Abstract
Cytolysin A (ClyA) of Escherichia coli is a pore-forming hemolytic protein encoded by the clyA (hlyE, sheA) gene that was first identified in E. coli K-12. In this study we examined various clinical E. coli isolates with regard to the presence and integrity of clyA. PCR and DNA sequence analyses demonstrated that 19 of 23 tested Shiga toxin-producing E. coli (STEC) strains, all 7 tested enteroinvasive E. coli (EIEC) strains, 6 of 8 enteroaggregative E. coli (EAEC) strains, and 4 of 7 tested enterotoxigenic E. coli (ETEC) strains possess a complete clyA gene. The remaining STEC, EAEC, and ETEC strains and 9 of the 17 tested enteropathogenic E. coli (EPEC) strains were shown to harbor mutant clyA derivatives containing 1-bp frameshift mutations that cause premature termination of the coding sequence. The other eight EPEC strains and all tested uropathogenic and new-born meningitis-associated E. coli strains (n = 14 and 3, respectively) carried only nonfunctional clyA fragments due to the deletion of two sequences of 493 bp and 204 or 217 bp at the clyA locus. Expression of clyA from clinical E. coli isolates proved to be positively controlled by the transcriptional regulator SlyA. Several tested E. coli strains harboring a functional clyA gene produced basal amounts of ClyA when grown under standard laboratory conditions, but most of them showed a clyA-dependent hemolytic phenotype only when SlyA was overexpressed. The presented data indicate that cytolysin A can play a role only for some of the pathogenic E. coli strains.
Many bacterial pathogens produce toxins that kill and lyse host cells by interacting with the plasma membrane and by disrupting the function of this membrane as a permeability barrier. The majority of these cytolytic toxins are pore-forming proteins, and several of them have been shown to represent important virulence factors of the corresponding bacteria (2).
In Escherichia coli several different pore-forming cytolysins have been identified. The one most extensively studied is α-hemolysin (HlyA), which is produced by many uropathogenic E. coli (UPEC) strains and which contributes to their virulence as shown in several animal models (14, 48). E. coli α-hemolysin is encoded by the hlyCABD operon and belongs to the family of RTX (repeats-in-toxin) toxins that are widespread among gram-negative pathogens (12, 26). Several UPEC strains have been shown to carry the hly gene cluster within unique chromosomal inserts called pathogenicity islands that are absent from the nonpathogenic E. coli laboratory strain K-12 (15).
A toxin related to α-hemolysin, enterohemorrhagic E. coli (EHEC) hemolysin (EHEC-HlyA), has been identified in EHEC strains of serotype O157:H7, which represent the major etiological agents of the hemolytic-uremic syndrome and of hemorrhagic colitis worldwide (4, 28, 38). The EHEC hemolysin operon, EHEC-hlyCABD (ehxCABD), is located on a large plasmid that is present in almost all clinical E. coli O157:H7 isolates (4, 38). Recent studies revealed that EHEC-hlyA is also present in most EHEC strains belonging to less prevalent serotypes, such as O157:H−, O26:H11/H−, and O103:H2 (7, 20, 40).
A novel pore-forming hemolysin not related to HlyA, cytolysin A (ClyA), has recently been detected in E. coli K-12. ClyA is a 34-kDa protein that is encoded by a chromosomal gene denoted clyA (also referred to as hlyE and sheA) (3, 8, 13, 25, 30, 31). The ClyA protein is not produced at phenotypically detectable levels when E. coli K-12 is grown under standard conditions on blood agar. This is apparently due to repression of the transcription of clyA by the nucleoid protein H-NS (49). Nevertheless, the expression of clyA in E. coli K-12 can be activated to a level that suffices to evoke a hemolytic phenotype when certain transcriptional regulators, such as SlyA from E. coli or Salmonella enterica serovar Typhimurium (24, 25, 30), MprA (EmrR) from E. coli (8), HlyX from Actinobacillus pleuropneumoniae (13), or FnrP from Pasteurella haemolytica (43) are overproduced in this strain.
Lipid bilayer experiments and electron microscopic studies have shown that ClyA forms stable pores in target membranes by assembling into ring-shaped toxin oligomers (25, 47). Due to this pore-forming activity, ClyA lyses erythrocytes from several mammalian species. In addition, it has been reported that ClyA is cytotoxic towards cultured mammalian cells and that it induces macrophage apoptosis (22, 31), which suggests that this toxin might contribute to the virulence of pathogenic E. coli strains. Consistent with this, some EHEC strains of serotype O157:H7 have recently been shown to harbor a complete clyA gene whose predicted product is almost identical in amino acid sequence to ClyA from E. coli K-12 (ClyAK-12) (9, 17, 36). Apart from that, however, the presence of clyA in the different pathogroups of E. coli has not yet been systematically studied. Interestingly, functional clyA homologues have recently been identified in S. enterica serovar Typhi and serovar Paratyphi A, demonstrating that ClyAK-12 represents the prototype of a novel family of bacterial cytolysins (33, 35, 47).
In this study, we analyzed various E. coli wild-type strains belonging to different pathogroups with regard to the presence and sequence characteristics of clyA. In addition, we investigated the expression of clyA from several of these strains and studied the influence of SlyA on clyA regulation. The data presented show that only some of the tested strains harbor a functional clyA gene, which in turn indicates that ClyA can play a role only for a subset of the pathogenic E. coli strains. The incidence of functional copies of clyA particularly showed a correlation with several E. coli pathogroups causing enteric diseases, while all E. coli strains isolated from extraintestinal infections merely harbored nonfunctional clyA fragments.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
The E. coli wild-type strains used in this study are listed in Table 1. E. coli DH5α [F− φ80 dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 phoA hsdR17 (rK− mK+) supE44 λ− thi-1 gyrA96 relA1] was employed as cloning host and for the propagation of all plasmids except pCVD442 and pCVD442 derivatives, which were propagated in E. coli SY327λpir and E. coli SM10λpir (29). The plasmids used in this work are listed in Table 2. All E. coli strains were grown aerobically at 37°C in 2× yeast extract-tryptone (2×YT) broth (yeast extract, 10 g/liter; tryptone, 16 g/liter; NaCl, 10 g/liter) or on YT broth solidified with 1.5% (wt/vol) agar. For the preparation of blood agar plates, the YT agar was supplemented with 4% defibrinated horse blood (Oxoid). When appropriate, antibiotic selection was carried out using ampicillin (Amp), chloramphenicol (Cm), and streptomycin (Str) at final concentrations of 100, 30, and 30 μg/ml, respectively.
TABLE 1.
E. coli wild-type strains used in this study
| Strain | Serogroup or serotype | Clinical origina | clyAb | Source of supplyc | Reference |
|---|---|---|---|---|---|
| STEC strainsd | |||||
| 0136/96 | O157:H7 | HUS | + | I | |
| 0296/96 | O157:H− | HUS | + | I | |
| 1104/96 | O157:H− | HUS | + | I | |
| 1885/96 | O157:H− | HUS | + | I | |
| 1938/96 | O157:H− | D | + | I | |
| 1994/96 | O157:H− | AC | + | I | |
| 2313/96 | O157:H− | AC | + | I | |
| 2471/96 | O157:H− | HUS | + | I | |
| 2711/96 | O157:H− | AC | + | I | |
| 3010/96 | O157:H7 | D | + | I | 5 |
| 3172/96 | O157:H− | HUS | + | I | |
| 3232/96 | O157:H7 | HUS | + | I | |
| 3817/96 | O157:H− | HUS | (+) | I | 5 |
| 4011/96 | O157:H7 | D | + | I | |
| 4049/96 | O157:H7 | HUS | + | I | |
| 4299/96 | O157:H− | D | (+) | I | |
| 4304/96 | O157:H− | D | (+) | I | |
| 5869/96 | O157:H− | HUS | + | I | |
| 16110/96 | O157:H− | D | + | I | |
| 86-24 | O157:H7 | HC | + | II | 11 |
| ST73/01 | O157:H7 | HUS | + | III | |
| ST2415/01 | O26:H11 | HUS | + | III | |
| ST3494/03 | O128 | HUS | (+) | III | |
| EIEC strains | |||||
| 12860 | O124 | D | + | I | 39 |
| 78-5 | O124 | D | + | I | |
| W7062 | O124:H− | D | + | II | |
| 76-5 | O143 | D | + | I | |
| 107-11 | O143 | D | + | I | |
| 4608-58 | O143 | D | + | IV | 16 |
| 309-36/85 | O145 | D | + | I | |
| EAEC strains | |||||
| 17-2 | O3:H2 | D | + | I | 45 |
| D4140-86 | O44 | D | + | I | 39 |
| DEF40 | O78 | D | + | I | |
| 5477/94 | O86 | D | + | I | |
| 4185/95 | O86 | D | + | I | |
| DEF53 | O111 | D | (+) | I | 39 |
| OPA065 | O119 | D | + | I | |
| DEF52 | O126 | D | (+) | I | |
| ETEC strains | |||||
| 117/86 | O6:H− | D | + | I | |
| 297/87 | O25:H42 | D | (+) | I | |
| 147/1 | O128:H− | D | + | I | |
| G1253 | O147:H19:K88 | D | + | I | |
| 164/82 | O148:H28 | D | (+) | I | |
| 284/97 | O149:H− | PD | (+) | I | |
| ST3135B/01 | O167:H6 | D | + | III | |
| EPEC strains | |||||
| 700-36/85 | O55 | ID | Del. | I | |
| 22CH | O55 | ID | Del. | I | |
| 273-4 | O55 | ID | Del. | I | |
| 111/87 | O111 | ID | (+) | I | |
| 212/87 | O111 | ID | (+) | I | |
| 402/87 | O111 | ID | (+) | I | |
| 227/63 | O114 | ID | (+) | I | |
| 315/60 | O114 | ID | (+) | I | |
| 12810 | O114 | ID | (+) | I | |
| 12-1 | O119 | ID | Del. | I | |
| 16-2 | O119 | ID | (+) | I | |
| 1104/80 | O127 | ID | Del. | I | |
| 3715/67 | O127 | ID | Del. | I | |
| E2348/69 | O127:K63:H6 | ID | Del. | II | 23 |
| 6447/89 | O128 | ID | (+) | I | |
| 6587/85 | O128 | ID | (+) | I | |
| 1083-36/91 | O157:H45 | ID | Del. | I | 5 |
| UPEC strains | |||||
| J96 | O4:K6 | UTI | Del. | II | 18 |
| AD110 | O6:K2:H1 | UTI | Del. | II | 44 |
| RZ460 | O6:K2:H− | UTI | Del. | II | 50 |
| RZ485 | O6:K2:H1 | UTI | Del. | II | 50 |
| RZ486 | O6:K2:H1 | UTI | Del. | II | 50 |
| RZ439 | O6:K5:H1 | UTI | Del. | II | 51 |
| RZ440 | O6:K5:H1 | UTI | Del. | II | 51 |
| RZ442 | O6:K5:H1 | UTI | Del. | II | 51 |
| RZ443 | O6:K5:H− | UTI | Del. | II | 51 |
| RZ495 | O6:K5:H− | UTI | Del. | II | 51 |
| RZ498 | O6:K5:H1 | UTI | Del. | II | 51 |
| RZ513 | O6:K5:H− | UTI | Del. | II | 51 |
| RZ533 | O6:K5:H− | UTI | Del. | II | 51 |
| 536 | O6:K15:H31 | UTI | Del. | II | 6 |
| NMEC strains | |||||
| IHE3034 | O18ac:K1:H7 | NBM | Del. | II | 1 |
| IHE3036 | O18ac:K1:H7 | NBM | Del. | II | 1 |
| RS218 | O18ac:K1:H7 | NBM | Del. | II | 1 |
| Additional E. coli strains | |||||
| RS226 | O18ac:K1:H7 | AC | Del. | II | 1 |
| 764 | O18:K5:H− | AC | Del. | II | 34 |
HUS, hemolytic-uremic syndrome; D, diarrhea; AC, asymptomatic carrier; HC, hemorrhagic colitis; PD, pig diarrhea; ID, infant diarrhea; UTI, urinary tract infection; NBM, new-born meningitis.
Results obtained by DNA sequencing (this study). +, functional clyA gene; (+), clyA present but containing a 1-bp frameshift mutation (two 1-bp frameshift mutations were detected in the clyA sequences of STEC ST3494/03 and ETEC 297/87); Del., extensive deletions (deletion I and deletion II) at the clyA locus.
I, Helge Karch, Institut für Hygiene, Universitätsklinikum Münster, Germany. II, Jörg Hacker, Institut für Molekulare Infektionsbiologie, Universität Würzburg, Germany. III, strains isolated between 2001 and 2003 at the Institut für Medizinische Mikrobiologie, Klinikum der J. W. Goethe-Universität, Frankfurt am Main, Germany. IV, Philippe J. Sansonetti, Institut Pasteur, Paris, France.
STEC strains causing the hemolytic-uremic syndrome or hemorrhagic colitis are also referred to as EHEC.
TABLE 2.
Plasmids used in this work
| Plasmid(s) | Relevant characteristicsa | Source or reference |
|---|---|---|
| pUC18, pUC19 | Cloning vectors; Ampr | New England Biolabs |
| pAL105 | pBluescript II SK(+) carrying slyAK-12; Ampr | 24 |
| pAL108 | pACYC184 carrying slyAK-12; Cmr | 25 |
| pAL115 | pUC18 carrying slyAK-12; Ampr | This work |
| pAL201 | pUC18 carrying clyAK-12 under control of lacZp; Ampr | 25 |
| pAL202 | pUC19 carrying clyAK-12; Ampr | 25 |
| pCLYA3232/96 | pUC18 carrying clyA from STEC strain 3232/96; Ampr | This work |
| pCLYA12860 | pUC18 carrying clyA from EIEC strain 12860; Ampr | This work |
| pCLYA5477/94 | pUC18 carrying clyA from EAEC strain 5477/94; Ampr | This work |
| pCLYAG1253 | pUC19 carrying clyA from ETEC strain G1253; Ampr | This work |
| pCLYA284/97 | pUC19 carrying clyA from ETEC strain 284/97; Ampr | This work |
| pCLYA284/97A | pUC18 carrying clyA from ETEC strain 284/97 under control of lacZp; Ampr | This work |
| pCLYA297/87 | pUC18 carrying clyA from ETEC strain 297/87; Ampr | This work |
| pCLYA212/87 | pUC18 carrying clyA from EPEC strain 212/87; Ampr | This work |
| pCVD442 | Positive-selection suicide vector containing the pir-dependent R6K replicon and sacB of Bacillus subtilis; Ampr | 10 |
| pCHΔclyA | pCVD442 carrying clyAK-12-flanking sequences but lacking clyAK-12; Ampr | This work |
| pANN202-812 | pBR322 carrying the E. coli α-hemolysin gene cluster (hlyCABD) from plasmid pHly152; Ampr | 46 |
The slyAK-12 gene in pAL105, pAL108, and pAL115 and the clyA genes in pAL202, pCLYA3232/96, pCLYA12860, pCLYA5477/94, pCLYAG1253, pCLYA284/97, pCLYA297/87, and pCLYA212/87 are under control of their native promoter regions.
DNA manipulations.
DNA manipulations were performed with standard protocols (37). PCR was conducted either with Taq DNA polymerase (Eppendorf), Deep Vent DNA polymerase (New England Biolabs), or Phusion High Fidelity DNA polymerase (Finnzymes). Only PCR products synthesized by one of the latter two DNA polymerases were cloned and sequenced. Nucleotide sequences of DNA fragments were determined by automated cycle sequencing with fluorescence dye terminator technology, using either the ABI PRISM 377 DNA Sequencer or the ABI PRISM 3700 DNA Analyzer (Applied Biosystems). Southern blot hybridizations were performed with the ECL direct nucleic acid labeling and detection system (Amersham Biosciences) following the recommendations of the manufacturer.
Cloning of clyA from E. coli wild-type strains.
The clyA genes of the E. coli strains 3232/96, 12860, 5477/94, G1253, 284/97, 297/87, and 212/87 were amplified by PCR, using several forward primers designed according to DNA sequences present 244 to 563 bp upstream from clyA of E. coli K-12 (clyAK-12) and using several reverse primers corresponding to DNA sequences present 13 to 55 bp downstream from clyAK-12. The PCR products were cloned into pUC18 or pUC19. Recombinant plasmids carrying the clyA genes of the above strains in opposite orientation relative to lacZα of the vector were selected and named pCLYA3232/96, pCLYA12860, pCLYA5477/94, pCLYAG1253, pCLYA284/97, pCLYA297/87, and pCLYA212/87. In all these plasmids the inserted clyA gene is controlled only by its native 5′-flanking regulatory region. An additionally isolated plasmid, pCLYA284/97A, carries clyA from E. coli 284/97 in the same orientation as lacZα. In this case, the clyA gene is consequently not only under control of its native promoter region but also under control of the lacZ promoter (lacZp).
Construction of plasmid pAL115.
The slyA gene of E. coli K-12 (slyAK-12) was amplified by PCR from strain CC118 (27) using the forward primer 5′-GAAGCAGGCGGTCGACGACAAGCC-3′, which was designed to introduce a SalI restriction site (underlined) 316 bp upstream from slyA, and the reverse primer 5′-GTTTCTCCGCGCTGGATCCGTTTGCGTGTG-3′, which introduced a BamHI site 43 bp downstream from the slyA stop codon. The 0.83-kb PCR product was cleaved with SalI and BamHI, and the generated 0.8-kb SalI-BamHI fragment was subsequently cloned into pUC18, resulting in pAL115. The authenticity of the insert of pAL115 was established by sequencing.
Construction of clyA knockout mutants of E. coli strains.
To delete clyA in the enteroinvasive E. coli (EIEC) strains 12860 and 4608-58, a 0.9-kb SalI-BamHI fragment comprising the DNA sequence present 1.04 to 0.14 kb upstream from clyA was isolated by PCR with the forward primer 5′-TAGCTCTTCCAGCGTCGACATCACCCG-3′ and the reverse primer 5′-TATCAAACAGGATCCAATGTCATTATGGCG-3′. Furthermore, a 1.03-kb BglII-SacI fragment representing the DNA sequence immediately downstream from clyA was amplified by PCR with the primers 5′-GTACCTGAAAGATCTTAAGCGATTATTCTC-3′ and 5′-GCGTTTGAGAGCTCTTGTCCGCTTTCC-3′. The restriction sites at the ends of these DNA fragments were introduced by the PCR primers (see underlined sequences; only the SalI site was naturally present). The two DNA fragments were fused with each other by ligation of the BamHI and BglII sites and then were inserted between the SalI and SacI sites of the suicide vector pCVD442, resulting in plasmid pCHΔclyA. pCHΔclyA was transferred by conjugation from E. coli SM10λpir into in vitro-selected Strr derivatives of E. coli 12860 and E. coli 4608-58. Transconjugant clones were selected on Amp/Str agar plates and were analyzed by Southern blot hybridization of EcoRV-digested genomic DNA using the insert of pCHΔclyA as probe. In both cases a clone was identified in which pCHΔclyA was inserted into the chromosomal DNA sequence downstream from clyA. These clones were then grown in the absence of antibiotics to allow for excision of the suicide plasmid by a second event of homologous recombination. Descendants that had lost the plasmid (including the plasmid-encoded sacB gene) were selected on YT agar plates supplemented with 1% (wt/vol) sucrose, and those descendants in which the plasmid was excised by recombination between the SalI-BamHI fragment of pCHΔclyA and the corresponding chromosomal DNA fragment were identified by Southern blot analysis, which was performed as described above. In the latter clones, the excision of the suicide plasmid concomitantly caused the deletion of the chromosomal clyA gene. This was confirmed for two of these clones, E. coli 12860ΔclyA and E. coli 4608-58ΔclyA, by sequencing of the clyA locus. In both clyA knockout mutants we detected the expected deletion of a 1.06-kb fragment spanning the entire clyA gene and the 149-bp sequence preceding clyA, which carries the clyA promoter region.
Isolation and analysis of proteins.
The periplasmic proteins of E. coli strains were isolated by osmotic shock as described previously (25), precipitated by addition of ice-cold trichloroacetic acid (final concentration, 10%), pelleted by centrifugation at 12,000 × g, washed with acetone, dried under vacuum, and dissolved in sample buffer (50 mM Tris-HCl [pH 6.8], 10% glycerol, 5% β-mercaptoethanol, 2% sodium dodecyl sulfate [SDS], 0.05% bromophenol blue). Extracellular proteins of E. coli strains were precipitated from cell-free culture supernatants by addition of 10% trichloroacetic acid and then were processed as described above for the periplasmic proteins. To analyze the proteins of whole cells, bacteria grown in 2×YT broth were harvested by centrifugation, washed with phosphate-buffered saline, pH 7.4, (PBS) (37), resuspended in PBS, and broken by ultrasonic treatment (10 times for 15 s each) at 4°C. Subsequently, the cell lysate was mixed with an equal volume of 2× sample buffer. Protein samples were neutralized, if required, by addition of saturated Tris solution and were boiled at 99°C prior to loading onto SDS-polyacrylamide gels. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described (21). Western immunoblotting was conducted according to Towbin et al. (42) with the modification that the proteins were blotted onto polyvinylidene difluoride membrane using a Tris-glycine transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol [pH 8.3]). Immunoblot analysis was performed either with a polyclonal antiserum raised in a rabbit against purified ClyAK-12 (see below) or with a polyclonal rabbit anti-HlyA antiserum (19). Horseradish peroxidase-conjugated anti-rabbit secondary antibodies were used to detect the immunoreactive protein bands. These bands were finally visualized by using either the ECL plus Western Blotting Detection System (Amersham Biosciences) (in this case, the first and secondary antibodies were used at final dilutions of 1:10,000 and 1:50,000, respectively) or by using 0.02% 4-chloro-1-naphthol/0.01% H2O2 in Tris-buffered saline (TBS) (10 mM Tris-HCl [pH 7.5], 150 mM NaCl) for chromogenic detection (in this case, both antibodies were used at final dilutions of 1:1,000 or 1:2,000).
Preparation of a polyclonal rabbit anti-ClyA antiserum.
ClyA was overexpressed in E. coli DH5α from plasmid pAL201 and was isolated by osmotic shock from the periplasm of bacteria grown to the stationary phase. The periplasmic proteins were mixed with 1 volume of 2× sample buffer lacking β-mercaptoethanol and bromophenol blue (100 mM Tris-HCl [pH 6.8], 20% glycerol, 4% SDS) and separated without previous boiling by SDS-PAGE. The predominant 34-kDa ClyA protein band was excised from the gel, and ClyA was eluted by diffusion at 4°C into PBS. About 0.5 mg of the purified ClyA protein was used for rabbit immunization. The serum taken 11 days after the third injection was used as polyclonal anti-ClyA antiserum. This serum reacted strongly with ClyA as established by Western blot analysis, while the preimmune serum did not recognize ClyA (data not shown).
Determination of hemolytic activity.
Quantitative hemolytic activity assays were performed with bacterial cell lysates which were prepared as described above by ultrasonic treatment of bacteria suspended in PBS. Different volumes (0.5 to 50 μl) of the lysates were mixed with 600 μl of a suspension of horse erythrocytes in 0.9% NaCl, containing about 7.0 × 108 red blood cells per ml. After incubation at 37°C for 30 min, the erythrocytes were pelleted by centrifugation. The amount of hemoglobin released into the supernatant was measured spectrophotometrically at 543 nm.
Nucleotide sequence accession numbers.
The clyA sequences of the following strains have been submitted to the EMBL/GenBank/DDBJ databases (accession numbers are given in parentheses): Shiga toxin-producing E. coli (STEC) strain 3232/96 (AY576656); EIEC strain 4608-58 (AY576657); EIEC strain 12860 (AY576658); enteroaggregative E. coli (EAEC) strain 5477/94 (AY576659); enterotoxigenic E. coli (ETEC) strain G1253 (AY576660); ETEC strain 284/97 (AY576661); ETEC strain 297/87 (AY576662); enteropathogenic E. coli (EPEC) strain 212/87 (AY576663); EPEC strain E2348/69 (AY576664); uropathogenic E. coli (UPEC) strain 536 (AY576665); UPEC strain RZ443 (AY576666); new-born meningitis-associated E. coli (NMEC) strain IHE3034 (AY576667).
RESULTS
PCR analysis of E. coli strains.
A significant number of clinical E. coli isolates belonging to the most representative groups of pathogenic E. coli was analyzed by PCR with regard to the presence of the clyA gene, using primers designed according to clyAK-12 and flanking DNA sequences. The tested strains included 23 STEC, 7 EIEC, 8 EAEC, 7 ETEC, 17 EPEC, 14 UPEC, and 3 NMEC strains (Table 1). Two additional strains which were isolated from the stool of healthy individuals, E. coli 764 and E. coli RS226, have not been assigned to a specific pathogroup but belong to serotypes that are frequently encountered among UPEC and NMEC strains, respectively.
By using several primer combinations, DNA fragments could be amplified from all STEC, EIEC, and EAEC strains, from 5 of the 7 ETEC strains (117/86, 147/1, G1253, 164/82, and ST3135B/01), and from 9 of the 17 EPEC strains (111/87, 212/87, 402/87, 227/63, 315/60, 12810, 16-2, 6447/89, and 6587/85) that were indistinguishable in size, when analyzed by agarose gel electrophoresis, from the PCR products obtained under the same conditions from E. coli K-12 (Fig. 1). Slightly shorter DNA fragments were amplified from the ETEC strains 284/97 and 297/87, indicating the presence of small deletions in the clyA genes of these strains. DNA fragments about 0.7 kb shorter than expected were amplified from the remaining eight EPEC strains, from all UPEC and NMEC strains, and from E. coli 764 and E. coli RS226 when the PCR was conducted with primers binding more than 0.16 kb upstream and immediately downstream from clyAK-12. In addition, no PCR products were obtained from these strains when primers binding to the 5′-terminal two-thirds of clyAK-12 or to the DNA region preceding clyAK-12 were used, suggesting that the latter strains have chromosomal deletions of about 0.7 kb affecting clyA and the 5′-flanking DNA sequence.
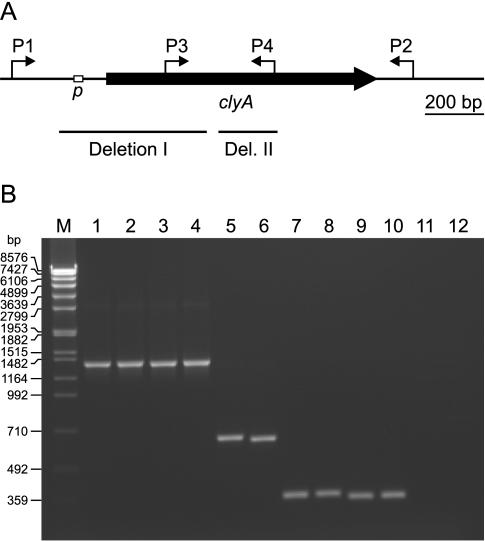
FIG. 1.
(A) Schematic presentation of the clyA gene from E. coli K-12. The position of the SlyA-controlled clyA promoter (p) (25) is indicated by an open box, and the binding sites of several primers used for PCR are shown by arrows (P1, 5′-GCCAGCAGATCAATACTG-3′; P2, 5′-CATAATGAGAGTTACCCGATACC-3′; P3, 5′-CTTATGGATAGCCAGGATAAG-3′; P4, 5′-CAAATGGACCGTCGACGACACC-3′). The position of deletion I and deletion II at the clyA locus of UPEC, NMEC, and several EPEC strains is indicated by solid bars. (B) Agarose gel electrophoresis of the PCR products obtained with the primer combinations P1-P2 (lanes 1 to 6) and P3-P4 (lanes 7 to 12) from E. coli K-12 (lanes 1 and 7), STEC (EHEC) strain 3232/96 (lanes 2 and 8), ETEC strain 284/97 (lanes 3 and 9), ETEC strain 297/87 (lanes 4 and 10), UPEC strain J96 (lanes 5 and 11), and UPEC strain 536 (lanes 6 and 12). Lane M, DNA size markers (SPP1 DNA cleaved with EcoRI).
Characterization of the clyA sequences of E. coli wild-type strains.
Sequencing of the PCR-amplified clyA-carrying or clyA-related DNA fragments from all 81 E. coli strains listed in Table 1 yielded the following results.
(i) Identification of an intact clyA gene in STEC, EIEC, EAEC, and ETEC strains
A complete clyA gene encoding, like clyAK-12, a protein of 303 amino acid residues was found in 19 of the 23 tested STEC strains, in all 7 tested EIEC strains, in 6 of 8 EAEC strains, and in 4 of the 7 tested ETEC strains (Tables 1 and 3). The clyA genes of some of these strains (EAEC strains 17-2, 5477/94, and OPA065 and ETEC strain 117/86) were identical in sequence to clyAK-12, but in most cases several nucleotide substitutions (between 5 and 16) were detected. Some of these substitutions proved to be highly conserved in strains belonging to the same E. coli pathogroup or even in members of different pathogroups.
TABLE 3.
Characteristics of the clyA gene in E. coli wild-type strains analyzed in this study
| E. coli pathotype | No. of strains tested | No. containing functional clyAa | No. containing mutant clyA sequencesa
|
|
|---|---|---|---|---|
| Strains with 1-bp frameshift mutations | Strains with large deletions (deletions I and II) | |||
| STEC | 23 | 19 | 4 | 0 |
| EIEC | 7 | 7 | 0 | 0 |
| EAEC | 8 | 6 | 2 | 0 |
| ETEC | 7 | 4 | 3 | 0 |
| EPEC | 17 | 0 | 9 | 8 |
| UPEC | 14 | 0 | 0 | 14 |
| NMEC | 3 | 0 | 0 | 3 |
Results obtained by DNA sequencing.
The amino acid sequences predicted for the clyA gene products of the above strains were either identical to that of ClyAK-12 (EAEC strains 17-2, 5477/94, and OPA065; ETEC strain 117/86; serogroup O26 STEC strain ST2415/01) or contained between one and three amino acid exchanges, which corresponds to a sequence identity of >99%. In particular, compared to ClyAK-12 the following amino acid substitutions were found: a single N220→S exchange in the putative ClyA proteins of several EIEC (78-5, 107-11), EAEC (D4140-86, 4185/95), and ETEC strains (147/1, G1253, ST3135B/01); K75→N/A199→T in ClyA of the EIEC strains 76-5, 4608-58, and 309-36/85; L99→F/V185→I in ClyA of the EIEC strains 12860 and W7062 and of EAEC strain DEF40; D64→N/N220→S/K279→N in the ClyA proteins of all 18 serogroup O157 STEC strains harboring an intact clyA gene.
The DNA sequences flanking the intact clyA genes of the different E. coli strains proved to be very similar to those flanking clyAK-12. In several cases (STEC strain ST2415/01; EIEC strains 12860, W7062, 76-5, 4608/58, and 309-36/85; EAEC strains 17-2, 5477/94, and OPA065; ETEC strains 117/86 and G1253) at least the first 185 bp preceding clyA were the same as those in E. coli K-12. This DNA region carries the SlyA-controlled promoter of clyA (25). The remaining strains exhibited a few nucleotide substitutions in the clyA 5′-flanking sequence, but the −10 and −35 signals of the clyA promoter (78 to 84 and 102 to 107 bp upstream from the translational start codon of clyA) were generally not affected. Most of the latter strains, including all serogroup O157 STEC strains, exhibited a C→T and a G→T exchange in the spacer between the −10 and −35 signals and a T→C substitution 61 bp upstream from the clyA start codon.
(ii) Detection of frameshift mutations in clyA of several STEC, ETEC, and EPEC strains.
Several E. coli wild-type strains were shown by DNA sequencing to harbor mutant clyA derivatives containing 1-bp frameshift mutations that cause premature truncation of the clyA open reading frame (ORF) (Tables 1 and 3, Fig. 2). In three serogroup O157 STEC strains (3817/96, 4299/96, and 4304/96) we found, for example, a clyA derivative exhibiting a unique 1-bp deletion in codon 248. Interestingly, this clyA derivative was otherwise identical in sequence to the intact clyA gene found in all other tested O157 STEC strains. In the clyA genes of the ETEC strains 284/97 and 297/87, which were already predicted from the PCR data to contain small deletions, we detected not only an in-frame deletion of the codons 179 to 182 but also an identical 1-bp insertion in codon 163. Furthermore, a unique 1-bp deletion was found in codon 15 of clyA from strain 297/87. All nine EPEC strains that did not exhibit noticeable clyA defects upon PCR analysis (111/87, 212/87, 402/87, 227/63, 315/60, 12810, 16-2, 6447/89, and 6587/85) were shown by DNA sequencing to harbor a mutant clyA gene exhibiting a specific 1-bp deletion in codon 165. The same deletion was also found in the EAEC strains DEF52 and DEF53, in ETEC strain 164/82, and in serogroup O128 STEC strain ST3494/03. In clyA of strain ST3494/03 we detected, in addition, a unique 1-bp deletion in codon 278. The clyA genes exhibiting the 1-bp deletion in codon 165 generally proved to be very similar or even identical in sequence. All of them encode an identical C-terminally truncated ClyA derivative with a predicted molecular mass of 19.03 kDa.
FIG. 2.
Alignment of the amino acid sequences predicted for the truncated clyA gene products of several clinical E. coli isolates with the amino acid sequence of ClyA from E. coli K-12. Amino acid substitutions in the ClyA derivatives of the clinical E. coli isolates are indicated by boldface type. The underlined C-terminal amino acid sequences of the truncated ClyA derivatives resulted from frameshift mutations in the corresponding clyA genes. The asterisk behind the last amino acid residue indicates the presence of a stop codon in the DNA sequence.
The promoter regions of the mutant clyA genes from the above strains were either identical in sequence to that of clyAK-12 (ETEC strains 284/97, 297/87, and 164/82; all mentioned EPEC strains; EAEC strains DEF52 and DEF53; STEC strain ST3494/03) or corresponded to the clyA promoter regions of the O157 STEC strains harboring an intact clyA gene (STEC strains 3817/96, 4299/96, and 4304/96), suggesting that all these clyA derivatives may be expressed under appropriate conditions.
(iii) Detection of deletions at the clyA locus in UPEC, NMEC, and several EPEC strains.
Sequencing of the strikingly short clyA-related PCR products obtained from all tested UPEC and NMEC strains as well as from E. coli 764, E. coli RS226, and eight EPEC strains (700-36/85, 22CH, 273-4, 12-1, 1104/80, 3715/67, E2348/69, and 1083-36/91) demonstrated that all these strains harbor only DNA sequences corresponding to an internal fragment and to the 3′-terminal region of clyA. The sequence data further indicated that these clyA-related sequences are left from two deletions at the clyA locus which we refer to as deletion I and deletion II (Fig. 1, Tables 1 and 3). Deletion I generally comprised the 493-bp fragment spanning the 160 bp preceding clyA and the first 333 bp of clyA. Deletion II was found in two versions: in the UPEC strains 536, RZ460, and RZ485 it comprised the 217-bp fragment spanning the nucleotides 377 to 593 of clyA, while in all other strains it comprised the 204-bp fragment from nucleotides 382 to 585 of clyA (codons 128 to 195).
The residual clyA sequences of the above-mentioned E. coli strains were at least 96% identical to the corresponding fragments of clyAK-12. Several nucleotide substitutions were found in all or in most of these strains, whereas others could be detected only in strains belonging to the same pathotype. The following groups of strains harbored identical residual clyA sequences: (i) the EPEC strains 700-36/85, 22CH, 273-4, 1104/80, 3715/67, and E2348/69, UPEC strain RZ533, and E. coli 764; (ii) the three UPEC strains exhibiting the larger version of deletion II (536, RZ460, and RZ485); (iii) all tested UPEC strains containing the smaller version of deletion II, except J96 and RZ533; (iv) all tested NMEC strains (IHE3034, IHE3036, and RS218) and E. coli RS226. The clyA sequences of UPEC strain J96 differed from those of the NMEC strains only at a single nucleotide position.
It is unlikely that these residual clyA sequences are expressed, because they lack a translational start codon and a fortuitous TAA stop codon is present 22 bp upstream from deletion I, in frame with the clyA coding sequence. Furthermore, the clyA promoter region is completely removed by deletion I.
Analysis of the stability of clyA in E. coli wild-type strains.
In order to test the stability of the clyA sequence in E. coli wild-type strains, four randomly selected clyA+ strains (STEC 3232/96, EIEC 4608-58, EAEC 5477/94, and ETEC G1253) were grown for 7 days in 2×YT broth with daily dilution of the cultures (1:100) into fresh medium. Subsequently, the clyA gene was amplified by PCR, using in each case the bacteria from 1 μl of the final culture as template. Sequencing of the PCR products yielded definite clyA sequences identical to those originally determined for the corresponding strains, indicating that the clyA genes of these strains are quite stable upon prolonged subculturing.
Expression of clyA from clinical E. coli isolates in E. coli K-12.
The clyA genes of several clinical E. coli isolates were cloned into pUC18 and pUC19 as described in Materials and Methods. Four of the resulting plasmids, pCLYA3232/96, pCLYA12860, pCLYA5477/94, and pCLYAG1253, carrying the functional clyA genes from STEC 3232/96, EIEC 12860, EAEC 5477/94, and ETEC G1253 under control of their native promoter regions, caused a hemolytic phenotype when introduced into the E. coli K-12 strain DH5α. The hemolytic activity on blood agar resembled in each case that of DH5α carrying clyAK-12 on plasmid pAL202 (Fig. 3). As shown in Fig. 4, these recombinant DH5α clones also produced amounts of the 34-kDa ClyA protein similar to amounts produced by E. coli DH5α/pAL202. Transformation of a slyAK-12-carrying plasmid (pAL108) into the DH5α clones harboring pAL202, pCLYA3232/96, pCLYA12860, pCLYA5477/94, and pCLYAG1253 resulted in each case in enhanced production of ClyA and in a significantly stronger hemolytic phenotype on blood agar, demonstrating that the clyA genes of the corresponding clinical E. coli isolates are positively controlled by SlyA, like clyAK-12 (Fig. 3 and 4). It should be pointed out that the stronger hemolytic phenotype of the DH5α double transformants does not completely reflect the enhancement of clyA expression, because ClyA overproduced in E. coli accumulates in the periplasmic space and only small amounts of it are released from the bacteria (25 and data not shown).
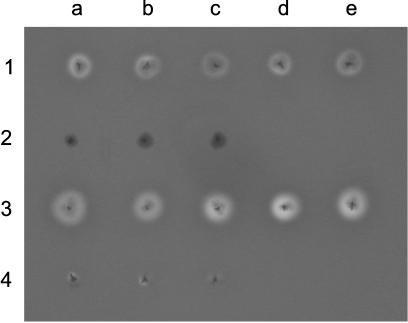
FIG. 3.
Hemolytic phenotypes of E. coli DH5α clones harboring the following plasmids: pAL202 (1a), pCLYA3232/96 (1b), pCLYA12860 (1c), pCLYA5477/94 (1d), pCLYAG1253 (1e), pCLYA212/87 (2a), pCLYA284/97 (2b), pCLYA297/87 (2c), pAL202 and pAL108 (3a), pCLYA3232/96 and pAL108 (3b), pCLYA12860 and pAL108 (3c), pCLYA5477/94 and pAL108 (3d), pCLYAG1253 and pAL108 (3e), pCLYA212/87 and pAL108 (4a), pCLYA284/97 and pAL108 (4b), and pCLYA297/87 and pAL108 (4c). A single colony of each strain was picked onto blood agar containing 100 μg of ampicillin/ml. The agar plate was photographed after overnight incubation at 37°C. Phenotypes identical to those shown here were observed in at least three independent experiments.
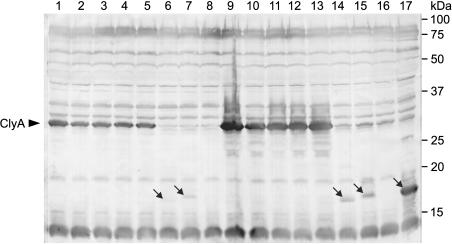
FIG. 4.
Western blot analysis of the expression of cloned clyA genes from clinical E. coli isolates in E. coli DH5α, using a polyclonal anti-ClyA antiserum. The production of ClyA was determined in E. coli DH5α clones carrying the following plasmids: pAL202 (lane 1), pCLYA3232/96 (2), pCLYA12860 (3), pCLYA5477/94 (4), pCLYAG1253 (5), pCLYA212/87 (6), pCLYA284/97 (7), pCLYA297/87 (8), pAL202 and pAL108 (9), pCLYA3232/96 and pAL108 (10), pCLYA12860 and pAL108 (11), pCLYA5477/94 and pAL108 (12), pCLYAG1253 and pAL108 (13), pCLYA212/87 and pAL108 (14), pCLYA284/97 and pAL108 (15), pCLYA297/87 and pAL108 (16), and pCLYA284/97A (17). Whole-cell proteins from 25 μl of an overnight culture (grown for 18 h) were analyzed in each case. The immunoreactive bands were visualized by chromogenic detection. The arrows in lanes 6, 7, 14, 15, and 17 point to truncated ClyA derivatives. Positions of marker proteins are shown on the right. The data presented are representative of three separate experiments.
By using the method of quantitative real-time reverse transcription-PCR we recently observed that transcription of clyA in exponentially growing E. coli DH5α/pAL202/pAL108 is 5- to 10-fold stronger than in E. coli DH5α carrying pAL202 only in combination with the vector pACYC184 (C. von Rhein and A. Ludwig, unpublished data). Similar results would be expected for corresponding experiments performed with isogenic E. coli DH5α clones carrying pCLYA3232/96, pCLYA12860, pCLYA5477/94, or pCLYAG1253 instead of pAL202.
The plasmids pCLYA212/87, pCLYA284/97, and pCLYA297/87, carrying the mutant clyA genes of EPEC 212/87 (clyA212/87), ETEC 284/97 (clyA284/97), and ETEC 297/87 (clyA297/87) under control of their own promoter sequences, did not cause a hemolytic phenotype when introduced into E. coli DH5α. Furthermore, transformation of pAL108 into the DH5α clones harboring these plasmids caused in each case only very weak hemolytic activity on blood agar due to the SlyA-mediated induction of the chromosomal clyAK-12 gene (Fig. 3 and 4). Proteins corresponding in size to the predicted products of clyA212/87 (19.03 kDa) and clyA284/97 (19.28 kDa) were specifically detected by Western blot analysis in cell lysates of E. coli DH5α harboring pCLYA212/87 and pCLYA284/97, respectively. The corresponding DH5α double transformants carrying pAL108 as well produced markedly larger amounts of these ClyA derivatives, confirming that clyA212/87 and clyA284/97 are positively controlled by SlyA (Fig. 4). Nevertheless, in the absence as well as in the presence of pAL108 the cellular levels of ClyA212/87 and ClyA284/97 were significantly lower than those of complete, functional ClyA proteins expressed under identical conditions, which suggests that these truncated ClyA derivatives are more unstable. E. coli DH5α transformed with pCLYA284/97A, a pUC18 derivative carrying clyA284/97 under control of the lacZ promoter, produced rather large amounts of ClyA284/97 (Fig. 4) but was also nonhemolytic on blood agar. In addition, no significant hemolytic activity could be detected in cell lysates of this strain by a quantitative hemolytic activity assay. The product of clyA297/87 (predicted molecular mass, 2.63 kDa) could be detected neither in lysates of E. coli DH5α/pCLYA297/87 nor in those of DH5α carrying both pCLYA297/87 and pAL108.
Analysis of the expression of clyA in clinical E. coli isolates.
Several E. coli strains possessing a functional clyA gene, such as STEC (EHEC) 3232/96, EIEC 12860, EIEC 4608-58, and ETEC G1253 showed a nonhemolytic phenotype when grown overnight on blood agar containing horse erythrocytes (the weak enterohemolytic phenotype of STEC strain 3232/96 caused by the production of EHEC-HlyA was visible only on sheep blood agar). Nevertheless, the colonies of EIEC strain 12860 developed a hemolytic phenotype on horse blood agar when the agar plate was stored for several days at 4°C after the initial overnight incubation at 37°C. A clyA knockout mutant of strain 12860 (E. coli 12860ΔclyA) remained nonhemolytic under the same conditions, demonstrating that this hemolytic phenotype is clyA dependent (Fig. 5A).
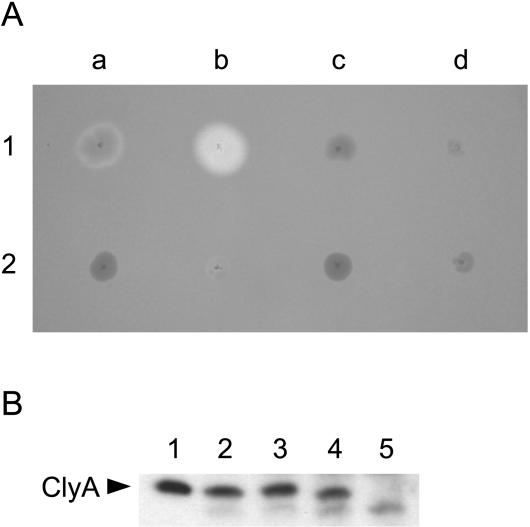
FIG. 5.
(A) Phenotypes of the EIEC strains 12860 and 4608-58 and of derivatives of these strains on blood agar. Shown are E. coli 12860 (1a), E. coli 12860/pAL115 (1b), E. coli12860ΔclyA (1c), E. coli 12860ΔclyA/pAL115 (1d), E. coli 4608-58 (2a), E. coli 4608-58/pAL115 (2b), E. coli 4608-58ΔclyA (2c), and E. coli 4608-58ΔclyA/pAL115 (2d). A single colony of each strain was picked onto a blood agar plate. The agar plate was incubated overnight at 37°C and then was stored for 2 weeks at 4°C prior to taking the photograph. (B) Analysis of the production of ClyA protein in different E. coli strains by immunoblotting using a polyclonal anti-ClyA antiserum. Lane 1, ClyA purified from the periplasmic protein fraction of E. coli DH5α/pAL201 employing the Model 491 Prep Cell (Bio-Rad); lane 2, E. coli DH5α; lane 3, E. coli 4608-58; lane 4, E. coli 12860; lane 5, E. coli12860ΔclyA. In lanes 2 to 5, whole-cell proteins of approximately 107 bacteria harvested from overnight cultures of the specified strains were analyzed. The immunoreactive bands were visualized with the ECL plus Western Blotting Detection System (Amersham Biosciences). The data presented are representative of three independent experiments.
To further study the expression of clyA in E. coli strains possessing a functional chromosomal clyA gene, we analyzed the cellular ClyA levels in stationary-phase cultures by immunoblotting with a polyclonal anti-ClyA antiserum. When a highly sensitive Western blotting detection system was employed, a protein of about 34 kDa corresponding to ClyA could be specifically detected in cell lysates of all tested E. coli strains harboring an intact clyA gene, such as DH5α, 4608-58, and 12860, but not in lysates of E. coli 12860ΔclyA (Fig. 5B). The amounts of ClyA found in the different clyA+ strains were very similar to each other, indicating that all these strains expressed clyA at similar low, basal levels. According to these data, the hemolytic phenotype of older colonies of EIEC strain 12860 is apparently not due to a stronger expression of clyA in this strain compared to that in the other strains but most likely is due to enhanced release of the toxin from the bacteria.
Introduction of a slyAK-12-carrying plasmid (pAL105 or pAL115) into the E. coli strains 3232/96, 12860, 4608-58, and G1253 by electroporation caused in each case a hemolytic phenotype, in line with the finding that the functional clyA genes of these strains are positively controlled by SlyA. Consistent with this, it was recently shown at the protein level that overexpression of SlyA in EIEC strain 12860 causes enhanced production of ClyA (41). As shown in Fig. 5A, E. coli 12860/pAL115 exhibited clearly stronger hemolytic activity on blood agar than E. coli 4608-58/pAL115, again suggesting that strain 12860 releases ClyA more readily than other clyA+ E. coli strains. clyA knockout mutants of E. coli 12860 and E. coli 4608-58 (12860ΔclyA and 4608-58ΔclyA) remained nonhemolytic after introduction of pAL115, confirming that the hemolytic activity of the SlyA-overproducing wild-type strains is dependent on clyA.
ETEC strain 297/87 and EPEC strain 212/87 (Ampr) were nonhemolytic on blood agar and retained this phenotype after introduction of slyAK-12-carrying plasmids (pAL105 and pAL108, respectively), consistent with the finding that the clyA genes of both strains encode only truncated, obviously nonhemolytic ClyA derivatives. ETEC strain 284/97 (Cmr), also harboring a defective clyA gene (see above), exhibited a strongly hemolytic phenotype that was not affected by introduction of pAL105. Southern blot analysis of genomic DNA from E. coli 284/97 using an E. coli α-hemolysin-specific DNA probe isolated from plasmid pANN202-812 revealed a single DNA fragment that hybridized with this probe. In addition, a protein possessing a molecular mass similar to that of HlyA (approximately 110 kDa) was specifically detected in culture supernatants of E. coli 284/97 by Western blot analysis using a polyclonal anti-HlyA antiserum (data not shown). These findings indicated that the hemolytic activity of E. coli 284/97 is most likely due to the production and secretion of α-hemolysin or of a closely related toxin.
DISCUSSION
Recent studies have shown that the ClyA protein of E. coli K-12 is a pore-forming toxin which lyses erythrocytes from various species and which exhibits cytotoxic and apoptotic activity towards cultured mammalian cells (22, 25, 30, 31, 47). Based on these findings the questions arose whether E. coli strains are generally able to produce ClyA and whether this toxin is involved in the virulence of strains causing intestinal or extraintestinal infections.
The data presented in this study demonstrate that only part of the pathogenic E. coli strains possess a functional clyA gene, while others harbor mutant clyA derivatives or even only clyA fragments. In particular, an intact clyA gene was found in all tested EIEC strains, in most of the tested STEC and EAEC strains, and also in several ETEC strains, but it was not detected in any of the tested EPEC, UPEC, and NMEC strains. Some STEC, EAEC, and ETEC strains and about half of the tested EPEC strains were shown to harbor clyA derivatives containing 1-bp frameshift mutations that cause premature truncation of the clyA ORF. In the remaining EPEC strains and in all tested UPEC and NMEC strains we found only nonfunctional clyA fragments that are apparently left from two deletions at the clyA locus. One of these deletions, denoted here as deletion I, generally comprised a 493-bp fragment spanning the 160-bp sequence preceding clyA and the 5′-terminal 111 clyA codons. The other deletion (deletion II) proved to be slightly heterogeneous in size, because in three of the tested UPEC strains it comprised a 217-bp fragment spanning the nucleotides 377 to 593 of clyA while in all other strains it comprised only the 204-bp fragment from nucleotides 382 to 585 of clyA (codons 128 to 195).
Interestingly, deletion I and deletion II were found only in combination, but we do not know whether these deletions occurred simultaneously or separately. It is also unclear whether or to what extent deletions I and II occurred independently in different E. coli strains. In any case, vertical DNA transfer most likely played an important role in the spreading of these deletions, even if they occurred in several strains. It is remarkable in this context that we did not observe the appearance of these deletions (nor of any other clyA mutations) upon prolonged cultivation of several E. coli strains harboring a functional clyA gene, which suggests that the clyA sequence is quite stable and not particularly prone to mutations.
Regarding deletion I, it is interesting that in E. coli K-12 and other clyA+ E. coli strains two very similar sequence motifs of 22 and 23 bp (AAGCATTCGCCATAATGACATT and AAGCATCCGCCCAGAAAGACATT) are centered 160 bp upstream and 333 bp downstream, respectively, from the 5′ end of clyA (i.e., at the end points of the sequence that is removed by deletion I). This suggests that deletion I occurred by homologous recombination between these related sequences. The processes that resulted in deletion II are, however, less clear because the corresponding deleted clyA fragment is not flanked by obvious direct repeats. Nevertheless, it is noteworthy that the 48-bp (or 43-bp) fragment left between deletion I and deletion II includes an imperfect palindromic sequence.
Results from immunoblot analyses demonstrated that E. coli K-12 and several tested clinical clyA+ E. coli isolates produce ClyA at similar low, basal levels when grown in rich medium. Consistent with this, a basal-level expression of ClyA in E. coli K-12 has recently also been observed by Oscarsson et al. (32). Thus, clyA is not totally silent in E. coli strains under in vitro cultivation conditions. The amounts of ClyA produced in E. coli K-12 and several other clyA+ E. coli strains are, however, apparently below the threshold that has to be passed to cause detectable hemolysis on blood agar. Nevertheless, in the case of EIEC strain 12860 we observed that the colonies grown on blood agar develop a clyA-dependent hemolytic phenotype when the agar plate is stored for several days at 4°C. To our knowledge, E. coli 12860 is the first reported phenotypically hemolytic E. coli wild-type strain in which the clyA gene has been identified as the genetic determinant of the hemolytic activity.
Western blot analyses of recombinant E. coli DH5α clones revealed that the clyA genes from clinical E. coli isolates are positively controlled, like clyAK-12, by the transcriptional regulator SlyA. In line with this finding, several tested E. coli wild-type strains possessing a functional clyA gene showed a clyA-dependent hemolytic phenotype when SlyA was overexpressed. This in turn indicates that these strains are able to release substantial amounts of ClyA under environmental conditions that cause increased cellular levels of SlyA and/or other factors involved in the positive regulation of clyA. It is tempting to speculate that such conditions might exist during the infection of host organisms.
Given the data presented in this work, it appears to be quite possible that cytolysin A contributes to the virulence of several STEC, EIEC, EAEC, and ETEC strains. In line with this, it was recently observed that EIEC strain 4608-58 exhibits significantly stronger cytotoxic activity towards J774 macrophage-like cells than a clyA knockout mutant of this strain (C. Hüttinger, W. Goebel, and A. Ludwig, unpublished data). The finding that all tested UPEC, NMEC, and EPEC strains are unable to produce functional cytolysin A suggests, on the other hand, that this toxin is not an important virulence factor for the latter groups of strains.
Acknowledgments
We thank Helge Karch, Jörg Hacker, and Philippe J. Sansonetti for providing most of the E. coli wild-type strains used in this study.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 176/B10 and LU 842/1-1).
REFERENCES
- 1.Achtman, M., A. Mercer, B. Kusecek, A. Pohl, M. Heuzenroeder, W. Aaronson, A. Sutton, and R. P. Silver. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alouf, J. E. 2001. Pore-forming bacterial protein toxins: an overview. Curr. Top. Microbiol. Immunol. 257:1-14. [PubMed] [Google Scholar]
- 3.Atkins, A., N. R. Wyborn, A. J. Wallace, T. J. Stillman, L. K. Black, A. B. Fielding, M. Hisakado, P. J. Artymiuk, and J. Green. 2000. Structure-function relationships of a novel bacterial toxin, hemolysin E. J. Biol. Chem. 275:41150-41155. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, M. E., and R. A. Welch. 1996. Characterization of an RTX toxin from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 64:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielaszewska, M., H. Schmidt, M. A. Karmali, R. Khakhria, J. Janda, K. Blahova, and H. Karch. 1998. Isolation and characterization of sorbitol-fermenting Shiga toxin (verocytotoxin)-producing Escherichia coli O157:H− strains in the Czech republic. J. Clin. Microbiol. 36:2135-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschäpe, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 8.del Castillo, F. J., S. C. Leal, F. Moreno, and I. del Castillo. 1997. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol. Microbiol. 25:107-115. [DOI] [PubMed] [Google Scholar]
- 9.del Castillo, F. J., F. Moreno, and I. del Castillo. 2000. Characterization of the genes encoding the SheA haemolysin in Escherichia coli O157:H7 and Shigella flexneri 2a. Res. Microbiol. 151:229-230. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., S. Tzipori, M. L. McKee, A. D. O'Brien, J. Alroy, and J. B. Kaper. 1993. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Investig. 92:1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felmlee, T., S. Pellett, and R. A. Welch. 1985. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J. Bacteriol. 163:94-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, J., and M. L. Baldwin. 1997. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coli by FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology 143:3785-3793. [DOI] [PubMed] [Google Scholar]
- 14.Hacker, J., C. Hughes, H. Hof, and W. Goebel. 1983. Cloned haemolysin genes from Escherichia coli that cause urinary tract infection determine different levels of toxicity in mice. Infect. Immun. 42:57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacker, J., G. Blum-Oehler, I. Mühldorfer, and H. Tschäpe. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 16.Hale, T. L., P. J. Sansonetti, P. A. Schad, S. Austin, and S. B. Formal. 1983. Characterization of virulence plasmids and plasmid-associated outer membrane proteins in Shigella flexneri, Shigella sonnei, and Escherichia coli. Infect. Immun. 40:340-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C.-G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 18.Hull, R. A., R. E. Gill, P. Hsu, B. H. Minshew, and S. Falkow. 1981. Construction and expression of recombinant plasmids encoding type 1 or D-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 33:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarchau, T., T. Chakraborty, F. Garcia, and W. Goebel. 1994. Selection for transport competence of C-terminal polypeptides derived from Escherichia coli hemolysin: the shortest peptide capable of autonomous HlyB/HlyD-dependent secretion comprises the C-terminal 62 amino acids of HlyA. Mol. Gen. Genet. 245:53-60. [DOI] [PubMed] [Google Scholar]
- 20.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Ölschläger, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 67:5994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lai, X.-H., I. Arencibia, A. Johansson, S. N. Wai, J. Oscarsson, S. Kalfas, K.-G. Sundqvist, Y. Mizunoe, A. Sjöstedt, and B. E. Uhlin. 2000. Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect. Immun. 68:4363-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, S. Sotman, and B. Rowe. 1978. Escherichia coli strains that cause diarrhea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119-1122. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, A., C. Tengel, S. Bauer, A. Bubert, R. Benz, H.-J. Mollenkopf, and W. Goebel. 1995. SlyA, a regulatory protein of Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol. Gen. Genet. 249:474-486. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, A., S. Bauer, R. Benz, B. Bergmann, and W. Goebel. 1999. Analysis of the SlyA-controlled expression, subcellular localization and pore-forming activity of a 34 kDa haemolysin (ClyA) from Escherichia coli K-12. Mol. Microbiol. 31:557-567. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig, A., and W. Goebel. 1999. The family of the multigenic encoded RTX toxins, p. 330-348. In J. E. Alouf and J. H. Freer (ed.), The comprehensive sourcebook of bacterial protein toxins, 2nd ed. Academic Press, London, United Kingdom.
- 27.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead, P. S., and P. M. Griffin. 1998. Escherichia coli O157:H7. Lancet 352:1207-1212. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 31.Oscarsson, J., Y. Mizunoe, L. Li, X.-H. Lai, A. Wieslander, and B. E. Uhlin. 1999. Molecular analysis of the cytolytic protein ClyA (SheA) from Escherichia coli. Mol. Microbiol. 32:1226-1238. [DOI] [PubMed] [Google Scholar]
- 32.Oscarsson, J., M. Westermark, L. Beutin, and B. E. Uhlin. 2002. The bacteriophage-associated Ehly1 and Ehly2 determinants from Escherichia coli O26:H- strains do not encode enterohemolysins per se but cause release of the ClyA cytolysin. Int. J. Med. Microbiol. 291:625-631. [DOI] [PubMed] [Google Scholar]
- 33.Oscarsson, J., M. Westermark, S. Löfdahl, B. Olsen, H. Palmgren, Y. Mizunoe, S. N. Wai, and B. E. Uhlin. 2002. Characterization of a pore-forming cytotoxin expressed by Salmonella enterica serovars Typhi and Paratyphi A. Infect. Immun. 70:5759-5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott, M., J. Hacker, T. Schmoll, T. Jarchau, T. K. Korhonen, and W. Goebel. 1986. Analysis of the genetic determinants coding for the S-fimbrial adhesin (sfa) in different Escherichia coli strains causing meningitis or urinary tract infections. Infect. Immun. 54:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. G. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schmidt, H., C. Kernbach, and H. Karch. 1996. Analysis of the EHEC hly operon and its location in the physical map of the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:907-914. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt, H., C. Geitz, P. I. Tarr, M. Frosch, and H. Karch. 1999. Non-O157:H7 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J. Infect. Dis. 179:115-123. [DOI] [PubMed] [Google Scholar]
- 41.Spory, A., A. Bosserhoff, C. von Rhein, W. Goebel, and A. Ludwig. 2002. Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J. Bacteriol. 184:3549-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uhlich, G. A., P. J. McNamara, J. J. Iandolo, and D. A. Mosier. 1999. Cloning and characterization of the gene encoding Pasteurella haemolytica FnrP, a regulator of the Escherichia coli silent hemolysin SheA. J. Bacteriol. 181:3845-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Bosch, J. F., U. Verboom-Sohmer, P. Postma, J. de Graaff, and D. M. MacLaren. 1980. Mannose-sensitive and mannose-resistant adherence to human uroepithelial cells and urinary virulence of Escherichia coli. Infect. Immun. 29:226-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vial, P. A., R. Robins-Browne, H. Lior, V. Prado, J. B. Kaper, J. P. Nataro, D. Maneval, A. Elsayed, and M. M. Levine. 1988. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J. Infect. Dis. 158:70-79. [DOI] [PubMed] [Google Scholar]
- 46.Vogel, M., J. Hess, I. Then, A. Juarez, and W. Goebel. 1988. Characterization of a sequence (hlyR) which enhances synthesis and secretion of hemolysin in Escherichia coli. Mol. Gen. Genet. 212:76-84. [DOI] [PubMed] [Google Scholar]
- 47.Wallace, A. J., T. J. Stillman, A. Atkins, S. J. Jamieson, P. A. Bullough, J. Green, and P. J. Artymiuk. 2000. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell 100:265-276. [DOI] [PubMed] [Google Scholar]
- 48.Welch, R. A., E. P. Dellinger, B. Minshew, and S. Falkow. 1981. Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature 294:665-667. [DOI] [PubMed] [Google Scholar]
- 49.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zingler, G., M. Ott, G. Blum, U. Falkenhagen, G. Naumann, W. Sokolowska-Köhler, and J. Hacker. 1992. Clonal analysis of Escherichia coli serotype O6 strains from urinary tract infections. Microb. Pathog. 12:299-310. [DOI] [PubMed] [Google Scholar]
- 51.Zingler, G., G. Blum, U. Falkenhagen, I. Orskov, F. Orskov, J. Hacker, and M. Ott. 1993. Clonal differentiation of uropathogenic Escherichia coli isolates of serotype O6:K5 by fimbrial antigen typing and DNA long-range mapping techniques. Med. Microbiol. Immunol. 182:13-24. [DOI] [PubMed] [Google Scholar]