Abstract
Introduction/Objective:
Pulmonary function tests (PFTs) are objective estimates of lung function, but are not reliably stored within the Veteran Health Affairs data systems as structured data. The aim of this study was to validate the natural language processing (NLP) tool we developed—which extracts spirometric values and responses to bronchodilator administration—against expert review, and to estimate the number of additional spirometric tests identified beyond the structured data.
Methods:
All patients at seven Veteran Affairs Medical Centers with a diagnostic code for asthma Jan 1, 2006–Dec 31, 2012 were included. Evidence of spirometry with a bronchodilator challenge (BDC) was extracted from structured data as well as clinical documents. NLP’s performance was compared against a human reference standard using a random sample of 1,001 documents.
Results:
In the validation set NLP demonstrated a precision of 98.9 percent (95 percent confidence intervals (CI): 93.9 percent, 99.7 percent), recall of 97.8 percent (95 percent CI: 92.2 percent, 99.7 percent), and an F-measure of 98.3 percent for the forced vital capacity pre- and post pairs and precision of 100 percent (95 percent CI: 96.6 percent, 100 percent), recall of 100 percent (95 percent CI: 96.6 percent, 100 percent), and an F-measure of 100 percent for the forced expiratory volume in one second pre- and post pairs for bronchodilator administration. Application of the NLP increased the proportion identified with complete bronchodilator challenge by 25 percent.
Discussion/Conclusion:
This technology can improve identification of PFTs for epidemiologic research. Caution must be taken in assuming that a single domain of clinical data can completely capture the scope of a disease, treatment, or clinical test.
Keywords: asthma, bronchodilator challenge, natural language processing, pulmonary function
Keywords: Data Use and Quality, Outcomes Assessment, Informatics, Data Reuse, Electronic Health Record (EHR), Natural Language Processing, Pulmonary Disease
Introduction
As the 10th leading cause of disability in the United States, asthma is a significant health problem.1 Pulmonary function testing (PFT) provides objective, quantifiable measurements of lung function and is a cornerstone of diagnosis and monitoring. Spirometry, the most commonly used test, is the measurement of the movement of air into and out of the lungs during breathing. It includes the measured forced expiratory volume in one second (FEV1), forced vital capacity (FVC), and bronchodilator responsiveness (change in FEV1 or FVC after administration of an inhaled bronchodilator).2
The interpretation of lung function tests involves two tasks: (1) the classification of the measured FEV1, FVC, and bronchodilator response with respect to a reference population based on the patient’s age, height and gender; and (2) the integration of the obtained values into the diagnosis.3 While the second task requires the interpretation of the measured values within the context of clinical presentation and patient care, the first task relies solely on the synthesis of data obtained by laboratory spirometry testing.
In order to identify a population of asthmatic patients with bronchodilator responsiveness, we aimed to identify spirometric values and bronchodilator response, referred to as the “Bronchodilator Challenge” (BDC). The Veteran Health Affairs (VHA) collects PFT data directly from spirometry equipment that is electronically connected to Veterans Health Information Systems and Technology Architecture (VistA). Nevertheless, preliminary analysis found approximately three times more procedure codes for BDCs than computer-generated BDC reports in the structured PFT data domain, suggesting that a large number of values are missing from the structured data. In searching the medical notes from the “back end” using the Corporate Data Warehouse (CDW), we found many cases where the computer-generated report appeared to be electronically copied and pasted into the clinicians’ medical note. Physician interpretation of the BDC study was also found in medical notes. Patients without computer-generated spirometry were also found to have Portable Document Format (PDF) and image files uploaded into the computerized patient record system—meaning the provider printed the report, then scanned and attached it to the patient’s medical record. This type of scanned file is not available in the CDW and cannot easily be used for research.
We developed a natural language processing (NLP) tool to extract FEV1 and FVC pre- and post BDC from medical notes in order to complement data obtained from the electronically captured BDC reports, to more completely assess bronchodilator response in our asthma population from the Rocky Mountain Network of the Veterans Affairs Health (VHA) System. The goal of this study was to validate the NLP against expert review for accuracy in identifying spirometric values and bronchodilator responsiveness, and to estimate the number of additional values identified with NLP plus structured data compared to the structured data alone.
Methods
Settings and Ethics
The study was conducted in the VHA Care System using data from the Rocky Mountain Network, a referral center for a large patient base of veterans for Utah, Colorado, Wyoming, and Montana. Seven VA Medical Centers in the region provide pulmonary function testing services. Documents were extracted from the Veterans Informatics, Information, and Computing Infrastructure, which provides centralized access to the VA CDW in a secure computing environment.4 This study was approved by the University of Utah Institutional Review Board (IRB#00062528) and the VA Salt Lake City Health Care System Research and Development Committee.
Study Population
We identified all patients between January 1, 2006 and December 31, 2012 who contained International Classification of Disease-Edition 9 (ICD-9) codes for asthma (493.xx) and excluded all patients with a concomitant diagnosis of chronic obstructive pulmonary disease (491.2, 493.2, 496, 506.4), emphysema (492.x), cystic fibrosis (277.0x), and bronchiectasis (494.xx).
Measurement
Identification of Bronchodilator Responsiveness
Evidence of spirometry with a BDC was extracted from structured data as well as clinical documents in two ways: presence of BDC in structured PFT tables, and presence of a Current Procedural Terminology (CPT) code 94060 during the study period. We linked patients’ clinical notes to visits that generated the CPT code for BDC and allowed for a 10 day window on each side of the visit date that generated the CPT BDC code. Response to the BDC was identified if a computer-generated BDC result was available or if NLP was able to extract spirometry data or provider interpretation from the medical notes. BDC response was defined by Equation 1 below. All complete BDC tests were classified into three clinically relevant categories in accordance with standard spirometry interpretation:5 change in FEV1 or FVC of 200mL, plus the following:
No significant response <12 percent improvement;
Significant response 12–19 percent improvement; or
Highly significant response ≥ 20 percent improvement.
Personalized medicine, a recent paradigm shift in health care, would suggest that asthma patients may respond to treatment differently, depending upon phenotype. A recent randomized, placebo-controlled study examining Brodalumab, a human anti-IL 17 receptor monoclonal antibody, reported a suggestive treatment effect only among study participants with asthma classified as having high bronchodilator reversibility.6 High bronchodilator reversibility may serve as a clinical phenotype or marker for people suffering from severe asthma that is often uncontrolled. Thus, this literature, coupled with the recent paradigm shift in health care, encouraged us to utilize the three analytic categories that are clinically interpretable in order to differentiate between potential asthma population phenotypes.
Identification of Note Patterns for Natural Language Processing (NLP)
The NLP algorithms were developed to identify three primary note patterns to extract spirometry information from, which included extraction of spirometry results from semistructured (tables) notes, extraction of spirometry results documented in unstructured narrative, and extraction of physician interpretation from narrative data.
Semistructured notes contained tables that appeared to be a copy and paste from the computer-generated spirometry report.
Unstructured narrative of spirometry results included actual FEV1 and FVC values documented by the provider.
Physician Interpretation means that the provider made a qualitative assessment based on patient response. For example, “Following the inhalation of the bronchodilator, there is no significant improvement in the spirometric numbers.”
| Equation 1. Percent Change in FEV1 and FVC |
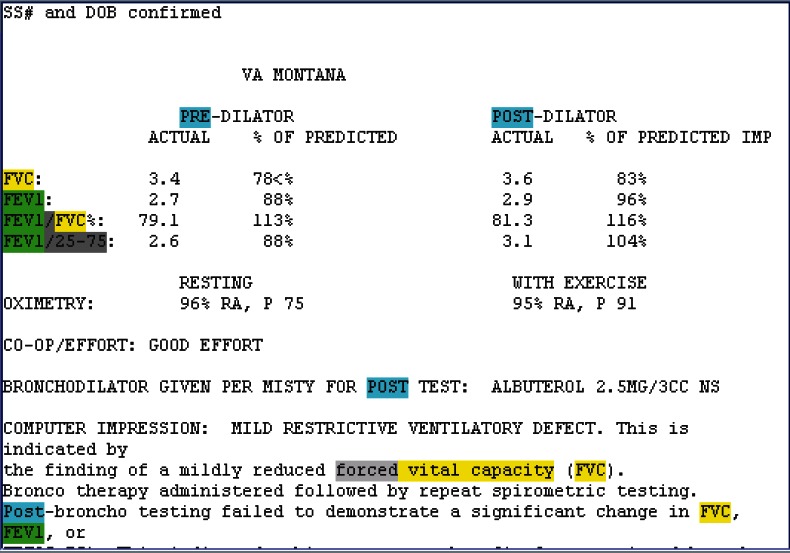
Semistructured Notes (Figure 1)
Figure 1.
Semistructured Pulmonary Function Test (PFT) Note
To determine the percent of FVC or FEV1 change, actual FVC and FEV1 values need to be identified and extracted for both pre- and postbronchodilator measurements. The JAVA software program we developed was used to generate regular expressions to identify and extract FEV1 and FVC values. Experts reviewed notes and collected patterns to build extraction rules. Extraction rules were composed of regular expressions that describe patterns and that tell the program when to capture information and how to classify that information, described in detail in Appendix A.
Unstructured Notes
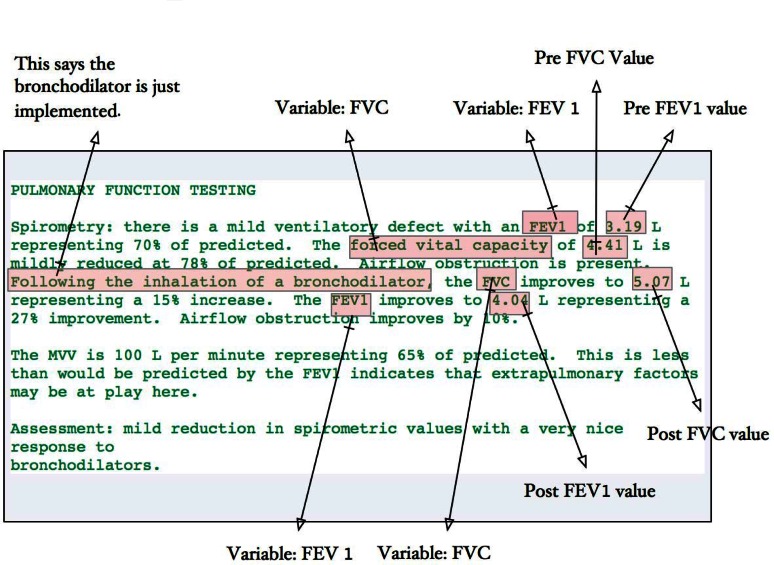
Clinicians also document spirometry findings during their clinical assessment, including pasted semistructured spirometry notes and free-text formats. Extracting spirometry data from unstructured text is more complex than extracting it from semistructured notes because the variability of unstructured text is infinitely greater than the variability of semistructured notes and unstructured text does not have a fixed pattern to describe each variable. The most difficult problem is correct assignment of study findings to pre and post statuses. The NLP required heavy use of verb and span position to differentiate the pre- and post variables. Figure 2 demonstrates the use of verb or span position to discriminate the pre- and post variables in addition to phrases that indicate a change, such as “improves to” or “reduced to.”
Figure 2.
Example of Unstructured Note with Description of Spirometry Findings
Physician Interpretation
The primary goal of our NLP program was to extract the actual FEV1 and FVC values required to compute percent change in FEV1. Even though physician interpretation was not quantifiable, it was deemed an important source of information since some facilities did not appear to have the ability to paste computer-generated reports directly into their clinical notes. For example, the clinician may have written the following: “Following the inhalation of the bronchodilator, there is no significant improvement in the spirometric numbers.”
This interpretation provides evidence that the change in FEV1 is likely less than 12 percent as it mentioned that there is no significant improvement on the FEV1 after inhaling the bronchodilator. The working assumption is that clinicians use the guidelines of 12 percent change in FEV1 to indicate a significant change. We thus developed an approach to capture clinical interpretation of the BDC to classify study results into significant versus nonsignificant results when actual values were not reported. Appendix B provides example text representing physician interpretation.
NLP Software and Training Procedures
Patients meeting inclusion criteria in Veterans Integrated Service Networks (VISN) 19 with a BDC identified by CPT code 94060 were eligible for NLP processing. NLP was limited to patients with CPT for BDC, since this would be a reasonable strategy for an epidemiological study and limiting to people with known tests would reduce false positives from other types of spirometry measures. Clinical notes generated 10 days before the visit date for BDC identified by CPT and 10 days after were included to reduce false negative findings. This process identified 12,156 notes with 2,468 having mention of FEV. Four hundred of the 2,468 notes were used to collect patterns and develop the NLP extraction procedures. After development, 1,001 notes were used to evaluate the software performance (the goal was to use 1,000 notes for validation but our counter started at 0).
The software customized for this project integrates NLP and performance evaluation into a single JAVA-based standalone program running under the Veterans Informatics, Information, and Computing Infrastructure research environment.7 A simple rule-based system was built to extract and standardize PFT results and interpretations. In the training stage, experts reviewed notes and collected patterns to design extraction rules using regular expression.8 These extraction rules are specific to terms and patterns identified in the note structure, and they operationalize knowledge about the relationships between concepts (FEV1) and values within the data. The current literature adequately supports utilizing regular expressions for NLP extraction.9–10 Several studies have utilized VHA data sources and followed an approach similar to ours in extracting information from unstructured data with adequate performance. For example, a similar NLP system was able to effectively extract left ventricular ejection fraction from free-text echocardiogram reports, which is a vital component to measure in ensuring that those at risk for heart failure are being cared for adequately.11 F-Measures at both the concept and document levels were >90 percent. Furthermore, an additional hybrid regular expression and NLP solution was utilized in processing blood culture microbiology reports.12 Sensitivity and positive predictive values represented superior NLP performance, further emphasizing the importance of utilizing regular expressions and NLP solutions within the VHA for health and surveillance outcomes research.
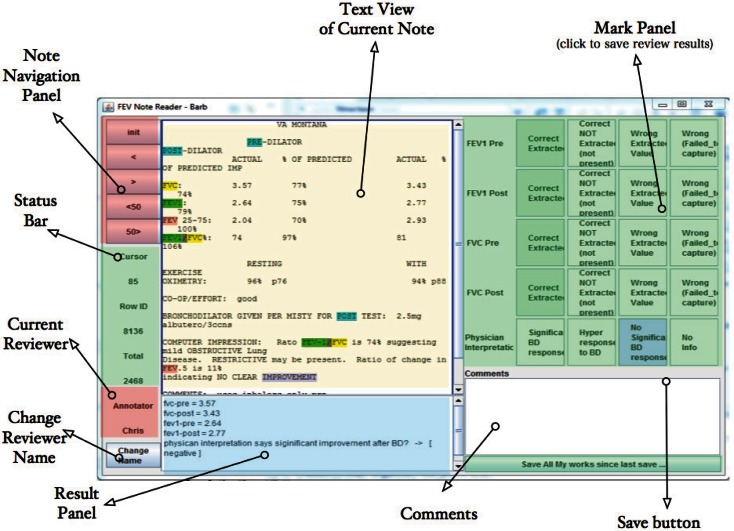
Our NLP extraction system runs in the background and lists FEV1 NLP results in the result area at the bottom of the screen for each note (Figure 3). An expert programmer reviewed 400 notes from VISN 19 with a pulmonologist. During this review process the programmer collected patterns and composed extraction rules for each pattern. The 400 notes were intentionally selected (sampled by facility) to represent variability among stations and facilities. The NLP algorithms were revised until the system could correctly extract FEV1 and FVC information from at least 95 percent of the 400 notes. This confirmation-based approach worked well for our extraction task because there was little clinical judgment required for assessing the accuracy of the concept extraction and we targeted notes based on CPT tests for BDCs. This approach, however, may not be ideal for more complex annotation and extraction tasks where information may be found in a variety of locations within the medical note.
Figure 3.
User Interface for NLP and Evaluation Software with Labeled Features
NLP Review Tool
The software developed for this project has two main panels: the left panel provided the text and extraction results, and the right panel provided the reviewer decision buttons. The lower left text box displays the data extracted and standardized from each note, such as “fvc-pre = 3.4, fvc-post = 3.6, fev1-pre = 2.7, fev1-post=2.9” and physician interpretation.
The upper text panel lists the text content from the clinical note being reviewed. Keywords and concepts are highlighted to show how the computer annotated and standardized content. Buttons on the far left were designed for navigation. They allow the reviewer and adjudicator to advance through different notes or batches of notes. Buttons on the right side were designed to capture reviewer judgment regarding NLP performance for each note. This software supports rapid confirmation of NLP extraction and quick identification of problems when the text extraction failed to capture the correct information (Figure 3).
NLP Evaluation
After the training phase was complete, the NLP software was run on all remaining medical notes for the study population. One reviewer was trained by the pulmonologist (Jones) to use the evaluation software to confirm that the NLP correctly extracted PFT data. A checklist was provided along with a brief help manual to improve consistency during evaluation. Each document was reviewed by the reviewers and pulmonologist, and was adjudicated by the two reviewers (the reviewers discussed and came to a consensus) when discrepancies were found.
Reviewers were given 10 batches of 100 notes each. After adjudication, each batch was sent to the statistical team to generate a report on the accuracy of the NLP software for that batch and all batches up to that point. Specifically, precision, recall, f-measure, overall accuracy, and 95 percent CI were computed. The evaluation stop rule was very conservative. The criterion was to stop evaluation if overall accuracy was at least 95 percent after reviewing 1,000 notes.
Statistical Analysis
Using the adjudicated human review results as the reference standard, NLP performance was evaluated using Accuracy, Recall, Precision, and F-measure13 for the each concept separately—i.e., FVC-pre, FVC-post, FEV1-pre, FEV1-post—and bronchodilator responsiveness among completed BDCs (paired FEV1 or FVC) and physician interpretations. The contingency Table 1 is a traditional way to report performance of information extraction software. Precision is the fraction of extracted instances that are correct (i.e., positive predictive value). Recall is the fraction of instances observed in the reference standard that are extracted by the software (sensitivity). The F-measure is the harmonic mean of the combined precision and recall. Accuracy is the proportion of extracted instances that are correctly classified among all possible classifications from the reference standard. Equation 2 contains the formulas used to compute the information extraction performance measures discussed.
Table 1.
Contingency Table Showing the Relationship Between Extraction Software and the Reference Standard
| FEV NLP PERFORMANCE | ADJUDICATED HUMAN REVIEWER RESULTS AS REFERENCE STANDARD | ||
|---|---|---|---|
| FEV EXTRACTION | NO FEV EXTRACTION | ||
| NLP | FEV Extraction | TP True Positive | FP False Positive |
| No FEV Extraction | FN False negative | TN True negative | |
Results
Performance of the NLP Software Against Human Review
Among 1,001 documents reviewed by clinical reviewers, adjudication was required for 40 notes. Table 2 describes the performance of the NLP tool to extract FVC and FEV1 values for pre- and postbronchodilator tests separately. Table 3 presents the overall performance for complete BDC information, i.e., complete pairs were identified for FEV1 and FVC tests. Table 4 presents the performance of the provider interpretation of BDC spirometry.
Table 2.
Performance of NLP to Extract FEV and FVC Values
| FVC-PRE | FVC-POST | FEV1-PRE | FEV1-POST | |
|---|---|---|---|---|
| True Positive | 153 | 88 | 154 | 105 |
| True Negative | 838 | 910 | 847 | 896 |
| False Positive | 0 | 1 | 0 | 0 |
| False Negative | 0 | 2 | 0 | 0 |
| Accuracy | 100% [99.63%, 100%] | 99.70% [99.13%, 99.94%] | 100% [98.48%, 100%] | 100% [99.63%, 100%] |
| Precision | 100% [97.62%, 100%] | 98.88% [93.90%, 99.70%] | 100% [97.63%, 100%] | 100% [96.55%, 100%] |
| Recall | 100% [97.62%, 100%] | 97.78% [92.20%, 99.73%] | 100% [97.63%, 100%] | 100% [96.55%, 100%] |
| F-measure | 100% | 98.32% | 100% | 100% |
Table 3.
Performance of NLP Extracted FEV and FVC for Pre- and Post Pairs
| FVC-PRE AND POST PAIR | FEV1-PRE AND POST PAIR | |
|---|---|---|
| True Positive | 88 | 105 |
| True Negative | 910 | 896 |
| False Positive | 1 | 0 |
| False Negative | 2 | 0 |
| Accuracy | 99.70% [99.13%, 99.94%] | 100% [99.63%, 100%] |
| Precision | 98.88% [93.90%, 99.70%] | 100% [96.55%, 100%] |
| Recall | 97.78% [92.20%, 99.73%] | 100% [96.55%, 100%] |
| F-measure | 98.32% | 100% |
Table 4.
Performance of Physician Interpretation of BDC Results
| SIGNIFICANT RESPONSE | NONSIGNIFICANT RESPONSE | |
|---|---|---|
| True Positive | 146 | 232 |
| True Negative | 600 | 600 |
| False Positive | 13 | 13 |
| False Negative | 4 | 6 |
| Accuracy | 97.77% [96.46%, 98.70%] | 97.77% [96.54%, 98.65%] |
| Precision | 91.82% [86.41%, 95.57%] | 94.69% [91.10%, 97.14%] |
| Recall | 97.33% [93.31%, 99.27%] | 97.48% [94.59%, 99.07%] |
| F-measure | 94.50% | 96.07% |
| Equation 2. Measures of NLP Performance |
Impact of NLP on Measurement of BDC in the Asthma Population
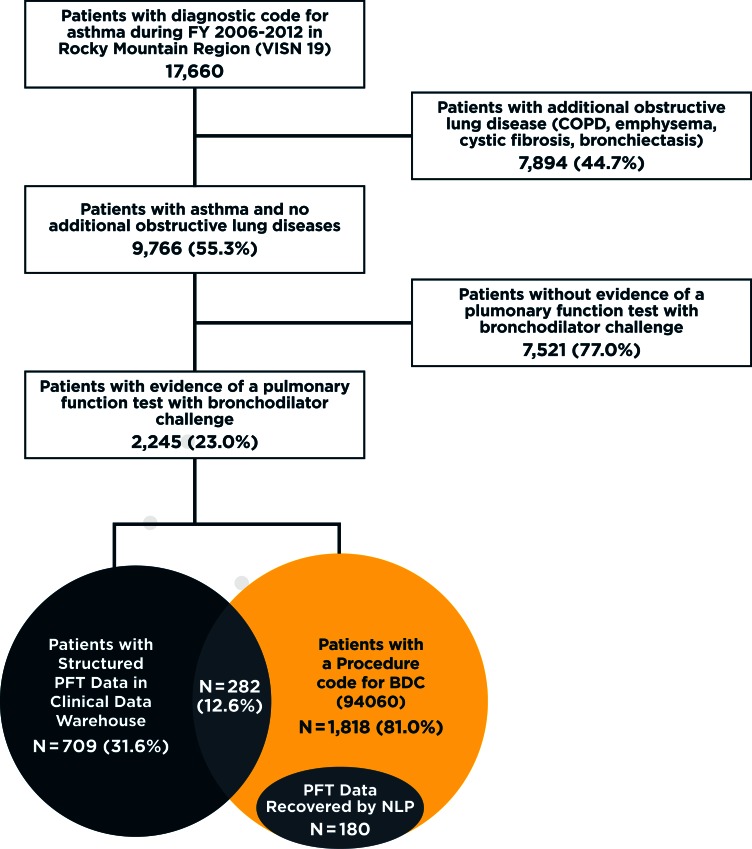
The attrition figure describing the asthma population, evidence of BDC tests, and proportion with measurement of BDC response is provided in Figure 4. Among 9,766 patients in the study population we identified, a CPT code for BDC or a computer-generated PFT report for a BDC test were observed in 23 percent (2,245 of the 9,766). Of these patients, 68.42 percent (n=1,536) had evidence of a CPT code only, 19 percent (n=427) had structured PFT data only, and 12.6 percent (n=282) had evidence of both structured PFT BDC data and a CPT code for a BDC.
Figure 4.
Attrition Figure for Asthma Population
Note: The total number of patients with complete BDC data increased from 709 to 889 after implementation of NLP on clinical notes—a 25% increase in the total number of patients with complete BDC data. The large number of patients with procedure codes for BDC but no available structured data or NLP extracted data indicates other techniques are needed to identify and extract BDC from the data warehouse. Chart review indicated most missing BDCs are scanned into the medical notes as image files, which are currently unavailable in the data wharehouse established for operational activities and research.
Among the 1,818 patients with a CPT code for BDC, we retrieved 2,464 notes containing the FEV1 search term. Applying the NLP resulted in 180 additional patients in our population, which was a 25.38 percent increase (709 to 889) in the number of patients in our cohort with a measure of reversibility. Nevertheless, the fraction of veterans in the cohort with complete BDCs who had evidence of receiving only a BDC increased from 31.6 percent to 39.6 percent. Among the 699 asthma patients in VISN 19 with complete PFT BDC results, there are only 272 (38.91 percent) patients who also had a CPT code for a BDC (CPT:94060).
NLP Impact on Classification of Reversibility in VISN 19 Asthma Population
Table 5 presents the highest level of reversibility for all asthma patients meeting inclusion criteria in VISN 19 when using only computer-generated BDC reports. Among the known reversibility group at the index date, 0.56 percent of the population had high reversibility, 1.22 percent of the population had significant reversibility, and 5.38 percent of the population had a nonsignificant reversibility response. The reversibility group characterized as unknown, with CPT BDC only, was 15.83 percent of the population, while those with no evidence of BDC studies were an estimated 77.01 percent of the population.
Table 5.
Distribution of BDC Reversibility in VISN 19 PFTs
| REVERSIBILITY GROUP AT INDEX DATE | REVERSIBILITY CRITERIA | NO VOL. CRITERIA | WITH VOL. CRITERIA | % POPULATION |
|---|---|---|---|---|
| Known | High reversibility | 55 | 55 | 0.56% |
| Significant reversibility | 124 | 119 | 1.22% | |
| Nonsignificant | 525 | 525 | 5.38% | |
| Unknown | CPT BDC only | 1,546 | – | 15.83% |
| No evidence of BDC studies | 7,521 | – | 77.01% |
Table 6 incorporates NLP findings into the classification of reversibility. Inclusion of NLP data increased the total number of people available for reversibility review from 699 to 879, thus increasing our ability to classify level of reversibility by 25.75 percent. The number of people classified as highly reversible doubled from 55 to 106 (1.09 percent of population). Asthma patients classified with significant reversibility rose from 119 to 158 (1.62 percent of population), and the number of people classified as having no significant response increased from 525 to 615 (6.30 percent of population).
Table 6.
Distribution of Reversibility in VISN 19 Based on Computer-Generated PFTs for BDC and NLP Results
| REVERSIBILITY GROUP AT INDEX DATE | REVERSIBILITY CRITERIA | NO VOL. CRITERIA | WITH VOL. CRITERIA | % POPULATION |
|---|---|---|---|---|
| Known | Highly | 106 | 106 | 1.09% |
| Significant | 164 | 158 | 1.62% | |
| Nonsignificant | 615 | 615 | 6.30% | |
| Unknown | CPT BDC Only | 1,366 | – | 13.99% |
| No evidence of BDC studies | 7,521 | – | 77.01% |
Discussion
Our study demonstrates that our natural language processing tool for PFT data was accurate in extracting spirometry values and BDC results from clinical documents. The NLP increased the ability to classify patients’ reversibility status by 25 percent in our study population, thus improving characterization of our asthma population for epidemiological evaluation. The NLP software extracted information from both semistructured and unstructured documents to extract FEV1 and FVC values before and after administration of a bronchodilator.
The overall performance of the NLP software, especially for FEV1 pre- and post pairs, appears to be better than that of other studies of NLPs designed to extract other clinical data, possibly due to the relatively small amount of variability in semistructured spirometry notes and unstructured notes within our study population and our document selection criteria.14–16 For example, a similarly designed study utilized NLP to identify patients in the VHA with systemic sclerosis that were on prednisone, which can potentially lead to scleroderma renal crisis.14 Overall, their NLP performed substantially less than what we were able to achieve (precision 81 percent, recall 97 percent, F-measure 87 percent). Furthermore, we recently developed NLP software in support of identifying outpatient infusions in the VHA; similarly, precision and recall were not as high as what we have observed in abstracting PFT reports.15
Even with the addition of the NLP, there nevertheless remained a large number of patients with codes for a BDC that remained uncharacterized. Notes were retrieved based on evidence of CPT codes for BDC, which is a reasonable strategy to reduce the NLP processing time. However, broadening our clinical document selection criteria by retrieving documents with terms such as FEV regardless of evidence of CPT codes indicating BDC may increase the yield of spirometric values identified, at the cost of some reduced accuracy. As our software is scaled up to the national VA or other data systems, future work will be needed to increase the ability of our NLP to capture more information while maintaining accuracy on a large scale.
Veteran patients with evidence of a BDC from CPT codes who did not have a computer-generated report or NLP evidence of a BDC were reviewed using the VA’s Computerized Patient Record System. An ad hoc review found many of these patients had a PDF image of their BDC in their medical record. These images are readily available for clinical care but the CDW has not built a system that supports easy retrieval of image files from VistA imaging systems.
Currently, the only way to access these files is to identify patients of interest and use the Compensation and Pension Records Interchange or VistAWeb, which is accessible through the computerized patient record system, to review each person.4 Nevertheless, there appears to be interest within the VA to make VistA image files available for research, but the system currently lacks a standard note title—making it difficult to retrieve specific information from the system. For example, there is no way to request all PFTs that are available as scanned PDF files from VistA imaging systems. The only way to retrieve scanned PFT and BDC images would be to identify CPT codes for BDCs, then attempt to link VistA imaging using visit dates or visit identifiers. Unfortunately, a review of CPT codes for BDCs in patients with computer-generated BDC reports suggests that this approach would not be very sensitive.
The current NLP system could be applied to PDF image files, but it would first require integration of optical character recognition software to electronically convert text from the image to machine-encoded text. Future efforts in the VA will focus on obtaining and processing scanned image files to develop a more complete assessment of lung function in patients with asthma.
An analysis of the national-level PFT table found 39 facilities out of 152 with evidence of computer-generated BDCs. Eleven stations had computer-generated BDCs for 95 percent or more of the CPT codes for BDC. Epidemiological studies evaluating the relationship between medication exposure and changes in PFT over time may be biased if a station is also associated with medication selection. VHA researchers should be careful using computer-generated BDCs for epidemiological evaluation until a more complete capture of PFTs is achieved in the CDW.
Conclusion
In this study we had access to structured laboratory spirometry data but soon discovered it to be incomplete. In order to address this, we developed an NLP program that extracted FEV findings in medical notes. However, this improved our overall capture to only 39.6 percent of all identified CPT codes for BDCs in our population. Complete capture of BDCs would require access to VistA image, but this is not viably accessible at this point for database research. From our investigation, we can conclude that several complexities exist when working with clinical data found within large health enterprise systems. How the data are captured and subsequently stored can dramatically influence the completeness and accuracy of epidemiologic findings. Research investigators need to be cautious in assuming a single domain of clinical data can completely capture the scope of a disease, treatment, or clinical test.
Acknowledgments
This work was supported by Amgen Inc. Gary Globe is an employee and stockholder of Amgen Inc. Brian C. Sauer and Patrick Sullivan are recipients of research grants from Amgen Inc. We have no other conflicts of interest to disclose. The authors would like to thank Saundra Duffy-Hawkins for her project management and administrative support, and Dr. Zach Burningham for his editorial support. We also would like to thank the data architects for the Corporate Data Warehouse at the Veterans Health Administration for providing technical support and database expertise.
Abbreviations
- BDC
Bronchodilator Challenge
- CDW
Corporate Data Warehouse
- CPT
Current Procedural Terminology
- FEV1
Forced Expiratory Volume in 1 second
- FVC
Forced Vital Capacity
- NLP
Natural Language Processing
- PFT
Pulmonary Function Test
- VA
Veteran Affairs
- VISN
Veterans Integrated Service Networks
- VistA
Veterans Health Information Systems and Technology Architecture
Appendix A.
Table A1. Rules and Regular Expressions to Extract FEV1 and FVCs from Semi-structured Notes
Appendix B.
Table B1. Physician Interpretation Examples
References
- 1.Murray CJL. The State of US Health, 1990–2010. JAMA. 2013;310(6):591–18. doi: 10.1001/jama.2013.13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crapo RO. Pulmonary-function testing. N Engl J Med. 1994;331(1):25–30. doi: 10.1056/NEJM199407073310107. [DOI] [PubMed] [Google Scholar]
- 3.Miller MR, Crapo R, Hankinson J, et al. General considerations for lung function testing. European Respiratory Journal. 2005;26(1):153–161. doi: 10.1183/09031936.05.00034505. [DOI] [PubMed] [Google Scholar]
- 4.Fihn SD, Francis J, Clancy C, et al. Insights From Advanced Analytics At The Veterans Health Administration. Health Affairs. 2014;33(7):1203–1211. doi: 10.1377/hlthaff.2014.0054. [DOI] [PubMed] [Google Scholar]
- 5.Global Initiative for Asthma. wwwginaasthmaorg. 2015:1–148. http://www.ginasthma.org/local/uploads/files/GINA_Report_2014_Aug12.pdf. Accessed February 21, 2015. [Google Scholar]
- 6.Busse WW, Holgate S, Kerwin E, Chon Y, Feng J, Lin J, Lin SL. Randomized, double-blind, placebo-controlled study of brodalumab, a human anti-IL-17 receptor monoclonal antibody, in moderate to severe asthma. Am J Respir Crit Care Med. 2013;188(11):1294–1302. doi: 10.1164/rccm.201212-2318OC. [DOI] [PubMed] [Google Scholar]
- 7.Nelson SD, Lu C-C, Teng C-C, et al. The use of natural language processing of infusion notes to identify outpatient infusions. Pharmacoepidemiol Drug Saf. 2014 doi: 10.1002/pds.3720. [DOI] [PubMed] [Google Scholar]
- 8.Turchin A, Kolatkar NS, Grant RW, Makhni EC, Pendergrass ML, Einbinder JS. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. J Am Med Inform Assoc. 2006;13(6):691–695. doi: 10.1197/jamia.M2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bui DD, Zeng-Treitler Q. Learning regular expressions for clinical text classification. J Am Med Inform Assoc. 2014;21(5):850–857. doi: 10.1136/amiajnl-2013-002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC, Miller RA, Waitman LR, Arrieta MA, Peterson JF. “Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor.”. Int J Med Inform. 2009;78(Suppl 1):S34–42. doi: 10.1016/j.ijmedinf.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garvin JH, DuVall SL, South BR, Bray BE, Bolton D, Heavirland J, Pickard S, Heidenreich P, Shen S, Weir C, Samore M, Goldstein MK. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J Am Med Inform Assoc. 2012;19(5):859–866. doi: 10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matheny ME, Fitzhenry F, Speroff T, Hathaway J, Murff HJ, Brown SH, Fielstein EM, Dittus RS, Elkin PL. “Detection of blood culture bacterial contamination using natural language processing.”. AMIA Annu Symp Proc. 2009;2009:411–415. [PMC free article] [PubMed] [Google Scholar]
- 13.Goutte C, Gaussier E. Advances in Information Retrieval. Vol. 3408. Berlin, Heidelberg: Springer Berlin Heidelberg; 2005. A Probabilistic Interpretation of Precision, Recall and F-Score, with Implication for Evaluation; pp. 345–359. Lecture Notes in Computer Science. [DOI] [Google Scholar]
- 14.Redd D, Frech TM, Murtaugh MA, Rhiannon J, Zeng QT. Informatics can identify systemic sclerosis (SSc) patients at risk for scleroderma renal crisis. Comput Biol Med. 2014;53:203–205. doi: 10.1016/j.compbiomed.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson SD, Lu CC, Teng CC, Leng J, Cannon GW, He T, Zeng Q, Halwani A, Sauer B. The use of natural language processing of infusion notes to identify outpatient infusions. Pharmacoepidemiol Drug Saf. 2015;24(1):86–92. doi: 10.1002/pds.3720. [DOI] [PubMed] [Google Scholar]
- 16.Al-Haddad MA, Friedlin J, Kesterson J, Waters JA, Aguilar-Saavedra JR, Schmidt CM. Natural language processing for the development of a clinical registry: a validation study in intraductal papillary mucinous neoplasms. HPB (Oxford) 2010;12(10):688–695. doi: 10.1111/j.1477-2574.2010.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]