ABSTRACT
Protein synthesis and its regulatory signaling pathways play essential roles in the initiation and maintenance of the cancer phenotype. Insight obtained over the last 3 decades on the mechanisms regulating translation in normal and transformed cells have revealed that perturbed control in cancer cells may offer an Achilles' heel for the development of novel anti-neoplastic agents. Several small molecule inhibitors have been identified and characterized that target translation initiation – more specifically, the rate-limiting step where ribosomes are recruited to mRNA templates. Among these, hippuristanol, a polyhydroxysteroid from the gorgonian Isis hippuris has been found to inhibit translation initiation by blocking the activity of eukaryotic initiation factor (eIF) 4A, an essential RNA helicase involved in this process. Herein, we highlight the biological properties of this compound, its potential development as an anti-cancer agent, and its use to validate eIF4A as an anti-neoplastic target.
KEYWORDS: chemical biology, eIF4A, hippuristanol, translational control
Introduction
Perturbed translational control has been implicated in the initiation and maintenance of the cancer phenotype, supporting angiogenesis, and modulating drug response.1-3 The energetically demanding process of translation is highly regulated predominantly at the level of initiation, with control exerted by 2 factors – eukaryotic initiation factor (eIF) 2 and eIF4F.4,5 The role of eIF2 in translation initiation under normal physiological or stress conditions has been extensively reviewed4,5 and regulation imposed at this step profoundly inhibits translation of a majority of mRNAs.4,5 Since perturbation of eIF2 activity in cancer biology is less characterized and understood than the role of eIF4F, this review will focus solely on the latter.
The eIF4F complex consists of 3 subunits that function to recruit ribosomes to mRNAs (Fig. 1A). The eIF4E subunit recognizes 5′ cap structures (m7GpppN, where N is any nucleotide), eIF4A is an RNA helicase required to remodel secondary structure proximal to the cap structure, and eIF4G provides a platform for subunit association, participates in RNA binding, and recruits the 40S ribosome (with associated factors) through bridging interactions with ribosome-bound eIF3.6-8 The eIF4F complex does not bind all mRNAs equally, but rather appears to favor templates with an accessible cap structure9-12 and reduced cap-proximal secondary structure.12-16 Assembly of the eIF4F complex is regulated by mTOR via phosphorylation of eIF4E-Binding Proteins (4E-BPs – of which there are 3 genes with 4E-BP1 being the best characterized). Stimulation of mTOR signaling results in phosphorylation of 4E-BPs, dissociating 4E-BP:eIF4E complexes and enabling eIF4E to associate with eIF4G.17
Figure 1.

A. Simplified model of eIF4F-dependent initiation displaying cap recognition and subsequent destabilization of local secondary structure. eIF4H and eIF4B share a common binding site on eIF4A and these interactions are mutually exclusive.111 Not shown is mRNA circularization mediated through poly(A) tail:poly(A)-binding protein (PABP) and eIF4G. B. Schematic diagram illustrating targets of various small molecules and anti-sense oligonucleotides (ASO) that target eIF4E-cap interaction (4Ei-1), prevent synthesis of the eIF4E subunit (4E ASO), inhibit eIF4E:eIF4G interaction (4E1RCat, 4E2RCat, 4E3RCat (unpublished data), 4EGI-1), and interfere with eIF4A helicase activity (silvestrol, hippuristanol, pateamine A). (Note that although ribavirin has been claimed to inhibit eIF4E-cap interaction,112 this has been questioned.113,114).
eIF4E appears to be the least abundant of all the initiation factors (0.2–0.3 molcules/ribosome)18,19 implying that mRNAs must compete for the limiting amounts of eIF4F during translation initiation with the outcome dictated, in part, by structural elements in the 5′ untranslated region (UTR). This point has been challenged since reductions in eIF4E levels in eIF4E+/− mice show little effect on global translation (at least in MEFs) and has been interpreted to indicate that eIF4E is not rate-limiting in vivo.20 However, similar observations had been previously documented in cells treated with anti-sense oligonucleotides21 or shRNAs22,23 targeting eIF4E. eIF4E is under homeostatic control via regulation of 4E-BP levels and reductions in eIF4E levels leads to increased degradation of 4E-BP1, buffering against changes in eIF4F levels.24
Changes in eIF4F activity is not expected to impact all mRNAs equally but rather cause a small change in global translation with a disproportionate, selective alteration in translation of a subset of mRNAs.1-3 As it turns out, several of these eIF4F-dependent mRNAs fuel tumorigenesis and thus one way to impact on tumor maintenance is to block eIF4F activity.1-3 There has therefore been intense interest in identifying small molecule inhibitors of eIF4F. Several HTS assays and directed synthetic efforts have identified compounds that target eIF4E-cap interaction,25 prevent synthesis of the eIF4E subunit,21,26 inhibit eIF4E:eIF4G interaction,27-29 and interfere with eIF4A helicase activity30-32 (Fig. 1B). Although these have been reviewed,1-3 here we update the current development status of hippuristanol, a polyhydroxysteroid first isolated from the coral Isis hippuris,33 and for which until recently, biological characterization was delayed due to limitations in supply.
eIF4A and translation initiation
eIF4A is a prototypical member of the DEAD-box family of RNA helicases and one of the more abundant translation factors (approximately 3 copies/ribosome).34,35 Given that eIF4E levels are approximately 10-fold lower,34,35 the majority (approximately 90%) of eIF4A exists as free form while only a small fraction is present in the eIF4F subunit.36-38 The higher abundance of eIF4A, relative to eIF4F, as well as the ability to crosslink (in a cap-dependent manner) eIF4A to sites located 52 nucleotides downstream of the cap structure, has lead to models invoking recycling of eIF4A through the eIF4F complex during translation initiation (Fig. 1A). 39,40 Mammals have 2 eIF4A proteins (eIF4AI [DDX2A] and eIF4AII [DDX2B]) which share approximately 90% sequence identity at the amino acid level.41 Although eIF4AI and eIF4AII are functionally interchangeable in vitro,42 they do not appear to be equivalent in vivo. eIF4AI, but not eIF4AII, is required for cell viability19,43 and eIF4AII is not capable of rescuing the inhibition of translation that ensues following suppression of eIF4AI.19 In general, eIF4AI is the more abundant protein41,44,45 and the majority of biochemical studies assessing eIF4A activity have been performed with eIF4AI. eIF4AI and eIF4AII possess both RNA-stimulated ATPase and ATP-stimulated RNA-binding activity. The eIF4A helicase and ATPase activities are strongly stimulated when eIF4A is part of the eIF4F complex or is associated with either of 2 RNA binding proteins, eIF4B or eIF4H46 47 (Fig. 1A).
Dominant-negative (dn) mutants of eIF4A have highlighted the critical role that this factor plays in translation.48,49 One such mutant is capable of assembling into the eIF4F complex and prevents cap recognition.49 Supplementing translation extracts with dn eIF4A mutants has revealed that translation inhibition is directly related to the degree of 5′ UTR secondary structure. Analysis of transcripts sensitive to eIF4A inhibition by silvestrol (another eIF4A inhibitor) is consistent with the concept that structural barriers within the 5′ UTR are a key determinant of eIF4A dependency.50-52
The activity of eIF4A can be negatively regulated by PDCD4, a tumor suppressor gene product.53-55 PDCD4 associates with eIF4A, displacing eIF4G and RNA,54 and resulting in preferential suppression of translation of mRNAs with structured 5′ UTRs.53,56 The association between PDCD4 and eIF4A is regulated by the PI3K/mTOR signaling pathway through the downstream S6K branch.57 Phosphorylation of PDCD4 by S6K1 leads to its ubiquitin-mediated degradation, freeing eIF4A for assembly into the eIF4F complex.57
Data implicating a direct role for dysregulated eIF4A levels contributing to tumor initiation or maintenance is sparse. This might be expected for an abundant protein, if its critical functional role is one mediated through a rate-limiting complex. On the other hand, there is a significant body of work indicating that eIF4F activity or eIF4E levels will drive tumor initiation, support tumor cell maintenance, and contribute to chemoresistance.1-3 Ex vivo experiments have shown the efficacy of targeting eIF4A using ASOs58 or ectopic PDCD4 expression to block transformation59 and delay tumor onset and progression in a chemically-induced skin tumor model.60 As well, another eIF4A inhibitor, silvestrol, has shown activity in a variety of pre-clinical cancer models.31,50,51,61-63 Presumably, these physiological responses are due to inhibition of eIF4F activity. Given the difficulty in “translating” biologicals such as proteins and ASOs into therapeutics, there was excitement when hippuristanol emerged from a high throughput screen aimed at identifying novel inhibitors of cap-dependent translation.32,64
Hippuristanol – A selective inhibitor of eIF4A
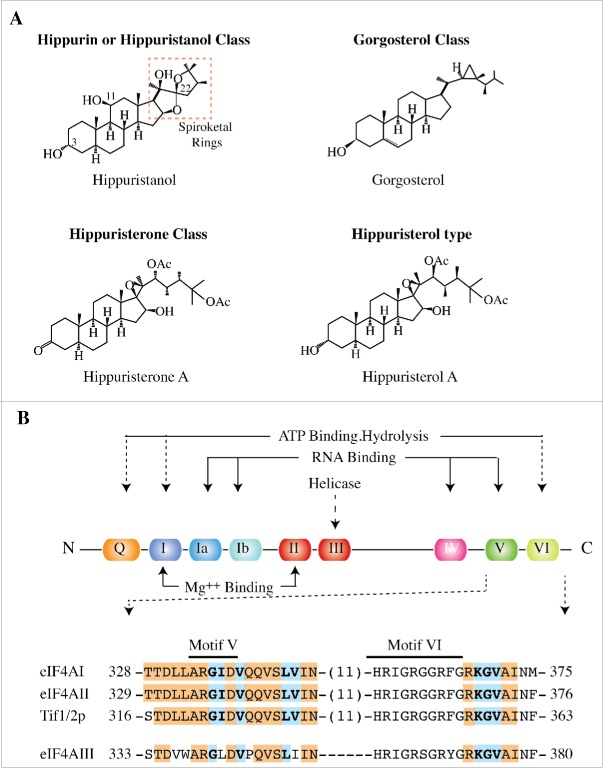
Hippuristanol is a member of one of 4 classes of polyoxygenated steroids. These include (i) the hippurin or hippuristanol type containing a spiroketal ring, (ii) the gorgosterol type containing a cyclopropane residue; (iii) the hippuristerone type possessing a 3-keto functionality, and (iv) the hippuristerol type (Fig. 2A). Although a large number of compounds from the various groups have been isolated and characterized, those belonging to the hippurin or hippuristanol class exhibit the most potent cytotoxic activity ex vivo against tumor cell lines, underscoring the importance of the spiroketal group for activity.32,33,65-67 Some members of the gorgosterol class exhibit moderate cytotoxicity68,69 (IC50 approximately 2 μM against NBT-T2 rat bladder epithelial cells and approximately 15 μM [for presumably a 4 day exposure period70]) and have shown activity against human epidermoid carcinoma KB drug-resistant cells expressing the drug transporter, ABCB1/P-glycoprotein (approximately 6–10 greater activity than against cells not expressing ABCB1), but less so against cells expressing multi-drug resistance protein-1 (MRP1).68 Tumor cell cytotoxicity has been reported for compounds of the hippuristerone family66 but these are far less potent than hippurin or hippuristanol-like compounds. There is also one report describing moderate activity of a hippuristerone against human cytomegalovirus (HCMV) (EC50 approximately 10 μM), while (together with 4 other hippuristerones) the same compound showed no activity against P-388 mouse lymphocytic leukemia, HT-29 human colon carcinoma, or human embryonic lung cells.71 Acknowledging the fact that not all gorgosterols, hippuristerones, and hippuristanols isolated to date have been tested for cytotoxicity,72-75 in general it is the hippurin/hippuristanol class of compounds that exhibit the highest level of activity against tumor cells in culture (e.g. IC50 approximately 700 nM against HeLa cells for a 24 hrs exposure32).
Figure 2.
A. Structure of representative molecules from the 4 polyoxygenated steroid classes. B. Schematic diagram illustrating characteristic domains that comprise DEAD-box RNA helicases with functions of the motifs indicated. Primary amino acid sequence of conserved motifs V and VI (indicated by overline) and neighboring amino acids of the indicated helicases. Direct protein-hippuristanol NOEs are highlighted in bold and light blue and those within ∼5Å are in orange shading.
Hippuristanol's mechanism of action differs significantly from that of 2 other eIF4A inhibitors, pateamine A and rocaglates. Whereas pateamine A and rocaglates appear to act as chemical inducers of dimerization by stimulating eIF4A:RNA binding, hippuristanol prevents both free eIF4A and eIF4F-bound eIF4A from interacting with RNA.32 Hippuristanol does not inhibit ATP binding.32 Single molecule FRET experiments have shown that hippuristanol locks eIF4AI in a closed conformation preventing its transition from a closed to an open state, an event essential to eIF4AI's helicase activity.76 Since eIF4A does not normally sample the closed state (transition to the closed conformation is eIF4G- and eIF4B-stimulated) and the eIF4A closed conformation is normally RNA bound,77 hippuristanol may be locking eIF4A in an aberrant closed complex that can no longer participate in initiation.
NMR and site-directed mutagenesis studies have revealed that hippuristanol interacts with the C-terminal domain of eIF4A in a pocket involving amino acids within and spanning conserved motifs V and VI (Fig. 2B).32,78 These studies provided valuable insight into some of the key residues involved in the interaction of eIF4A with hippuristanol.32,78 Hippuristanol is thought to make direct contacts with mouse eIF4AI residues G335I336, V338, L343V344, as well as K369 - V371 whereas residues T328 - N346 and R368 - I373 lie within 5Å of hippuristanol (Fig. 2B). Residues making direct contacts with hippuristanol are conserved among eIF4AI, eIF4AII, as well as the yeast TIF1 and TIF 2 homologues (Fig. 2B).78 These results have been validated by mutagenesis studies of the hippuristanol binding site which led to altered sensitivity (RNA binding and helicase activity of recombinant proteins) to hippuristanol in vitro.78 Hippuristanol-resistant eIF4A alleles were able to rescue in vitro inhibition of translation by hippuristanol, consistent with the effects of this small molecule on translation being mediated through eIF4A inhibition.78 Hippuristanol inhibits the RNA-stimulated ATPase activity of both eIF4AI and eIF4AII to similar extents.78
Among all members of the DEAD-box helicase family, eIF4AIII [DDX48]) [implicated in nonsense-mediated decay (NMD)79-82] has the most related hippuristanol binding site - differing by 6 amino acids compared to the eIF4AI site (Figs. 2B and 3). Consequently ten times more hippuristanol is required to inhibit the ATPase activity of eIF4AIII compared to eIF4AI or eIF4AII.78 Increased sensitivity to hippuristanol has been engineered into eIF4AIII by grafting the eIF4AI hippuristanol binding site into eIF4AIII.78 Other members of the DEAD-box helicase family display higher degeneracy within the hippuristanol-binding site (Fig. 3) and are thus not expected to be as sensitive to hippuristanol as eIF4AI or eIF4AII. Indeed, to date none have been found to be responsive to hippuristanol concentrations as high as 50 μM.78 Whether hippuristanol targets other cellular proteins is unknown.
Figure 3.
Cladogram displaying the multiple sequence alignment obtained with murine DEAD-box RNA helicase family members using Clustal Omega (http://www.ebi.ac.uk/clustalw/). Alignments were performed with sequences spanning and immediately flanking conserved motifs V and VI only. For example, this would correspond to 328-TTDLLARGIDVQQVSLVIN—-HRIGRGGRFGRKGVAINM-375 for eIF4AI. A complete list of sequences compared can be found in Figure S2 of Ref.78 The Entrez Protein IDs are in parenthesis. eIF4AI and eIF4AII are highlighted in yellow for easy reference.
The selectivity of hippuristanol for eIF4A has provided a powerful tool by which to probe for eIF4A-dependent processes. Hippuristanol has been used to characterize viral and cellular IRESes in vitro and in vivo32,83-86 and to probe the eIF4A-dependency of cellular mRNAs.87-91 As well, it has been used to investigate the effects of Herpes Simplex Virus 1 (HSV 1) virion host shutoff (vhs) protein on cell type specific translation of viral late RNAs,92 the effect of HSV 1 host translation shutoff on nuclear envelope-derived autophagy,93 and the dependency of Influenza virus A polymerase on eIF4F.94 As might be expected, hippuristanol exhibits anti-viral activity,29 but whether it can be used as an anti-viral therapeutic remains untested. Hippuristanol has also been used to characterize the eIF4A dependency of translational events required for long-term synaptic plasticity and potentiation.95,96
Anti-neoplastic activity of hippuristanol
Hippuristanol as a single agent has shown promising anti-neoplastic activity. In 1981, Higa et al.33 reported that hippuristanol inhibited the growth of DBA/MC fibrosarcoma cells and exhibited in vivo activity against lymphocytic leukemia P-388 tumors in mice. More recently, hippuristanol has shown activity against human adult T-cell leukemia (ATL) in vitro and in vivo in a xenograft model.97 Additionally, hippuristanol is active against primary effusion lymphoma (PEL), causing cell cycle arrest and caspase activation followed by apoptosis.98 What is currently required is a comprehensive understanding of the pharmacokinetic/pharmacodynamics properties of hippuristanol so that an optimal dosing schedule and route of administration can be determined for testing in a larger number of xenograft and genetically engineered mouse models (GEMMs) of cancer.
Elevated eIF4E levels and eIF4F activity have also been linked to acquired resistance of PI3K and MAPK pathway targeted therapies (reviewed in Refs.1-3). Increased eIF4E levels are associated with resistance to PI3K/mTOR targeted therapies in cell based models,99,100 as well as doxorubicin101 and rapamycin102 resistance in the Eμ-Myc lymphoma model.101 In addition, elevated eIF4F levels have been associated with resistance to anti-BRAF and anti-MEK therapies103 and targeting eIF4F synergizes with anti-BRAF therapy.103 Targeting eIF4A has also been shown to be a viable approach for overcoming some of this acquired resistance. More specifically, in a Myc-driven lymphoma model (the Eμ-Myc mouse) hippuristanol is capable of resensitizing tumor cells to DNA damaging agents (doxorubicin).104 As well, hippuristanol is capable of synergizing with the Bcl-2 family inhibitor, ABT-737, to induce a potent synergistic response that triggers cell death in mouse and human lymphoma and leukemia cells.104 Multiple myeloma cells are sensitive to hippuristanol (IC50 approximately 50 nM for a 48 h exposure)105 and hippuristanol synergizes with dexamethasone ex vivo, a frontline glucocorticoid used in the management of this disease.105 These studies demonstrate a role for eIF4F in contributing to drug resistance and proof-of-concept for overcoming this with eIF4A inhibitors.
Structure-activity relationships of hippuristanol
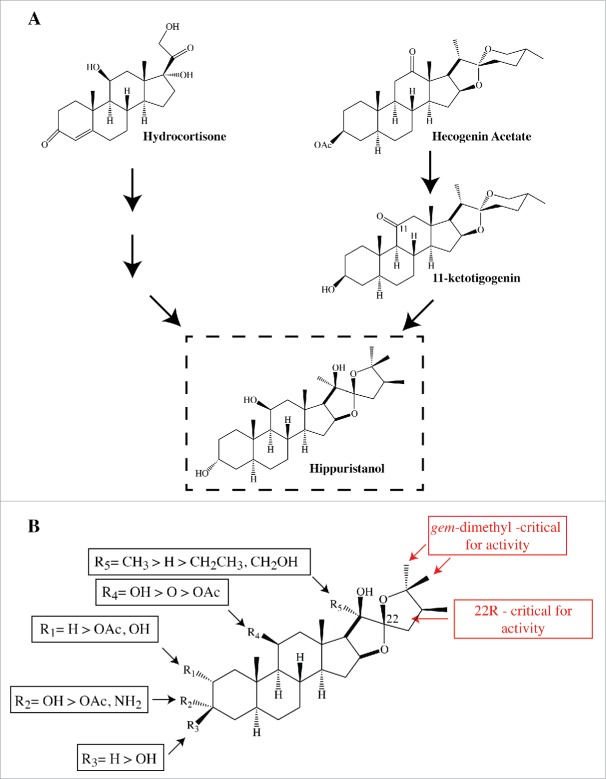
Hippuristanol is a rare natural product and it was critical to develop synthetic routes to obtain sufficient material for biological studies as well as undertaking structure-activity relationship (SAR) studies. Four synthetic routes to hippuristanol have been published and used as starting material either hydrocortisone or hecogenin acetate/11-ketotigigenin106-109 (Fig. 4A). These routes were employed to generate a number of analogs that have been tested for translation inhibition activity106,109 and inhibition of cell proliferation against HeLa cells.106 Taken together with data assessing the activity of several naturally occurring hippuristanol congeners in translation and eIF4A helicase activity,32 we have a fairly good understanding of the essential features required for optimal hippuristanol activity (Fig. 4B). The R stereochemistry at C22 is essential for activity32 as are the gem-dimethyl substitutions on the F ring.110 Appending functionalities onto R1 results in a 3-fold decrease in activity, whereas altering the R2 OH leads to a 25-fold decrease in activity.32,109 Converting the R4 OH to a ketone or acetate diminishes activity approximately 25- and >2000 -fold, respectively.32 Eliminating the R5 CH3 group decreases activity approximately 5–8 fold, whereas increasing the bulkiness at R5 decreases activity >15-fold.109 The rank order of inhibition obtained in in vitro translation assays was similar to when several of the same congeners were assessed for direct inhibition of eIF4A RNA helicase activity32 - consistent with eIF4A being the target through which hippuristanol exerts its inhibitory effects on protein synthesis. These results demonstrate that a large surface area of hippuristanol likely participates in binding to eIF4A.
Figure 4.
A. Several synthetic routes to hippuristanol have been elaborated, involving hydrocortisone,106,109 hecogenin acetate,108 or 11-ketotigogenin107 as starting material. B. Structure-activity relationship for hippuristanol. See text for details.
Future perspectives
There are several issues that will need to be resolved as hippuristanol is developed for clinical assessment. A detailed assessment of the pharmacological properties is urgently needed. In the past this information could not be obtained due to limitations in compound availability, but this has recently been overcome by synthetic routes allowing access to sufficient quantities of material for pharmacological studies. A better understanding of the types of tumor cells that are most likely to respond to eIF4A inhibition is required. The identification of surrogate biomarkers that can report on eIF4A/eIF4F inhibition will also be important to ensure that target inhibition is maintained in vivo. As challenging as these are, they are essential to optimizing the chances of success for eIF4A inhibitors as they move forward from being powerful tools used in the laboratory to potential drugs for blocking eIF4A/eIF4F function in tumor cells. We look forward helping extend the paradigm of targeting eIF4F for the treatment of cancer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Work in the author's laboratory on targeting translation using small molecule inhibitors is supported by grants from the Canadian Institutes of Health Research (MOP-115126 and MOP-106530).
References
- [1].Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res 2015; 75:250-63; PMID:25593033; http://dx.doi.org/ 10.1158/0008-5472.CAN-14-2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov 2015; 14:261-78; PMID:25743081; http://dx.doi.org/ 10.1038/nrd4505 [DOI] [PubMed] [Google Scholar]
- [3].Chu J, Pelletier J. Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim Biophys Acta 2015; 1849:781-91; PMID:25234619; http://dx.doi.org/ 10.1016/j.bbagrm.2014.09.006 [DOI] [PubMed] [Google Scholar]
- [4].Pestova TV, Lorsch JR, Hellen CU. The Mechanism of Translation Initiation in Eukaryotes In: Mathews MB, Sonenberg N, Hershey JWB, eds. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2007:87-128. [Google Scholar]
- [5].Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 2011; 75:434-67; PMID:21885680; http://dx.doi.org/ 10.1128/MMBR.00008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Villa N, Do A, Hershey JW, Fraser CS. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 2013; 288:32932-40; PMID:24092755; http://dx.doi.org/ 10.1074/jbc.M113.517011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J Biol Chem 2000; 275:41369-76; PMID:11022043; http://dx.doi.org/ 10.1074/jbc.M007525200 [DOI] [PubMed] [Google Scholar]
- [8].Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem 1995; 270:21975-83; PMID:7665619; http://dx.doi.org/ 10.1074/jbc.270.37.21975 [DOI] [PubMed] [Google Scholar]
- [9].Godefroy-Colburn T, Ravelonandro M, Pinck L. Cap accessibility correlates with the initiation efficiency of alfalfa mosaic virus RNAs. Eur J Biochem 1985; 147:549-52; PMID:2983983; http://dx.doi.org/ 10.1111/j.0014-2956.1985.00549.x [DOI] [PubMed] [Google Scholar]
- [10].Lawson TG, Cladaras MH, Ray BK, Lee KA, Abramson RD, Merrick WC, Thach RE. Discriminatory interaction of purified eukaryotic initiation factors 4F plus 4A with the 5′ ends of reovirus messenger RNAs. J Biol Chem 1988; 263:7266-76; PMID:3366779 [PubMed] [Google Scholar]
- [11].Parkin NT, Cohen EA, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J 1988; 7:2831-7; PMID:3181141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. Rna 2006; 12:851-61; PMID:16540693; http://dx.doi.org/ 10.1261/rna.2309906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pelletier J, Sonenberg N. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: effect of mRNA 5′ secondary structure. Mol Cell Biol 1985; 5:3222-30; PMID:3837842; http://dx.doi.org/ 10.1128/MCB.5.11.3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Svitkin YV, Evdokimova VM, Brasey A, Pestova TV, Fantus D, Yanagiya A, Imataka H, Skabkin MA, Ovchinnikov LP, Merrick WC, et al.. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J 2009; 28:58-68; PMID:19078965; http://dx.doi.org/ 10.1038/emboj.2008.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Svitkin YV, Ovchinnikov LP, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. Embo J 1996; 15:7147-55; PMID:9003790 [PMC free article] [PubMed] [Google Scholar]
- [16].Goossen B, Caughman SW, Harford JB, Klausner RD, Hentze MW. Translational repression by a complex between the iron-responsive element of ferritin mRNA and its specific cytoplasmic binding protein is position-dependent in vivo. Embo J 1990; 9:4127-33; PMID:1701143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gingras AC RB, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 2001; 15:2852-64; PMID:11691836; http://dx.doi.org/ 10.1101/gad.887201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Duncan R, Hershey JW. Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J Biol Chem 1983; 258:7228-35; PMID:6853516 [PubMed] [Google Scholar]
- [19].Galicia-Vazquez G, Cencic R, Robert F, Agenor AQ, Pelletier J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA 2012; 18:1373-84; PMID:22589333; http://dx.doi.org/ 10.1261/rna.033209.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 2015; 162:59-71; PMID:26095252; http://dx.doi.org/ 10.1016/j.cell.2015.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, et al.. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest 2007; 117:2638-48; PMID:17786246; http://dx.doi.org/ 10.1172/JCI32044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lin CJ, Nasr Z, Premsrirut PK, Porco JA Jr., Hippo Y, Lowe SW, Pelletier J. Targeting Synthetic Lethal Interactions between Myc and the eIF4F Complex Impedes Tumorigenesis. Cell Reports 2012; 1:325-33; PMID:22573234; http://dx.doi.org/ 10.1016/j.celrep.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yanagiya A, Suyama E, Adachi H, Svitkin YV, Aza-Blanc P, Imataka H, Mikami S, Martineau Y, Ronai ZA, Sonenberg N. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell 2012; 46:847-58; PMID:22578813; http://dx.doi.org/ 10.1016/j.molcel.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, et al.. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res 2012; 72:6468-76; PMID:23100465; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-2395 [DOI] [PubMed] [Google Scholar]
- [25].Ghosh B, Benyumov AO, Ghosh P, Jia Y, Avdulov S, Dahlberg PS, Peterson M, Smith K, Polunovsky VA, Bitterman PB, et al.. Nontoxic chemical interdiction of the epithelial-to-mesenchymal transition by targeting cap-dependent translation. ACS Chem Biol 2009; 4:367-77; PMID:19351181; http://dx.doi.org/ 10.1021/cb9000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hong DS, Kurzrock R, Oh Y, Wheler J, Naing A, Brail L, Callies S, Andre V, Kadam SK, Nasir A, et al.. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin Cancer Res 2011; 17:6582-91; PMID:21831956; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-0430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al.. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 2007; 128:257-67; PMID:17254965; http://dx.doi.org/ 10.1016/j.cell.2006.11.046 [DOI] [PubMed] [Google Scholar]
- [28].Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, Qui M, Lewis I, Kurtkaya S, Dingledine R, et al.. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proceedings of the National Academy of Sciences of the United States of America 2011; 108:1046-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, Dingledine R, Fu H, Vajda S, Talbot PJ, et al.. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. Journal of virology 2011; 85:6381-9; PMID:21507972; http://dx.doi.org/ 10.1128/JVI.00078-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc Natl Acad Sci U S A 2005; 102:10460-5; PMID:16030146; http://dx.doi.org/ 10.1073/pnas.0504249102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA Jr., et al.. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest 2008; 118:2651-60; PMID:18551192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2006; 2:213-20; PMID:16532013; http://dx.doi.org/ 10.1038/nchembio776 [DOI] [PubMed] [Google Scholar]
- [33].Higa T, Tanaka J, Yasumasa T, Hiroyuki K. Hippuristanols, cytotoxic polyoxygenated steroids from the gorgonian Isis hippuris. Chem Lett 1981; 11:1647-50; http://dx.doi.org/ 10.1246/cl.1981.1647 [DOI] [Google Scholar]
- [34].Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem 1987; 262:380-8; PMID:3793730 [PubMed] [Google Scholar]
- [35].Rogers GW Jr., Komar AA, Merrick WC. eIF4A: The godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 2002; 72:307-31; PMID:12206455; http://dx.doi.org/ 10.1016/S0079-6603(02)72073-4 [DOI] [PubMed] [Google Scholar]
- [36].Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. New initiation factor activity required for globin mRNA translation. J Biol Chem 1983; 258:5804-10; PMID:6853548 [PubMed] [Google Scholar]
- [37].Edery I, Humbelin M, Darveau A, Lee KA, Milburn S, Hershey JW, Trachsel H, Sonenberg N. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J Biol Chem 1983; 258:11398-403; PMID:6604056 [PubMed] [Google Scholar]
- [38].Thomas A, Goumans H, Amesz H, Benne R, Voorma HO. A comparison of the initiation factors of eukaryotic protein synthesis from ribosomes and from the postribosomal supernatant. Eur J Biochem 1979; 98:329-37; PMID:488105; http://dx.doi.org/ 10.1111/j.1432-1033.1979.tb13192.x [DOI] [PubMed] [Google Scholar]
- [39].Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog Nucleic Acid Res Mol Biol 1988; 35:173-207; PMID:3065823; http://dx.doi.org/ 10.1016/S0079-6603(08)60614-5 [DOI] [PubMed] [Google Scholar]
- [40].Lindqvist L, Imataka H, Pelletier J. Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. RNA 2008; 14:960-9; PMID:18367715; http://dx.doi.org/ 10.1261/rna.971208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nielsen PJ, Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. Embo J 1988; 7:2097-105; PMID:3046931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yoder-Hill J, Pause A, Sonenberg N, Merrick WC. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J Biol Chem 1993; 268:5566-73; PMID:8449919 [PubMed] [Google Scholar]
- [43].Galicia-Vazquez G, Chu J, Pelletier J. eIF4AII is dispensable for miRNA-mediated gene silencing. RNA 2015; 21:1826-33; PMID:26286746; http://dx.doi.org/ 10.1261/rna.052225.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nielsen PJ, McMaster GK, Trachsel H. Cloning of eukaryotic protein synthesis initiation factor genes: isolation and characterization of cDNA clones encoding factor eIF-4A. Nucl Acids Res 1985; 13:6867-80; PMID:3840589; http://dx.doi.org/ 10.1093/nar/13.19.6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sudo K, Takahashi E, Nakamura Y. Isolation and mapping of the human EIF4A2 gene homologous to the murine protein synthesis initiation factor 4A-II gene Eif4a2. Cytogenet Cell Genet 1995; 71:385-8; PMID:8521730; http://dx.doi.org/ 10.1159/000134145 [DOI] [PubMed] [Google Scholar]
- [46].Rogers GW Jr., Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 2002; 72:307-31; PMID:12206455; http://dx.doi.org/ 10.1016/S0079-6603(02)72073-4 [DOI] [PubMed] [Google Scholar]
- [47].Garcia-Garcia C, Frieda KL, Feoktistova K, Fraser CS, Block SM. RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science 2015; 348:1486-8; PMID:26113725; http://dx.doi.org/ 10.1126/science.aaa5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 1994; 13:1205-15; PMID:8131750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 2001; 7:382-94; PMID:11333019; http://dx.doi.org/ 10.1017/S135583820100108X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, et al.. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One 2009; 4:e5223; PMID:19401772; http://dx.doi.org/ 10.1371/journal.pone.0005223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al.. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014; 513:65-70; PMID:25079319; http://dx.doi.org/ 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, Fanidi A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol 2014; 15:476; PMID:25273840; http://dx.doi.org/ 10.1186/s13059-014-0476-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 2003; 23:26-37; PMID:12482958; http://dx.doi.org/ 10.1128/MCB.23.1.26-37.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Suzuki C, Garces RG, Edmonds KA, Hiller S, Hyberts SG, Marintchev A, Wagner G. PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc Natl Acad Sci U S A 2008; 105:3274-9; PMID:18296639; http://dx.doi.org/ 10.1073/pnas.0712235105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zakowicz H, Yang HS, Stark C, Wlodawer A, Laronde-Leblanc N, Colburn NH. Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. Rna 2005; 11:261-74; PMID:15661843; http://dx.doi.org/ 10.1261/rna.7191905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol 2004; 24:3894-906; PMID:15082783; http://dx.doi.org/ 10.1128/MCB.24.9.3894-3906.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006; 314:467-71; PMID:17053147; http://dx.doi.org/ 10.1126/science.1130276 [DOI] [PubMed] [Google Scholar]
- [58].Eberle J, Fecker LF, Bittner JU, Orfanos CE, Geilen CC. Decreased proliferation of human melanoma cell lines caused by antisense RNA against translation factor eIF-4A1. Br J Cancer 2002; 86:1957-62; PMID:12085193; http://dx.doi.org/ 10.1038/sj.bjc.6600351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang HS, Knies JL, Stark C, Colburn NH. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene 2003; 22:3712-20; PMID:12802278; http://dx.doi.org/ 10.1038/sj.onc.1206433 [DOI] [PubMed] [Google Scholar]
- [60].Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res 2005; 65:6034-41; PMID:16024603; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-2119 [DOI] [PubMed] [Google Scholar]
- [61].Kogure T, Kinghorn AD, Yan I, Bolon B, Lucas DM, Grever MR, Patel T. Therapeutic potential of the translation inhibitor silvestrol in hepatocellular cancer. PLoS One 2013; 8:e76136; PMID:24086701; http://dx.doi.org/ 10.1371/journal.pone.0076136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lucas DM, Edwards RB, Lozanski G, West DA, Shin JD, Vargo MA, Davis ME, Rozewski DM, Johnson AJ, Su BN, et al.. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood 2009; 113:4656-66; PMID:19190247; http://dx.doi.org/ 10.1182/blood-2008-09-175430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, Shi W, Zhang Z, Rajasekhar VK, Pagano NC, et al.. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med 2011; 208:1799-807; PMID:21859846; http://dx.doi.org/ 10.1084/jem.20110846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Novac O, Guenier AS, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res 2004; 32:902-15; PMID:14769948; http://dx.doi.org/ 10.1093/nar/gkh235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Longley RE, McConnell OJ, Essich E, Harmody D. Evaluation of marine sponge metabolites for cytotoxicity and signal transduction activity. J Nat Prod 1993; 56:915-20; PMID:8350092; http://dx.doi.org/ 10.1021/np50096a015 [DOI] [PubMed] [Google Scholar]
- [66].Gonzalez N, Barral MA, Rodriguez J, Jimenez C. New cytotoxic steroids from the gorgonian Isis hippuris. Structure-activity studies. Tetrahedron 2001; 57:3487-97; http://dx.doi.org/ 10.1016/S0040-4020(01)00223-X [DOI] [Google Scholar]
- [67].Chao CH, Huang LF, Yang YL, Su JH, Wang GH, Chiang MY, Wu YC, Dai CF, Sheu JH. Polyoxygenated steroids from the gorgonian Isis hippuris. J Nat Prod 2005; 68:880-5; PMID:15974612; http://dx.doi.org/ 10.1021/np050033y [DOI] [PubMed] [Google Scholar]
- [68].Tanaka J, Trianto A, Musman M, Issa HH, Ohtani II, Ichiba T, Higa T, Yoshida WY, Scheuer PJ. New polyoxygenated steroids exhibiting reversal of multidrug resistance from the gorgonian Isis hippuris. Tetrahed Lett 2002; 58:6259-66; http://dx.doi.org/ 10.1016/S0040-4020(02)00625-7 [DOI] [Google Scholar]
- [69].Uddin MH, Hanif N, Trianto A, Agarie Y, Higa T, Tanaka J. Four new polyoxygenated gorgosterols from the gorgonian Isis hippuris. Nat Prod Res 2011; 25:585-91; PMID:21409719; http://dx.doi.org/ 10.1080/14786419.2010.485303 [DOI] [PubMed] [Google Scholar]
- [70].Aoki S, Chen ZS, Higasiyama K, Setiawan A, Akiyama S, Kobayashi M. Reversing effect of agosterol A, a spongean sterol acetate, on multidrug resistance in human carcinoma cells. Jpn J Cancer Res 2001; 92:886-95; PMID:11509122; http://dx.doi.org/ 10.1111/j.1349-7006.2001.tb01177.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen WH, Wang SK, Duh CY. Polyhydroxylated steroids from the bamboo coral Isis hippuris. Mar Drugs 2011; 9:1829-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rao CB, Ramana KV, Rao DV, Fahy E, Faulkner JD. Metabolites of the gorgonian Isis hippuris from India. J Nat Products 1988; 51:954-8; http://dx.doi.org/ 10.1021/np50059a023 [DOI] [Google Scholar]
- [73].Sheu J-H, Chen S-P, Sung P-J, Chiang MY, Dai C-F. Hippuristerone A, a novel polyoxygenated steroid from the gorgonian Isis hippuris. Tetrahed Lett 2000; 41:7885-8; http://dx.doi.org/ 10.1016/S0040-4039(00)01348-4 [DOI] [Google Scholar]
- [74].Shen Y-C, Prakash CVS, Chang Y-T. Two new polyhydroxysteroids from the gorgonian Isis hippuris. Steroids 2001; 66:721-5; PMID:11546560; http://dx.doi.org/ 10.1016/S0039-128X(01)00098-8 [DOI] [PubMed] [Google Scholar]
- [75].Chao CH, Huang LF, Wu SL, Su JH, Huang HC, Sheu JH. Steroids from the gorgonian Isis hippuris. J Nat Prod 2005; 68:1366-70; PMID:16180815; http://dx.doi.org/ 10.1021/np050200u [DOI] [PubMed] [Google Scholar]
- [76].Sun Y, Atas E, Lindqvist LM, Sonenberg N, Pelletier J, Meller A. Single-Molecule Kinetics of the Eukaryotic Initiation Factor 4AI upon RNA Unwinding. Structure 2014; 22:941-48 [DOI] [PubMed] [Google Scholar]
- [77].Harms U, Andreou AZ, Gubaev A, Klostermeier D. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res 2014; 42:7911-22; PMID:24848014; http://dx.doi.org/ 10.1093/nar/gku440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, Vogt E, Marintchev A, Tanaka J, Fagotto F, Altmann M, et al.. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS ONE 2008; 3:e158; http://dx.doi.org/ 10.1371/journal.pone.0001583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol 2004; 11:346-51; PMID:15034551; http://dx.doi.org/ 10.1038/nsmb750 [DOI] [PubMed] [Google Scholar]
- [80].Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natil Acad Sci U S A 2004; 101:4118-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 2004; 427:753-7; PMID:14973490; http://dx.doi.org/ 10.1038/nature02351 [DOI] [PubMed] [Google Scholar]
- [82].Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. Rna 2004; 10:200-9; PMID:14730019; http://dx.doi.org/ 10.1261/rna.5230104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Chard LS, Bordeleau ME, Pelletier J, Tanaka J, Belsham GJ. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J Gen Virol 2006; 87:927-36; PMID:16528042; http://dx.doi.org/ 10.1099/vir.0.81546-0 [DOI] [PubMed] [Google Scholar]
- [84].Bakhshesh M, Groppelli E, Willcocks MM, Royall E, Belsham GJ, Roberts LO. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J Virol 2008; 82:1993-2003; PMID:18077729; http://dx.doi.org/ 10.1128/JVI.01957-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Belsham GJ, Nielsen I, Normann P, Royall E, Roberts LO. Monocistronic mRNAs containing defective hepatitis C virus-like picornavirus internal ribosome entry site elements in their 5′ untranslated regions are efficiently translated in cells by a cap-dependent mechanism. Rna 2008; 14:1671-80; PMID:18567818; http://dx.doi.org/ 10.1261/rna.1039708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Spriggs KA, Cobbold LC, Jopling CL, Cooper RE, Wilson LA, Stoneley M, Coldwell MJ, Poncet D, Shen YC, Morley SJ, et al.. Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Mol Cell Biol 2009; 29:1565-74; PMID:19124605; http://dx.doi.org/ 10.1128/MCB.01283-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Fred RG, Sandberg M, Pelletier J, Welsh N. The human insulin mRNA is partly translated via a cap- and eIF4A-independent mechanism. Biochem Biophys Res Commun 2011; 412:693-8; PMID:21867683; http://dx.doi.org/ 10.1016/j.bbrc.2011.08.030 [DOI] [PubMed] [Google Scholar]
- [88].Tsai BP, Wang X, Huang L, Waterman ML. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol Cell Proteomics 2011; 10:M110 007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Olson CM, Donovan MR, Spellberg MJ, Marr MT 2nd. The insulin receptor cellular IRES confers resistance to eIF4A inhibition. eLife 2013; 2:e00542; PMID:23878722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tsai BP, Jimenez J, Lim S, Fitzgerald KD, Zhang M, Chuah CT, Axelrod H, Wilson L, Ong ST, Semler BL, et al.. A novel Bcr-Abl-mTOR-eIF4A axis regulates IRES-mediated translation of LEF-1. Open Biol 2014; 4:140180; PMID:25392452; http://dx.doi.org/ 10.1098/rsob.140180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Webb TE, Hughes A, Smalley DS, Spriggs KA. An internal ribosome entry site in the 5′ untranslated region of epidermal growth factor receptor allows hypoxic expression. Oncogenesis 2015; 4:e134; PMID:25622307; http://dx.doi.org/ 10.1038/oncsis.2014.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Dauber B, Pelletier J, Smiley JR. The herpes simplex virus 1 vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J Virol 2011; 85:5363-73; PMID:21430045; http://dx.doi.org/ 10.1128/JVI.00115-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Radtke K, English L, Rondeau C, Leib D, Lippe R, Desjardins M. Inhibition of the host translation shutoff response by herpes simplex virus 1 triggers nuclear envelope-derived autophagy. J Virol 2013; 87:3990-7; PMID:23365427; http://dx.doi.org/ 10.1128/JVI.02974-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Yanguez E, Castello A, Welnowska E, Carrasco L, Goodfellow I, Nieto A. Functional impairment of eIF4A and eIF4G factors correlates with inhibition of influenza virus mRNA translation. Virology 2011; 413:93-102; PMID:21377182; http://dx.doi.org/ 10.1016/j.virol.2011.02.012 [DOI] [PubMed] [Google Scholar]
- [95].Ran I, Laplante I, Bourgeois C, Pepin J, Lacaille P, Costa-Mattioli M, Pelletier J, Sonenberg N, Lacaille JC. Persistent transcription- and translation-dependent long-term potentiation induced by mGluR1 in hippocampal interneurons. J Neurosci 2009; 29:5605-15; PMID:19403827; http://dx.doi.org/ 10.1523/JNEUROSCI.5355-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hoeffer CA, Santini E, Ma T, Arnold EC, Whelan AM, Wong H, Pierre P, Pelletier J, Klann E. Multiple components of eIF4F are required for protein synthesis-dependent hippocampal long-term potentiation. J Neurophysiol 2013; 109:68-76; PMID:23054596; http://dx.doi.org/ 10.1152/jn.00342.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Tsumuraya T, Ishikawa C, Machijima Y, Nakachi S, Senba M, Tanaka J, Mori N. Effects of hippuristanol, an inhibitor of eIF4A, on adult T-cell leukemia. Biochem Pharmacol 2011; 81:713-22; PMID:21219881; http://dx.doi.org/ 10.1016/j.bcp.2010.12.025 [DOI] [PubMed] [Google Scholar]
- [98].Ishikawa C, Tanaka J, Katano H, Senba M, Mori N. Hippuristanol reduces the viability of primary effusion lymphoma cells both in vitro and in vivo. Mar Drugs 2013; 11:3410-24; PMID:24018901; http://dx.doi.org/ 10.3390/md11093410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A 2011; 108:E699-708; PMID:21876152; http://dx.doi.org/ 10.1073/pnas.1108237108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Cope CL, Gilley R, Balmanno K, Sale MJ, Howarth KD, Hampson M, Smith PD, Guichard SM, Cook SJ. Adaptation to mTOR kinase inhibitors by amplification of eIF4E to maintain cap-dependent translation. J Cell Sci 2014; 127:788-800; PMID:24363449; http://dx.doi.org/ 10.1242/jcs.137588 [DOI] [PubMed] [Google Scholar]
- [101].Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 2004; 428:332-7; PMID:15029198; http://dx.doi.org/ 10.1038/nature02369 [DOI] [PubMed] [Google Scholar]
- [102].Wendel HG, Malina A, Zhao Z, Zender L, Kogan SC, Cordon-Cardo C, Pelletier J, Lowe SW. Determinants of sensitivity and resistance to rapamycin-chemotherapy drug combinations in vivo. Cancer Res 2006; 66:7639-46; PMID:16885364; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, Thomas M, Basmadjian C, Ribeiro N, Thuaud F, et al.. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature 2014; 513:105-9; PMID:25079330; http://dx.doi.org/ 10.1038/nature13572 [DOI] [PubMed] [Google Scholar]
- [104].Cencic R, Robert F, Galicia-Vazquez G, Malina A, Ravindar K, Somaiah R, Pierre P, Tanaka J, Deslongchamps P, Pelletier J. Modifying chemotherapy response by targeted inhibition of eukaryotic initiation factor 4A. Blood Cancer J 2013; 3:e128; PMID:23872707; http://dx.doi.org/ 10.1038/bcj.2013.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Robert F, Roman W, Bramoulle A, Fellmann C, Roulston A, Shustik C, Porco JA Jr., Shore GC, Sebag M, Pelletier J. Translation initiation factor eIF4F modifies the dexamethasone response in multiple myeloma. Proc Natl Acad Sci U S A 2014; 111:13421-6; PMID:25197055; http://dx.doi.org/ 10.1073/pnas.1402650111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Li W, Dang Y, Liu JO, Yu B. Expeditious synthesis of hippuristanol and congeners with potent antiproliferative activities. Chemistry 2009; 15:10356-9; PMID:19746484; http://dx.doi.org/ 10.1002/chem.200901732 [DOI] [PubMed] [Google Scholar]
- [107].Ravindar K, Reddy MS, Lindqvist L, Pelletier J, Deslongchamps P. Efficient Synthetic Approach to Potent Antiproliferative Agent Hippuristanol via Hg(II)-Catalyzed Spiroketalization. Org Lett 2010; 12:4420-3; PMID:20828120; http://dx.doi.org/ 10.1021/ol1019663 [DOI] [PubMed] [Google Scholar]
- [108].Ravindar K, Reddy MS, Lindqvist L, Pelletier J, Deslongchamps P. Synthesis of the antiproliferative agent hippuristanol and its analogues via Suarez cyclizations and Hg(II)-catalyzed spiroketalizations. J Org Chem 2011; 76:1269-84; PMID:21268618; http://dx.doi.org/ 10.1021/jo102054r [DOI] [PubMed] [Google Scholar]
- [109].Somaiah R, Ravindar K, Cencic R, Pelletier J, Deslongchamps P. Synthesis of the antiproliferative agent hippuristanol and its analogues from hydrocortisone via Hg(II)-catalyzed spiroketalization: structure-activity relationship. J Med Chem 2014; 57:2511-23; PMID:24588834; http://dx.doi.org/ 10.1021/jm401799j [DOI] [PubMed] [Google Scholar]
- [110].Li W, Dang Y, Liu JO, Yu B. Structural and stereochemical requirements of the spiroketal group of hippuristanol for antiproliferative activity. Bioorg Med Chem Lett 2010; 20:3112-5; PMID:20409710; http://dx.doi.org/ 10.1016/j.bmcl.2010.03.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Rozovsky N, Butterworth AC, Moore MJ. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA 2008; 14:2136-48; PMID:18719248; http://dx.doi.org/ 10.1261/rna.1049608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A 2004; 101:18105-10; PMID:15601771; http://dx.doi.org/ 10.1073/pnas.0406927102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yan Y, Svitkin Y, Lee JM, Bisaillon M, Pelletier J. Ribavirin is not a functional mimic of the 7-methyl guanosine mRNA cap. Rna 2005; 11:1238-44; PMID:16043507; http://dx.doi.org/ 10.1261/rna.2930805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Westman B, Beeren L, Grudzien E, Stepinski J, Worch R, Zuberek J, Jemielity J, Stolarski R, Darzynkiewicz E, Rhoads RE, et al.. The antiviral drug ribavirin does not mimic the 7-methylguanosine moiety of the mRNA cap structure in vitro. Rna 2005; 11:1505-13; PMID:16131589; http://dx.doi.org/ 10.1261/rna.2132505 [DOI] [PMC free article] [PubMed] [Google Scholar]