Abstract
We used Porphyromonas gingivalis gene microarrays to compare the total gene contents of the virulent strain W83 and the avirulent type strain, ATCC 33277. Signal ratios and scatter plots indicated that the chromosomes were very similar, with approximately 93% of the predicted genes in common, while at least 7% of them showed very low or no signals in ATCC 33277. Verification of the array results by PCR indicated that several of the disparate genes were either absent from or variant in ATCC 33277. Divergent features included already reported insertion sequences and ragB, as well as additional hypothetical and functionally assigned genes. Several of the latter were organized in a putative operon in W83 and encoded enzymes involved in capsular polysaccharide synthesis. Another cluster was associated with two paralogous regions of the chromosome with a low G+C content, at 41%, compared to that of the whole genome, at 48%. These regions also contained conserved and species-specific hypothetical genes, transposons, insertion sequences, and integrases and were located adjacent to tRNA genes; thus, they had several characteristics of pathogenicity islands. While this global comparative analysis showed the close relationship between W83 and ATCC 33277, the clustering of genes that are present in W83 but divergent in or absent from ATCC 33277 is suggestive of chromosomal islands that may have been acquired by lateral gene transfer.
The identification of virulent strains of pathogenic bacteria, and consequently their virulence genes, is a basic doctrine of the microbial pathogenesis field. Historically, identification has depended on phenotypic properties, biochemical activities, and immunological classifications. Increasingly, these tests have been replaced by genomic DNA-based analyses that can be successfully adapted to identify species, strains, and even mutants within strains. The availability of complete genome sequences for many bacterial pathogens has further increased the accuracy and specificity of such tests. A new addition to the existing repertoire of DNA analyses is comparative genome profiling using DNA microarrays, and this technology has been adapted to identify genes associated with pandemic strains of Vibrio cholerae (9) and to distinguish virulent strains of group A Streptococcus (30), Helicobacter pylori (4), and Salmonella species (6).
Porphyromonas gingivalis is a gram-negative oral anaerobe associated with periodontal disease in adults. The organism is the most-studied oral pathogen, partly because it produces several virulence factors that can be isolated and studied biochemically (reviewed in reference 16) and partly because it is relatively easy to grow and manipulate genetically. According to animal models of disease, strains are classified as virulent and avirulent, and studies with bacterial strains and defined mutants have validated both the models and putative virulence factors (2, 3, 11). Strains of P. gingivalis have been differentiated by restriction fragment length polymorphism analysis of insertion sequences (8) and by heteroduplex and PCR analysis of the ribosomal intergenic spacer region (13, 21). The genome sequence of P. gingivalis was recently completed (26), and DNA microarrays were prepared from PCR amplicons derived from the annotated open reading frames. We compared a virulent and an avirulent strain of P. gingivalis by microarray analysis to identify genetic differences. The microarray results identified over 150 divergent genes, with several organized in clusters associated with low-G+C genomic regions. This suggests that they were relatively recent additions to the genome and were possibly acquired by lateral gene transfer.
MATERIALS AND METHODS
Bacterial strains and genomic DNA preparation.
P. gingivalis strains W83, W50, ATCC 33277, and 381 were cultured anaerobically on blood agar as described previously (7). Two-day-old cultures were washed once in phosphate-buffered saline, and genomic DNAs were prepared with MasterPure DNA purification kits (Epicentre Technologies, Madison, Wis.).
P. gingivalis microarrays.
P. gingivalis microarrays were manufactured by The Institute for Genomic Research (TIGR) and were based on the genome sequence of the virulent strain W83. PCR amplicons were generated from open reading frames (ORFs) predicted by TIGR GLIMMER automated annotation software. Amplicons in 50% dimethyl sulfoxide buffer were spotted at least twice for each ORF onto aminosilane-coated glass microscope slides (CMT-GAPS, Corning, N.Y.) by a microarray robot (Intelligent Automation Systems, Cambridge, Mass.). The mean and median sizes of the amplicons were 486 and 461 bp, respectively, and represented 2,558 ORFs identified in the genome. Due to a high number of repeat elements such as insertion sequences, only 1,990 ORFs were unique. Detailed array information, e.g., grid formation, PCR primer and amplicon sequences, and annotation, can be viewed at the web site described below.
Competitive DNA-DNA hybridizations and microarray data acquisition.
Genomic DNAs were labeled by a two-step protocol. Briefly, at least 3 μg of DNA was digested with Sau3A1 (New England Biolabs, Beverly, Mass.), concentrated by ethanol precipitation, and dissolved in 10 mM Tris-HCl, pH 8.5. The DNA was combined with 3 μg of random hexamers (Invitrogen Life Technologies, Carlsbad, Calif.) in a 30-μl reaction volume, heated at >95°C for 5 min, and then chilled on ice. The rest of the reaction components, in a total volume of 50 μl, were as follows: 5 μl of 10× E. coli DNA polymerase I buffer (NEB); 6 μl each of 2.5 mM dATP, dGTP, and dCTP (Perkin-Elmer, Wellesley, Mass.); 6 μl of 2.5 mM amino allyl-dUTP (Sigma Chemical Company, St. Louis, Mo.); and 3 μl of Klenow enzyme (New England Biolabs). The reaction was carried out at 37°C for 2 h, and the products were removed from unincorporated amino allyl-dUTP by precipitation with ethanol. The dried pellet was dissolved in 5 μl of 2× coupling buffer (0.2 M NaHCO3, pH 9.0), and 5 μl of 0.5 mM Cy3 or Cy5 was added; the coupling reaction was incubated for 30 min to 1 h at room temperature in the dark. Dye-coupled DNA samples were purified with a PCR purification kit (Qiagen, Valencia, Calif.). Hybridization and stringency washes were performed as described previously (9). Arrays were scanned in a GenePix 4000B microarray scanner, and amplicon spot intensities were read with GenePix Pro software (Axon Instruments, Inc., Union City, Calif.). Spots that could not be identified by both automated and human visual inspection were discarded.
Data normalization.
The normalization of array data was performed with Statistics for Microarray Analysis (SMA) software, an R add-on package for cDNA microarray data processing (17) available at http://stat-www.berkeley.edu/users/terry/zarray/Software/smacode.html. Data within the same slide were normalized by locally weighted scatter-plot smoothing (LOWESS) and scaled print-tip group normalization under the premise that the majority of genes in the two DNA samples would have similar overall signal intensities. This method combined multiple approaches that considered both the overall signal ratio and the distribution of signal ratios. Data between slides were normalized similarly before the comparative analysis described below.
EPP analysis.
Normalized array data were subjected to estimation of the probability of presence (EPP) with the GACK genomotyping analysis software at http://falkow.stanford.edu/whatwedo/software/software.html (19). Each amplicon was assigned a value between 0.5 and −0.5 based on the graded assignment algorithm provided by the software.
Microarray data visualization and storage.
Microarray data visualization was carried out with GenomeViewer software (http://genome.oralgen.org), in which P. gingivalis PCR amplicons and genome annotations from the TIGR Comprehensive Microbial Resource (CMR) (http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl) and the Los Alamos Oral Pathogen Sequence Database (http://www.oralgen.lanl.gov) were linked for side-by-side comparisons. Whole-genome heat-map comparison images were created with the same software.
Verification of highly divergent genes.
Sequences of primer pairs for 16 highly divergent genes were obtained through links provided by the GenomeViewer software. The primer sequences were identical to those used by TIGR to generate amplicons for the microarrays and are listed in Table 1. PCRs were performed in a PTC-200 Peltier thermal cycler (MJ Research, Inc., Watertown, Mass.) in 50-μl reaction volumes that contained 1 μM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, a 0.2 μM concentration of each primer, 5 ng of genomic DNA template, and 1.25 U of AmpliTaq Gold (Applied Biosystems, Foster City, Calif.). The cycling conditions were as follows: 10 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 55°C, and 1.5 min at 72°C; and 5 min at 72°C.
TABLE 1.
Primer sequences used for PCR verification of selected amplicons with low EPPs and negative graded scoresb
| ORF | Encoded protein | Primera | Primer sequence (5′-3′) | Amplicon length (bp) |
|---|---|---|---|---|
| PG0019 | ISPg4, transposase | F | AGCCACAGGTAACCTCAACC | 847 |
| R | CCACCGATATTTGGCGATAC | |||
| PG0110 | Glycosyl transferase, group 1 family protein | F | CGGAGTCGTTCTAAGCCTTG | 670 |
| R | AGTCCACAATGACTCCTGGG | |||
| PG0111 | Capsular polysaccharide biosynthesis gene, putative | F | GCTATCGCCCTCCAATATGA | 711 |
| R | TGTGTCACAACAACGACCCT | |||
| PG0117 | Polysaccharide transport protein, putative | F | TCAATATTCGAGGGGCGTAG | 824 |
| R | AGGAGCGCAAATAGCAAAAA | |||
| PG0683 | ABC transporter, permease protein, putative | F | ACTATCTGCTCAAAGCCGGA | 795 |
| R | CCAATTCGGCACGAAGTATT | |||
| PG0742 | Antigen PgaA | F | CATTCTGCTCCGAGCTTAGG | 699 |
| R | ATCACGAATTAGCGGTGGTC | |||
| PG0826 | Transcriptional regulator, AraC family | F | AAGCGTTGGAGAAACTCCTG | 800 |
| R | GTTCGCAACTCACCGATTTT | |||
| PG0827 | MATE efflux family protein | F | CATCGCAATGCTGATTATGG | 1,162 |
| R | TCCGTTCAATCCCCAATATG | |||
| PG0828 | RteC protein, truncation | F | CTTTCAGATCGCTTTCCACC | 294 |
| R | AGGGACTTCTTCCTGCATTG | |||
| PG1445 | RteC protein, truncation | F | TGCCAGTCAGACCTGCTAAG | 271 |
| R | AGATCGCTTTCCACCATACG | |||
| PG1446 | MATE efflux family protein | F | TTCTTGGGATAGGGCTGATG | 1,067 |
| R | CGGTTGTGCATAAAGCACTC | |||
| PG1447 | Transcriptional regulator, AraC family | F | CAAATCCCAAACCTTGTGCT | 755 |
| R | GCCAATCCATAGAAGTTCGC | |||
| PG1454 | Integrase | F | TTATGGAATCCCCGTGAGAG | 884 |
| R | TCCTTTATGTCGGCGAGAAC | |||
| PG1644 | ISPg5, transposase Orf2 | F | AGACCTGGGGAACTCCTTGT | 501 |
| R | CGGATTTTTAGACTCTGGCG | |||
| PG1645 | ISPg5, transposase Orf1 | F | GAGGATTACCTCTCGGGGTC | 228 |
| R | TCGCTTGAGACGACTCTTGA | |||
| PG2100 | Immunoreactive 63-kDa antigen PG102 | F | TATACGTAATGGCCCGGGTA | 855 |
| R | TTACAAGATGGCTGTGGCAG |
F, forward; R, reverse.
Genomic DNAs from P. gingivalis strains were used as PCR templates. Primer sequences were the same as those used to generate microarray amplicons. ORF and protein names were from the TIGR CMR.
Oral Pathogens Microarray Database.
The original microarray images, the raw data generated by GenePix software, and the relevant minimum information about a microarray experiment can be accessed at the Oral Pathogens Microarray Database (http://array.oralgen.org). The complete list of EPP values and graded divergent scores can be viewed and downloaded by using the GenomeViewer software at the same web site.
RESULTS
Genomotyping by microarray analysis.
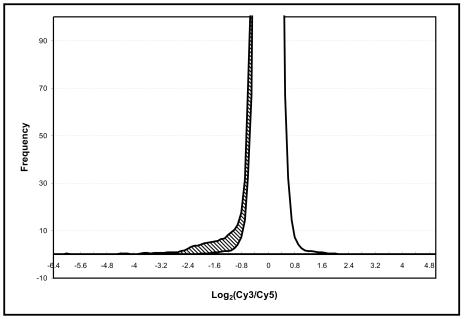
Microarray-based competitive hybridizations with labeled genomic DNAs from control (W83) and tester (ATCC 33277) strains were performed with a total of six slides. Independently isolated and labeled genomic DNA samples were used for each slide. Each slide contained two identical sets of amplicons representing P. gingivalis ORFs, and thus there were two duplicate arrays per slide. Data from a total of 12 repeats were normalized first within the slides and then between slides by a combined approach that included print-tip group normalization, LOWESS, and the scaled normalization schemes that were provided in the SMA package. Normalized data were used as input for the GACK program to evaluate and rank genes that diverged between strains W83 and ATCC 33277. Figure 1 shows the skewed frequency distribution of logarithm signal ratios between the two strains. The skewed effect of signal ratios on one tail of the normal distribution curve was anticipated in these experiments since the probes (amplicons on the slides) were from the control strain (W83) and the normalized signals of the tester strain (ATCC 33277) were seldom higher than those of the control, except for genes that were present in higher copy numbers in the tester strain. Based on GACK analysis, each gene was assigned an EPP score and a graded assessment of divergence (graded mean score). A total of 154 ORFs predicted by the TIGR annotation (7%) had EPP scores of <100% and negative graded mean scores and were considered slightly (EPP near 100) to highly (EPP close to 0) divergent between strains W83 and ATCC 33277, i.e., they were present in W83 but not detected in ATCC 33277. In Table 2, we present selected genes with the lowest EPP scores (the cutoff was 20%).
FIG. 1.
Distribution of log ratios of signal intensities between strains W83 (Cy5) and ATCC 33277 (Cy3). The difference (hatched area) between the mapped normal curve (inner curve) and the raw frequency curve (outer curve) represents the skewed frequency distribution due to absent or variant counterpart sequences in strain ATCC 33277.
TABLE 2.
Strain W83 genes that are hightly divergent in ATCC 33277 with an EPP cutoff of <20%
| Gene no. | Encoded protein | ORF | Sample size | EPP (%) | Mean score | Standard deviation | Variance |
|---|---|---|---|---|---|---|---|
| 1 | ISPg4, transposase | PG0019 | 13 | 1 | −0.49 | 0.04 | 0.00 |
| 2 | ISPg4, transposase | PG0050 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 3 | Null | PG0051 (ORF00091) | 1 | 15 | −0.35 | 0.00 | 0.00 |
| 4 | Hypothetical protein | PG0089 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 5 | Glycosyl transferase, group 1 family protein | PG0110 | 14 | 12 | −0.38 | 0.16 | 0.03 |
| 6 | Capsular polysaccharide biosynthesis gene putative | PG0111 | 14 | 18 | −0.32 | 0.28 | 0.08 |
| 7 | Null, conserved hypothetical protein, authentic frameshift | PG0112 (ORF00191) | 14 | 18 | −0.32 | 0.27 | 0.07 |
| 8 | Hypothetical protein | PG0113 | 14 | 15 | −0.35 | 0.17 | 0.03 |
| 9 | Hypothetical protein | PG0114 | 13 | 8 | −0.42 | 0.11 | 0.01 |
| 10 | Polysaccharide transport protein, putative | PG0117 | 14 | 9 | −0.41 | 0.13 | 0.02 |
| 11 | Glycosyl transferase, group 2 family protein | PG0118 | 11 | 10 | −0.40 | 0.18 | 0.03 |
| 12 | Lipoprotein RagB | PG0186 | 14 | 13 | −0.37 | 0.18 | 0.03 |
| 13 | Hypothetical protein | PG0244 | 1 | 15 | −0.35 | 0.00 | 0.00 |
| 14 | Secretion activator protein, putative | PG0293 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 15 | Hypothetical protein | PG0410 | 4 | 18 | −0.33 | 0.10 | 0.01 |
| 16 | Hypothetical protein | PG0421 | 2 | 18 | −0.33 | 0.03 | 0.00 |
| 17 | ISPg5, transposase Orf1 | PG0427 | 2 | 18 | −0.33 | 0.03 | 0.00 |
| 18 | ISPg5, transposase Orf1 | PG0459 | 14 | 4 | −0.46 | 0.09 | 0.01 |
| 19 | Type I restriction modification system, M subunit, putative | PG0544 | 13 | 19 | −0.31 | 0.16 | 0.03 |
| 20 | Hypothetical protein | PG0565 | 13 | 15 | −0.35 | 0.14 | 0.02 |
| 21 | Hypothetical protein | PG0626 | 14 | 13 | −0.38 | 0.14 | 0.02 |
| 22 | ABC transporter, permease protein, putative | PG0683 | 14 | 12 | −0.38 | 0.16 | 0.03 |
| 23 | Hypothetical protein | PG0717 | 14 | 11 | −0.39 | 0.14 | 0.02 |
| 24 | Antigen PgaA | PG0742 | 14 | 11 | −0.39 | 0.15 | 0.02 |
| 25 | Integrase | PG0820 | 14 | 1 | −0.49 | 0.02 | 0.00 |
| 26 | Hypothetical protein | PG0821 | 13 | 11 | −0.39 | 0.26 | 0.07 |
| 27 | MATE efflux family protein | PG0827 | 14 | 12 | −0.38 | 0.14 | 0.02 |
| 28 | RteC protein, truncation | PG0828 | 11 | 19 | −0.31 | 0.28 | 0.08 |
| 29 | Integrase | PG0838 | 13 | 8 | −0.42 | 0.13 | 0.02 |
| 30 | Conserved hypothetical protein | PG0839 | 14 | 7 | −0.43 | 0.08 | 0.01 |
| 31 | Hypothetical protein | PG0848 | 13 | 18 | −0.32 | 0.16 | 0.03 |
| 32 | Conserved hypothetical protein | PG0859 | 14 | 15 | −0.35 | 0.18 | 0.03 |
| 33 | Hypothetical protein | PG0892 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 34 | ISPg1, transposase, authentic frameshift | PG0939 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 35 | ISPg4, transposase | PG0970 | 2 | 18 | −0.33 | 0.03 | 0.00 |
| 36 | Conserved hypothetical protein | PG1057 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 37 | Hypothetical protein | PG1059 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 38 | Hypothetical protein | PG1102 | 14 | 1 | −0.49 | 0.04 | 0.00 |
| 39 | Hypothetical protein | PG1107 | 14 | 15 | −0.35 | 0.16 | 0.02 |
| 40 | Null (PG1275 hypothetical) | PG1275 (ORF02037a) | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 41 | RteC protein, truncation | PG1445 | 8 | 19 | −0.31 | 0.29 | 0.08 |
| 42 | Conserved hypothetical protein | PG1449 | 13 | 15 | −0.35 | 0.17 | 0.03 |
| 43 | Conserved hypothetical protein | PG1450 | 2 | 15 | −0.35 | 0.15 | 0.02 |
| 44 | Integrase | PG1454 | 14 | 2 | −0.48 | 0.05 | 0.00 |
| 45 | Conserved domain protein | PG1512 | 11 | 5 | −0.45 | 0.10 | 0.01 |
| 46 | O-Succinylbenzoic acid-coenzyme A ligase | PG1521 | 4 | 11 | −0.39 | 0.11 | 0.01 |
| 47 | Toprim domain protein | PG1533 | 2 | 8 | −0.43 | 0.08 | 0.01 |
| 48 | HDIG domain protein | PG1592 | 1 | 15 | −0.35 | 0.00 | 0.00 |
| 49 | Isoleucyl-tRNA synthetase | PG1596 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 50 | ISPg5, transposase Orf2 | PG1644 | 14 | 2 | −0.48 | 0.04 | 0.00 |
| 51 | ISPg5, transposase Orf1 | PG1645 | 5 | 14 | −0.36 | 0.26 | 0.07 |
| 52 | Hypothetical protein | PG1685 | 4 | 1 | −0.49 | 0.02 | 0.00 |
| 53 | ISPg5, transposase Orf1 | PG1709 | 4 | 1 | −0.49 | 0.02 | 0.00 |
| 54 | Null, hypothetical protein | PG1740 (ORF02751) | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 55 | Hypothetical protein | PG1988 | 4 | 10 | −0.40 | 0.12 | 0.02 |
| 56 | Hypothetical protein | PG2008 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 57 | Hypothetical protein | PG2018 | 14 | 17 | −0.33 | 0.18 | 0.03 |
| 58 | ISPg5, transposase Orf1 | PG2058 | 4 | 6 | −0.44 | 0.08 | 0.01 |
| 59 | Hypothetical protein | PG2095 | 1 | 0 | −0.50 | 0.00 | 0.00 |
| 60 | Immunoreactive 63-kDa antigen PG102 | PG2100 | 13 | 19 | −0.31 | 0.17 | 0.03 |
| 61 | ISPg5, transposase Orf1 | PG2129 | 13 | 12 | −0.38 | 0.15 | 0.02 |
| 62 | Hypothetical protein | PG2136 | 14 | 15 | −0.35 | 0.15 | 0.02 |
| 63 | Hypothetical protein | PG2203 | 1 | 5 | −0.45 | 0.00 | 0.00 |
| 64 | Hypothetical protein | PG2204 | 2 | 0 | −0.50 | 0.00 | 0.00 |
Verification of microarray results.
For further study, we selected 16 genes that were highly divergent according to the microarray results, i.e., the data indicated that they were present in W83 but not in ATCC 33277. Of these, PG0019 (ISPg4) and PG01644 and PG01645 (the two ORFs of ISPg5) were previously shown to be absent from ATCC 33277 (5, 29). ORFs PG0110, PG0111, and PG0117 were from a cluster of genes involved in capsular polysaccharide biosynthesis; PG0826, PG0827, PG0828, PG1446, and PG1447 were from two paralogous regions of the genome with characteristics of pathogenicity islands. The absence of these genes in ATCC 33277 was tested by PCR amplification with the W83 sequence-derived primer pairs that were used to generate the respective amplicons for the microarrays. Two close relatives of strains W83 and ATCC 33277, strains W50 and 381, respectively, were also included in the PCR analysis. Amplicons of the predicted sizes were detected for all 16 genes in strains W83 and W50 (Fig. 2), confirming the strong similarity between these two strains. However, for ATCC 33277 and 381, no or very weak amplification was obtained for 15 of the genes (Fig. 2), indicating either that the templates were absent from these strains or that the W83-derived primer sequences were so dissimilar that amplicons could not be generated; both possibilities support gene divergence between strain W83 and strains ATCC 33277 and 381. Despite the low EPP and mean scores predicted for PG1446 (MATE efflux family protein), amplicons were found in all four strains (also confirmed by Southern blot analysis [data not shown]). However, since the surrounding ORFs were all highly divergent or absent, PG1446 may encode an essential protein in both virulent and avirulent strains.
FIG. 2.
Verification of microarray data by PCR. Genomic DNAs from four P. gingivalis strains were used as PCR templates with primer pairs for 16 ORFs that were predicted to be highly variant between strains W83 and ATCC 33277. Amplicons were visualized after agarose gel electrophoresis and ethidium bromide staining.
Survey of divergent genes in W83 genome.
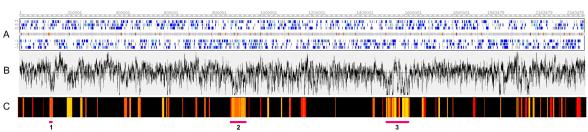
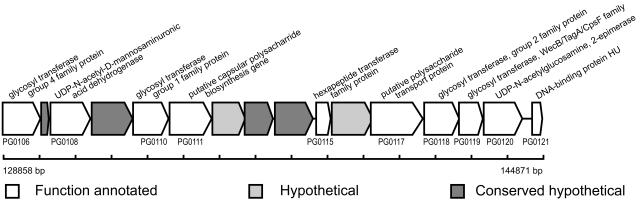
To determine the distribution of divergent genes in the W83 genome, we plotted graded mean scores of all the genes across the length of the complete genome (Fig. 3). At least two regions contained a high density of divergent genes, and these “hot spots” are shown in Fig. 3C. Interestingly, the hot spots coincided with regions of lower G+C ratios (Fig. 3B). According to the P. gingivalis annotation in the CMR database (TIGR), the genes encoding PG0106, -0108, -0117, -0118, -0119, -0120, and -0121 are predicted to be part of an operon based on comparisons of similar genes in different microbial genomes. As yet, we have no experimental evidence that these genes are cotranscribed in strain W83. The complete region consists of up to 14 genes encoding enzymes that may be involved in polysaccharide capsule synthesis (Fig. 4). The genomotyping results obtained in this study reveal eight genes in the cluster: they are PG0109, -0110, -0111, -0112, -0113, -0114, -0117, and -0118, and they are highly divergent in or absent from strain ATCC 33277. Furthermore, the coding sequences of the genes have the lowest G+C content (mean, 40.1%; range, 36.2% to 47.7%) within the region, suggesting that they may be new additions to the genome and possibly were acquired by lateral gene transfer.
FIG. 3.
Distribution of divergent genes in genome of strain W83. (A) W83 genome and six-frame distribution of predicted genes. Dark blue, genes annotated with names or function; light blue, conserved hypothetical genes; gray, hypothetical genes and intergenic regions; red, tRNAs; yellow, rRNA. ORF coordinates were obtained from the CMR at TIGR (http://www.oralgen.lanl.gov); rRNA and tRNA coordinates were obtained from the Oral Pathogen Sequence Databases at the Los Alamos National Laboratory (http://www.oralgen.lanl.gov). (B) GC distribution based on the G+C content of a 500-bp window. (C) Simulated heat map of the distribution of genes categorized from slightly (yellow) to highly (red) divergent between strains W83 and ATCC 33277. The black background indicates areas where genes are present in both genomes. Images were compiled with GenomeViewer software (http://www.oralgen.org).
FIG. 4.
Putative polysaccharide capsule synthesis operon in P. gingivalis W83 genome. A region of 16,014 bp containing 16 ORFs is shown with exact nucleotide positions in the complete genome.
Two paralogous regions, one of approximately 28 kb (PG0819 to PG0844) and a deleted version of approximately 18 kb (PG1435 to PG1454), were also identified by microarray analysis as being present in W83 and divergent in ATCC 33277. It is probable that the paralogs were generated by duplication and intrachromosomal recombination. With an average G+C composition of 41%, compared to 48% for the whole genome, the regions are bounded on one side by homologs of the Bacteroides transposon Tn5520 and on the other by either a serine or aspartate tRNA; thus, these regions have characteristics of pathogenicity islands. Half of the genes encode homologs of transcription regulators, mobilization and transfer functions of Bacteroides conjugative transposons, excisases, integrases, ISPg1, and an efflux pump family protein; the rest of the genes are either conserved hypothetical or species-specific hypothetical ORFs.
DISCUSSION
We used microarrays to compare the genomes of P. gingivalis strains and to identify genes that were present in a virulent strain but absent from or divergent in an avirulent strain, potentially representing a repertoire of functions associated with pathogenicity. The goals of this initial study were to detect differences in the total gene complement of the two strains, rather than identifying sequence differences in specific genes. However, since microarray data readouts are based on signals generated from DNA-DNA hybridizations, very low EPP scores were presumed to be due to extensive sequence variations in genes rather than to minor nucleotide polymorphisms.
Strains W83 and ATCC 33277 were selected as the virulent and avirulent strains, respectively, since many previous reports have compared their virulence-associated activities and disease-promoting characteristics in vitro and in vivo (12, 20, 31). To determine the degree of divergence, we used an approach that determines and ranks gene variations between the two strains based on the shape of the signal ratio distribution (19), thus alleviating the empirical determination of a cutoff. The cutoff was determined independently for each array repeat and thus compensated for the variation in hybridization. Also, this algorithm calculates an estimate of the probability of the W83 genes being present in ATCC 33277 (EPP), providing an additional measure of confidence in the divergence assignment.
Our results indicated many genetic differences between the two strains, and several divergent genes encoded activities that putatively contribute to virulence. The microarray analyses identified genes in W83 that previously were shown to be absent from ATCC 33277, including an insertion sequence renamed ISPg4 (29), ISPg5 (5), and the ragB gene (10, 15). These results gave credence to the rest of the microarray data showing that approximately 7% of the W83 genes were divergent to various degrees in ATCC 33277.
A cluster of ORFs involved in the synthesis of capsular polysaccharide that was present in W83 was not found in strain ATCC 33277. There are six serotypes of P. gingivalis based on capsular polysaccharide (K) antigens, and the severity of disease was correlated with the presence of the capsule and with the capsule serotype in a mouse infection model (20). The capsule of strain W83 (K1 type) was associated with the severest form of infection, while strain 381, a close relative of strain ATCC 33277, which does not possess a capsule (K−), caused minimal infection. The animal infection study indicated a role for the capsule in virulence, which suggests that the genes identified in the present work may be involved in pathogenesis.
Interestingly, many of the divergent genes were located in low-G+C regions, suggesting that they may be relatively recent additions to the genome. DNA-based assays have shown that the majority of virulent bacterial strains or clones differ from their avirulent counterparts because of the acquisition of virulence genes by lateral transfer on mobile genetic elements such as plasmids, transposons, and conjugative transposons. Clues that suggest a gene may have been acquired by lateral gene transfer include a different GC content and/or different codon usage from the other host genes, antibiotic resistance functions, activities associated with virulence, and genetic linkage with known moveable DNA elements. Many of these criteria are fulfilled by pathogenicity islands, so called because they contain genes for virulence factors in microorganisms that cause disease (reviewed in reference 14). Ranging in size from 10 to 200 kb, pathogenicity islands often carry genes encoding integrases and transposases that are involved in DNA mobility, and they may be associated with tRNA genes, which are favored sites for the integration of foreign DNA. These are properties of two regions of the genome (ORFs PG0819 to PG0844 and PG1435 to PG1454) that were identified in this study. Many genes in these regions are hypothetical ORFs, and their functional identification will determine whether they are virulence factors in true pathogenicity islands. The existence of these atypical islands prompts the question of how they got there. Over 40% of the protein sequences in these regions show the highest homology to proteins of Bacteroides thetaiotaomicron, an enteric commensal (34), and it is conceivable that a gram-negative oral anaerobe may act as an intermediary in transfer. The close and constant bacterial associations in dental plaque present favorable conditions for the transfer of conjugative transposons by cell-to-cell contact (27, 33), and recently it was shown that natural competence for DNA uptake increases when bacteria are grown in plaque-like biofilms (22, 23, 32).
To validate the microarray results, we used PCR to confirm the divergence of specific genes in both W83 and ATCC 33277, as well as in two additional strains, W50 (virulent) and 381 (avirulent). Although strains ATCC 33277 and 381 have sequence differences in ribosomal intergenic spacer regions (28) and different vitamin K requirements (12), genomic and proteomic studies have revealed strong similarities, even between distantly situated genes, that could suggest that they are the same strain or sequence type (10, 24, 25). These studies also showed strong similarities between W83 and W50 but placed them in a different group from that of ATCC 33277 and 381. Evidence that the four strains may be independent comes from analyses of the protein compositions of their outer membranes, from which subtle differences could be observed (18).
Frandsen et al. (10) reported both genotypic and phenotypic diversity in a study of 132 P. gingivalis strains. A sequence analysis of four genes from disparate genomic loci in 57 strains yielded 41 genotypes, providing evidence for a predominantly nonclonal population structure and prompting the hypothesis that recombination dominates over mutations in P. gingivalis. However, six strains from different geographic locations showed close genetic relatedness and may constitute a clone. The inclusion of strains W83 and W50 in this clone and their association with periodontal disease (1, 13) suggested that this genotype had the capacity to spread through the population (10). Microarray-based whole-genomic profiling studies may uncover many genetic differences that determine virulence and provide further evidence of a clonal identity.
Acknowledgments
We thank the Harvard Center for Genomic Research for the use of their scanning facilities.
This work was supported by NIDCR grant R01DE10510 (M.J.D.) and grant K22 DE14742 (T.C.), and array construction was supported by grant R01 DE12082.
REFERENCES
- 1.Amano, A., M. Kuboniwa, I. Nakagawa, S. Akiyama, I. Morisaki, and S. Hamada. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 79:1664-1668. [DOI] [PubMed] [Google Scholar]
- 2.Baker, P. J., M. Dixon, R. T. Evans, and D. C. Roopenian. 2000. Heterogeneity of Porphyromonas gingivalis strains in the induction of alveolar bone loss in mice. Oral Microbiol. Immunol. 15:27-32. [DOI] [PubMed] [Google Scholar]
- 3.Baker, P. J., R. T. Evans, and D. C. Roopenian. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035-1040. [DOI] [PubMed] [Google Scholar]
- 4.Bjorkholm, B., A. Lundin, A. Sillen, K. Guillemin, N. Salama, C. Rubio, J. I. Gordon, P. Falk, and L. Engstrand. 2001. Comparison of genetic divergence and fitness between two subclones of Helicobacter pylori. Infect. Immun. 69:7832-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Califano, J. V., T. Kitten, J. P. Lewis, F. L. Macrina, R. D. Fleischmann, C. M. Fraser, M. J. Duncan, and F. E. Dewhirst. 2000. Characterization of Porphyromonas gingivalis insertion sequence-like element ISPg5. Infect. Immun. 68:5247-5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, T., K. Nakayama, L. Belliveau, and M. J. Duncan. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect. Immun. 69:3048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, H., T. Chen, F. E. Dewhirst, R. D. Fleischmann, C. M. Fraser, and M. J. Duncan. 1999. Genomic loci of the Porphyromonas gingivalis insertion element IS1126. Infect. Immun. 67:3416-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frandsen, E. V., K. Poulsen, M. A. Curtis, and M. Kilian. 2001. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect. Immun. 69:4479-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genco, C. A., D. R. Kapczynski, C. W. Cutler, R. J. Arko, and R. R. Arnold. 1992. Influence of immunization on Porphyromonas gingivalis colonization and invasion in the mouse chamber model. Infect. Immun. 60:1447-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenier, D., and D. Mayrand. 1987. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J. Clin. Microbiol. 25:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffen, A. L., S. R. Lyons, M. R. Becker, M. L. Moeschberger, and E. J. Leys. 1999. Porphyromonas gingivalis strain variability and periodontitis. J. Clin. Microbiol. 37:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 15.Hanley, S. A., J. Aduse-Opoku, and M. A. Curtis. 1999. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect. Immun. 67:1157-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 17.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299-314. [Google Scholar]
- 18.Kennell, W., and S. C. Holt. 1990. Comparative studies of the outer membranes of Bacteroides gingivalis, strains ATCC 33277, W50, W83, 381. Oral Microbiol. Immunol. 5:121-130. [DOI] [PubMed] [Google Scholar]
- 19.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laine, M. L., and A. J. van Winkelhoff. 1998. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 13:322-325. [DOI] [PubMed] [Google Scholar]
- 21.Leys, E. J., J. H. Smith, S. R. Lyons, and A. L. Griffen. 1999. Identification of Porphyromonas gingivalis strains by heteroduplex analysis and detection of multiple strains. J. Clin. Microbiol. 37:3906-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loos, B. G., D. W. Dyer, T. S. Whittam, and R. K. Selander. 1993. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect. Immun. 61:204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menard, C., and C. Mouton. 1995. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect. Immun. 63:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts, A. P., J. Pratten, M. Wilson, and P. Mullany. 1999. Transfer of a conjugative transposon, Tn5397, in a model oral biofilm. FEMS Microbiol. Lett. 177:63-66. [DOI] [PubMed] [Google Scholar]
- 28.Rumpf, R. W., A. L. Griffen, B. G. Wen, and E. J. Leys. 1999. Sequencing of the ribosomal intergenic spacer region for strain identification of Porphyromonas gingivalis. J. Clin. Microbiol. 37:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawada, K., S. Kokeguchi, H. Hongyo, S. Sawada, M. Miyamoto, H. Maeda, F. Nishimura, S. Takashiba, and Y. Murayama. 1999. Identification by subtractive hybridization of a novel insertion sequence specific for virulent strains of Porphyromonas gingivalis. Infect. Immun. 67:5621-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundqvist, G., D. Figdor, L. Hanstrom, S. Sorlin, and G. Sandstrom. 1991. Phagocytosis and virulence of different strains of Porphyromonas gingivalis. Scand. J. Dent. Res. 99:117-129. [DOI] [PubMed] [Google Scholar]
- 32.Wang, B. Y., B. Chi, and H. K. Kuramitsu. 2002. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol. Immunol. 17:108-112. [DOI] [PubMed] [Google Scholar]
- 33.Waters, V. L. 1999. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 4:D433-D456. [DOI] [PubMed] [Google Scholar]
- 34.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]