a product with a bright yellow color and an absorbance maximum at 375 nm (25). White and light yellow clones, representing deficient CDO production, were selected after 5 min of color development, patched to new agar plates, rescreened, and compared to pTKepNM (nonmutated eis promoter) and pTKmx isolates. True-white and light-yellow colonies were used to inoculate liquid media or were patched onto agar to prepare crude extracts for PCR amplification of the mutated eis promoter insert. Quantitative CDO assays were performed, similar to those previously described, with slight modifications (28, 29). M. smegmatis and M. tuberculosis H37Ra cultures were grown in their respective liquid media to stationary phase. One-milliliter aliquots were centrifuged at 13,000 rpm for 3 min and washed once with 0.5 ml of 50 mM potassium phosphate buffer (pH 7.5). Cells were suspended in 0.5 ml of 50 mM potassium phosphate buffer (pH 7.5) and lysed as described above for RNA extraction with the addition of 1 mM final phenylmethanesulfonyl fluoride (Sigma). Bicinchoninic acid assays (Pierce) were used to determine protein concentrations of lysates, according to the manufacturer's protocol. One hundred microliters of lysate was then mixed with 0.9 ml of 0.3 mM catechol in 50 mM potassium phosphate buffer (pH 7.5), and the OD375 was recorded over a 2-min period. A change in the OD375 of 0.0147 is equal to 1 mU of specific activity at 24°C (25). Results presented are from experiments performed in triplicate using three independent cultures for each sample tested. Values are expressed as milliunits/milligram/minute.
DNA sequencing.
Sequencing reactions were performed by the Arizona Research Laboratories Genetic Analysis and Technology Core using an Applied Biosystems 3730xl DNA analyzer.
Statistical analysis.
Results of flow cytometry and CDO activity experiments are expressed as the mean ± standard error of the mean. Differences in fluorescence or CDO activity between three or more constructs were assessed using one-way analysis of variance (ANOVA). When two constructs were compared to one another, a paired Student's t test analysis was used to determine the statistical difference.
RESULTS
Transcriptional start point mapping of eis.
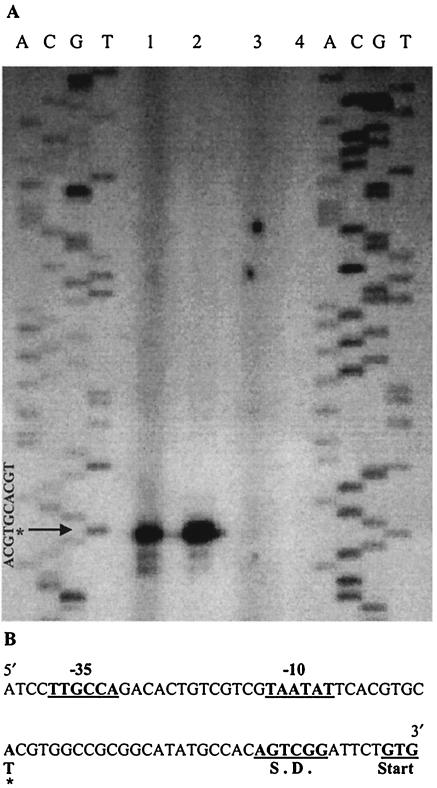
The transcriptional start point (TSP) of the eis gene was determined by primer extension analyses on total RNA from M. smegmatis(p69), M. tuberculosis H37Ra(p69), and wild-type M. tuberculosis H37Ra. In both M. smegmatis and M. tuberculosis H37Ra, the TSP mapped to an A nucleotide 33 bp upstream from the start codon of eis (Fig. 1A and B). The wild-type M. tuberculosis H37Ra product was very faint compared to M. tuberculosis H37Ra harboring p69, indicating low levels of chromosomal eis transcript production during logarithmic growth. A putative −10 region matching the consensus E. coli σ70 sequence at 4 of 6 bases was found 8 bp upstream from the TSP, and a putative −35 region matching the E. coli σ70 consensus sequence at 5 of 6 positions was located 13 bp upstream from the putative −10 region (Fig. 1B). The consensus sequence data for the putative −35 and −10 regions place the eis promoter within the group A mycobacterial promoters (15).
FIG. 1.
Primer extension analysis of the eis transcript in mycobacteria. (A) Primer extension analysis of M. smegmatis(p69) (lane 1), M. tuberculosis H37Ra(p69) (lane 2), and M. tuberculosis H37Ra wild type (lane 3), showing the TSP mapping to an A nucleotide in both species. Labeled primer was run as a negative control (lane 4). (B) Sequence upstream from the published eis start codon. Putative Shine-Dalgarno (S.D.), −35, and −10 regions are underlined and boldfaced, and the TSP is indicated by an asterisk.
Flow cytometry of site-directed core eis promoter mutants in M. smegmatis and M. tuberculosis.
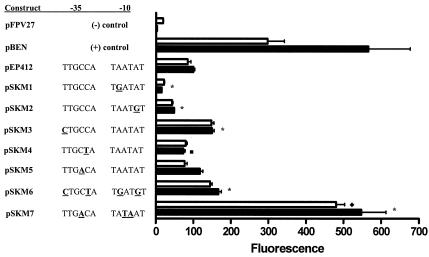
We tested whether the putative −35 and −10 regions of the eis promoter were important for the transcriptional activity of gfp. Figure 2 shows the specific mutations that were made in these regions. Two substitutions were made in the −10 region, A→G at −12 (pSKM1) and A→G at −9 (pSKM2) (Fig. 2). pSKM1 exhibited a fourfold and sixfold decrease in fluorescence in M. smegmatis and M. tuberculosis H37Ra, respectively, compared to pEP412 (P < 0.05). pSKM2 showed a twofold decrease in fluorescence compared to pEP412 in both mycobacteria (P < 0.05). These results indicate that the −10 region is critical for the expression of eis in mycobacteria.
FIG. 2.
Flow-cytometric analysis of gfp expression in mycobacteria by site-directed mutants (pSKM series) in the core promoter of eis. Open bars represent pSKM constructs assayed with M. smegmatis and solid bars represent pSKM constructs assayed with M. tuberculosis H37Ra. Mutations are underlined and boldfaced. Asterisks indicate significant differences at P values of <0.05 using one-way ANOVA between pEP412 and pSKM constructs with both M. smegmatis and M. tuberculosis H37Ra. The diamond indicates a significant difference using a paired t test between pBEN and pSKM7 in M. smegmatis at P values of <0.05. The square represents a significant difference using a paired t test between pSKM4 and pEP412 in M. tuberculosis H37Ra at P values of <0.05. The −35 and −10 sequences of pSKM7 represent the E. coli σ70 consensus sequences.
Three mutations were made in the −35 region to examine its role in eis expression. A single change from T to C at −32 (Fig. 2, pSKM3) enhanced expression in both M. smegmatis and M. tuberculosis H37Ra by 1.75- and 1.5-fold, respectively. A C→T change at −28 (pSKM4) had no significant effect in M. smegmatis but caused a 1.36-fold decrease (P < 0.05) in expression in M. tuberculosis H37Ra. A change from C to A at −29 (pSKM5) had no significant effect on expression in either mycobacteria (Fig. 2). These results suggest that the putative −35 region may be subtly involved in the recognition of the eis promoter by the mycobacterial transcriptional machinery but is not essential for expression.
We then changed the eis promoter sequences further from (pSKM6) and closer to (pSKM7) the E. coli σ70 consensus sequence. We discovered a 1.7-fold increase (P < 0.05) in transcriptional activity in pSKM6 in both mycobacteria, indicating slightly better promoter recognition. Interestingly, an enormous increase in activity occurred when the eis promoter was changed to the E. coli σ70 consensus sequence (pSKM7). In M. smegmatis, pSKM7 exhibited a 5.7-fold increase in activity over that of pEP412 and a 1.84-fold increase in activity over that of pBEN. In M. tuberculosis H37Ra, pSKM7 exhibited a 5.5-fold increase in activity over that of pEP412 and displayed levels of fluorescence similar to that of pBEN. pSKM7 also showed a twofold increase in gfp expression in E. coli compared to that of pEP412 in logarithmic-phase cells (data not shown). The results from the site-directed mutagenesis of the putative −35 and −10 regions indicate that these regions comprise the core promoter of eis. In addition, these results confirm that the eis promoter is a group A/SigA-like mycobacterial promoter.
Flow cytometry of M. smegmatis containing 5′ eis promoter deletion constructs.
To identify the region of the eis promoter required for maximal expression in the heterologous host M. smegmatis, 5′ eis promoter deletions were cloned upstream of promoterless gfp in pFPV27 to create the pEP vector series (Fig. 3A). These plasmids were transformed into M. smegmatis 1-2c, and cells harboring these constructs were grown to log phase and diluted for flow cytometry. The negative control carrying pFPV27 displayed minimal fluorescence (Fig. 3B). The heat shock promoter control, pBEN, produced a 120-fold higher level of fluorescence than the negative control, pFPV27 (Fig. 3B). pBEN produced a fourfold higher level of fluorescence than the largest eis promoter construct, pEP922 (Fig. 3B). When the pEP series constructs were compared to each other, there was no significant difference in the levels of fluorescence from pEP922 to pEP200 (Fig. 3B). However, pEP112 showed a twofold-lower level of fluorescence than pEP200. pEP112 conferred more than 14-fold more fluorescence than the negative control, indicating that while it was weaker than pEP200, it was still positive (Fig. 3B). These data indicate that at least 200 bp of the eis promoter are required for maximal eis expression in log-phase M. smegmatis.
Flow cytometry of M. tuberculosis containing 5′ eis promoter deletion constructs.
To determine the region of the eis promoter required for maximal expression in M. tuberculosis, M. tuberculosis H37Ra was transformed with the pEP vector series, grown to log phase, and diluted for flow cytometry. The negative control, pFPV27, showed minimal fluorescence, and the positive control, pBEN, produced a fluorescence signal 215-fold higher than that of the negative control, indicating that the heat shock promoter functions at a nearly twofold-higher level in M. tuberculosis H37Ra than in M. smegmatis (Fig. 3B and C). This difference could be due to variability in plasmid copy number between the two species. In M. tuberculosis H37Ra, the positive control produced fivefold more fluorescence than the largest eis promoter construct, pEP922 (Fig. 3C). There was no significant difference in the level of fluorescence from pEP922 to pEP412 (Fig. 3C). Unlike the case with M. smegmatis, a decrease in expression was observed when the promoter region was shortened to 300 bp. Compared to pEP412, pEP300 and pEP200 displayed approximately twofold less fluorescence. In addition, pEP112 was fourfold less fluorescent than pEP412 (Fig. 3C). These data suggest that the 412-bp region of the eis promoter is required for maximal expression of eis in log-phase M. tuberculosis H37Ra. The data also indicate that different cis elements may be involved in eis expression between log-phase M. smegmatis and M. tuberculosis H37Ra.
Quantitative analysis of random eis promoter mutants in M. smegmatis and M. tuberculosis.
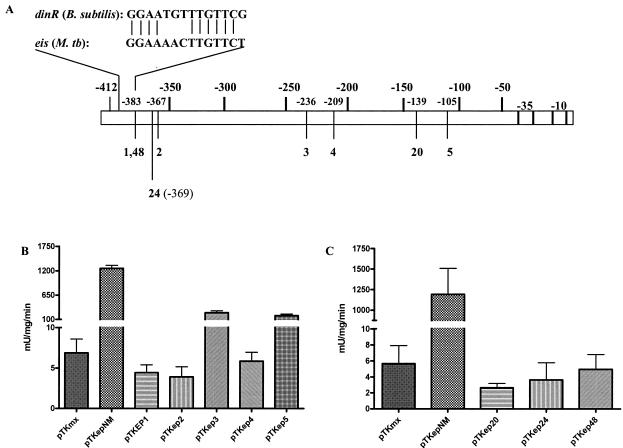
To identify regions required for eis transcriptional activity, we analyzed plasmid clones containing random mutations in the 412-bp eis promoter that were isolated from xylE reporter screens in M. smegmatis and M. tuberculosis H37Ra. For each species, 10,000 independent clones were screened for diminished CDO activity based on yellow/white screening. For M. smegmatis, 48 clones were isolated as PCR positive for promoter inserts. Of these clones, only five (pTKep1, -2, -3, -4, and -5) contained single nucleotide changes in the 412-bp eis promoter (Fig. 4A). pTKep1 and -2 were separated by only 16 bp. pTKep1 contained a T→C mutation at −383. pTKep2 was located at −367 and had mutated from C to T. pTKep3 and 4 were within 27 bp of one another. pTKep3 contained an A→G substitution at −236, while pTKep4 contained a T→C substitution at −209. pTKep5 contained an A→G substitution at −105. These clones were assayed for quantitative CDO activity in M. smegmatis. pTKep1, pTKep2, and pTKep4 displayed CDO activity comparable to that of the negative control, pTKmx, indicating the complete loss of eis promoter activity (Fig. 4B). pTKep3 and pTKep5 showed a 5- and 6.7-fold (P < 0.05) decrease of transcriptional activity, respectively (Fig. 4B).
FIG. 4.
CDO assay of random eis promoter mutants in mycobacteria. (A) Map of pTKep mutants isolated in qualitative CDO screening in relation to the 412-bp eis promoter. An alignment between the B. subtilis dinR element and the putative dinR element of the eis promoter (−397 to −383) is also shown. Quantitative analysis of the CDO activity of pTKep mutants assayed with M. smegmatis (B) and M. tuberculosis H37Ra (C) compared to the nonmutated eis promoter, pTKepNM, and the negative control, pTKmx. All pTKep constructs were significantly different from pTKepNM in both M. smegmatis and M. tuberculosis H37Ra using paired t tests at P values of <0.05.
For M. tuberculosis H37Ra, only 14 clones were PCR positive for promoter inserts, and of those only three (pTKep20, -24, and -48) contained single-nucleotide changes in the eis promoter. All three mutants displayed CDO activity at or below levels for the negative control, pTKmx (Fig. 4C). The mutation in pTKep20, a T→C substitution at −139, was 34 bp from the mutation in pTKep5 found in the M. smegmatis screen. However, unlike the case with pTKep5 in M. smegmatis, this mutation caused complete loss of CDO activity. The mutation in pTKep24, a T→G transversion at −369, was only 2 bp upstream from the mutation in pTKep2 and caused a complete loss of CDO activity similar to that with pTKep2. The single-nucleotide change in pTKep48 was identical to that in pTKep1 of the M. smegmatis assay. In addition, pTKep1 and pTKep48 map to a region that contains a putative site for DinR (Fig. 4A), a negative regulator of DNA-damage inducible genes found in both gram-negative and gram-positive organisms (21, 33).
DISCUSSION
The intracellular survival of M. tuberculosis within macrophages is central to its success as a human pathogen, yet little is known of the factors that allow its survival within this inhospitable environment. Even less is known about the promoter function of these factors. Very few promoters of mycobacterial genes involved in its pathogenesis have been characterized. These include the promoters of sigE, sigH, oxyR, ahpC, katG, mas, and fadD28 (8, 10, 12, 17, 28, 34). Several other genes have been shown to be involved in the survival of mycobacteria during the infection of macrophages, including eis (32) and the isocitrate lyase gene (11, 30). However, the promoters for these genes have not been characterized.
Primer extension analysis (Fig. 1) determined that the TSP of the eis gene, in both M. smegmatis and M. tuberculosis H37Ra, maps to the same A nucleotide. The level of signal for eis transcript from wild-type M. tuberculosis H37Ra was very low compared to that for mycobacteria containing p69 (Fig. 1, lane 3). This indicated either that low levels of eis expression from the M. tuberculosis H37Ra chromosome occur during logarithmic growth or that the eis mRNA transcript has a short half-life. This made it necessary to use a plasmid-borne copy of eis in M. tuberculosis H37Ra to ensure sufficient transcript yield to identify the TSP. The most striking feature of the eis promoter region is the high similarity to the canonical −35 and −10 sequences for E. coli σ70 promoters.
We used site-directed mutagenesis (Fig. 2) to determine the importance of the putative −35 and −10 regions. Single mutations in the putative −10 region significantly reduced the transcription of gfp in both M. smegmatis and M. tuberculosis H37Ra (Fig. 2). Loss of function upon alteration of −10 regions is common for a variety of bacteria. For example, base substitutions in the −10 region of the rpsL promoter of M. smegmatis caused dramatic defects in transcriptional activity (19). Interestingly, changes in the −35 region of the eis promoter had variable and low-level effects on transcriptional activity. Mutations in the putative −35 region either increased transcriptional activity (pSKM3) or had no effect (pSKM4 in M. smegmatis), with the exception of a slight decrease in fluorescence from pSKM4 in M. tuberculosis H37Ra (Fig. 2). These results suggest that while the −35 region may be involved in eis expression, it is not essential for promoter activity.
We moved the eis promoter away from the E. coli σ70 consensus by incorporating a combination of mutations in pSKM6 (Fig. 2). Surprisingly, fluorescence from pSKM6 was higher than that of its parent, pEP412, in both mycobacterial species. We expected fluorescence to decrease since single mutations caused decreased fluorescence in pSKM1, -2, and -4. However, the −32 T→C mutation in pSKM3 was also incorporated into pSKM6, which showed increased levels of fluorescence in both mycobacteria. It is possible that the single alteration in the −35 region was able to overcome single mutations known to negatively influence eis expression. However, we cannot be certain of this, since multiple mutations in the −10 region in a single construct were not tested.
E. coli promoters generally function very poorly in mycobacteria (3). Alteration of the core promoter of eis toward consensus E. coli σ70 promoter elements (Fig. 2, pSKM7) greatly enhanced eis transcriptional activity in both mycobacterial species. These results show that the eis promoter can be changed into a strong, heat shock-like promoter. Although it would be expected that conversion of promoter elements to consensus elements would cause better promoter recognition and therefore greater activity, this is the first report that alteration of mycobacterial promoter elements to E. coli consensus elements positively influences gene expression in mycobacteria. Our results suggest that it is perhaps the spacing between the −35 and −10 regions in E. coli promoters and not the consensus elements themselves that are critical to the lack of recognition of these promoters in mycobacteria.
To identify upstream regions necessary for the maximal expression of eis, 5′promoter deletions were assayed with mycobacteria using GFP, which has been used widely to study mycobacterial gene expression (2, 6, 9). Flow-cytometric analysis of M. smegmatis harboring the pEP vector series suggested that at least 200 bp of the upstream eis promoter region are necessary for the maximal expression of eis in M. smegmatis(p69). During the initial characterization of eis, it was not noted that there was a decrease in the production of the Eis protein in the deletion derivatives p69-97 and p69-96 compared to the intact clone p69 (32). These derivatives contain a promoter region identical to the pEP112 construct. Although the survival phenotype was retained when using these deletion derivatives in M. smegmatis, the promoter region necessary for the maximum expression of eis was not present. In contrast to M. smegmatis, flow-cytometric analysis of M. tuberculosis H37Ra harboring the pEP vector series indicated that a 412-bp region upstream of eis is required for maximum expression in M. tuberculosis H37Ra. The difference in the region required for maximum expression between the two species may be attributed to the fact that the eis promoter is not in its native host when analyzed in M. smegmatis. The results from M. tuberculosis H37Ra, therefore, present a more relevant analysis of eis promoter activity.
Because the flow cytometry data from the pEP series showed that a 200-bp and 412-bp region of the eis promoter was necessary for maximum expression of eis in M. smegmatis and M. tuberculosis H37Ra, respectively, we were confident that more than just the core promoter was required for activity. Random mutagenesis of the 412-bp eis promoter was employed to delineate other regions involved in transcriptional activity. Interestingly, we did not recover mutants that mapped to the core promoter region of eis, which suggests that mutations in the core promoter did not fully inhibit the production of CDO or that the core promoter is not essential for expression in stationary-phase cells, therefore allowing qualitatively screened colonies to retain a yellow color upon exposure to catechol. With M. smegmatis, five mutants were localized to regions far upstream from the core promoter of eis (Fig. 4A and B). pTKep1, -2, and -4 showed a complete loss of CDO activity, while pTKep3 and -5 displayed significantly reduced levels of CDO activity. With M. tuberculosis H37Ra, only three single-nucleotide mutants (pTKep20, -24, and -48) were recovered, and all showed complete loss of CDO production. The results from the CDO activities strongly suggest that the upstream region is essential for the expression of eis in stationary mycobacteria.
The mutations found in pTKep1 and -48 mapped within a region with high homology to a DinR cis element from Bacillus subtilis (Fig. 4A). The match was 71% identical when the 412-bp eis promoter was used to query the recently updated transcriptional regulatory network database, DBTBS, for B. subtilis (22). DinR is a negative regulator of genes inducible by DNA damage and is the gram-positive equivalent of the LexA SOS repressor in gram-negative bacteria (21). LexA (DinR) is a transcriptional repressor that binds as a dimer to a consensus sequence presenting dyad symmetry commonly known as the SOS box or Cheo box (33). LexA in M. tuberculosis has been shown to bind to a mycobacterial version of the B. subtilis Cheo box (24). It is unlikely that this putative DinR cis element actually represents a site for negative regulation, because our data support the requirement of the upstream region for eis promoter activity in stationary mycobacteria. The data presented here suggest that there is a transcriptional activator binding site in the upstream region of the eis promoter with similarity to the DinR binding site. Studies employing gel retardation analysis to determine if this region is a binding site for an activator or repressor protein will be needed before hypotheses can be formulated on its role in eis expression.
A comparison of the data from the flow-cytometric analysis and the CDO analysis of the 412-bp eis promoter revealed a difference in the region necessary for expression in M. smegmatis. The flow cytometry data show that a 200-bp fragment is required for maximum expression, whereas mutations upstream of −200 cause loss of expression. In addition, if the proposed DinR-like site is important for expression, then its exclusion in the pEP300 construct should have caused a loss in gfp transcription in both mycobacteria. The mycobacteria analyzed for flow cytometry were grown to logarithmic phase, while those analyzed using the CDO activity assay were grown to stationary phase. We believe that the discrepancies in the data are due to the difference in growth phase of the cells and not the region being analyzed. We have preliminary evidence, using real-time PCR, that eis is up-regulated in stationary-phase M. smegmatis carrying an integrated copy of the p69 insert (unpublished results). It is possible that the putative DinR element is required for eis expression in stationary-phase M. smegmatis but not for expression during log phase. This possibility remains to be determined for both M. smegmatis and M. tuberculosis H37Ra. In conclusion, we show that the promoter of the putative virulence gene eis of M. tuberculosis is a group A/SigA-like mycobacterial promoter that contains both a core promoter region and an upstream region required for transcriptional activity. Future studies to identify the putative trans-activating factors involved in the expression of eis will enhance our understanding of how potential virulence genes may be regulated in M. tuberculosis and will perhaps provide clues to the function of Eis during mycobacterial infection.
Acknowledgments
We thank Norma Seaver and Debbie Sakiestewa from the Flow Cytometry Shared Services of the Arizona Cancer Center, University of Arizona, for expert technical assistance with the flow cytometry studies. Special thanks are extended to the laboratory of Stanley Falkow at Stanford University for providing the vectors pFPV27 and pBEN. A special thanks to Gordon Churchward of Emory University is also in order for providing the vector pTKmx. We also thank Janet Hatt, James Moulder, Amy Windley, Linoj Samuel, and Chris Alteri for critical review of the paper.
This work was supported by National Institutes of Health grant AI45537-01A2 to R.L.F., the American Society for Microbiology Robert D. Watkins Minority Fellowship to E.A.R., and funding through the Undergraduate Biology Research Program of the University of Arizona to S.K.M.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2001. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Barker, L. P., S. F. Porcella, R. G. Wyatt, and P. L. Small. 1999. The Mycobacterium marinum G13 promoter is a strong sigma 70-like promoter that is expressed in Escherichia coli and mycobacteria species. FEMS Microbiol. Lett. 175:79-85. [DOI] [PubMed] [Google Scholar]
- 3.Bashyam, M. D., D. Kaushal, S. K. Dasgupta, and A. K. Tyagi. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J. Bacteriol. 178:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cappelli, G., P. Volpe, A. Sanduzzi, A. Sacchi, V. Colizzi, and F. Mariani. 2001. Human macrophage gamma interferon decreases gene expression but not replication of Mycobacterium tuberculosis: analysis of the host-pathogen reciprocal influence on transcription in a comparison of strains H37Rv and CMT97. Infect. Immun. 69:7262-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. (Erratum, 396:190, 1998.) [DOI] [PubMed] [Google Scholar]
- 6.Cowley, S. C., and Y. Av-Gay. 2001. Monitoring promoter activity and protein localization in Mycobacterium spp. using green fluorescent protein. Gene 264:225-231. [DOI] [PubMed] [Google Scholar]
- 7.Dahl, J. L., J. Wei, J. W. Moulder, S. Laal, and R. L. Friedman. 2001. Subcellular localization of the intracellular survival-enhancing Eis protein of Mycobacterium tuberculosis. Infect. Immun. 69:4295-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhandayuthapani, S., M. Mudd, and V. Deretic. 1997. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J. Bacteriol. 179:2401-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhandayuthapani, S., L. E. Via, C. A. Thomas, P. M. Horowitz, D. Deretic, and V. Deretic. 1995. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol. Microbiol. 17:901-912. [DOI] [PubMed] [Google Scholar]
- 10.Dhandayuthapani, S., Y. Zhang, M. H. Mudd, and V. Deretic. 1996. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J. Bacteriol. 178:3641-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubnau, E., P. Fontan, R. Manganelli, S. Soares-Appel, and I. Smith. 2002. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect. Immun. 70:2787-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes, N. D., Q. L. Wu, D. Kong, X. Puyang, S. Garg, and R. N. Husson. 1999. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181:4266-4274. (Erratum, 181:6222, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez, M., and I. Smith. 2000. Determinants of mycobacterial gene expression, p. 111-129. In G. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 16.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen-Cain, D. M., and F. D. Quinn. 2001. Differential expression of sigE by Mycobacterium tuberculosis during intracellular growth. Microb. Pathog. 30:271-278. [DOI] [PubMed] [Google Scholar]
- 18.Kaufmann, S. H. 2002. Protection against tuberculosis: cytokines, T cells, and macrophages. Ann. Rheum. Dis. 61(Suppl. 2):ii54-ii58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenney, T. J., and G. Churchward. 1996. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J. Bacteriol. 178:3564-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung, D. W., E. Chen, and D. V. Goeddel. 1989. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11-15. [Google Scholar]
- 21.Makarova, K. S., A. A. Mironov, and M. S. Gelfand. March 2001, posting date. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2:RESEARCH0013. http://genomebiology.com/2001/2/4/research/0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makita, Y., M. Nakao, N. Ogasawara, and K. Nakai. 2004. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 32:D75-D77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAdam, R. A., S. Quan, D. A. Smith, S. Bardarov, J. C. Betts, F. C. Cook, E. U. Hooker, A. P. Lewis, P. Woollard, M. J. Everett, P. T. Lukey, G. J. Bancroft, W. R. Jacobs Jr., and K. Duncan. 2002. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 148:2975-2986. [DOI] [PubMed] [Google Scholar]
- 24.Movahedzadeh, F., M. J. Colston, and E. O. Davis. 1997. Characterization of Mycobacterium tuberculosis LexA: recognition of a Cheo (Bacillus-type SOS) box. Microbiology 143:929-936. [DOI] [PubMed] [Google Scholar]
- 25.Nozaki, M. 1970. Metapyrocatechase (Pseudomonas). Methods Enzymol. 17A:522-525. [Google Scholar]
- 26.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saviola, B., S. C. Woolwine, and W. R. Bishai. 2003. Isolation of acid-inducible genes of Mycobacterium tuberculosis with the use of recombinase-based in vivo expression technology. Infect. Immun. 71:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sirakova, T. D., A. M. Fitzmaurice, and P. Kolattukudy. 2002. Regulation of expression of mas and fadD28, two genes involved in production of dimycocerosyl phthiocerol, a virulence factor of Mycobacterium tuberculosis. J. Bacteriol. 184:6796-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stolt, P., Q. Zhang, and S. Ehlers. 1999. Identification of promoter elements in mycobacteria: mutational analysis of a highly symmetric dual promoter directing the expression of replication genes of the Mycobacterium plasmid pAL5000. Nucleic Acids Res. 27:396-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Triccas, J. A., W. J. Britton, and B. Gicquel. 2001. Isolation of strong expression signals of Mycobacterium tuberculosis. Microbiology 147:1253-1258. [DOI] [PubMed] [Google Scholar]
- 31.Wards, B. J., and D. M. Collins. 1996. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol. Lett. 145:101-105. [DOI] [PubMed] [Google Scholar]
- 32.Wei, J., J. L. Dahl, J. W. Moulder, E. A. Roberts, P. O'Gaora, D. B. Young, and R. L. Friedman. 2000. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 182:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winterling, K. W., A. S. Levine, R. E. Yasbin, and R. Woodgate. 1997. Characterization of DinR, the Bacillus subtilis SOS repressor. J. Bacteriol. 179:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, Q. L., D. Kong, K. Lam, and R. N. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Y., R. Lathigra, T. Garbe, D. Catty, and D. Young. 1991. Genetic analysis of superoxide dismutase, the 23 kilodalton antigen of Mycobacterium tuberculosis. Mol. Microbiol. 5:381-391. [DOI] [PubMed] [Google Scholar]