Abstract
When grown under conditions of potassium limitation or high osmolality, Escherichia coli synthesizes the K+-translocating KdpFABC complex. The KdpA subunit, which has sequence homology to potassium channels of the KcsA type, has been shown to be important for potassium binding and transport. Replacement of the glycine residues in KdpA at positions 345 and 470, members of putative selectivity filter regions III and IV, alters the ion selectivity of the KdpFABC complex.
The KdpFABC complex of Escherichia coli, which belongs to the group of P-type ATPases, is an inducible, high-affinity K+ uptake system (reviewed in references 1 and 2). The KdpB catalytic subunit is homologous to the large subunits of other P-type ATPases (11); it contains an ATP binding site and forms a phosphointermediate during the catalytic cycle (13, 15). Mutations affecting ion binding and selectivity are clustered in defined regions within the KdpA subunit (3, 4, 14, 18). While some authors have concluded that KdpA has two (3, 4) KcsA-like selectivity filter regions (5) at the periplasmic side and one cytoplasmic binding site for K+, where the ion becomes occluded, others have proposed that KdpA has four selectivity filter regions analogous to a KcsA tetramer (5), thereby forming by itself a functional potassium channel (6, 7). The latter suggestions were drawn mainly from sequence comparisons with other potassium channels and symporters, like KcsA, KtrB, HKT, rand TrkH (6). In order to provide experimental evidence for the four-filter-region hypothesis, site-directed mutagenesis of glycine residue 345 (in putative filter region III) and glycine residue 470 and serine residue 471 (both in putative filter region IV) was performed. These residues have been aligned with the K+ channel filter motif (TCGYG) (6, 10). In the present study we focused on the conserved glycine residues within filter regions III and IV, because the topology of KdpA in that part of the protein has been a topic of controversy (3, 6).
Site-directed mutagenesis.
All strains and plasmids used are listed in Table 1. All kdpA cassettes containing the different substitutions were cloned into the pSMCHis10 vector (based on pSM2 [18], but containing in addition 10 histidine codons downstream of kdpC). The pSM vector series carries the wild-type kdpFABC operon under control of its native kdp promoter. Therefore, expression of the kdpFABC operon is regulated via the KdpDE proteins. The pSMCHis10 derivatives were transformed into TKW3205, which carries the chromosomal kdpDE operon but from which the kdpFABC operon has been deleted. Cassettes carrying the codons for the G345 mutants were cloned into the NsiI and NcoI sites, while cassettes with the altered codons for G470 and S471 were introduced into the NsiI and AflII sites of pSMCHis10.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype and derivation | Source or reference |
|---|---|---|
| E. coli strains | ||
| TKW3205 | ΔkdpFABC Δatp706 thi rha lacZ nagA trkA405 trkD1 | 12 |
| DH5α | kdp+recA1 endA1 gyrA96 thi-1 relA1 hsdR17 supE44 Δ(lac-proAB) F′ (traD36 laqIqlacZΔM15 proA+proB+) | 9 |
| Plasmids | ||
| pUC19 | 19 | |
| pSMCHis10 | kdpFABC(KdpFABC-His10),a derivative of pSM2 (18) | This study |
| pSMCHis10-KdpA(G345A) | kdpA(G345A), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(G345D) | kdpA(G345D), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(G345S) | kdpA(G345S), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(G470A) | kdpA(G470A), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(G470D) | kdpA(G470D), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(G470K) | kdpA(G470K), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(G470S) | kdpA(G470S), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(S471D) | kdpA(S471D), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(S471G) | kdpA(S471G), derivative of pSMCHis10 | This study |
| pSMCHis10-KdpA(S471K) | kdpA(S471K), derivative of pSMCHis10 | This study |
Decahistidinyl motif at C terminus of KdpC.
In vivo characterization of the mutants.
In order to test the effects of the different mutations on the K+ affinity of the KdpFABC complexes in vivo, strain TKW3205, which contains no functional K+ uptake system, was transformed with the different pSMCHis10 derivatives listed in Table 1. Cells were grown as described previously (15). Carbenicillin-resistant single colonies were selected and transferred to minimal medium agar plates containing 0 to 115 mM KCl (according to the method described in reference 8). TKW3205/pSMCHis10 (here referred to as the wild type) was able to grow on medium with all of the different KCl concentrations (K0 agar plates contain up to 20 μM K+ due to impurities, enabling the growth of cells synthesizing the wild-type KdpFABC complex). TKW3205/pSMCHis10-KdpA(G345A) (the G345A mutant) did not grow on K+ concentrations below 1.5 mM, and TKW3205/pSMCHis10-KdpA(G345S) needed 0.7 mM K+, while TKW3205/pSMCHis10-KdpA(G345D) was able to grow on 0.3 mM K+. Alterations at position G470 even more strongly impaired growth on medium with low potassium concentrations. TKW3205/pSMCHis10-KdpA(G470A) and TKW3205/pSMCHis10-KdpA(G470S) were able to grow on 2.3 mM K+, TKW3205/pSMCHis10-KdpA(G470D) needed 1.6 mM K+, and TKW3205/pSMCHis10-KdpA(G470K) grew on 1.8 mM K+. Less-severe effects were observed for alterations at position S471. TKW3205/pSMCHis10-KdpA(S471D) and TKW3205/pSMCHis10-KdpA(S471G) grew like the wild type. Only the strain with the extreme alteration, TKW3205/pSMCHis10-KdpA(S471K), was unable to grow on the medium with K+ at concentrations below 10 mM. It should be noted that all of the KdpFABC variants were membrane bound, as determined by immunoblot analysis using the poly-His motif as an epitope (data not shown). Phenotypic alterations are therefore due to changes in binding and transport capacity and not to misrouting of the protein complexes. Although these data do not allow a distinction between Km or Vmax effects, the phenotypic characterization provides valuable information about effects on K+ affinity and transport per se.
In vitro characterization of purified wild-type and mutant KdpFABC complexes.
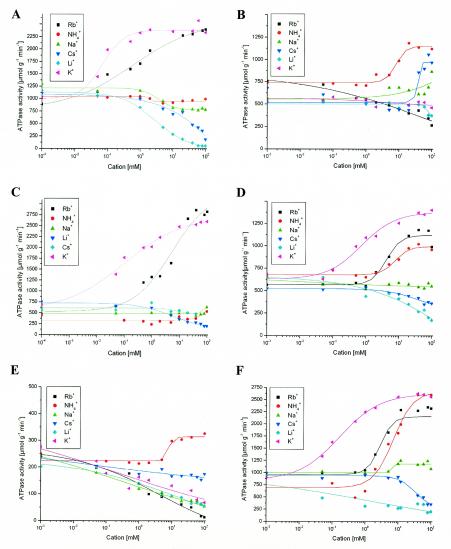
Induction of wild-type and mutant kdpFABC operons was carried out as described previously (18). Membranes were prepared as described elsewhere (16), with the exception that buffer A (20 mM HEPES-Tris [pH 8.0], 0.5 mM MgCl2, 10 mM imidazole, 0.2% aminoxide, 0.5 mM phenylmethylsulfonyl fluoride) was used. The KdpFABC complexes were solubilized using 1% (vol/vol) aminoxide. The solubilized proteins were applied to Ni-nitrilotriacetic acid (NTA), equilibrated with buffer A, and incubated for 60 min with constant shaking on ice. Subsequently, the Ni-NTA resin was transferred into a glass column and connected to a fast protein liquid chromatography device (Amersham Pharmacia Biotech, Freiburg, Germany). Unbound proteins were removed with 10 column volumes of buffer A, and subsequently a 10-column-volume gradient of 0 to 100% buffer B (20 mM HEPES-Tris [pH 8.0], 0.5 mM MgCl2, 250 mM imidazole, 0.2% aminoxide, 300 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride) was used for elution of the KdpFABC-His10 complexes. The pooled fractions were concentrated and analyzed by size-exclusion chromatography with a Superdex 75 column (Amersham Pharmacia Biotech) in 20 mM HEPES-Tris (pH 8.0)-0.5 mM MgCl2-0.2% aminoxide. The fractions containing KdpFABC-His10 were stored in liquid nitrogen until use. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the purified KdpFABC complexes with a substitution mutation in KdpA revealed that the KdpA/KdpB/KdpC ratio was the same in all cases (data not shown) and, therefore, that the stoichiometry of the complexes was not altered upon mutation. By applying metal affinity chromatography, protein purity was significantly increased compared to that of the previously described procedure (15). Furthermore, the use of a polyhistidine tag at KdpC for purification demonstrated that complex formation and interactions between the subunits were not impaired by the mutations. Cation-dependent ATPase activity, which is a common feature of P-type ATPases, was measured as described previously (2, 18). The cations tested in the present study were K+, Rb+, Na+, Li+, Cs+, and NH4+. The factors by which ATPase activity was stimulated or inhibited, as well as the cation concentrations needed for half-maximal stimulation or inhibition (K0.5), are summarized in Table 2. For the wild-type KdpFABC complex (Fig. 1A), K+ caused a stimulation factor of 2.3 with a K0.5 of 70 μM. The stimulation by ammonium which was reported previously (18) was not observed (Fig. 1A). However, some of the mutant complexes (the G345A, G345S, G470S, and S471G mutants) did show NH4+-stimulated ATPase activity, ruling out the possibility that the histidine motif, which was not present in the complex used in the previous report, might have abolished the ammonium effect. The purification procedure described here leads to a protein preparation that is much more homogeneous than the previous one. In addition, an NH4Cl gradient for the elution of the protein has been omitted. Therefore, it is quite conceivable that these changes are responsible for the differences observed. The G345A mutant (Fig. 1B) exhibited effects similar to those reported for the G232A substitution mutant: NH4+ induced a 1.5-fold stimulation, while K+ had an inhibitory effect (1.2-fold). Replacement of G345 with serine had a less severe effect than the G232S (18) or G470S substitution. However, the G345S mutant was stimulated 1.5-fold by NH4+ (Fig. 1D). The G345D mutant had a cation-stimulated ATPase activity similar to that of the wild-type KdpFABC-His10 complex; stimulation of the ATPase activity with K+ and Rb+ was significantly higher than that of the wild-type enzyme. Furthermore, the inhibitory effects of Li+ and Cs+, as observed for the wild-type KdpFABC-His10 complex, were absent (Fig. 1C). A similar observation has been reported for the G232D mutant (18). The G470S mutant exhibited an ion-dependent ATPase activity almost identical to that described for the G232S mutant (18). Except for NH4+, all cations tested were inhibitory (Fig. 1E). The S471G mutant was stimulated in its ATPase activity by K+, Rb+, and NH4+ (Fig. 1F), indicating that changes at position S471 might be more tolerable than those at position G470, although an effect on ion selectivity was still observed.
TABLE 2.
Cation-dependent ATPase activities of purified KdpFABC-His10 complexesa
| Complex | Fold change in ATPase activity (K0.5 [mM]) withb:
|
|||||
|---|---|---|---|---|---|---|
| K+ | Rb+ | NH4+ | Na+ | Li+ | Cs+ | |
| KdpFABC-His10 (wild type) | 2.3 (0.07) | 2.2 (1) | 1.0 (—) | 1.3 (5) | 18.6 (2) | 1.1 (10) |
| G345A mutant | 1.2 (10) | 2.9 (1) | 1.5 (6) | 1.5 (11) | 1.4 (80) | 1.4 (50) |
| G345D mutant | 3.8 (0.5) | 5.9 (3) | 1.0 (—) | 1.0 (—) | 2.9 (10) | 1.0 (—) |
| G345S mutant | 2.0 (1) | 1.8 (3) | 1.5 (15) | 1.0 (—) | 3.7 (8) | 1.5 (30) |
| G470S mutant | 4.0 (5) | 18.0 (2) | 1.3 (9) | 1.3 (0.9) | 3.6 (1) | 4.7 (0.5) |
| S471G mutant | 2.9 (0.2) | 1.9 (2.5) | 1.9 (3) | 1.1 (10) | 4.7 (1) | 2.8 (30) |
Data represent average values obtained in at least three independent experiments.
Values for stimulating cations are in boldface, and values for inhibiting cations are in lightface; change was measured in the presence of saturating concentrations of the different cations. Values of 1 represent no stimulation or inhibition. K0.5 values for half-maximal stimulation or inhibition are given in parentheses. —, K0.5 = 0.
FIG. 1.
Cation-dependent ATPase activities of different KdpFABC-His10 variants. KdpFABC-His10 (wild type) (A), G345A mutant (B), G345D mutant (C), G345S mutant (D), G470S mutant (E), S471G mutant (F). KdpFABC-His10 variants were purified and ATPase activity assays were performed as described previously (18).
Conclusions.
Sequence alignment (6, 7) and mutagenesis studies (3, 4, 18) lend support to the notion that KdpA is the K+-binding and -translocating subunit of the KdpFABC complex. Beside the general agreement that KdpA has selectivity filter regions similar to those found in KcsA (5), the number and localization of the filter regions in KdpA remained unclear. Sequence comparisons between E. coli KdpA and KcsA revealed that three out of four regions of KdpA (I, 112-NTNWQ-116; II, 230-TNGGG-234; III, 343-SCGAV-347; IV, 468-NNGSA-472) show some similarity to the selectivity filter motif (TCGYG) of KcsA. Although there is no obvious similarity between the first putative filter region (region I) and the K+ selectivity filter motif, it should be mentioned that the Q116R mutant exhibits a drastic change in K+ affinity from 2 μM to 6 mM (3). In the present study we focused on filter regions III and IV, because the existence of two selectivity filter regions in this part of the protein (the N-terminal part) would support the topology model suggested by Durell and coworkers (6). Interestingly, only the N-terminal glycine residues of filter regions II to IV are highly conserved. In other K+-translocating systems, such as KtrB, TrkH, and HKT, a similar situation is found. Furthermore, in the case of Arabidopsis thaliana HKT, the N-terminal glycine residue within the first selectivity filter is replaced by serine, a change that is perhaps correlated with the shift in selectivity from K+ to Na+ (17). The second putative filter region of KdpA (residues G232, G233, and G234) has been well characterized (3, 4, 14, 18). In particular, residue G232 was of fundamental importance for ion selectivity (14, 18), whereas residues G233 and G234 had only a minor effect (18). Based on these observations, we have replaced the conserved glycine residues in putative selectivity filter regions III (G345) and IV (G470) in KdpA and tested the ion-stimulated ATPase activity. It is shown here that ion selectivity is impaired by substitutions at G345 and G470. Interestingly, a change from glycine to aspartate at position 345 has a less severe effect on ion selectivity than expected, similar to the results reported for the G232D mutant (18). Replacement of glycine 470 is not tolerated and resulted in almost no ion stimulation of ATPase activity. Although several substitution mutants (the G470A, G470D, G470S, G470K mutants) were constructed, so far only the G470S mutant complex could be purified. In the other cases, only KdpC, containing the histidine motif, was eluted from Ni-NTA. This observation emphasizes the possibility that the stability of the complex is tightly coupled with correctly folded subunits and might be one explanation for the finding in previous studies that these residues could not be changed (4). The substitutions at S471 did not affect K+- and Rb+-stimulated ATPase activity, but ammonium was now effective. However, the substitution of charged residues at position S471 resulted in an unstable protein (data not shown), indicating the necessity of a properly folded pore region for correct membrane insertion and assembly. The results presented here strongly support the idea that KdpA contains four selectivity filter motifs, as previously proposed (6, 7), and might therefore be evolutionarily derived from a KcsA-like potassium channel by gene duplication and fusion, making the existence of an internal K+ binding site unlikely (3).
Acknowledgments
We thank Michael Gaβel for constructing the plasmid pSMCHis10. We are grateful to Brigitte Herkenhoff-Hesselmann for excellent technical assistance and to E. P. Bakker for helpful discussion.
Support for this study was provided by the Deutsche Forschungsgemeinschaft (SFB 431) and the Fonds der Chemischen Industrie (fellowship to M.B.).
REFERENCES
- 1.Altendorf, K., and W. Epstein. 1996. The Kdp-ATPase of Escherichia coli, p. 403-420. In A. G. Lee (ed.), Biomembranes (ATPases), vol. 5. JAI Press Inc., London, England.
- 2.Altendorf, K., M. Gaβel, W. Puppe, T. Möllenkamp, A. Zeeck, C. Boddien, K. Fendler, E. Bamberg, and S. Dröse. 1998. Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. 163:137-146. [PubMed] [Google Scholar]
- 3.Buurman, E. T., K.-T. Kim, and W. Epstein. 1995. Genetic evidence of two sequentially occupied K+ binding sites in the Kdp transport ATPase. J. Biol. Chem. 270:6678-6685. [DOI] [PubMed] [Google Scholar]
- 4.Dorus, S., H. Mimura, and W. Epstein. 2001. Substrate-binding clusters of the K+-transporting Kdp ATPase of Escherichia coli investigated by amber suppression scanning mutagenesis. J. Biol. Chem. 276:9590-9598. [DOI] [PubMed] [Google Scholar]
- 5.Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280:69-77. [DOI] [PubMed] [Google Scholar]
- 6.Durell, S. R., E. P. Bakker, and H. R. Guy. 2000. Does the KdpA subunit from the high affinity K+-translocating P-type Kdp-ATPase have a structure similar to that of K+ channels? Biophys. J. 78:188-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durell, S. R., Y. Hao, T. Nakamura, E. P. Bakker, and H. R. Guy. 1999. Evolutionary relationship between K+ channels and symporters. Biophys. J. 77:775-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein, W., and B. S. Kim. 1971. Potassium transport loci in Escherichia coli K-12. J. Bacteriol. 108:639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Jan, L. Y., and Y. N. Jan. 1997. Cloned potassium channels from eukaryotes and prokaryotes. Annu. Rev. Neurosci. 20:91-123. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan, J. 2002. Biochemistry of Na, K-ATPase. Annu. Rev. Biochem. 71:511-535. [DOI] [PubMed] [Google Scholar]
- 12.Puppe, W. 1991. Kalium-Transport bei Escherichia coli: molekulargenetische und biochemische Untersuchungen zu funktionellen Domänen der Kdp-ATPase. Dissertation. Universität Osnabrück, Osnabrück, Germany.
- 13.Puppe, W., A. Siebers, and K. Altendorf. 1992. The phosphorylation site of the Kdp-ATPase of Escherichia coli: site-directed mutagenesis of the aspartic acid residues 300 and 307 of the KdpB subunit. Mol. Microbiol. 6:3511-3520. [DOI] [PubMed] [Google Scholar]
- 14.Schrader, M., K. Fendler, E. Bamberg, M. Gassel, W. Epstein, K. Altendorf, and S. Dröse. 2000. Replacement of glycine 232 by aspartic acid in the KdpA subunit broadens the ion specificity of the K+-translocating KdpFABC complex. Biophys. J. 79:602-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siebers, A., and K. Altendorf. 1988. The K+-translocating Kdp-ATPase from Escherichia coli. Purification, enzymatic properties and production of complex- and subunit-specific antisera. Eur. J. Biochem. 178:131-140. [DOI] [PubMed] [Google Scholar]
- 16.Siebers, A., R. Kollmann, G. Dirkes, and K. Altendorf. 1992. Rapid, high-yield purification and characterization of the K+-translocating Kdp-ATPase from Escherichia coli. J. Biol. Chem. 267:12717-12721. [PubMed] [Google Scholar]
- 17.Uozumi, N., E. J. Kim, F. Rubio, T. Yamaguchi, S. Muto, A. Tsuboi, E. P. Bakker, T. Nakanmura, and J. I. Schroeder. 2000. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant. Physiol. 122:1249-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Laan, M., M. Gaβel, and K. Altendorf. 2002. Characterization of amino acid substitutions in KdpA, the K+-binding and -translocating subunit of the KdpFABC complex of Escherichia coli. J. Bacteriol. 184:5491-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]