Abstract
The icosahedral bacteriophage PM2 has a circular double-stranded DNA (dsDNA) genome and an internal lipid membrane. It is the only representative of the Corticoviridae family. How the circular supercoiled genome residing inside the viral membrane is translocated into the gram-negative marine Pseudoalteromonas host has been an intriguing question. Here we demonstrate that after binding of the virus to an abundant cell surface receptor, the protein coat is most probably dissociated. During the infection process, the host cell outer membrane becomes transiently permeable to lipophilic gramicidin D molecules proposing fusion with the viral membrane. One of the components of the internal viral lipid core particle is the integral membrane protein P7, with muralytic activity that apparently aids the process of peptidoglycan penetration. Entry of the virion also causes a limited depolarization of the cytoplasmic membrane. These phenomena differ considerably from those observed in the entry process of bacteriophage PRD1, a dsDNA virus, which uses its internal membrane to make a cell envelope-penetrating tubular structure.
The key step in the life cycle of most viruses is the successful interaction with an appropriate host via a receptor on the cell surface. Several obstacles must be overcome for successful entry of a virus into the host cell. In the case of gram-negative bacteria, the envelope contains three layers: two membranes and a rigid peptidoglycan layer. In addition, nucleases in the periplasmic space form a challenge to bacteriophage entry. Binding to a specific outer membrane (OM) receptor has been well described for several phages (44, 45), but the mechanism of DNA entry into the bacterial cell is still a poorly understood process.
Bacteriophages use diverse strategies for genome entry (for reviews, see references 30, 59, 60, and 77). Tailed double-stranded-DNA (dsDNA) bacteriophages of Escherichia coli, such as T4, T5, T7, and lambda, have stable icosahedral capsids with a proteinaceous tail associated with the portal vertex. This unique vertex is used for genome packaging and delivery into the host bacterium. Receptor binding by the tail leads to structural rearrangements, and then the linear DNA is injected with the aid of the tail. The empty capsid is left on the cell surface. Mechanisms of penetration vary among tailed phages: the genomes of T5 and T7 are transported across the envelope through a protein-rich channel partially formed by phage proteins (35, 59, 67, 68), whereas the channel for the transfer of the lambda genome is formed by host-derived proteins (11, 79). Translocation of the T7 genome occurs simultaneously with its transcription by the host and T7 polymerases (36, 94), while phage T4 induces fusion between the OM and cytoplasmic membrane (CM) (88, 89). However, in the case of bacterial viruses that contain a membrane and no tail, the lipid component is used for cell entry. For example, the enveloped dsRNA phage φ6 fuses its external membrane with the host OM (5, 76) and the dsDNA phage PRD1 uses its internal membrane to form a tubular structure that penetrates the cell envelope (4, 38, 61).
Transport of viral DNA across the host cell membranes is a highly efficient energy-dependent process. It has been demonstrated that some phages utilize the proton motive force (Δp), consisting of membrane voltage (ΔΨ) and pH difference across the CM (ΔpH) for DNA delivery. The injection of phage T4 DNA into the host is strictly dependent on the metabolic state of the cell (50). A minimal CM voltage of ∼90 mV is needed for the proper delivery of the genome (56). However, other tailed phages such as T5 and lambda can transfer their genomes across the envelope of deenergized cells (57, 79). Changes in the CM permeability and the depolarization of the membrane indicate that the genomes of phages T4, T5, and lambda are injected through a transient channel formed in the CM (14, 15, 41). Surprisingly, during the penetration of phage PRD1 DNA, the host CM is not depolarized but the membrane voltage increases (27).
Measurements of ion gradients across the cell envelope during bacteriophage infection have been used to study DNA entry processes (27, 59, 60). The OM of gram-negative bacteria forms a relatively permeable barrier allowing small hydrophilic metabolites and inorganic ions to pass through porins (72). However, the highly charged lipopolysaccharide (LPS) and proteins in the outer leaflet of the OM make a permeability barrier to lipophilic compounds (73). The bacterial CM is a highly selective barrier. It is permeable to lipophilic ions like tetraphenylphosphonium (TPP+) and ionophoric antibiotics like gramicidin D (GD), while inorganic ions like K+ do not cross the CM. The efflux of intracellular potassium is used as an indicator of increased CM permeability (for details, see references 27 and 59). Measuring the distribution of the lipophilic membrane voltage indicator TPP+ between cells and medium or binding of phenyldicarbaundecaborane (PCB−) to the cellular membranes has been used to study the energy state of the CM and the permeability of the OM. The ion fluxes across the cell envelope can be observed by using selective electrodes that monitor the concentrations of indicator ions in the media.
Bacteriophage PM2 (31) is a lipid-containing dsDNA virus that infects two marine pseudoalteromonads. Pseudoalteromonas espejiana BAL-31 and the virus were isolated from coastal seawater in Chile (32). Another host, ER72M2, was isolated from the East River, New York City, N.Y.. Based on its ribosomal 16S sequence, ER72M2 also belongs to the pseudoalteromonads (53). Gram-negative pseudoalteromonads are common in the marine environment. They are strictly aerobic, polarly flagellated, heterotrophic Pseudomonas-like bacteria originally classified as alteromonads (9). Later, the genus Alteromonas was divided into Pseudoalteromonas and Alteromonas (37).
PM2 is the only known isolate of the Corticoviridae family (1, 7). The virion (∼65 nm in diameter) consists of an icosahedral capsid surrounding a protein-rich membrane vesicle, the lipid core (LC), which encloses the circular dsDNA genome of 10,079 bp (43, 48, 54, 62). The highly supercoiled phage genome replicates in association with the host CM, using the rolling-circle mechanism initiated by the virally encoded replication initiation protein P12 (18, 62). Among its 21 putative genes, ten (I to X) have been shown to encode structural components, proteins P1 to P10 (48, 53, 54, 62). All structural proteins, except P9, are encoded by the late operon, which is regulated by two phage-encoded transcription factors, P13 and P14 (63). For transcription, PM2 uses the host RNA polymerase (95). Virus-derived endolysin activity induced during PM2 infection has been described (90).
The mass of the virion is distributed among protein (72%), lipid (14%), and nucleic acid (14%) (22, 23, 24). Cryo-electron microscopy-based three-dimensional image analysis of native PM2 showed that trimers of the major capsid protein P2 are organized on an icosahedral pseudo-T = 21 lattice (48). There are pentameric spike complexes composed of protein P1 at the fivefold vertices. The proximal N-terminal domains of P1 are connected to the P2 capsid, and the distal C-terminal domains are exposed to the medium (54). The PM2 virion is a metastable structure stabilized by Ca2+ ions. In the absence of calcium, the coat dissociates revealing the spherical hydrophobic LC particle (54). LC-associated proteins (P3 to P10) occupy about one-third of the PM2 membrane, and the rest are lipids derived from the host CM during virus maturation (33, 43). The capsid and the underlying LC are linked by 60 transmembrane anchors near the twofold symmetry axes (48).
The process of entry of the viral genome into its host in general, and into a gram-negative cell in particular, is poorly understood. The internal membrane component in the PM2 virion suggests its involvement in this process. Currently, there is no information describing the early stages in the PM2 life cycle. In this investigation, we characterized the adsorption parameters and measured cellular energy changes during PM2 entry. We also identified a virion protein with murolytic activity that potentially is involved in locally digesting the peptidoglycan during virus entry.
MATERIALS AND METHODS
Bacterial strains, virus, and plasmids.
Bacterial strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Phage PM2 (31) and Pseudoalteromonas cells were cultured in SB broth (53) or in a marine defined rich medium (63, 70) at 28°C. E. coli strains were propagated in Luria-Bertani broth (LB) (82) at 37°C. When HB101 was used in adsorption tests, LB medium was supplemented with 10 mM CaCl2 to maintain phage viability.
TABLE 1.
Bacteria used in this study
| Bacteria | Relevant property or usage | Reference |
|---|---|---|
| Pseudoalteromonas | ||
| espejiana BAL-31 | PM2 host | 32, 37 |
| sp. strain ER72M2 | PM2 host | 53 |
| sp. strain A28 | Adsorption test, transfection | 52 |
| haloplanktis subsp. haloplanktis | Transfection | 37, 96 |
| haloplanktis subsp. tetraodonis | Transfection | 37, 87 |
| undina | Transfection | 26, 37 |
| nigrifaciens | Transfection | 37, 93 |
| Alteromonas macleodie | Transfection | 9, 37 |
| PM2-resistant Pseudo- alteromonas sp. strain ER72M2 mutants | Adsorption test | This study |
| Escherichia coli | ||
| HB101 | Adsorption test | 16 |
| DH5α | Molecular cloning and source of peptidoglycan | 82 |
| DH5α(pDMI)a | Protein expression | 25, 82 |
| HMS174(DE3) | Protein expression | 87 |
Plasmid pDMI has a p15A replicon encodes resistance to kanamycin as the selective marker, and contains the gene for the lac repressor.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Relevant property or usage | Reference |
|---|---|---|---|
| pDS12 | Expression vector, colE1 replicon, Apr | 19 | |
| pJJ2 | Expression vector, colE1 replicon, Apr | 74 | |
| pET24 | Expression vector, pMB1 replicon, Kmr | Novagen | |
| pRM203 | pDS12 Δ(EcoRI-BamHI) Ω(PM2 6333-6659) | Expression of P3 | This study |
| pRM206 | pDS12 Δ(EcoRI-BamHI) Ω(PM2 7903-8928) | Expression of P1 | This study |
| pRM301 | pJJ2 Δ(EcoRI-BamHI) Ω(PM2 9388-9783) | Expression of P6 | This study |
| pRM315 | pJJ2 Δ(EcoRI-BamHI) Ω(PM2 6646-6781) | Expression of P4 | This study |
| pRM318 | pJJ2 Δ(EcoRI-BamHI) Ω(PM2 5393-5510) | Expression of P7 | This study |
| pRM319 | pJJ2 Δ(EcoRI-BamHI) Ω(PM2 8911-9407) | Expression of P5 | This study |
| pRM325 | pJJ2 Δ(EcoRI-BamHI) Ω(PM2 6766-7008) | Expression of P8 | This study |
| pRM607 | pET24 Δ(EcoRI-BamHI) Ω(PM2 5508-6335) | Expression of P2 | This study |
Transfection of Pseudoalteromonas cells by phage DNA.
PM2 DNA was isolated as described by Männistö et al. (62) and transferred into Pseudoalteromonas cells by transfection according to van der Schans et al. (91). Transfected spheroplasts were grown for 2 h followed by plating on a lawn of PM2-sensitive host cells.
Isolation of phage-resistant mutants.
For the isolation of spontaneous PM2-resistant ER72M2 cells, confluent plates were prepared. After overnight growth at 28°C, PM2-resistant colonies were picked, and five subsequent single-colony purifications were carried out. The sensitivity of the resulting strains to phage was tested, and the total protein compositions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to detect possible contaminating species.
Growth and purification of the phage.
Radioactive labeling of the PM2 virion was carried out in marine defined rich medium (see above) not including methionine. PM2 was labeled either with l-[35S]methionine (11 μCi/ml; Amersham SJ204) added 10 min after infection or with 33P (11 μCi/ml; Amersham BF1003) added 3 min prior to infection of a BAL-31 culture (7.5 × 108 CFU/ml), using a multiplicity of infection (MOI) of 10. The virus particles from the lysate were concentrated with polyethylene glycol 6000 and purified by rate zonal centrifugation (53, 54). The fresh virus zones were stored on ice and used in the adsorption assays within 2 days. In other cases the analyses were carried out using radioactive virus preparation stored at −20°C. The specific activities were 1.3 × 10−6 cpm/PFU and 1.8 × 1013 PFU/mg of protein for fresh l-[35S]methionine-labeled PM2, and 2.0 × 10−6 cpm/PFU and 2.2 × 1013 PFU/mg of protein for fresh 33P-labeled PM2.
Distribution of the incorporated l-[35S]methionine in the PM2 virion was analyzed by separating the proteins by SDS-PAGE and measuring the radioactivity associated with the Coomassie-blue stained protein bands by scintillation counting. Incorporation of 33P label into the lipid membrane and nucleic acid was analyzed by extracting the phospholipids (12, 54) and nucleic acid (62) components from the virus preparation and measuring the radioactivity. The viral DNA was also digested with PstI and analyzed by 0.7% agarose gel electrophoresis, confirming the association of 33P label with phage DNA.
The nonradioactive virus was grown on ER72M2 in SB broth and purified as described previously (53, 54). The 1× purified virus was concentrated by differential centrifugation and resuspended in 20 mM Tris-HCl (pH 7.2)-100 mM NaCl-5 mM CaCl2. The specific infectivity of this virus material was 6 × 1012 to 1 × 1013 PFU/mg of protein. The nonradioactive virus material was stored on ice and used within 3 days.
Phage adsorption.
The phage adsorption tests were performed in SB broth (except for HB101, see above) by infecting cells grown to a density of 7 × 108 CFU/ml with an MOI of ∼0.3 using a fresh filtered virus agar stock. After different incubation times (at 28°C with aeration), adsorption was stopped by diluting the cells 100-fold in ice-cold SB buffer (50 mM Tris-HCl [pH 8.0], 450 mM NaCl, 50 mM MgSO4, 10 mM KCl, 10 mM CaCl2). The cells were removed (Heraeus Biofuge; 13,000 rpm, 5 min, 4°C), and the PFU in the supernatant was determined by plating on lawns of PM2-sensitive cells. The adsorption rate constants were calculated as described by Adams (2). For the receptor saturation assay, a constant number of BAL-31 or ER72M2 cells was infected with MOI values between 1 and 850. After a 15-min incubation at 28°C with aeration, the cells were collected and washed once with SB buffer. The reduction in the number of PFU in the supernatant fractions was determined by plating on the lawns of phage-sensitive cells. The adsorption test using the labeled virus preparations was performed in SB medium, infecting ER72M2 cells using MOIs of 1, 2, 5, and 10. After a 15-min incubation at 28°C with aeration the cells were separated by centrifugation, washed once with SB buffer, and resuspended in the original volume. We measured the levels of radioactivity in the cell and supernatant fractions and determined the reduction in the number of PFU in the supernatant fraction. The cell and supernatant fractions were also analyzed by tricine-SDS-PAGE followed by autoradiography. The adsorption of the labeled virus to ER72M2 cells was also analyzed by rate zonal centrifugation. After a 15-min adsorption the cells were layered on a linear 5 to 17% sucrose gradient (SB buffer; Sorvall TH641 rotor; 24,000 rpm, 60 min, 15°C). A similar amount of virus particles alone was used as a control. The gradients were fractionated, and the amount of radioactivity was measured by scintillation counting. The number of infective centers and viable cells were determined after a 10-min adsorption, using an MOI of 10, by separating the nonadsorbed virus particles from the cells by centrifugation. The cells were washed once and plated on SB plates (viable cell count) and on SB soft-agar lawns of sensitive cells (infective centers).
Adsorption in the presence of isolated LPS.
LPSs were isolated from ER72M2, BAL-31, and HB101 cells according to the procedure of Westphal and Jann (92). A 250-ml overnight culture was harvested, washed with 200 mM NaCl, and suspended in 5 ml of 67°C deionized water. Five milliliters of phenol (67°C; Merck) was added, and the extraction was incubated for 15 min at 67°C and cooled to ∼10°C on ice. The water and phenol phases were separated by centrifugation (Sorvall SS34 rotor; 3,000 rpm, 45 min, 10°C). The resulting phenol layer was extracted with another 5 ml of water. The two water phases were combined and dialyzed against deionized water at 4°C for five nights. After dialysis, the mixture (∼10 ml) was concentrated threefold under vacuum, stored overnight at 4°C, and used in the adsorption test as described above. Increasing amounts of the isolated LPS fraction were mixed with a constant number of exponentially growing ER72M2 cells and incubated for 15 min at 28°C with aeration. After this, the cells were infected with PM2 (MOI, ∼0.4). The amount of nonadsorbed phages was determined after a 10-min adsorption as described above.
Adsorption in the presence of isolated cell fractions.
Twenty milliliters of exponentially growing ER72M2, BAL-31, and HB101 cells (∼2 × 108 CFU/ml) was collected and resuspended in 2 ml of 10 mM Tris-HCl, pH 8.0, containing 1 mM EDTA. Cells were disrupted by sonication on ice (four 5-s pulses), and intact cells were removed by centrifugation (Heraeus Biofuge; 10,000 rpm, 30 s, 4°C). The broken cells were fractionated by centrifugation (Heraeus Biofuge; 13,000 rpm, 10 min, 4°C), and the resulting pellet was washed with the above-mentioned buffer and centrifuged again. The two supernatant fractions were combined (soluble fraction), and the pellet (insoluble fraction) was resuspended in deionized water. After overnight storage at 4°C, the fractions were used in the adsorption test as described above. Increasing amounts of these fractions were mixed with exponentially growing ER72M2 cells and incubated for 15 min at 28°C. The cells were infected with PM2 (MOI, ∼0.4), and the number of nonadsorbed particles was determined.
Adsorption to protease-treated cells.
ER72M2 and HB101 cells were grown to a density of ∼5 × 108 CFU/ml. HB101 cultures were transferred to 28°C, and tetracycline was added to both cultures at a final concentration of 100 μg/ml to prevent protein synthesis. After 30 min, pronase, trypsin, and proteinase K were added at final concentrations of 2 mg/ml, 100 μg/ml, and 400 μg/ml, respectively, followed by incubation at 28°C for 10, 30, or 60 min. The cells were washed twice to remove the proteases and resuspended in SB medium containing 100 μg of tetracycline/ml. Treated cells were incubated for 15 min at 28°C before usage in the adsorption test as described above (10-min adsorption time). Cell densities and viabilities were determined by plating the cells on SB or LB plates. All proteases were purchased from Boehringer Mannheim.
Electron microscopy.
Bacteria were grown to a cell density of ∼7 × 108 CFU/ml and infected using fresh virus stock to obtain an MOI of 160. Samples were taken at 2, 5, and 10 min after infection, and the cells were fixed with 3% (vol/vol) glutaraldehyde for 20 min at 22°C. Fixed cells were collected by centrifugation, washed twice, and prepared for transmission electron microscopy as previously described (3). The micrographs were taken using a JEOL 1200 EX electron microscope operating at 60 kV (Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki).
Measurements of ion fluxes and determination of membrane voltage.
Ion flux measurements were performed as described by Daugelavičius et al. (27). Briefly, cells were grown to a density of 7 × 108 CFU/ml, collected by centrifugation, concentrated 200-fold, and dissolved in 50 mM Tris-HCl, pH 8.0, containing 450 mM NaCl. For measurements, the concentrated cell suspension was diluted in an appropriate buffer to obtain a final concentration of 2 × 109 CFU/ml and added to a 5-ml reaction vessel at 28°C with a thermostat. The cells were infected with nonradioactive virus. The concentrations of TPP+, PCB−, and K+ ions in the reaction vessel medium were monitored by selective electrodes while stirring. The characteristics of TPP+- and PCB−-selective electrodes, connected to Orion 520A pH/ISE meters, have been described previously (29, 40). The K+-selective electrode was from Orion Research Inc. (model 93-19). The Ag/AgCl reference electrodes (Orion Research Inc.; model 9001 or 9002) were indirectly connected to the measuring vessels through an agar salt bridge. TPP+ chloride was obtained from Sigma, and the potassium salt of PCB− was provided by A. Beganskiene, Department of Inorganic Chemistry, Vilnius University. Polymyxin B sulfate (PMB; 7,730 U of PMB base/mg) and GD were purchased from Sigma.
The K+ content of the cells and the nonspecific binding of TPP+ were measured by adding GD and PMB to the cell suspension in the reaction vessel. The internal K+ and TPP+ concentrations of the cells were calculated from the external concentrations, assuming that 1 × 1010 cells correspond to an intracellular water volume of 5 μl. The volume of P. espejiana BAL-31 and Pseudoalteromonas sp. strain ER72M2 cells was 60 to 70% that of E. coli (1.1 ml/g of dry mass) (14) as determined by comparing their cell dimensions to E. coli HB101, using phase-contrast light microscopy. The membrane voltage (ΔΨ) values were calculated using a modified Nernst equation, as previously described (27). The measurements of ion fluxes were carried out simultaneously in three reaction vessels, and a typical registration course of ion movements is presented. The concentrated cell suspension was kept on ice until used (maximally 5 h).
Determination of cellular ATP content.
The ATP content of the cells was determined by the luciferin-luciferase method (BioOrbit). The cells were incubated as described for the ion flux measurements. Fifty microliters of the cell suspension was withdrawn and mixed with 750 μl of 100 mM Tris-acetate (pH 7.75), 2 mM EDTA buffer, and 200 μl of ATP Monitoring Reagent (BioOrbit). The amount of light produced was measured by using a model 1250 Luminometer (LKB-Wallac). The total ATP content of the cells was measured by adding 200 μl of ATP Releasing Reagent (BioOrbit) to the bacterial suspension. The amount of ATP present in the incubation buffer was calculated from a calibration curve. The intracellular ATP concentration of the cells was calculated from the external concentrations, assuming that 1 × 1010 cells correspond to an intracellular water volume of 5 μl (see above).
Expression and purification of PM2 proteins P1 and P2.
The E. coli DH5α(pDMI)(pRM206) strain was used for expression of protein P1 (see Tables 1 and 2). Standard molecular cloning techniques were used (82). The cells were grown overnight in LB supplemented with 150 μg of ampicillin/ml and 25 μg of kanamycin/ml. The culture was diluted ∼30-fold in the same medium and grown to a density of 2 × 108 CFU/ml at 28°C. Expression was induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM, and the culture was transferred to 18°C. After 16 to18 h, the cells were collected (Sorvall SLA3000 rotor; 5,000 rpm, 10 min, 5°C), resuspended in 20 mM Tris-HCl (pH 8.0) buffer containing 5% (vol/vol) glycerol (1/100 of the original volume), and stored at −80°C. The cells were disrupted by two passages through a precooled French pressure cell (∼105 MPa, 35-ml cell), and the cell debris was removed by centrifugation (Sorvall SS34 rotor; 12,000 rpm, 20 min, 5°C). The supernatant was cleared by centrifugation (Beckman Ti 50 rotor; 40,000 rpm, 1 h and 40 min, 5°C). Protein P1 was precipitated from the supernatant by 35% (vol/vol) (NH4)2SO4, resuspended in 20 mM Tris-HCl, pH 8.0, containing 150 mM NaCl, and purified by gel filtration (Superdex 200 16/60 [Amersham Pharmacia Biotech] equilibrated with 20 mM Tris-HCl [pH 8.0]-150 mM NaCl). The peak fractions containing protein P1 were pooled and used in the adsorption inhibition tests. It was estimated from SDS-PAGE that this fraction contained ∼80% P1.
For the expression of protein P2, the E. coli HMS174(DE3)(pRM607) strain was used (see Tables 1 and 2). The cells were grown in LB supplemented with 25 μg of kanamycin/ml and stored and disrupted as described above for protein P1 except that the pH of the resuspension buffer was 7.5. Protein P2 was precipitated with 42.5% (vol/vol) (NH4)2SO4, resuspended in 20 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, and purified by size exclusion chromatography (Superdex 200 26/60 [Amersham Pharmacia Biotech] equilibrated with 20 mM Tris-HCl[pH 7.5]-150 mM NaCl). The fractions containing P2 were pooled, dialyzed against 20 mM Tris-HCl (pH 7.5) buffer containing 30 mM NaCl (Spectra/Por membrane; cutoff, 12 to14 kDa), and loaded onto 1-ml HiTrap Q Sepharose columns (Amersham Pharmacia Biotech). Flowthrough containing protein P2 was collected, stored at 4°C, and used in the adsorption inhibition tests. With this technique, P2 was purified to near homogeneity.
The multimericity of recombinant proteins was determined by gel filtration as described above and by sedimentation in a linear 10 to 40% (wt/vol) sucrose gradient in 20 mM Tris-HCl, pH 7.5, containing 150 mM NaCl buffer (Sorvall TH641 rotor; 35,000 rpm, 42 h, 15°C). Protein P1 (37.5 kDa) monomers and P2 (90.6 kDa) trimers isolated from PM2 virus particles (54), protein P3 (120 kDa) and P5 (85 kDa) trimers isolated from PRD1 virus particles (4), bovine serum albumin (68 kDa,; Sigma), and lysozyme (14 kDa; Boehringer Mannheim) were used as molecular mass markers. Gradient fractions were analyzed by SDS-PAGE.
Adsorption tests and ion flux measurements in the presence of protein P1 or P2.
A constant number of ER72M2 cells (grown to 7 × 108 CFU/ml) was mixed with increasing amounts of purified P1 in equal volumes of SB broth (taking into account that the P1 specimen contained ∼20% impurities). After 15 min of incubation at 28°C with shaking, the cells were infected with the fresh virus agar stock (MOI, ∼0.3). After 10 min, adsorption was stopped and the number of nonadsorbed phages was determined as described for the phage adsorption test.
For the measurements of ion fluxes, ER72M2 cells in 50 mM Tris-HCl, pH 8.0, containing 450 mM NaCl and 10 mM CaCl2 were combined with a mixture of P1 and P2 proteins isolated from phage particles and purified recombinant P1 (see above) or purified recombinant P2 (see above). The cells were incubated for 5 min with protein prior to the addition of phages. The measurements were carried out as described above.
Lytic activity assayed by zymogram analysis.
The production of 2× purified PM2 virions and isolated lipid cores from freeze-thawed phage particles for zymogram assays were carried out as described previously (54). For the expression of PM2 proteins P3, P4, P5, P6, P7, and P8, the corresponding coding sequences were amplified by PCR and inserted into expression vectors (see Table 2). Strains DH5α(pDMI)(pRM203), HMS174(DE3)(pRM315), HMS174(DE3)(pRM319), HMS174(DE3)(pRM301), HMS174(DE3)(pRM318), and HMS174(DE3)(pRM325) were used for the expression of proteins P3, P4, P5, P6, P7, and P8, respectively. DH5α(pDMI)(pDS12) and HMS174(DE3)(pJJ2) were used as negative controls. Cells carrying different plasmids were grown in LB medium supplemented with appropriate selective antibiotics (ampicillin, 150 μg/ml; kanamycin, 25 μg/ml), and protein expression was carried out as described for protein P1. After expression, cells were collected and concentrated tenfold by resuspension in 20 mM Tris-HCl (pH 7.2) buffer. The cells were disrupted by sonication, and the soluble and insoluble fractions were separated (Heraeus Biofuge; 10,000 rpm, 4 min, 4°C). Bacteriophage PRD1 particles used as a control in zymogram assays were purified as previously described (8). All samples were stored at −20°C until used in zymogram analysis.
The peptidoglycan sacculus of E. coli DH5α was isolated as described by Hoyle and Beveridge (47) by boiling 500 ml of stationary-phase cells in 4% SDS followed by removal of the peptidoglycan-associated proteins with 2 M NaCl (10). The isolated peptidoglycan preparation was resuspended in 5 ml of water and stored at −20°C until used. Zymogram analysis was performed as previously described by Bernadsky et al. (10). SDS-16% polyacrylamide gels (101 by 82 by 0.5 mm) were cast as described by Olkkonen and Bamford (75) except that the peptidoglycan preparation homogenized by sonication was added at a final concentration of 7.5% (vol/vol) in the separation gel. After electrophoresis, gels first were soaked in deionized water and then in renaturation buffer (25 mM KPO4 [pH 7.2], 0.1% [vol/vol] Triton X-100) for 30 min. Following this, zymogram gels were transferred into fresh renaturation buffer and the incubation was continued for 40 to 60 h. After renaturation, gels were rinsed with water, stained with 0.1% methylene blue in 0.01% KOH for 1 h at 28°C, and destained with deionized water.
Analytical methods.
Protein concentration was measured by using the Coomassie blue method with bovine serum albumin as a standard (17). Proteins were separated by SDS-PAGE using either 14% polyacrylamide-tricine-SDS gels (53, 85) or 16% polyacrylamide-SDS gels (75). For amino-terminal amino acid sequencing, the proteins were transferred from the gel to a polyvinylidene difluoride membrane (Millipore) and stained with Coomassie blue (PhastGel Blue R; Pharmacia Biotech). Sequencing was performed by using a Procise 494A protein sequencer (Perkin-Elmer/Applied Biosystems, Foster City, Calif.) at the Protein Chemistry Laboratory, Institute of Biotechnology, University of Helsinki.
RESULTS
Large amounts of phage particles bind to the host cells, but no empty capsids are seen on the cell surface.
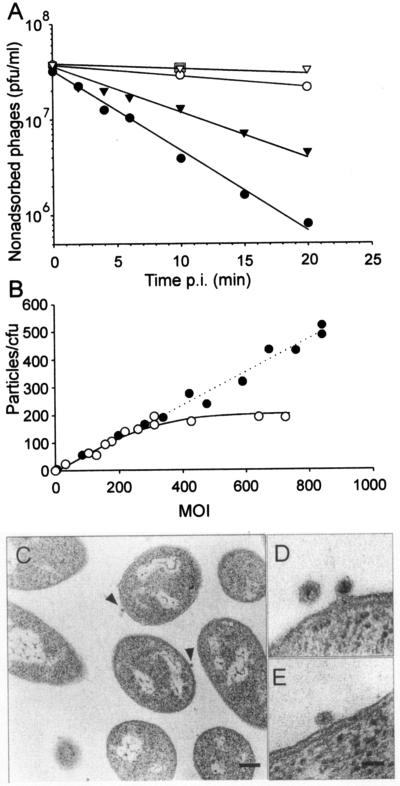
When a number of Pseudoalteromonas species were screened for PM2 sensitivity using the plaque assay, only one new isolate, ER72M2, in addition to BAL-31 cells, was able to plate PM2, indicating that the receptor is not a common Pseudoalteromonas surface structure (53). Also note that Pseudoalteromonas sp. strain A28 was unable to plate PM2 (see below). The adsorption of the phage to different cells was studied under vigorous aeration in normal growth medium resembling seawater. The adsorption was strictly aeration dependent. If the cells were not aerated, no virus binding was detected. The plating efficiency of PM2 on BAL-31 and ER72M2 is the same, but the binding of virus to the cell surface differed considerably (Fig. 1A and B). The adsorption rate constants calculated for the two hosts were 1.4 × 10−10 ml/min and 2.2 × 10−10 ml/min for ER72M2 and BAL-31, respectively (Fig. 1A). Virus adsorption to the sensitive strains was compared to that to PM2-resistant control cells (Pseudoalteromonas sp. strain A28 or E. coli HB101). In addition, 50 independent spontaneous PM2-resistant ER72M2 cell lines were isolated (see Materials and Methods). The electrochemical characteristics of the resistant mutants were the same as those of the sensitive ER72M2 strain, indicating that the envelopes of the resistant cells were not altered. All the tested cell lines showed no binding of the virus above the background level in the adsorption test (Fig. 1A), and they served as isogenic controls for the measurements of PM2-specific effects in the electrochemical measurements. The nonspecific binding of the virus to resistant E. coli (Fig. 1A) did not deviate from virus decay during the assay, which was demonstrated with virus material in the absence of cells.
FIG. 1.
Phage PM2 adsorption. (A) Time-dependent adsorption to BAL-31 (black circles), ER72M2 (black triangles), A28 (open circles), and E. coli HB101 (open triangles) cells. The amount of nonadsorbed phages was measured after different times p.i. Fifty PM2-resistant ER72M2 cell lines were tested using a 10-min adsorption time, and the results did not deviate from those obtained for E. coli, as indicated by a grey square. (B) The number of PM2 particles adsorbed to ER72M2 (open circles) and BAL-31 (black circles) cells at different MOIs (15-min adsorption time). (C to E) Thin sections of PM2-infected cells (MOI, 160). (C) Two DNA-containing virus particles in association with ER72M2 cells after 10 min of infection are denoted by arrowheads. Bar, 200 nm. (D) A higher magnification of a free virion and one adsorbed DNA-containing particle on an ER72M2 cell after 2 min of infection. (E) A cell-associated DNA-containing particle on a BAL-31 cell after 2 min of infection. Bar in panel E, 50 nm for panels D and E.
In one-step growth experiments (MOI, 10) the PM2-infected bacterial culture lyses effectively (53). Ten minutes postinfection (p.i.) ∼95% of cells were infected (MOI, 10). With an increase in the MOI, the fraction of infected cells increased (99% at an MOI of 140). To determine how many virus particles bind per CFU, receptor saturation experiments were carried out. The maximum number of bound particles was ∼180 per 1 ER72M2 CFU, achieved at an MOI of ∼300 (Fig. 1B). BAL-31 bound over 400 particles per CFU at the highest MOI used (850) with no signs of saturation. Thin-section electron microscopy of BAL-31 and ER72M2 cells during the first 10 min of infection (Fig. 1C to E) showed only a few DNA-containing virus particles associated with the cell surface when an MOI of 160 was used. Counts of visible virus particles on the cell surface revealed approximately 20 DNA-containing particles per 100 cell sections. The cell-associated particles had a mean diameter of ∼40 nm, while PM2 has a ∼51-nm diameter in thin-section electron microscopy (Fig. 1C to E) (54). In addition, no empty virus capsids were detected in association with the cells.
Binding inhibition assays were used to search for the PM2 receptor. Isolated LPSs from sensitive cells had no effect on PM2 adsorption (data not shown). Neither did the addition of the soluble or insoluble fractions from disrupted sensitive cells. In addition, extensive protease treatment of the host cells to digest the surface proteins did not alter the capacity of the cells to adsorb phages (data not shown).
Release of virion components follows phage adsorption.
Incorporation of l-[35S]methionine to capsid and LC-associated proteins was studied after separation of the labeled virion proteins in a polyacrylamide gel. About 60% of the radioactivity associated with the capsid (proteins P1 and P2), and the rest (40%) associated with the LC proteins. Extraction of the nucleic acid and lipids from the 33P-labeled virions showed that ∼87% of the radioactivity was associated with nucleic acid and the rest (13%) resided in the lipid fraction.
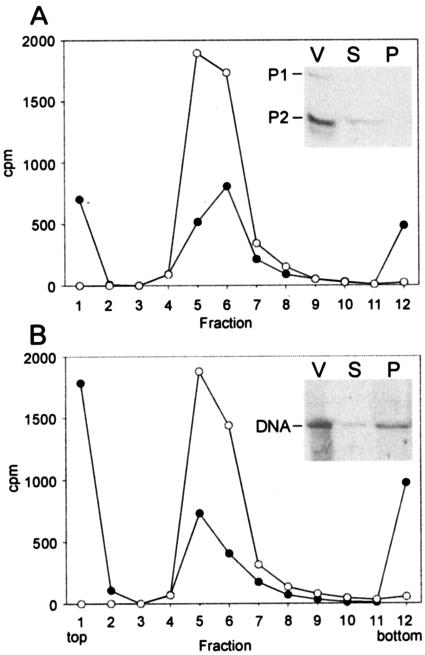
Association of the labeled viruses to ER72M2 cells was examined by rate zonal centrifugation (Fig. 2). This allowed the radioactivity associated with the nonadsorbed particles to be distinguished from that released or bound to host cells. The mixing of l-[35S]methionine-labeled PM2 (MOI, 10) with ER72M2 cells resulted in the formation of two separate fractions. Sixty percent of the radioactivity was found free at the top of the gradient, and 40% associated with the cells (Fig. 2A). In these conditions ∼75% of PM2 particles were bound, as detected by the adsorption assay or reduction of the virus peak (Fig. 2A). An autoradiogram of the supernatant and cell-associated fractions of the adsorption mixture separated by tricine-SDS-PAGE revealed that the major coat protein P2 was found in the soluble fraction (inset in Fig. 2A). Due to the low specific activity obtained, we could not demonstrate the association of the LC proteins with the cells. Sedimentation analysis of the 33P-labeled PM2 mixed with ER72M2 cells (MOI, 10) showed that ∼35% of the radioactivity associated with the cells while the majority (65%) was released (Fig. 2B). In this experiment ∼80% of the particles associated with the cells (see above). An autoradiogram of the supernatant and cell-associated fraction demonstrated that the viral DNA resided in both fractions (Fig. 2B).
FIG. 2.
Association of l-[35S]methionine- (A) and 33P- (B) labeled PM2 virions with ER72M2 cells. The distribution of the radioactivity was analyzed by rate zonal centrifugation after a 15-min adsorption period using an MOI of 10 (black circles). A corresponding amount of labeled virus particles alone was used as a control (open circles). The radioactivity on top of the control gradient (virus only) was subtracted from the adsorption experiment to correct the effect of spontaneously dissociated phage particles. The insets show the supernatant (S) and the cell fraction (P) of an adsorption mixture (MOI, 5) and the labeled virions (V) analyzed in a tricine-polyacrylamide gel followed by autoradiography. The positions of PM2 P1 and P2 proteins and DNA are indicated.
Receptorless Pseudoalteromonas sp. strain A28 supports virus reproduction.
Pseudoalteromonas sp. strain A28 harbors a plasmid, pAS28 (52), which shares significant sequence similarity with the early operon of PM2 (62, 63). The adsorption test revealed that PM2 could not bind to the A28 cell surface (Fig. 1A). Further, analysis of the effects of PM2 on A28 cells, as determined by measuring ion fluxes, revealed that the phage had no effect on envelope permeability of A28 cells (Fig. 3). Interestingly, transfection of purified PM2 genomes into A28 spheroplasts produced plaques if the transfected cells were plated on a lawn of PM2-sensitive cells. This shows that A28, a close relative to the two Pseudoalteromonas hosts, does not have the receptor for PM2, but after the genome has entered the cell, viruses are produced. No other tested pseudoalteromonads or Alteromonas macleodie (Table 1) was able to produce virus in this assay, except the host cells.
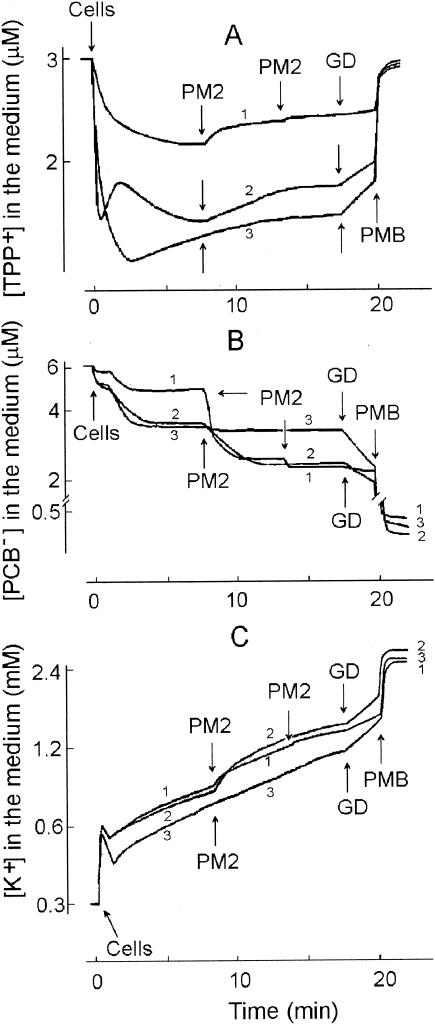
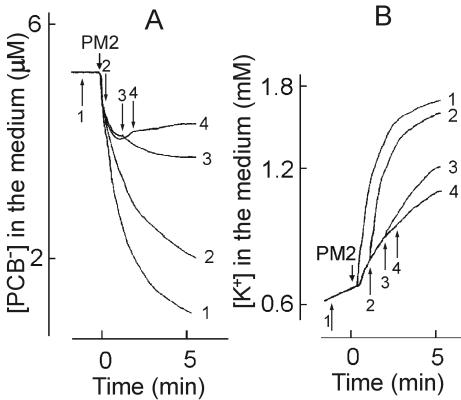
FIG. 3.
Effects of phage PM2 infection on TPP+ (A), PCB− (B), and K+ (C) ion uptake by different Pseudoalteromonas strains. The experiments were carried out at 28°C in 50 mM Tris-HCl (pH 8.0)-450 mM NaCl-10 mM CaCl2 with a cell concentration of 2 × 109 CFU/ml. ER72M2 (curve 1) cells were infected with an MOI of 10, and BAL-31 (curve 2) and A28 (curve 3) cells were infected with an MOI of 50. A second infection of ER72M2 cells (curve 1) was carried out using an MOI of 50. GD and PMB were added at final concentrations of 5 and 100 μg/ml, respectively.
The outer membrane of marine pseudoalteromonads is highly permeable.
Pseudoalteromonads are gram-negative cells. The OMs of the best-studied gram-negative bacteria, E. coli and Salmonella enterica, form a permeability barrier to lipophilic compounds such as TPP+ or ionophoric antibiotics (for a review, see reference 73). However, the OM permeability of pseudoalteromonads to TPP+ was rather high (Fig. 3A). Accumulation of TPP+ differed between the studied strains. BAL-31 and A28 accumulated considerably higher amounts of TPP+ compared to ER72M2, indicating a higher membrane voltage (ΔΨ). However, higher OM permeability to lipophilic compounds in the case of BAL-31 and A28 also is possible, as they accumulated a higher amount of PCB− and were more sensitive to GD compared to the ER72M2 strain (Fig. 3B).
In spite of the high OM permeability of the studied three Pseudoalteromonas strains, the CM-depolarizing and intracellular K+-releasing effect of GD was low. In addition, the depolarizing effect of the polycationic antibiotic PMB was rather low on ER72M2, but somewhat higher in the case of BAL-31 (data not shown). However, the combination of GD and PMB induced very effective depolarization of the CM and an efficient release of intracellular K+ (Fig. 3 and 4). The presence of TPP+ or PCB− ions had no effect on the viability of the cells or the amount of infective centers observed when BAL-31 and ER72M2 were infected with an MOI of 10.
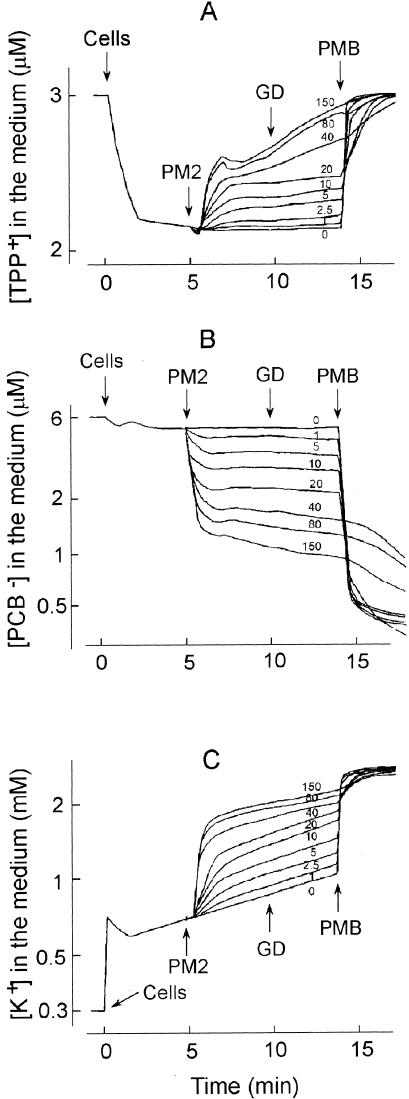
FIG. 4.
Effects of the MOI on amounts of TPP+ (A), PCB− (B), and K+ (C) accumulated by ER72M2 cells. The experiments were carried out as described for Fig. 3. PM2 phages were added to obtain MOIs as indicated by the numbers in each panel. GD and PMB were added at final concentrations of 5 and 100 μg/ml, respectively.
EDTA is known to chelate divalent cations away from the LPS layer, increase the OM permeability to lipophilic compounds, and induce the additional accumulation of lipophilic cations in gram-negative cells (71). However, in the case of Pseudoalteromonas strains, the addition of up to 500 μM EDTA to the cell suspension (in 50 mM Tris-HCl [pH 8.0] containing 450 mM NaCl) caused only partial release of accumulated TPP+ and K+ (data not shown).
PM2 induces depolarization of the CM.
Phage-induced effects on the CM can be detected by measuring the efflux of the membrane voltage indicator TPP+ ion from the infected cells. The release of TPP+ from infected BAL-31 and ER72M2 cells began 20 to 22 s after addition of phage particles and continued for approximately 2 min, indicating depolarization of the CM (Fig. 3A). The phage-induced leakage of accumulated TPP+ was MOI dose dependent (Fig. 4A). Approximately five-times-higher numbers of infectious particles induced effects with similar amplitudes on BAL-31 cells when compared to ER72M2 cells. Four-times-higher numbers of superinfecting phages, added 6 min after the initial infection, had no effect on the external concentrations of the three ions studied (Fig. 3, ER72M2 strain).
Since the combination of GD and PMB very effectively depolarized the CM, it was possible to calculate the membrane voltage. The Δψ of ER72M2 and BAL-31 cells before infection was ∼165 and ∼187 mV, respectively (calculated from Fig. 3A). After infection, before GD addition, Δψ was ∼153 mV for ER72M2 and ∼177 mV for BAL-31. At a high MOI (150), the CMs of ER72M2 cells were depolarized from ∼167 to ∼142 mV in 5 min (Fig. 4A). After infection, the membrane voltage stayed at the lower level and repolarization of the CM was not detected. The accuracy of the calculations depends on viable count measurements. Light microscopy of growing bacteria revealed that both strains did not separate the dividing cells, and the cells were seen predominantly as doublets. When this is taken into account, the effective Δψ is ∼18 mV lower than the calculated one.
PM2 induces strong binding of PCB− to cell membranes and K+ leakage.
In addition to depolarization of the CM, PM2 induced high accumulation of PCB− by infected BAL-31 and ER72M2 cells (Fig. 3B). The decrease of PCB− concentration in the medium started 12 to 15 s after the addition of phages and continued for approximately 1.5 min. The amplitude of the phage-induced effect on the accumulation of PCB− increased with the MOI (Fig. 4B). PM2 virus particles bound rather low amounts of PCB−. However, at high MOIs, the binding of PCB− to the viral membranes can be observed immediately after the phage addition as a fast uptake of this anion.
Cells initially accumulated potassium ions, but after approximately 1 min of incubation, a slow spontaneous leakage of intracellular K+ started (Fig. 3C). GD and PMB caused elimination of the K+ gradient on the CM, allowing us to calculate the intracellular K+ concentration during infection (calculated from Fig. 3C). The maximal calculated intracellular K+ concentration (just before the spontaneous leakage starts) was ∼2.0 and ∼2.3 M for ER72M2 and BAL-31, respectively. However, when the inaccuracy in viable count measurements (see Δψ calculations) is taken into account, the effective intracellular K+ concentration is about half of the calculated one. The addition of infectious PM2 particles stimulated the efflux of K+ ions starting ∼15 s after infection and lasting for ∼2 min (Fig. 3C). The amplitude of this efflux was proportional to the MOI used (Fig. 4C). After this K+ efflux stimulation, the rate of K+ leakage from infected ER72M2 and BAL-31 cells was close to the rate of spontaneous leakage, which was also observed for A28 cells (Fig. 3C). The calculated intracellular potassium ion concentrations of ER72M2 and BAL-31 during the first 2 min of infection decreased from ∼1.7 to ∼1.5 M and from ∼2.0 to ∼1.75 M, respectively (see also above).
Additional energy considerations.
In order to estimate the role of cell energetics in infection, the effects of energy transformation-modifying compounds were studied. The addition of glucose did not induce additional accumulation of either TPP+ or K+ ions, but some release of the accumulated TPP+ was registered. The addition of glycerol, lactose, or sucrose to the incubation medium had no effect on the accumulation of TPP+ or K+. NaN3 eliminates bacterial growth by blocking respiration and uncoupling oxidative phosphorylation due to inhibition of cytochrome oxidase and membrane H+-ATPase (42). In the presence of 20 mM NaN3, the amount of accumulated TPP+ was somewhat lower but the cells were able to maintain membrane voltage and keep the K+ gradient. In these conditions, the amplitude of PM2-induced TPP+ and especially PCB− and K+ fluxes was even higher than without NaN3 (data not shown).
The measurements of ATP-dependent light emission by the luciferin-luciferase system showed that the calculated intracellular concentration of ATP was rather high, ∼8 mM. However, the effective concentration was ∼4 mM (see Δψ calculations). The concentration of intracellular ATP in ER72M2 cells did not decrease during infection (calculated concentration 8.3 mM at 3.5 min p.i. and 9.3 mM at 10 min p.i.).
Purified PM2 protein P1 inhibits phage adsorption.
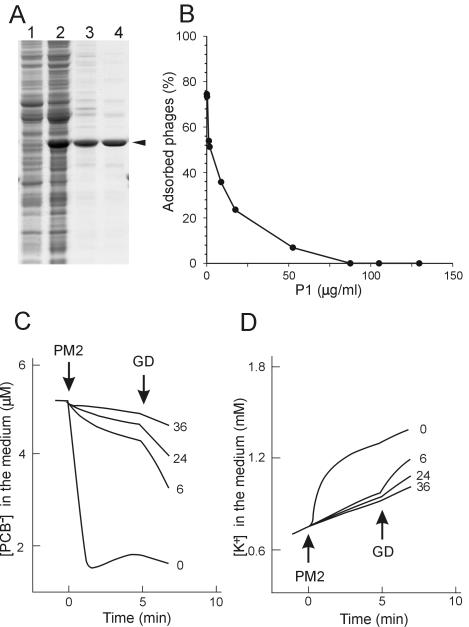
The outer layer of the PM2 virion consists of two major structural proteins, P1 and P2. To evaluate whether these capsid proteins could interfere with the interaction between the phages and the cells, the proteins were isolated from viral particles (see reference 54) and added to the adsorption mixture. Clear adsorption inhibition was observed (data not shown). To test which of the proteins was responsible for the effect, they were separately expressed from plasmids and purified (Fig. 5A for P1; for P2, data not shown). The recovery of purified proteins P1 and P2 was ∼1 and ∼4 mg/liter of culture, respectively. N-terminal amino acid sequence analysis of the purified proteins gave sequences MIVKKKLAA and MRSFLNLNSI, equivalent to the P1 and P2 sequences, respectively, as deduced from the virus genome. In addition to the information obtained during purification by size exclusion chromatography (P1, ∼40 kDa, and P2, ∼65 kDa), the multimericity of isolated proteins was determined by rate zonal centrifugation (P1, ∼55 kDa, and P2, ∼85kDa). Recombinant P1 and P2 behaved as the corresponding proteins from the virus particle (54), indicating that the recombinant P1 is a monomer and P2 is a trimer in solution.
FIG. 5.
Effects of PM2 protein P1 on phage adsorption. (A) Purification of PM2 protein P1. Samples were taken after different purification steps and analyzed by SDS-PAGE. Soluble fraction of disrupted DH5α(pDMI)(pDS12) (lane 1) and DH5α(pDMI)(pRM206) (lane 2) cells, proteins after ammonium sulfate precipitation (lane 3), and purified protein P1 after size exclusion chromatography (lane 4) are shown. The position of P1 is indicated by an arrowhead. (B) Phage adsorption to ER72M2 cells in the presence of increasing amounts of purified protein P1 (SB medium; MOI, 0.3; 7 × 108 CFU/ml). The percentage of adsorbed phages after a 10-min adsorption period is shown. (C and D) PM2-induced PCB− (C) and K+ (D) fluxes in the presence of different amounts of purified P1. The measurements were carried out as described for Fig. 2. The final concentrations of P1 protein (micrograms per milliliter) added to the ER72M2 cell suspension 5 min before the infection are indicated. PM2 was added to obtain an MOI of 40, and GD was added to a final concentration of 5 μg/ml.
The ability of these proteins to interfere with PM2 adsorption was analyzed by two different methods. In the adsorption test, the presence of increasing amounts of P1 blocked adsorption (Fig. 5B). In the presence of P1, phage-induced effects on the envelope permeability of ER72M2 cells with respect to PCB− and K+ ions also were prevented (Fig. 5C and D). Recombinant P2 did not have any inhibitory effects on PM2 adsorption. Binding of the monomeric protein P1 alone to the phage receptor had no electrochemical effects on ER72M2 cells (data not shown). We conclude that the first step in the PM2 infection cycle is the binding of the pentameric vertex protein P1 to the cellular receptor.
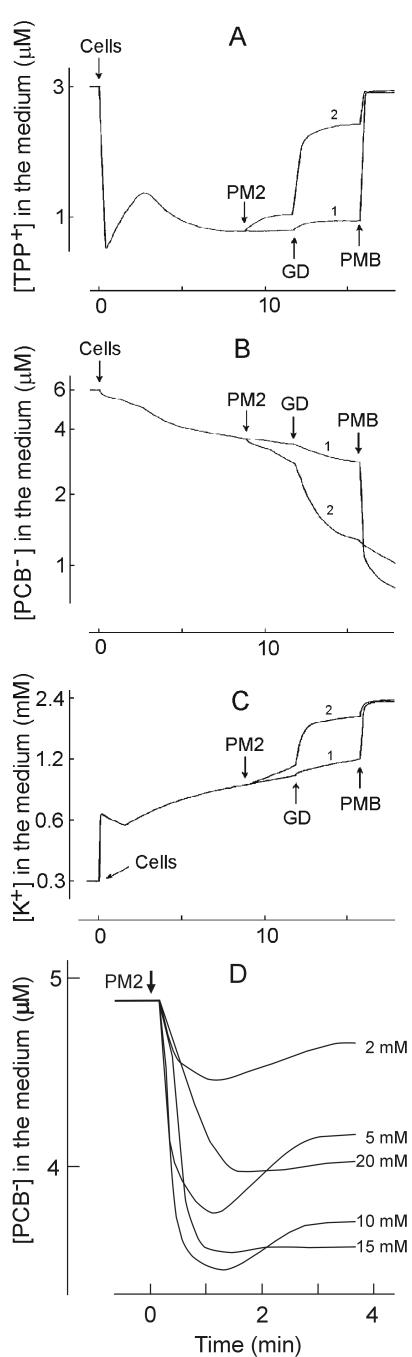
PM2 has a temporal effect on OM permeability.
The addition of GD at different time points before and after the addition of PM2 particles in the presence of calcium (Fig. 6) revealed that cells were sensitive to GD effects (PCB− binding and K+ leakage) only during the first 2 min of infection, but not after that time or before infection. This indicates the presence of a temporal virus-induced patch on the OM, which is permeable to lipophilic GD molecules.
FIG. 6.
Effects of GD on phage-induced PCB− (A) and K+ (B) fluxes in ER72M2 cells. The measurements were carried out as described for Fig. 2. Cells were infected with PM2 using an MOI of 5 (marked by PM2). GD was added to a final concentration of 5 μg/ml. The arrows (1 to 4) show the time points of GD addition, and the curves are numbered accordingly. GD was added 60 s before infection for curve 1 and 30, 90 or 130 s after phage addition for curves 2, 3, and 4, respectively.
In the absence of calcium, ER72M2 cells had considerably higher membrane voltage (∼212 mV; see above). Under these conditions, PM2 alone did not cause depolarization of the CM (∼209 mV), PCB− accumulation, or K+ leakage (Fig. 7A to C). However, GD added 3 min or later after addition of the phage particles induced strong depolarization of the CM, leakage of K+, and accumulation of PCB−, indicating that the transient nature of OM permeability was abolished (Fig. 7A to C). The amplitude of phage PM2-induced binding of PCB− depended on the concentration of CaCl2 at the beginning of infection (Fig. 7D). The maximal effect was achieved with 10 to 15 mM CaCl2. Higher concentrations of Ca2+ had deleterious effects on infection.
FIG.7.
Phage-induced effects on envelope permeability of ER72M2 cells with respect to TPP+ (A), PCB− (B), and K+ (C) ions in the absence of CaCl2. The measurements were carried out as described for Fig. 2 except that the incubation medium contained 50 mM Tris-HCl (pH 8.0) and 450 mM NaCl. Curve 1 shows noninfected cells, and curve 2 shows cells infected with an MOI of 50. GD and PMB were added at final concentrations of 5 and 100 μg/ml, respectively. (D) Phage PM2-induced accumulation of PCB− by ER72M2 cells in the presence of different CaCl2 concentrations as indicated. The measurements were carried out as described for Fig. 1. The cells were infected with an MOI of 50.
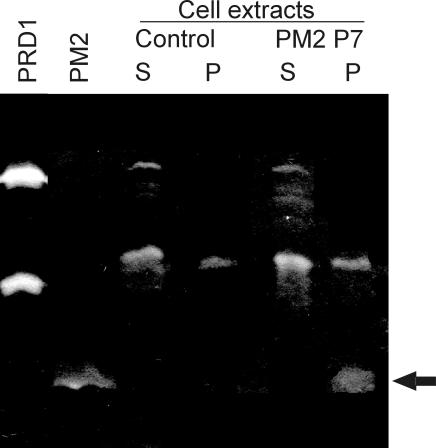
Small structural PM2 protein P7 has muralytic activity.
The lytic activities associated with bacteriophage PM2 particles were examined by analyzing highly purified virions by zymogram analysis. After separation of viral proteins by SDS-PAGE (16% polyacrylamide-SDS gel containing peptidoglycan), proteins were allowed to renature at different temperatures (4, 15, 28, and 37°C). After renaturation at 4 or 15°C, one clear PM2-derived zone was detected (Fig. 8). The molecular mass of the PM2 structural protein with lytic activity was below 17 kDa, which is the mass of the PRD1 lytic enzyme P15 (Fig. 8).
FIG. 8.
Zymogram analysis of purified PM2 virions and a cell extract containing recombinant PM2 protein P7. PM2 virions show a single protein with muralytic activity (arrow). A cell extract of E. coli with protein P7 produced similar activity. The control E. coli extract indicates the cell-derived lytic activities. Supernatant and pellet fractions of disrupted cells are indicated by S and P, respectively. As a control, purified bacteriophage PRD1 virions containing two proteins with muralytic activity (P7, 27 kDa; and P15, 17 kDa) are shown.
The internal LC particle contains proteins P3 to P10 (48, 53). Isolated LC particles were analyzed in zymograms, and the protein with muralytic activity was present in this preparation (data not shown). In order to determine which protein was responsible for this activity, the structural proteins P3, P4, P5, P6, P7, and P8 were expressed in E. coli (see Table 2) at two different temperatures, 18 and 37°C. After expression, the cells were disrupted and fractionated into soluble and insoluble fractions. Expression of proteins P3, P5, P6, and P7 could be detected in cell extracts with Coomassie blue staining. P5 was detected in the soluble fraction (18°C). P3 (37°C), P6 (18°C and 37°C), and P7 (18°C and 37°C) were found in the insoluble fraction. Expression of proteins P4 (4.4 kDa) and P8 (7.3 kDa) could not be detected by this method. The produced proteins were identified by N-terminal amino acid sequence analysis. The obtained sequences, MNTSVPTS, MKKAHMF, ANFLTKNF, and MINKTTIK, were identical to the sequences deduced from the genes for PM2 proteins P3, P5, P6, and P7, respectively. The first methionine of recombinant protein P6 is removed after translation, which is also the case with the virion-derived protein.
To analyze whether the cell extracts containing recombinant PM2 proteins could produce peptidoglycan-hydrolyzing activity similar to that observed in viral specimens, zymogram analysis was performed. Cell extracts of the bacteria carrying the cloning vectors without inserts were used as negative controls to reveal the cell-derived lytic activities. Screening of the cell extracts in zymogram analyses using different renaturation temperatures showed that protein P7 (expressed at 37°C) had similar enzymatic activity as that observed in PM2 virions and in the LC particles (Fig. 8). According to these data, we conclude that protein P7 is a lytic enzyme. It is the smallest characterized structural protein (3.7 kDa) in the PM2 virion, containing one putative membrane-spanning helix (53, 62). However, a data bank search of the PM2 proteins did not reveal any matches to known sequences for lytic activities (PROSITE) (34).
DISCUSSION
Only two Pseudoalteromonas isolates carrying a high number of receptors on the cell surface have been shown to act as hosts for PM2 (Fig. 1) (53). Pseudoalteromonas sp. strain A28 harboring a plasmid (pAS28) (52), but no receptor, was shown to support PM2 growth after purified phage genomes were transfected into the cell. This finding is consistent with the observation that the replication and regulatory regions of the PM2 genome and plasmid pAS28 share significant similarity (63). In this study, we showed that an abundant receptor molecule is nonproteinaceous or is a protein that is very resistant to proteolysis. Extracts of sensitive cells did not interfere with the virus, indicating that the receptor or receptor complex is functional only on the cell surface, not being extractable, as is that of phage PRD1 (28).
In the native virion, P1 monomers are grouped as pentamers at the fivefold vertices by the N-terminal domain, which connects the complex to the capsid formed from P2 trimers (48). Previously, it has been demonstrated that the lack of the C-terminal portion of P1 results in an inability of the virus to infect the host (54). We showed that the monomeric protein P1 readily associated with the cellular receptor, interfering with virus adsorption (Fig. 5). This demonstrates that the initial interaction between the host and PM2 is mediated by P1, which is known to build up the spike structure of the virion. The trimeric viral coat protein P2 did not interfere with binding of PM2 to its primary receptor.
The adsorption experiments (Fig. 1A and B) and thin-section electron microscopy (Fig. 1C to E) of infected cells gave contradicting results, as only very few DNA-containing particles were seen to be in contact with cells in spite of the high adsorption rate and high MOI used. Interestingly, the diameter of the cell-associated particles corresponds to the LC, not to the virion (Fig. 1D). In addition, no empty particles were detected in association with the cells, even 10 min p.i., when all the DNA entry-derived electrochemical events were over (Fig. 3). Moreover, the mixing of PM2 with its host cell leads to the dissociation of the particle (Fig. 2). Our previous results have demonstrated that the capsid is a metastable structure easily dissociated by calcium depletion, freezing and thawing of the particles, or by storage (53, 54). In contrast to these results, the empty viral capsid stays associated with the cell in the case of numerous dsDNA phages, including internal membrane-containing bacteriophages PRD1 and Bam35 (61, 78). The gross morphology of the PM2 virion resembles that of PRD1 (6, 39), which uses the internal membrane vesicle for DNA delivery by transforming it into a tube-like structure (38). No tubular membrane structures have been detected for PM2. Accordingly PM2 entry diverges considerably from those viruses.
The envelope permeability of Pseudoalteromonas cells deviates from that determined for the other well-studied enteric bacteria E. coli and S. enterica (73). In contrast to enterobacteria, the OM of pseudoalteromonads was permeable to TPP+ (Fig. 3). The intracellular concentration of solutes in bacteria has been shown to increase in conjunction with increasing external salt concentration (46, 49). High intracellular K+ concentrations observed in pseudoalteromonads presumably are a consequence of the extracellular osmolarity in its natural environment, seawater (and in the growth medium).
The dependence of the depolarizing effect of external GD on PM2 addition and its temporal nature (Fig. 6) indicate virus-induced changes in the OM. We envision that the LC interacts with the OM, creating a window for GD entry into the periplasm, where it causes depolarization in the CM. Presumably, due to the diffusion of lipids and movement of LPS molecules on the plane of the membrane, this permeability window closes within a few minutes. If there is a fusion between the virus membrane and the host OM, then the supercoiled DNA and its associated proteins, as well as the internal surface of the LC, will face the peptidoglycan. The interaction between protein P4 and the PM2 DNA has been proposed, and it seems likely that P4 is associated with the viral membrane inside the vesicle (54, 64, 83, 84). However, after entering the cytoplasm, no genome-associated proteins are needed for proper initiation of virus replication (62, 91).
Many bacteriophages have proteins with lytic activity (55, 58). Furthermore, PM2-derived endolysin activity has been characterized (90). In addition to playing a role in host cell lysis, lytic enzymes are often structural virion components. These proteins locally digest the murein sacculus during penetration into the cell. Examples of such proteins are the two murolytic enzymes, P7 and P15 of PRD1 (21, 80, 81), gp16 of bacteriophage T7 (65), base-plate protein gp5 of T4 (51, 69), P5 of phage φ6 (20), and the recently described terminal protein gp3 of bacteriophage φ29 (66). Here, we demonstrated that a small LC-associated integral membrane protein, P7, harbors lytic activity (Fig. 8). It is conceivable that P7 locally opens the murein layer, allowing the entering DNA to reach the host CM.
PM2-induced effects on the CM (TPP+ and K+ leakage and PCB− binding) started some 15 to 20 s after phage addition and were MOI dependent (Fig. 3 and 4). These effects lasted approximately 2 min. There was only a moderate decrease in the membrane voltage during DNA penetration and no decrease in the cellular ATP concentration. DNA penetration through the CM obviously causes only a temporal destabilization in the membrane with no severe adverse effects on the cellular energy balance. Similar to the OM, there is a well-controlled pore in the CM for the penetration of DNA comprised of virus or host proteins or both. The PM2-induced relatively strong binding of PCB− to cellular membranes is an indication of pore formation in the bacterial envelope, as has been shown for bacteriophages T4 and lambda (14, 29, 41). This PM2-induced pore is permeable to potassium ions but not to ATP. After DNA passage, the pore is sealed, indicating a gating phenomenon.
Calcium controls the infection process and stability of PM2. The LC-associated proteins P6 and P10 and the gene product of open reading frame h contain a putative calcium-binding site (48, 54, 62). In the absence of calcium, the infection process seems to stop at the CM stage, since the OM was permeabilized but depolarization of the CM did not take place. Bacteriophage T5 DNA penetration through the CM also is controlled by calcium ions (13), pointing to a similar mechanism.
Combining these and previous results allowed us to delineate an entry pathway for PM2. It deviates considerably from previously described phage DNA delivery mechanisms. The binding of the virion to the cell surface receptor triggers a cascade of events where the viral coat is dissociated and the LC most probably fuses with the bacterial OM. This event is rapid (∼2 min) but also occasionally leads to failure, as a fraction of the viral DNA is released. The murein-hydrolyzing enzyme in the phage membrane aids in peptidoglycan penetration. The penetration of the CM is a controlled gated event where a transient lowering of the membrane voltage is detected. Obviously, unique delivery and packaging mechanisms for the highly supercoiled circular PM2 genome have evolved.
Acknowledgments
The Helsinki Graduate School in Biotechnology and the Molecular Biology and Viikki Graduate School in Biosciences are acknowledged for fellowships to H.M.K. and R.H.H., respectively. This work was supported by research grants 1202108 and 1202855 from the Academy of Finland (to D.H.B.; Finnish Center of Excellence Program [2000-2005]) and grant 1201964 (to J.K.H.B.). R.D. is a Lithuanian State Fellowship holder.
Pia Rydman is thanked for the guidance with the zymogram analysis.
REFERENCES
- 1.Ackermann, H. W. 2000. Family Corticoviridae, p. 117-120. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, Inc., San Diego, Calif.
- 2.Adams, M. H. 1959. Bacteriophages. Interscience Publishers, Inc., New York, N.Y.
- 3.Bamford, D. H., and L. Mindich. 1980. Electron microscopy of cells infected with nonsense mutants of bacteriophage φ6. Virology 107:222-228. [DOI] [PubMed] [Google Scholar]
- 4.Bamford, D. H., and L. Mindich. 1982. Structure of the lipid-containing bacteriophage PRD1: disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J. Virol. 44:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamford, D. H., M. Romantschuk, and P. J. Somerharju. 1987. Membrane fusion in prokaryotes: bacteriophage φ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 6:1467-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bamford, D. H., J. Caldentey, and J. K. H. Bamford. 1995. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281-319. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, D. H., and J. K. H. Bamford. Lipid-containing bacteriophage PM2, the type-organism of Corticoviridae. In R. Calendar (ed.), The bacteriophages, in press. Oxford University Press, Oxford, United Kingdom.
- 8.Bamford, J. K. H., and D. H. Bamford. 1990. Capsomer proteins of bacteriophage PRD1, a bacterial virus with a membrane. Virology 177:445-451. [DOI] [PubMed] [Google Scholar]
- 9.Baumann, L., P. Baumann, M. Mandel, and R. D. Allen. 1972. Taxonomy of aerobic marine eubacteria. J. Bacteriol. 110:402-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernadsky, G., T. J. Beveridge, and A. J. Clarke. 1994. Analysis of the sodium dodecyl sulfate-stable peptidoglycan autolysins of select gram-negative pathogens by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 176:5225-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berrier, C., M. Bonhivers, L. Letellier, and A. Ghazi. 2000. High-conductance channel induced by the interaction of phage lambda with its receptor maltoporin. FEBS Lett. 476:129-133. [DOI] [PubMed] [Google Scholar]
- 12.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. 27:911-917. [DOI] [PubMed] [Google Scholar]
- 13.Bonhivers, M., and L. Letellier. 1995. Calcium controls phage T5 infection at the level of the Escherichia coli cytoplasmic membrane. FEBS Lett. 374:169-173. [DOI] [PubMed] [Google Scholar]
- 14.Boulanger, P., and L. Letellier. 1988. Characterization of ion channels involved in the penetration of phage T4 DNA into Escherichia coli cells. J. Biol. Chem. 263:9767-9775. [PubMed] [Google Scholar]
- 15.Boulanger, P., and L. Letellier. 1992. Ion channels are likely to be involved in the two steps of phage T5 DNA penetration into Escherichia coli cells. J. Biol. Chem. 267:3168-3172. [PubMed] [Google Scholar]
- 16.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification system of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 17.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 18.Brewer, G. J. 1978. Membrane-localized replication of bacteriophage PM2. Virology 84:242-245. [DOI] [PubMed] [Google Scholar]
- 19.Bujard, H., R. Gentz, M. Lanzer, D. Stueber, M. Mueller, I. Ibrahimi, M. T. Haeuptle, and B. Dobberstein. 1987. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 155:416-433. [DOI] [PubMed] [Google Scholar]
- 20.Caldentey, J., and D. H. Bamford. 1992. The lytic enzyme of the Pseudomonas phage φ6. Purification and biochemical characterization. Biochim. Biophys. Acta 1159:44-50. [DOI] [PubMed] [Google Scholar]
- 21.Caldentey, J., A. L. Hänninen, and D. H. Bamford. 1994. Gene XV of bacteriophage PRD1 encodes a lytic enzyme with muramidase activity. Eur. J. Biochem. 225:341-346. [DOI] [PubMed] [Google Scholar]
- 22.Camerini-Otero, R. D., and R. M. Franklin. 1972. Structure and synthesis of a lipid-containing bacteriophage. XII. The fatty acids and lipid content of bacteriophage PM2. Virology 49:385-393. [DOI] [PubMed] [Google Scholar]
- 23.Camerini-Otero, R. D., P. N. Pusey, D. E. Koppel, D. W. Schäfer, and R. M. Franklin. 1974. Intensity fluctuation spectroscopy of laser light scattered by solutions of spherical viruses: R17, Q beta, BSV, PM2, and T7. II. Diffusion coefficients, molecular weights, solvation, and particle dimensions. Biochemistry 13:960-970. [DOI] [PubMed] [Google Scholar]
- 24.Camerini-Otero, R. D., and R. M. Franklin. 1975. Structure and synthesis of a lipid-containing bacteriophage. The molecular weight and other physical properties of bacterophage PM2. Eur. J. Biochem. 53:343-348. [DOI] [PubMed] [Google Scholar]
- 25.Certa, U., W. Bannwarth, D. Stüber, R. Gentz, M. Lanzer, S. le Grice, F. Guillot, I. Wendler, G. Hunsmann, H. Bujard, and J. Mous. 1986. Subregions of a conserved part of the HIV gp41 transmembrane protein are differentially recognized by antibodies of infected individuals. EMBO J. 5:3051-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan, K. Y., L. Baumann, M. M. Garza, and P. Baumann. 1978. Two new species of Alteromonas: Alteromonas espeijiana and Alteromonas undida. Int. J. Syst. Bacteriol. 28:217-222. [Google Scholar]
- 27.Daugelavičius, R., J. K. H. Bamford, and D. H. Bamford. 1997. Changes in host cell energetics in response to bacteriophage PRD1 DNA entry. J. Bacteriol. 179:5203-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daugelavičius, R., J. K. H. Bamford, A. M. Grahn, E. Lanka, and D. H. Bamford. 1997. The IncP plasmid-encoded cell envelope-associated DNA transfer complex increases cell permeability. J. Bacteriol. 179:5195-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daugelavičius, R., E. Bakienė, J. Beržinskienė, and D. H. Bamford. 2001. Use of lipophilic anions for estimation of biomass and cell viability. Biotechnol. Bioeng. 71:208-216. [DOI] [PubMed] [Google Scholar]
- 30.Dreiseikelmann, B. 1994. Translocation of DNA across bacterial membranes. Microbiol. Rev. 58:293-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Espejo, R. T., and E. S. Canelo. 1968. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology 34:738-747. [DOI] [PubMed] [Google Scholar]
- 32.Espejo, R. T., and E. S. Canelo. 1968. Properties and characterization of the host bacterium of bacteriophage PM2. J. Bacteriol. 95:1887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espejo, R. T., and E. S. Canelo. 1968. Origin of phospholipid in bacteriophage PM2. J. Virol. 2:1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. A. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feucht, A., A. Schmid, R. Benz, H. Schwarz, and K. J. Heller. 1990. Pore formation associated with the tail-tip protein pb2 of bacteriophage T5. J. Biol. Chem. 265:18561-18567. [PubMed] [Google Scholar]
- 36.Garcia, L. R., and I. J. Molineux. 1995. Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J. Bacteriol. 177:4066-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gauthier, G., M. Gauthier, and R. Christen. 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 45:755-761. [DOI] [PubMed] [Google Scholar]
- 38.Grahn, A. M., R. Daugelavičius, and D. H. Bamford. 2002. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 46:1199-1209. [DOI] [PubMed] [Google Scholar]
- 39.Grahn, A. M., S. J. Butcher, J. K. H. Bamford, and D. H. Bamford. PRD1—dissecting the genome, structure and entry. In R. Calendar (ed.), The bacteriophages, in press. Oxford University Press, Oxford, United Kingdom.
- 40.Grinius, L., R. Daugelavičius, and G. Alkimavičius. 1981. Studies of the membrane potential of Bacillus subtilis and Escherichia coli cells by the method of penetrating ions. Biochimya 45:1222-1230. (English translation.) [Google Scholar]
- 41.Grinius, L., and D. Daugelavičius. 1988. Depolarization of Escherichia coli cytoplasmic membrane by bacteriophage T4 and lambda: evidence for induction of ion-permeable channels. Biochem. Bioenerg. 19:235-245. [Google Scholar]
- 42.Harold, F. M. 1972. Conservation and transformation of energy by bacterial membranes. Bacteriol. Rev. 36:172-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrison, S. C., D. L. Caspar, R. D. Camerini-Otero, and R. M. Franklin. 1971. Lipid and protein arrangement in bacteriophage PM2. Nat. New Biol. 229:197-201. [DOI] [PubMed] [Google Scholar]
- 44.Haywood, A. M. 1994. Virus receptors: binding, adhesion strengthening, and changes in viral structure. J. Virol 68:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heller, K. J. 1992. Molecular interaction between bacteriophage and the gram-negative cell envelope. Arch. Microbiol. 158:235-248. [DOI] [PubMed] [Google Scholar]
- 46.Houssin, C., N. Eynard, E. Shechter, and A. Ghazi. 1991. Effect of osmotic pressure on membrane energy-linked functions in Escherichia coli. Biochim. Biophys. Acta 1056:76-84. [DOI] [PubMed] [Google Scholar]
- 47.Hoyle, D. B., and T. J. Beveridge. 1984. Metal binding by the peptidoglycan sacculus of Echerichia coli K12. Can. J. Microbiol. 30:204-211. [DOI] [PubMed] [Google Scholar]
- 48.Huiskonen, J. T., H. M. Kivelä, D. H. Bamford, and S. J. Butcher. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat. Struct. Mol. Biol., in press. [DOI] [PubMed]
- 49.Ingraham, J. L., and A. G. Marr. 1996. Effect of temperature, pressure, pH, and osmotic stress on growth, p. 1570-1578. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 50.Kalasauskaitė, E. V., D. L. Kadišaite, R. J. Daugelavičius, L. L. Grinius, and A. A. Jasaitis. 1983. Studies on energy supply for genetic process. Requirement for membrane potential in Escherichia coli infection by phage T4. Eur. J. Biochem. 130:123-130. [PubMed] [Google Scholar]
- 51.Kao, S. H., and W. H. McClain. 1980. Baseplate protein of bacteriophage T4 with both structural and lytic functions. J. Virol. 34:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato, J., J. Amie, A. Kuroda, A. Mitsutani, and H. Ohtake. 1998. Development of a genetic transformation system for an alga-lysing bacterium. Appl. Environ. Microbiol. 64:2061-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kivelä, H. M., R. H. Männistö, N. Kalkkinen, and D. H. Bamford. 1999. Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:364-374. [DOI] [PubMed] [Google Scholar]
- 54.Kivelä, H. M., N. Kalkkinen, and D. H. Bamford. 2002. Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core. J. Virol. 76:8169-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koonin, E. V., and K. E. Rudd. 1994. A conserved domain in putative bacterial and bacteriophage transglycosylases. Trends Biochem. Sci. 19:106-107. [DOI] [PubMed] [Google Scholar]
- 56.Labedan, B., K. B. Heller, A. A. Jasaitis, T. H. Wilson, and E. B. Goldberg. 1980. A membrane potential threshold for phage T4 DNA injection. Biochem. Biophys. Res. Commun. 93:625-630. [DOI] [PubMed] [Google Scholar]
- 57.Lambert, O., L. Plancon, J. L. Rigaud, and L. Letellier. 1998. Protein-mediated DNA transfer into liposomes. Mol. Microbiol. 30:761-765. [DOI] [PubMed] [Google Scholar]
- 58.Lehnherr, H., A. M. Hansen, and T. Ilyina. 1998. Penetration of the bacterial cell wall: a family of lytic transglycosylases in bacteriophages and conjugative plasmids. Mol. Microbiol. 30:454-457. [DOI] [PubMed] [Google Scholar]
- 59.Letellier, L., L. Plancon, M. Bonhivers, and P. Boulanger. 1999. Phage DNA transport across membranes. Res. Microbiol. 150:499-505. [DOI] [PubMed] [Google Scholar]
- 60.Letellier, L., P. Boulanger, M. de Frutos, and P. Jacquot. 2003. Channeling phage DNA through membranes: from in vivo to in vitro. Res. Microbiol. 154:283-287. [DOI] [PubMed] [Google Scholar]
- 61.Lundström, K. H., D. H. Bamford, E. T. Palva, and K. Lounatmaa. 1979. Lipid-containing bacteriophage PR4: structure and life cycle. J. Gen. Virol. 43:583-592. [DOI] [PubMed] [Google Scholar]
- 62.Männistö, R. H., H. M. Kivelä, L. Paulin, D. H. Bamford, and J. K. H. Bamford. 1999. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:355-363. [DOI] [PubMed] [Google Scholar]
- 63.Männistö, R. H., A. M. Grahn, D. H. Bamford, and J. K. H. Bamford. 2003. Transcription of bacteriophage PM2 involves phage-encoded regulators of heterologous origin. J. Bacteriol. 185:3278-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marcoli, R., V. Pirrotta, and R. M. Franklin. 1979. Interaction between bacteriophage PM2 protein IV and DNA. J. Mol. Biol. 131:107-131. [DOI] [PubMed] [Google Scholar]
- 65.Moak, M., and I. J. Molineux. 2000. Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol. Microbiol. 37:345-355. [DOI] [PubMed] [Google Scholar]
- 66.Moak, M., and I. J. Molineux. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51:1169-1183. [DOI] [PubMed] [Google Scholar]
- 67.Moffatt, B. A., and F. W. Studier. 1988. Entry of bacteriophage T7 DNA into the cell and escape from host restriction. J. Bacteriol. 170:2095-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Molineux, I. J. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1-8. [DOI] [PubMed] [Google Scholar]
- 69.Nakagawa, H., F. Arisaka, and S. Ishii. 1985. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J. Virol. 54:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nikaido, H. 1994. Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem. 269:3905-3908. [PubMed] [Google Scholar]
- 73.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 74.Ojala, P. M., J. T. Juuti, and D. H. Bamford. 1993. Protein P4 of double-stranded RNA bacteriophage φ6 is accessible on the nucleocapsid surface: epitope mapping and orientation of the protein. J. Virol. 67:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olkkonen, V. M., and D. H. Bamford. 1989. Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection. Virology 171:229-238. [DOI] [PubMed] [Google Scholar]
- 76.Poranen, M. M., R. Daugelavičius, P. M. Ojala, M. W. Hess, and D. H. Bamford. 1999. A novel virus-host cell membrane interaction. Membrane voltage-dependent endocytic-like entry of bacteriophage φ6 nucleocapsid. J. Cell Biol. 147:671-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poranen, M. M., R. Daugelavičius, and D. H. Bamford. 2002. Common principles in viral entry. Annu. Rev. Microbiol. 56:521-538. [DOI] [PubMed] [Google Scholar]
- 78.Ravantti, J. J., A. Gaidelytė, D. H. Bamford, and J. K. H. Bamford. 2003. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrates a viral lineage. Virology 313:401-414. [DOI] [PubMed] [Google Scholar]
- 79.Roessner, C. A., and G. M. Ihler. 1986. Formation of transmembrane channels in liposomes during injection of lambda DNA. J. Biol. Chem. 261:386-390. [PubMed] [Google Scholar]
- 80.Rydman, P. S., and D. H. Bamford. 2000. Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol. Microbiol. 37:356-363. [DOI] [PubMed] [Google Scholar]
- 81.Rydman, P. S., and D. H. Bamford. 2002. The lytic enzyme of bacteriophage PRD1 is associated with the viral membrane. J. Bacteriol. 184:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 83.Satake, H., H. Akutsu, M. Kania, and R. M. Franklin. 1980. Structure and synthesis of a lipid-containing bacteriophage. Studies on the structure of the bacteriophage PM2 nucleocapsid. Eur. J. Biochem. 108:193-201. [DOI] [PubMed] [Google Scholar]
- 84.Satake, H., M. Kania, and R. M. Franklin. 1981. Structure and synthesis of a lipid-containing bacteriophage. Amphiphilic properties of protein IV of bacteriophage PM2. Eur. J. Biochem. 114:623-628. [DOI] [PubMed] [Google Scholar]
- 85.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 86.Simidu, U., K. Kita-Tsukamoto, T. Yasumoto, and M. Yotsu. 1990. Taxomony of four bacterial strains that produce tetrodotoxin. Int. J. Syst. Bacteriol. 40:331-336. [DOI] [PubMed] [Google Scholar]
- 87.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 88.Tarahovsky, Y. S., A. A. Khusainov, A. A. Deev, and Y. V. Kim. 1991. Membrane fusion during infection of Escherichia coli cells by phage T4. FEBS Lett. 289:18-22. [DOI] [PubMed] [Google Scholar]
- 89.Tarahovsky, Y. S., A. A. Khusainov, R. Daugelavičius, and E. Bakene. 1995. Structural changes in Escherichia coli membranes induced by bacteriophage T4 at different temperatures. Biophys. J. 68:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsukagoshi, N., R. Schäfer, and R. M. Franklin. 1977. Structure and synthesis of a lipid-containing bacteriophage. An endolysin activity associated with bacteriophage PM2. Eur. J. Biochem. 77:585-588. [DOI] [PubMed] [Google Scholar]
- 91.van der Schans, G. P., J. P. Weyermans, and J. F. Bleichrodt. 1971. Infection of spheroplasts of Pseudomonas with DNA of bacteriophage PM2. Mol. Gen. Genet. 110:263-271. [DOI] [PubMed] [Google Scholar]
- 92.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. Methods Cardohydr. Chem. 5:83-91. [Google Scholar]
- 93.White, A. H. 1940. A bacterial discoloration of print butter. Sci. Agric. 20:638-645. [Google Scholar]
- 94.Zavriev, S. K., and M. F. Shemyakin. 1982. RNA polymerase-dependent mechanism for the stepwise T7 phage DNA transport from the virion into E. coli. Nucleic Acids Res. 10:1635-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zimmer, S. G., and R. L. Millette. 1975. DNA-dependent RNA polymerase from Pseudomonas BAL-31. II. Transcription of the allomorphic forms of bacteriophage PM2 DNA. Biochemistry. 14:300-307. [DOI] [PubMed] [Google Scholar]
- 96.ZoBell, C. E., and H. C. Upham. 1944. A list of marine bacteria including description of sixty new species. Bull. Scripps Inst. Oceanogr. Univ. Calif. 5:239-292. [Google Scholar]