Abstract
A leading hypothesis of N-acyl ethanolamine (NAE) biosynthesis, including the endogenous cannabinoid anandamide (AEA), is that it depends on hydrolysis of N-acyl-phosphatidylethanolamines (NAPE) by a NAPE-specific phospholipase D (NAPE-PLD). Thus, deletion of NAPE-PLD should attenuate NAE levels. Previous analyses of two different NAPE-PLD knockout (KO) strains produced contradictory data on the importance of NAPE-PLD to AEA biosynthesis. Here, we examine this hypothesis with a strain of NAPE-PLD KO mice whose lipidome is uncharacterized. Using HPLC/MS/MS, over 70 lipids, including the AEA metabolite, N-arachidonoyl glycine (NAGly), the endocannabinoid 2-arachidonyl glycerol (2-AG) and prostaglandins (PGE2 and PGF2α), and over 60 lipoamines were analyzed in 8 brain regions of KO and wild-type (WT) mice.
Lipidomics analysis of this third NAPE-PLD KO strain shows a broad range of lipids that were differentially affected by lipid species and brain region. Importantly, all 6 NAEs measured were significantly reduced, though the magnitude of the effect varied by fatty acid saturation length and brain region. 2-AG levels were only impacted in the brainstem, where levels were significantly increased in KO mice. Correspondingly, levels of arachidonic acid were significantly decreased exclusively in brainstem. NAGly levels were significantly increased in 4 brain regions and levels of PGE2 increased in 6 of 8 brain regions in KO mice. These data indicate that deletion of NAPE-PLD has far broader effects on the lipidome than previously recognized. Therefore, behavioral characteristics of suppressing NAPE-PLD activity may be due to a myriad of effects on lipids and not simply due to reduced AEA biosynthesis.
Keywords: Lipidomics, NAPE-PLD, endogenous cannabinoid biosynthesis, anandamide, lipoamine
1. Introduction
Two of the most studied signaling molecules of the endogenous cannabinoid (eCB) system are N-arachidonoyl ethanolamine (AEA) [1], also known as anandamide, and 2-arachidonoyl glycerol (2-AG) [2, 3]. Both are derived from the polyunsaturated fatty acid arachidonic acid and are hypothesized to be produced “on demand” in mammalian cells [4, 5]. AEA and 2-AG engage the cannabinoid receptors CB1 and CB2 and these cannabinoid receptors are implicated in the regulation of appetite, pain perception, immune system function, inflammation and learning and memory [6, 7]. Biosynthesis and metabolism of each of these eCBs is of particular importance from a therapeutic standpoint as these ligand/receptor interactions are actively being studied in the context of pathophysiology of each of the above stated processes.
1.1 Biosynthesis of N-arachidonoyl ethanolamine
The metabolism of N-acyl phosphatidylethanolamine (NAPE) is central to the synthesis of anandamide. NAPE is formed when a calcium-dependent N-acyl transferase transfers arachidonic acid from the sn-1 position of a phospholipid to the primary amine group of phosphatidylethanolamine [8]. NAPE can be directly cleaved to synthesize AEA in-vitro by a NAPE-specific phospholipase D (NAPE-PLD), an enzyme found in the mammalian brain [9]. In situ hybridization of mouse brain tissue, with knockout (KO) controls, revealed high concentrations of NAPE-PLD mRNA in the dentate gyrus of the hippocampus (HIPP), and more moderate concentrations in several other brain areas. NAPE-PLD protein was detected throughout the brain, with higher levels of staining in the HIPP and the olfactory bulb, and lower levels in the midbrain (MID), brainstem (STEM), and cerebellum (CER) [10]. Electron microscopy with immunogold staining highlighted the presence of NAPE-PLD on dendrites, some axon terminals, in laminae I-II of the spinal dorsal horn, and in microglia and astrocytes [11].
Currently, NAPE-PLD is hypothesized to be a key enzyme in animal tissue that is confirmed to release AEA and its structural analogues, though alternative pathways have been suggested [12]. In one alternative pathway to synthesize AEA from its precursor NAPE, NAPE-specific phospholipases A1/A2 (PLA1/2) convert NAPE into lyso-NAPE. Lyso-NAPE can then be converted to AEA by a specific PLD, named lysophospholipase D [13]. Upon the finding that tissue expression of the NAPE-specific PLA2 was very low, other enzymes were hypothesized to contribute to the synthesis of lyso-NAPE in vivo. Simon and Cravatt reported that α/β-hydrolase domain 4 (ABHD4) was able to remove an acyl group from NAPE to synthesize lyso-NAPE [14]. In another ABHD4-dependent reaction, lyso-NAPE can lose another acyl group and be converted into glycerophosphoAEA, which can then be hydrolyzed into AEA by glycerophosphodiesterase 1 (GDE1) [15]. Another alternative pathway to yield AEA involves the generation of phospho-AEA from NAPE catalyzed by a NAPE-specific phospholipase C. A lipid phosphatase can then dephosphorylate phospho-AEA, yielding AEA [16]. Furthermore, it is possible to synthesize AEA from plasmalogen-type NAPE (pNAPE). Like NAPE, pNAPE can be converted into lyso-pNAPEs, which can then be converted into AEA by lysophospholipase D [17]. The relative importance of each of these pathways remains unknown [5].
1.2 Structural analogs of endogenous cannabinoids
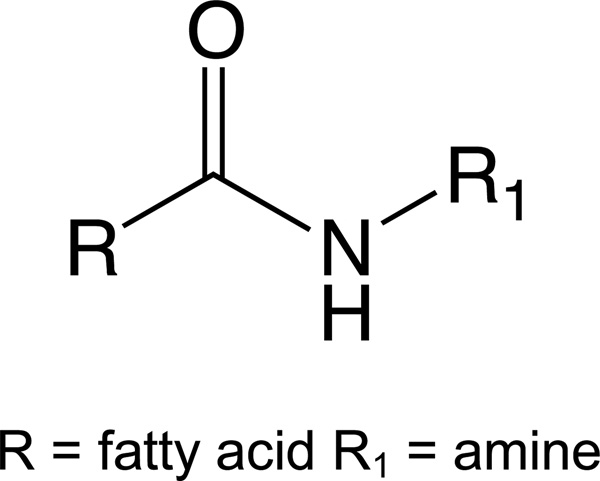
AEA belongs to a larger structural family of N-acyl ethanolamines (NAEs), which are part of a still larger chemical family of Lipoamines (also referred to as N-acyl amides, fatty acid amide, N-acyl amino acids, and lipo amino acids). Whereas AEA is constructed via an amide bond of ethanolamine with arachidonic acid, other NAEs are constructed via an amide bond of diverse fatty acids and ethanolamine. Nearly every endogenous fatty acid has been detected as an NAE conjugate in mammalian tissues [18]. N-acyl amides, as a group are all structurally analogous to the NAEs, in which amino acids or simple amines are conjugated with fatty acids (Figure 1).
Figure 1.
Generic structure of a Lipoamine
Panel A: Construct for Cravatt line; Panel B: Construct for Luquet line; Panel C: Construct for Deutsch line
A theoretical computation of seven common fatty acids in mammalian systems (arachidonic, stearic, docosahexaenoic, oleic, palmitic, linolenic, and linoleic) with twenty amino acids, and 4 common amines (ethanolamine, dopamine, gamma-amino butyric acid (GABA), and taurine), demonstrates that there are 168 possible N-acyl amide combinations that could be formed in mammals [19]. To date, at least 70 N-acyl amides have been endogenously identified in a variety of species, with a limiting factor being a lack of availability of all the synthetic standards needed for complete cataloging [20, 21]. The Bradshaw lab has produced and maintained a large collection of N-acyl amides and uses them in ongoing lipidomics projects.
1.3 NAPE-PLD knockout mice
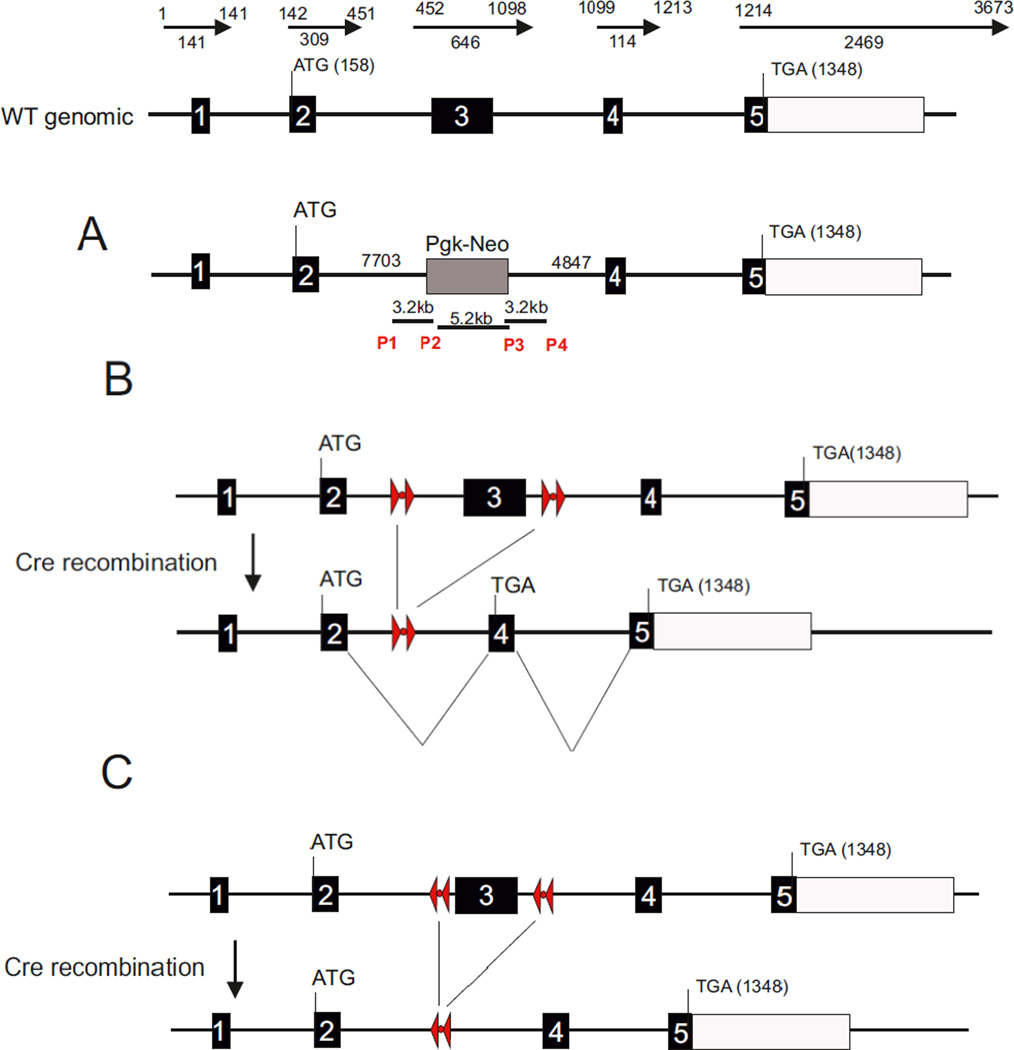
Given NAPE-PLD’s ability to synthesize AEA in-vitro, it was hypothesized that mice lacking NAPE-PLD would have lower brain levels of AEA and related NAEs than wild-type (WT) mice. To date, three variants of NAPE-PLD KO mice have been developed (Figure 2).
Figure 2.
Constructs published for each variant of NAPE-PLD mice.
In the first line of NAPE-PLD KO mice, developed in the laboratory of Ben Cravatt (hereafter referred to as the Cravatt line), no differences were detected in whole-brain levels of AEA compared to WT, despite the fact that brain homogenates from KO mice showed lower activity in synthesizing AEA from exogenously added NAPE. Neither did they find any difference in levels of N-docosahexaenoyl ethanolamine (DEA). Changes in NAE production were restricted to saturated and monounsaturated fatty acid conjugates, with significant decreases in N-palmitoyl ethanolamine (PEA), N-stearoyl ethanolamine (SEA), and N-oleoyl ethanolamine (OEA). In this line of NAPE-PLD KO mice, the larger decreases in NAE levels were in 20, 22 and 24 carbon saturated and monounsaturated fatty acid conjugates. N-linoleoyl ethanolamine (LEA) levels were not measured in the Cravatt line [22].
An additional strain of NAPE-PLD KO mice was later developed in the laboratory of Dale Deutsch (hereafter referred to as the Deutsch line) [17]. In this strain of NAPE-PLD KO mice, whole brain levels of AEA were significantly lower than WT. Additionally, levels of NAEs containing unsaturated conjugates, such as OEA, LEA, and DEA, were likewise significantly lower in the NAPE-PLD KO mice. Results were more varied for saturated conjugates, as these NAPE-PLD KO mice had significantly lower levels of PEA, whereas levels of SEA were not significantly different from WT mice. Additionally, the concentration of longer chain saturated NAEs with 20 and 22 carbons were not statistically different from WT in the NAPE-PLD KO mice [17].
A third NAPE-PLD KO mouse was developed by Serge Luquet when he was a post-doctoral fellow in Richard Palmiter’s lab (hereafter referred to as the Luquet line) wherein brain homogenates from the Luquet line showed reduced conversion of exogenous NAPE to AEA by about 75% compared to WT [12]. However, lipid levels have not yet been measured in the Luquet line of NAPE-PLD KO mice.
Here, we compare levels of NAEs and over 60 structurally analogous N-acyl amides, 2-AG and two structural analogs, and two species of prostaglandins, PGE2 and PGF2α, between NAPE-PLD KO mice from the Luquet line and age, sex, and strain-matched WT mice in a broad scale lipidomics screen in 8 distinct brain regions. Our data show that NAPE-PLD plays a broader role in lipid metabolism than first suspected, including, but not limited to, AEA metabolism. These data show that the NAPE-PLD deletion in the Luquet line also significantly modifies levels of 2-AG, prostaglandins, and a range of N-acyl amide lipids in a region specific manner.
2. Methods
2.1 Mice
Six WT and 6 NAPE-PLD KO 2 month old male mice from the Luquet line [12] were used in this study. WT mice were of the same strain to which the KO mice were derived from, the C57BL/6J (JAX) strain. While the NAPE-PLD KO colony is maintained as homozygous crosses, once a year they were backcrossed to C57BL/6J (JAX) mice to minimize genetic drift, and two heterozygotes from this cross were bred to yield homozygous KO’s. The Bloomington Institutional Animal Care and Use Committee of Indiana University reviewed and approved the procedures used here.
2.2 Tissue collection
All brain tissue was collected and dissected as previously described [23]. In brief, all mice were sacrificed via cervical dislocation on the same day; brains were immediately removed and placed in liquid nitrogen, and then stored in a −80°C freezer until dissections were performed. Brains were dissected while frozen on an ice-cold dissection plate into the following regions: striatum (STR), hippocampus (HIPP), cerebellum (CER), thalamus (THAL), cortex (CTX), hypothalamus (HYP), midbrain (MID), and brainstem (STEM) as previously described [20]. Each dissected area was immediately placed in liquid nitrogen and then stored at −80°C until used for lipid extraction.
2.3 Lipid extraction
Tissue extracts were performed as previously described [20]. In brief, samples from one brain area from 6 WT and 6 NAPE-PLD KO were placed in 50 volumes of HPLC-grade methanol then spiked with 500pmols deuterium-labeled N-arachidonoyl glycine (d8NAGly; Cayman Chemical, Ann Arbor, MI) as an internal standard to determine extraction efficiency. Samples were placed on ice in darkness for 2 hours then individually homogenized. Homogenates were then centrifuged at 19,000g for 20 minutes at 20°C. Supernatants were decanted and diluted with HPLC H2O to make a 75:25 water to supernatant solution. Partial purification was achieved using C-18 solid phase extraction columns (Agilent Technologies, Lake Forest, CA). A series of 4 elutions with 1.5 mL of 60%, 75%, 85%, and 100% methanol were collected for analysis, yielding 48 elutions per brain region. This was then repeated for the other 7 regions.
2.4 HPLC/MS/MS
Samples were analyzed using an Applied Biosystems API 3000 triple quadrupole mass spectrometer with electrospray ionization. Levels of each compound (Table 1) were determined by running each sample using a multiple reactions monitoring (MRM) method tailored for each amide family of compounds as previously described [21]. 20µL from each elution were chromatographed using an XDB-C18 reversed phase HPLC analytical column (Agilent) and optimized mobile phase gradients.
Table 1.
List of lipids screened in HPLC/MS/MS analysis of NAPE-PLD KO (Luquet line) and WT mouse brains.
| N-acyl alanine | [M − H]− | Fragment | N-acyl proline | [M − H]− | Fragment |
| N-palmitoyl alanine | 326.5 | 88.09 | N-palmitoyl proline | 352.53 | 114.12 |
| N-stearoyl alanine | 354.55 | 88.09 | N-stearoyl proline | 380.59 | 114.12 |
| N-oleoyl alanine | 352.53 | 88.09 | N-oleoyl proline | 378.31 | 114.12 |
| N-linoleoyl alanine | 350.52 | 88.09 | N-linoleoyl proline | 376.56 | 114.12 |
| N-arachidonoyl alanine | 374.5 | 88.09 | N-arachidonoyl proline | 400.58 | 114.12 |
| N-docosahexaenoyl alanine | 398.56 | 88.09 | N-docosahexaenoyl proline | 424.6 | 114.12 |
| N-acyl dopamine | [M − H]− | Fragment | N-acyl serine | [M − H]− | Fragment |
| N-oleoyl dopamine | 416.3 | 123.2 | N-palmitoyl serine | 342.3 | 74 |
| N-arachidonoyl dopamine | 438.4 | 123.2 | N-stearoyl serine | 370.3 | 74 |
| N-acyl ethanolamine | [M + H]+ | Fragment | N-oleoyl serine | 368.3 | 74 |
| N-palmitoyl ethanolamine | 300.29 | 62.1 | N-linoleoyl serine | 366.27 | 74 |
| N-stearoyl ethanolamine | 328.3 | 62.1 | N-arachidonoyl serine | 390.3 | 74 |
| N-oleoyl ethanolamine | 326.3 | 62.1 | N-docosahexaenoyl serine | 414.3 | 74 |
| N-linoleoyl ethanolamine | 324.3 | 62.1 | N-acyl taurine | [M − H]− | Fragment |
| N-arachidonoyl ethanolamine | 348.29 | 62.1 | N-arachidonoyl taurine | 410.6 | 124 |
| N-docosahexaenoyl ethanolamine | 372.6 | 62.1 | N-acyl tryptophan | [M − H]− | Fragment |
| N-acyl GABA | [M − H]− | Fragment | N-palmitoyl tryptophan | 441.63 | 203.1 |
| N-palmitoyl GABA | 340.54 | 102.1 | N-stearoyl tryptophan | 469.68 | 203.1 |
| N-stearoyl GABA | 368.58 | 102.1 | N-oleoyl tryptophan | 467.67 | 203.1 |
| N-oleoyl GABA | 366.57 | 102.1 | N-linoleoyl tryptophan | 465.65 | 203.1 |
| N-linoleoyl GABA | 364.54 | 102.1 | N-arachidonoyl tryptophan | 489.67 | 203.1 |
| N-arachidonoyl GABA | 388.57 | 102.1 | N-docosahexaenoyl tryptophan | 513.69 | 203.1 |
| N-docosahexaenoyl GABA | 412.59 | 102.1 | N-acyl tyrosine | [M − H]− | Fragment |
| N-acyl glycine | [M − H]− | Fragment | N-palmitoyl tyrosine | 418.59 | 180.18 |
| N-palmitoyl glycine | 312.26 | 74.2 | N-stearoyl tyrosine | 446.65 | 180.18 |
| N-stearoyl glycine | 340.3 | 74.2 | N-oleoyl tyrosine | 444.63 | 180.18 |
| N-oleoyl glycine | 338.3 | 74.2 | N-linoleoyl tyrosine | 442.61 | 180.18 |
| N-linoleoyl glycine | 336.3 | 74.2 | N-arachidonoyl tyrosine | 466 | 180.18 |
| N-arachidonoyl glycine | 360.3 | 74.2 | N-docosahexaenoyl tyrosine | 490.66 | 180.18 |
| N-docosahexaenoyl glycine | 384.3 | 74.2 | N-acyl valine | [M − H]− | Fragment |
| N-acyl leucine | [M − H]− | Fragment | N-palmitoyl valine | 354.31 | 116.31 |
| N-palmitoyl leucine | 368.58 | 130.1 | N-stearoyl valine | 382.6 | 116.14 |
| N-stearoyl leucine | 396.63 | 130.1 | N-oleoyl valine | 380.59 | 116.14 |
| N-oleoyl leucine | 394.61 | 130.1 | N-nervonoyl valine | 464.75 | 116.14 |
| N-linoleoyl leucine | 392.6 | 130.1 | N-linoleoyl valine | 378.58 | 116.14 |
| N-docosahexaenoyl leucine | 440.64 | 130.1 | N-docosahexaenoyl valine | 426.62 | 116.14 |
| N-acyl methionine | [M − H]− | Fragment | Free Fatty Acids | [M − H]− | Fragment |
| N-palmitoyl methionine | 386.62 | 148.2 | Linoleic Acid | 279.5 | 261 |
| N-stearoyl methionine | 414.64 | 148.2 | Arachidonic Acid | 303.5 | 285 |
| N-oleoyl methionine | 412.65 | 148.2 | 2-acyl-sn-glycerol | [M + H]+ | Fragment |
| N-linoleoyl methionine | 410.64 | 148.2 | 2-arachidonoyl-sn-glycerol | 379.3 | 287.5 |
| N-arachidonoyl methionine | 434.66 | 148.2 | 2-linoleoyl-sn-glycerol | 355.5 | 245 |
| N-docosahexaenoyl methionine | 458.68 | 148.2 | 2-oleoyl-sn-glycerol | 357.5 | 265.2 |
| N-acyl phenylalanine | [M − H]− | Fragment | Prostaglandins | [M − H]− | Fragment |
| N-palmitoyl phenylalanine | 402.59 | 164.1 | PGE2 | 351.2 | 315 |
| N-stearoyl phenylalanine | 430.65 | 164.1 | PGF2α | 353.3 | 309.2 |
| N-oleoyl phenylalanine | 428.63 | 164.1 | |||
| N-linoleoyl phenylalanine | 426.61 | 164.1 | |||
| N-arachidonoyl phenylalanine | 450.64 | 164.1 | |||
| N-docosahexaenoyl phenylalanine | 474.66 | 164.1 | |||
Lipids are grouped by amide family and all members of that lipid family are screened in a multiple reactions monitoring (MRM) method. The parent ion mass is also listed: negative ionization mode, resulting in a [M − H]− parent ion, is used for all methods except the N-acyl ethanolamine and 2-acyl glycerol methods, which used positive ionization and generates a parent ion with a mass of [M + H]+.
Mobile phase A: 20% / 80% (v / v) methanol / water and 1 mM ammonium acetate.
Mobile phase B: 100% methanol, 1 mM ammonium acetate.
2.5 Data Analysis
Analysis of the HPLC/MS/MS data was performed using Analyst software as previously described [21, 23, 24]. Standards of known concentrations were first run for each analyte. Then, peaks from the brain area samples were compared with the standards. Therefore, unknown lipids are matched to known standards according to retention time from the analytical column and according to their mass fingerprint. Concentrations in moles per gram adjusted for percent recovery from the KO animals were compared to WT concentrations using a one-way ANOVA. Percent recovery was calculated using deuterium-labeled standards as previously described [20, 21, 23, 24]. All statistical tests were carried out using SPSS Statistics 20 (IBM). Statistical significance was defined as p≤.05 and a trending effect was defined as .05<p≤.10.
Analysis of such large data sets continues to be problematic for both the implementation and the interpretation of such data. The hypothesis that we are testing is simply that the WT values for each individual lipid differs from the KO values in each brain region analyzed. There would be the expectation that the values would be evenly distributed and a T-type comparison would yield an analysis of the means by which this could be determined. However, theoretical constructs of using T-test analyses on large data sets suggest the potential to generate false positives with a series of T-tests; therefore, the use of corrective analyses allows for a more conservative approach to these types of calculations. To this end, we have grouped data for analyses by lipid species based on either the amine group for the lipoamines, free fatty acids, 2-acyl glycerols, or prostaglandins. This separation also extends to discrete analyses being performed on these categories only within a brain region. In this configuration there is no expectation that one specific lipid category has an influence on the other; therefore, the post-hoc adjustments for multiple comparisons are limited to the specific subcategories listed. We do recognize that there may be potential predictive analyses that can be performed between brain areas and categories of lipid species; however, those types of analyses fall into a realm of modeling that is beyond the scope of the questions being asked of these initial data sets.
An example of a categorical analysis is that of all N-acyl ethanolamine species (7 total) within a discrete brain region would be considered a selection of values with the most likelihood of having the same direction and magnitude of effects by the deletion of a gene given their strong structural homology and their regional specificity (i.e. a discreet brain region). Therefore, all of these lipids were analyzed as a unit with an overall ANOVA and then a corrected post-hoc analysis using Fishers Least Significant Difference was performed in the event that the overall ANOVA determined an interaction.
Analyzed data are represented in tabular format illustrating both the direction and magnitude of change. To determine the magnitude change and therefore the number of arrows to assign each significant difference, the mean level of a particular lipid in a specific region of the NAPE-PLD KO mice was divided by that same lipid’s mean level in the same brain region of the corresponding WT mice. For example, the average level of PGE2 in the hippocampus of NAPE-PLD KO mice was 8.67×10−10 moles per gram and the average level of PGE2 in the hippocampus of the WT was 4.33×10−10 moles per gram; 8.67×10−10 divided by 4.33×10−10 equals 2.002, meaning that PGE2 levels are twice as high in the hippocampus of the NAPE-PLD KO mice and assigning it 3 up arrows in the figures because the magnitude of change was between 2 and 3 times higher than WT. For decreases the process was very similar: the mean level in the KO was divided by the mean level in the WT; however, the reciprocal of the decimal was taken to express a fold decrease (if the level in the KO mouse is ½ of the WT level then that is a 2 fold decrease). As an example the mean level of AEA was 1.13×10−11 moles per gram in the hippocampus of the NAPE-PLD KO mice and 3.97×10−11 moles per gram in the corresponding WT hippocampus. 1.13×10−11 divided by 3.97×10−11 is 0.28 and the reciprocal of 0.28 is 3.57, meaning that the decrease is between 3 and 10 times WT levels and giving it 4 down arrows on our scale.
3. Results
3.1 Analysis of the targeted lipidome in WT and NAPE-PLD KO mice
Of the 73 N-acyl amides in our screening library (Table 1), over 50 were detected in most brain regions in both the WT and NAPE-PLD KO mice. The most lipids in the library were detected in larger brain regions such as the cortex (CTX), cerebellum (CER), and brainstem (STEM). The fewest were detected in smaller brain regions such as the hypothalamus (HYP). Each of the NAE, prostaglandin, free fatty acid, and 2-acyl glycerol species analyzed here were detected in all brain regions in WT and NAPE-PLD KO. In our analysis 426 post-hoc comparisons were made. This number is result of being able to analyze data from only those lipid detected (38–59 per region) in each brain region (8 regions) between WT (6 mice) and NAPE-PLD KO (6 mice). This level of analysis constitutes ~8000 data points; therefore, the detailed list of levels of analytes detected in each of the eight brain regions assayed from WT and NAPE-PLD and the statistical analyses of each are available in the Supplemental Tables.
3.2 Levels of all N-acyl ethanolamines in all brain regions are significantly reduced in this strain of NAPE-PLD KO mice
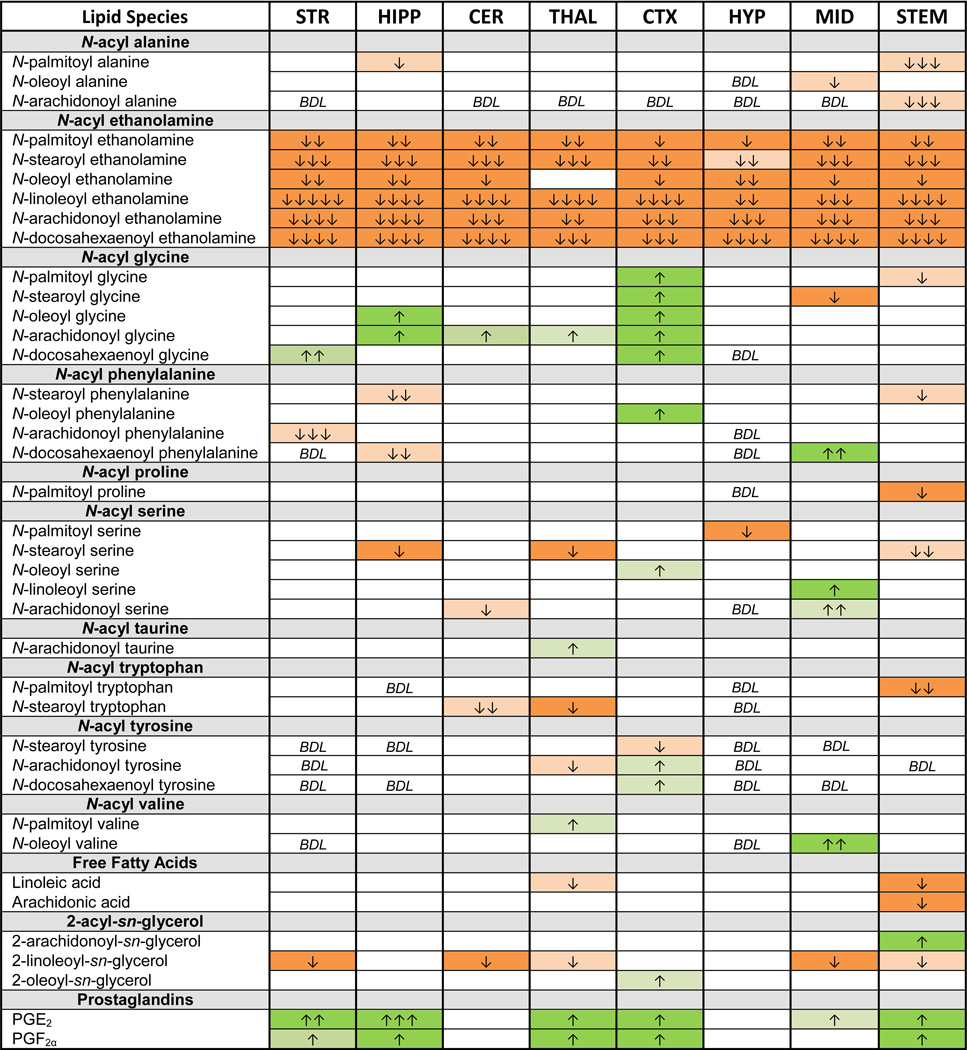
Of the six NAEs in the screening library, all were significantly lowered by NAPE-PLD deletion (Table 2). Importantly, polyunsaturated conjugates, LEA, AEA, and DEA, showed a greater magnitude change in concentration relative to WT than that of more saturated conjugates PEA, SEA or OEA. A notable exception was a complete lack of change in OEA that was restricted to the thalamus (THAL); whereas, OEA levels were significantly lower in all other brain regions. (This is a replicated finding from our pilot study with another set of the same strain of NAPE-PLD KO mice; therefore, this appears to be a reproducible phenomenon and not merely a statistical anomaly of this data set- data not shown).
Table 2.
Changes in lipidome of Luquet line NAPE-PLD KO compared to WT controls.
Boxes in green indicate significant increases in the lipid and the number of arrows in the box indicates the magnitude difference (1 arrow = 1–1.49 fold change; 2 arrows = 1.5–1.99 fold change; 3 arrows = 2–2.99 fold change; 4 arrows = 3–9.99 fold change; 5 arrows = 10 fold or more change relative to WT). Likewise, orange boxes indicate a significant decrease. Blank cells indicate no change between WT and NAPE-PLD KO, whereas BDL marks where lipids were present at concentrations below detection limits. See supplemental tables for means and statistical analyses.
3.3 Differential changes in the lipidome as a function of brain region suggests region-specific biosynthesis and metabolism
In contrast to the ubiquitous unidirectional change in levels of NAEs throughout the brain, changes in other lipids were restricted to specific brain regions and varied in directionality (i.e. increase or decrease; Table 2). Here, we will highlight the cases of the hippocampus (HIPP), CTX, and STEM. In each case, they share a commonality of an increase in levels of both prostaglandins, PGE2 and PGF2α. By contrast, levels of all six N-acyl glycines increase in the CTX, only two members of the N-acyl glycine family increase in HIPP, and yet none increase in STEM. This example suggests differential regulation of the synthesis of these lipid families in different brain areas.
3.4 Levels of arachidonic acid conjugates and metabolites are particularly sensitive to NAPE-PLD deletion
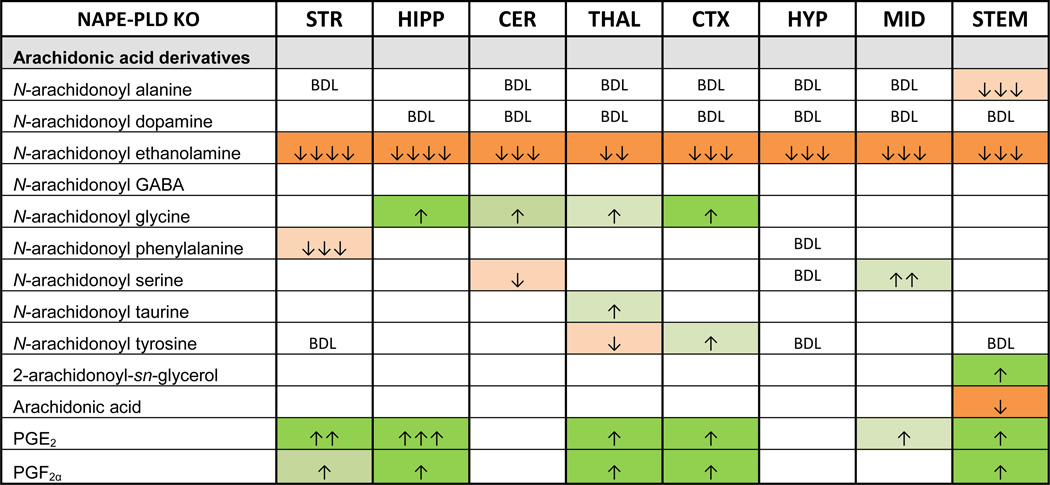
Of the lipids measured in this screen, those with arachidonic acid as part of their molecular structure showed greater variability of levels following NAPE-PLD deletion (Table 3). Although several N-arachidonoyl amides were detected throughout the brain in WT and NAPE-PLD KO mice, only AEA was significantly affected in all brain regions. Four N-arachidonoyl amides decreased in different areas of the brain. Conversely, NAGly increased in four regions – the most consistent change in any non-NAE lipoamine measured here. Surprisingly, 2-AG levels were significantly higher in the STEM of NAPE-PLD KO mice, a change that was accompanied by a significant decrease in the amount of free arachidonic acid. This was the only brain area where arachidonic acid levels were altered.
Table 3.
Summary of all arachidonic acid derived lipid species in the screen and their significance and magnitude of change in the Luquet line relative to WT controls.
Boxes in green indicate significant increases in the lipid and the number of arrows in the box indicates the magnitude difference (same scale as in Table 2). Likewise, orange boxes indicate a significant decrease. See supplemental tables for means and statistical analyses. Blank cells indicate no change between WT and NAPE-PLD KO, whereas BDL marks where lipids were present at concentrations below detection limits. See supplemental tables for means and statistical analyses.
Perhaps the most unexpected finding was that in 6 of the 8 brain regions tested, prostaglandin levels were significantly higher in NAPE-PLD KO mice. Levels of both PGE2 and PGF2α were higher in the striatum (STR), HIPP, THAL, CTX, and STEM of NAPE-PLD KO mice relative to WT. In the midbrain (MID) of NAPE-PLD KO mice, PGE2 was the only prostaglandin with differing concentrations compared to the WT MID. However, in the CER and HYP, levels of prostaglandins were equivalent in WT and NAPE-PLD KO. When levels of both prostaglandins did change, PGE2 levels increased more than those of PGF2α. The largest increase in PGE2 levels was in the HIPP of the NAPE-PLD KO mice relative to WT HIPP.
4. Discussion
That NAPE-PLD KO has broad, yet region-specific effects on the brain lipidome examined here speaks to the interconnectedness of lipid biosynthesis and metabolism as well as regional specificity. Previous findings that showed decreased production of specific NAEs in the Cravatt and Deutsch NAPE-PLD KO models were replicated here, with the caveat that all NAE levels screened here are reduced in the Luquet line of NAPE-PLD KO mice relative to WT. This is potentially meaningful for understanding the effects on lipid signaling pathways in multiple systems. Although AEA is the only NAE with significant affinity for CB1 and CB2, the other NAEs measured are also biologically active, often targeting non-classical cannabinoid receptors (e.g. the peroxisome proliferator-activated receptors (PPARs) or ion channels) [18, 20].
4.1 Comparison of different constructs among the 3 different NAPE-PLD KOs
The first line of NAPE-PLD KO, the Cravatt line, was described in a manuscript by Leung et al [22]. In this study the authors used homologous recombination approach targeted in 129Svj embryonic stem cells. Positive clones were used to create chimeric mice in a C57Bl/6 background. The construct strategy described the replacement of exon 4 by the phosphoglycerate kinase I promoter driving the neomycin resistance gene (Pgk-Neo). However, when the sequence of the primers used to generate the 5’ and 3’ homology arms are aligned with the genomic sequence of the NAPE-PLD gene, the two primers that were used to achieve homologous recombination span exon 3. The fact that exon 3 rather than 4 was removed is more in line with the expected phenotype, since exon 4 does not contain the essential catalytic activity of the enzyme. It is probable that the nomenclature for Nape-pld exon number changed after the manuscript was published. This might be at the origin of the misleading Figure 1A in Leung et al. If our assumption is correct, then the allele obtained lacks exon 3 and is therefore unable to produce a functional NAPE-PLD. It is possible that the surrounding genomic sequence might be impacted by the presence of the Pgk-Neo cassette that resides still in the null allele (Figure 2A).
The Deutsch and Luquet NAPE-PLD transgenic mice took advantage of the Cre/Lox P recombinase approach, which allows tissue specific invalidation of the gene. In both cases the manuscript by Liu et al. [12] and Tsuboi et al [17] described a strategy that result from the insertion of two LoxP sites on other side of exon 3. In Liu et al, for the Luquet line, the targeting construct was electroporated in embryonic 129S4/SvJ stem cells in order to eventually obtain chimeric mice. NAPE-PLD floxed (NAPE-PLDlox) animals were bred with Mox2-Cre mice [25] to generate heterozygous animals with one null Nape-pld allele. These mice were crossed with C57Bl/6 mice to remove the Mox2-Cre gene, and these offspring were then bred together to generate mice homozygous for the Nape-pld null allele. At this point mice were on a mixed 129S4 × C57Bl/6 genetic background. This line was further bred at least 8 times onto C57Bl/6 background in the Luquet lab (Figure 2B).
In the manuscript by Tsuboi et al [17] regarding the Deutsch line a similar strategy is described in which LoxP sites were inserted on both sides of exon 3. The targeting construct was electroporated into iTL 129 Sv embryonic stem cells and positive clones were used to obtain chimera that were mated onto C57Bl/6 background. The NAPE-PLD null allele was obtained by breeding NAPE-PLD floxed animals (NAPE-PLDlox) with B6.FVB-Tg(EIIa-cre)C5379Lmgd/J allowing for Cre expression in the early mouse embryo [26]. The resulting mice carrying a null NAPE-PLD allele (NAPE-PLD+/−) were bred backcrossed onto C57BL6/J more than 8 times and intercrossed to obtain KO animals (NAPE-PLD−/−). Noteworthy, in both strategies the removal of exon 3 does not preclude the initiation of a transcript from the starting ATG codon located in exon 2. In the Luquet line the strategy was designed so as to introduce a frameshift upon Cre-mediated deletion of the exon 3 in between the LoxP sites. This results in 19 amino acids after the initiation starts, before a stop sequence is encountered. The methodology described in Tsuboi et al does not provide sufficient details to determine if a frameshift mutation is present in the recombined allele. There are specific differences in the lipidomics results from each NAPE-PLD KO study, with decreases in AEA and DEA in the Deutsch and Luquet lines but not in the Cravatt lines, which may provide an insight into the overall functioning of this enzyme.
4.2 Differences in analytical methods among 3 lines of NAPE-PLD KO
Another source of the discrepancy in effects of deleting NAPE-PLD on AEA levels may be the analytical methods used to measure NAEs. The lipid extractions for analysis of the Cravatt line and the Deutsch line were both based on the chloroform liquid-liquid extraction [27]. Major differences exist in the internal standards that lipid extracts were spiked with. Two of the analyses used stable radiolabeled isotopes: Leung and colleagues used d4AEA and d4OEA spikes and we used d8NAGly. Tsuboi and colleagues used 1-Heptadecanoyl-2-hydroxy-glycero-3-phosphoethanolamine. In contrast to the tandem MS performed in this study and by Tsuboi and colleagues (4000 Q-TRAP), the studies on the Cravatt line used a single quadrupole benchtop MS (Agilent 1100 LC-MSD SL) [22, 28]. The Cravatt group quantified lipids by comparing the peak area of the NAE to the peak area of the d4 labeled internal standard that was added to each sample [29, 30]. For our analysis and for analysis of the Deutsch line [17], standard curves for each MRM method were generated by running purified standard lipids of known concentrations. Standards for NAEs were not mixed in with the samples from brain tissue in our analysis or Tsuboi and colleagues’ analysis
Eighty lipids were measured in analyzing the Luquet line, including lipoamines and prostaglandins that were measured in other lines of NAPE-PLD KO. However, a notable absence from our present analysis is comparing levels of precursor NAPEs. We would expect levels of NAPE to increase in NAPE-PLD KO, similar to that seen in Tsuboi and colleagues [17]. Levels of NAPEs correspond well with NAE levels: the analysis of the Cravatt line saw increases in levels of NAPEs whose conjugate NAEs showed decreases and saw no change in N-arachidonoyl phosphatidylethanolamine levels, corresponding with no change in AEA [22].
4.3 Lipoamines are broad targets for enzymes and receptors
One of the first NAEs to be identified was PEA, the conjugate of palmitic acid and ethanolamine. Kuehl and colleagues found it in soybeans, peanuts, and egg yolks and determined that PEA had anti-inflammatory effects [31]. PEA and OEA both bind to PPAR-α to have analgesic, anti-inflammatory, and anorexic effects. In addition, SEA also possesses anti-inflammatory and anorexic properties [32]. OEA is an effective agonist of GPR119, a receptor highly expressed in the pancreas and colon [33]. The stimulation of GPR119 in the small intestine releases glucagon-like peptide-1, which enhances satiety and reduces food intake [34]. LEA can also activate PPAR-α in the intestine, where it may have a role in prolonging feeding latency [33]. Levels of PEA, OEA, and LEA decrease after rats are fed a high fat diet, though what is happening to the broader lipidome of N-acyl amide lipids is unknown [34].
Non-NAE N-acyl amide endogenous lipids have also been shown to have biological significance. N-arachidonoyl taurine (A-Taur), activates transient receptor potential vanilloid 1 (TRPV1) and TRPV4 channels at micromolar concentrations [35]. Likewise, AEA and OEA were shown previously to also be agonists at TRPV1 [36, 37]. More recently, we identified 8 additional N-acyl amides that activate TRPV1; including LEA, DEA, and N-docosahexaenoyl glycine that are modified here in the Luquet NAPE-PLD mouse [20]. The only change in levels of A-Taur in NAPE-PLD KO mice was an increase in the thalamus; otherwise, A-Taur levels were the same as WT. Therefore, inactivating NAPE-PLD does not affect all endogenous TRPV1 ligands in the same manner or in the same region of the brain.
4.4 Anandamide-NAGly synergy is the opposite of what is predicted in the NAPE-PLD mouse
The N-acyl amide, N-arachidonoyl glycine (NAGly), is an agonist at GPR18 and drives migration in both microglial and human endometrial carcinoma cells [38, 39]. NAGly can be formed from AEA via two distinct pathways, both of which are found in mammalian cells. The first is a direct oxidation of AEA by alcohol dehydrogenase, and the second is via the conjugation of arachidonic acid and glycine using arachidonic acid released by the hydrolysis of AEA by fatty acid amide hydrolase (FAAH) [40]. NAGly possesses antinociceptive and anti-inflammatory properties in several animal models of pain. NAGly has also been shown to suppress the proliferation of murine macrophage cells [41]. Like AEA, NAGly can undergo oxygenation reactions via the enzyme cyclooxygenase 2 (COX-2) to produce a glycine conjugate of prostaglandins [33, 42]. These relationships would predict that if AEA were decreased in the cells then NAGly would, likewise, decrease. This, however, was not the case in 4 of the 8 brain regions where NAGly was actually increased in NAPE-PLD KO mice. Therefore, the biosynthetic mechanism that cell systems are using in the brain for lipid metabolism even between these two tightly-controlled lipids appears to be compromised when this upstream enzyme is eliminated.
4.5 Brainstem metabolism of 2-AG and arachidonic acid are unique in NAPE-PLD KO mice, though 2-acyl glycerols are more globally regulated
Any potential differences in 2-AG and arachidonic acid levels after NAPE-PLD deletion cannot be evaluated as these lipids were not reported for either the Cravatt or Deutsch NAPE-PLD KO strains [17, 22]. Changes observed for 2-AG and arachidonic acid in the Luquet mice were restricted to the brainstem, highlighting a region-dependent role for NAPE-PLD. Importantly, in 5 of the 8 brain regions analyzed here, levels of the 2-AG structural analog 2-linoleoyl glycerol (2-LG) were significantly reduced in the Luquet NAPE-PLD KO mouse, again, suggesting more biosynthetic and metabolic cross-talk among lipid species. Although 2-LG does not directly activate CB1 or CB2 [43], it acts as an agonist at some of the orphan GPCRs that are putative cannabinoid receptors. In COS-7 cells transfected with human GPR119, 2-LG stimulates cAMP formation [34]. Another potential mechanism of action of 2-LG is that it potentiates that binding of 2-AG to CB1 and inhibits 2-AG’s deactivation in neuronal cells. This effect has been called the “entourage effect” [43], though recent unpublished data from the Mackie lab presented at ICRS in 2014 demonstrated that 2-LG was acting as a negative allosteric modulator for 2-AG in hippocampal neurons, adding still more complexity to these signaling systems. Therefore some of the region-dependent decreases in 2-LG levels in the Luquet NAPE-PLD KO mice may be indirectly affecting cannabinoid signaling.
4.6 Prostaglandins are sensitive to changes in the arachidonic acid lipidome
Our finding of region dependent increases in prostaglandin levels is the first report of altered prostaglandin signaling in NAPE-PLD KO mice. Altering the expression of an enzyme that regulates eCB production can have effects on prostaglandins, as the two families of lipids are derived from arachidonic acid and share some precursors [42]. Prostaglandins are well known for their role in inflammation [44]. Interestingly, inflammatory challenges can acutely alter the expression of NAPE-PLD: in cultured RAW 264.7 macrophage cells, an inflammatory insult caused by treatment with LPS downregulated NAPE-PLD expression by altering transcription factors [45]. However, treatment of RAW macrophage cells with LPS increases levels of AEA compared to vehicle treatment, suggesting that NAPE-PLD is not the major enzyme responsible for LPS-induced up regulation of AEA [16]. Therefore, despite the fact that NAPE-PLD KO mice do not have an overt behavioral phenotype under baseline conditions [17, 22], deleting NAPE-PLD may impair the brain’s ability to respond to inflammation [46].
Possible mechanistic explanations for why prostaglandin levels are upregulated in some brain areas of NAPE-PLD KO involve alterations in expression or activity of enzymes that synthesize prostaglandins. PLA2 releases arachidonic acid from phospholipid pools. COX oxidizes arachidonic acid and forms the endoperoxide PGH2. Prostaglandin synthases then generate the prostaglandins (PGE2, PGD2, and PGF2α) and prostacyclin (PGI2) [44]. Stimuli, like growth factors or inflammatory agents, induce expression of COX-2. [47]. Several studies that reported increases in PGE2 due to pharmacological manipulations identified upregulation of COX-2 as a potential mechanism [48–50]. Therefore, a follow-up study will examine COX-2 expression in the brain regions of NAPE-PLD KO where prostaglandins are upregulated, predicting increases in COX-2 where prostaglandin levels are higher.
COX-2 dependent PGE2 synthesis has important implications for both psychopathology and neurodegeneration. For example, the post-mortem brains of patients with schizophrenia and bipolar disorder displayed higher expression of COX-2 [51, 52] and this is correlated with reports of higher PGE2 levels in plasma [53]. The anti-psychotic drug olanzapine reduced both COX-2 and PGE2 levels [54]. In Alzheimer’s disease, brains from patients demonstrated higher COX-2 levels, along with other markers of inflammation [55] that were not present in normal aging [56]. Indeed, the use of COX inhibitors can reduce amyloid beta buildup in animal models of Alzheimer’s disease [50, 57]. Studies examining inflammatory markers in humans with neuroinflammation did not measure NAPE-PLD activity. However, given that NAPE-PLD is required for inflammatory responses in cell systems [45], it is possible that dysregulation of NAPE-PLD may be contributing to increased prostaglandin signaling.
How are concentrations of prostaglandins and eCBs related to levels of free arachidonic acid? Illustrating the close connections of the arachidonic acid lipidome, Nomura and colleagues showed an integral relationship between 2-AG and prostaglandins in monoacylglycerol lipase (MAGL) KO mice: brain levels of 2-AG robustly increased, at the expense of levels of prostaglandins and arachidonic acid [58]. As prostaglandin production is regionally upregulated in NAPE-PLD KO, it was expected that free arachidonic acid levels would increase. However, in cases where prostaglandin production is augmented, free arachidonic acid levels do not necessarily change [59]. Furthermore, the relationship between AEA metabolism and arachidonic acid levels is not fully clarified. Despite the fact that inhibiting AEA hydrolysis prevents the formation of radiolabeled arachidonic acid from labeled AEA, levels of free arachidonic acid in the brains of FAAH KO mice have not been measured [30]. As arachidonic acid is a target for many oxidative enzymes [60], such as COX-2, increases in arachidonic acid may be so rapidly metabolized that a change cannot be detected in NAPE-PLD KO. The factors that drive sustained increases in arachidonic acid levels need to be further elucidated, as well as detailing the relationships between eCBs, arachidonic acid, and prostaglandins.
5. Concluding Remarks
NAPE-PLD is hypothesized to be responsible for the production of NAEs in the brain, yet, to rule out that it is involved in broader areas of lipid metabolism a wider variety of lipids must be screened in more discreet brain regions. Here, a targeted lipidomics screen of over 70 N-acyl amides illustrates that in the Luquet NAPE-PLD KO mouse not only were there significant decreases in levels of NAEs throughout the brain, they were fatty acid chain and area dependent. Levels of other N-acyl amides, 2-acyl glycerols and prostaglandins were also significantly impacted by NAPE-PLD deletion in a region-specific manner. This is the first report of changes in prostaglandin production in any NAPE-PLD KO mice. Therefore, deletion of NAPE-PLD impacts production of several cannabimimetic molecules, and is not just important for the synthesis of NAEs. Furthermore, the impact of NAPE-PLD deletion on the lipidome varies by brain region, which no doubt has functional implications. The widespread lipidomics shifts identified here in the Luquet NAPE-PLD KO mouse highlights the interconnectedness of the arachidonic acid lipidome and may explain some of the behavioral changes seen after deletion of this enzyme.
Supplementary Material
Highlights for NAPE-PLD KO lipidomics.
NAPE-PLD KO mice show a significant reduction in brain levels of Anandamide.
Modifying NAPE-PLD has broader effects on the lipidome than previously recognized.
NAPE-PLD KO has brain region-dependent effects on arachidonic acid derivatives.
Increased prostaglandin levels illustrate a dramatic shift in arachidonic acid metabolism
Acknowledgments
Funding
This study was funded by NIH grants EY024625, DA011322, and DA021696.
Abbreviations
- 2-AG
2-arachidonyl glycerol
- 2-LG
2-linoleoyl glycerol
- ABHD4
α/β-hydrolase domain 4
- AEA
N-arachidonoyl ethanolamine (anandamide)
- A-Taur
N-arachidonoyl taurine
- CER
cerebellum
- COX
cyclooxygenase
- CTX
cortex
- DEA
N-docosahexaenoyl ethanolamine
- eCB
endogenous cannabinoid
- FAAH
fatty acid amide hydrolase
- GABA
gamma-amino butyric acid
- GDE1
glycerophosphodiesterase 1
- HIPP
hippocampus
- HYP
hypothalamus
- KO
knockout
- LEA
N-linoleoyl ethanolamine
- MAGL
monoacylglycerol lipase
- MID
midbrain
- MRM
multiple reactions monitoring
- NAE
N-acyl ethanolamine
- NAGly
N-arachidonoyl glycine
- NAPE
N-acyl-phosphatidylethanolamine
- NAPE-PLD
NAPE-specific phospholipase D
- NAPE-PLDlox
NAPE-PLD floxed
- OEA
N-oleoyl ethanolamine
- PEA
N-palmitoyl ethanolamine
- PGK-neo
phosphoglycerate kinase I promoter driving neomycin resistance gene
- PLA2
phospholipase A2
- pNAPE
plasmalogen-type NAPE
- PPAR
peroxisome proliferator-activated receptor
- SEA
N-stearoyl ethanolamine
- STEM
brainstem
- STR
striatum
- THAL
thalamus
- TRPV1
transient receptor potential vanilloid 1
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The author, Heather B Bradshaw, of this manuscript is on the Advisory Board for Phytecs and consults on how endogenous cannabinoids function in the central nervous system. Phytecs had no financial contribution to the current work.
Ethical Approval
All procedures using animals were approved by the Bloomington Institutional Animal Care and Use Committee of Indiana University. This article does not contain any studies with human participants performed by any of the authors.
References
- 1.Devane WA, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258(5090):1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 2.Sugiura T, et al. 2-Arachidonoylglycerol: a possible endogenous cannabinoid receptor ligand in brain. Biochem Biophys Res Commun. 1995;215(1):89–97. doi: 10.1006/bbrc.1995.2437. [DOI] [PubMed] [Google Scholar]
- 3.Mechoulam R, et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem Pharmacol. 1995;50(1):83–90. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- 4.Maccarrone M, et al. Anandamide inhibits metabolism and physiological actions of 2-arachidonoylglycerol in the striatum. Nat Neurosci. 2008;11(2):152–159. doi: 10.1038/nn2042. [DOI] [PubMed] [Google Scholar]
- 5.Ueda N, Tsuboi K, Uyama T. Metabolism of endocannabinoids and related N-acylethanolamines: canonical and alternative pathways. FEBS J. 2013;280(9):1874–1894. doi: 10.1111/febs.12152. [DOI] [PubMed] [Google Scholar]
- 6.Di Marzo V. Targeting the endocannabinoid system: to enhance or reduce? Nat Rev Drug Discov. 2008;7(5):438–455. doi: 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- 7.Pertwee RG. Handbook of Cannabis. Oxford University Press; 2014. [Google Scholar]
- 8.Di Marzo V, et al. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372(6507):686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto Y, et al. Molecular characterization of a phospholipase D generating anandamide and its congeners. J Biol Chem. 2004;279(7):5298–5305. doi: 10.1074/jbc.M306642200. [DOI] [PubMed] [Google Scholar]
- 10.Egertova M, et al. Localization of N-acyl phosphatidylethanolamine phospholipase D (NAPE-PLD) expression in mouse brain: A new perspective on N-acylethanolamines as neural signaling molecules. J Comp Neurol. 2008;506(4):604–615. doi: 10.1002/cne.21568. [DOI] [PubMed] [Google Scholar]
- 11.Hegyi Z, et al. Differential distribution of diacylglycerol lipase-alpha and N-acylphosphatidylethanolamine- specific phospholipase d immunoreactivity in the superficial spinal dorsal horn of rats. Glia. 2012;60(9):1316–1329. doi: 10.1002/glia.22351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, et al. Multiple pathways involved in the biosynthesis of anandamide. Neuropharmacology. 2008;54(1):1–7. doi: 10.1016/j.neuropharm.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun YX, et al. Biosynthesis of anandamide and N-palmitoylethanolamine by sequential actions of phospholipase A2 and lysophospholipase D. Biochem J. 2004;380(Pt 3):749–756. doi: 10.1042/BJ20040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon GM, Cravatt BF. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway. J Biol Chem. 2006;281(36):26465–26472. doi: 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- 15.Simon GM, Cravatt BF. Anandamide biosynthesis catalyzed by the phosphodiesterase GDE1 and detection of glycerophospho-N-acyl ethanolamine precursors in mouse brain. J Biol Chem. 2008;283(14):9341–9349. doi: 10.1074/jbc.M707807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, et al. A biosynthetic pathway for anandamide. Proc Natl Acad Sci U S A. 2006;103(36):13345–13350. doi: 10.1073/pnas.0601832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuboi K, et al. Enzymatic formation of N-acylethanolamines from N-acylethanolamine plasmalogen through N-acylphosphatidylethanolamine-hydrolyzing phospholipase D-dependent and -independent pathways. Biochim Biophys Acta. 2011;1811(10):565–577. doi: 10.1016/j.bbalip.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Dumlao DS, et al. High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines. Biochim Biophys Acta. 2011;1811(11):724–736. doi: 10.1016/j.bbalip.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradshaw HB, Raboune S, Hollis JL. Opportunistic activation of TRP receptors by endogenous lipids: Exploiting lipidomics to understand TRP receptor cellular communication. Life Sci. 2012 doi: 10.1016/j.lfs.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raboune S, et al. Novel endogenous N-acyl amides activate TRPV1-4 receptors, BV-2 microglia, and are regulated in brain in an acute model of inflammation. Front Cell Neurosci. 2014;8:195. doi: 10.3389/fncel.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tortoriello G, et al. Targeted lipidomics in Drosophila melanogaster identifies novel 2-monoacylglycerols and N-acyl amides. PLoS One. 2013;8(7):e67865. doi: 10.1371/journal.pone.0067865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung D, et al. Inactivation of N-acyl phosphatidylethanolamine phospholipase D reveals multiple mechanisms for the biosynthesis of endocannabinoids. Biochemistry. 2006;45(15):4720–4726. doi: 10.1021/bi060163l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuart JM, et al. Brain levels of prostaglandins, endocannabinoids, and related lipids are affected by mating strategies. Int J Endocrinol. 2013;2013:436252. doi: 10.1155/2013/436252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradshaw HB, et al. Sex and hormonal cycle differences in rat brain levels of pain-related cannabimimetic lipid mediators. Am J Physiol Regul Integr Comp Physiol. 2006;291(2):R349–R358. doi: 10.1152/ajpregu.00933.2005. [DOI] [PubMed] [Google Scholar]
- 25.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26(2):113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 26.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93(12):5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 28.Saghatelian A, et al. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43(45):14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 29.Clement AB, et al. Increased seizure susceptibility and proconvulsant activity of anandamide in mice lacking fatty acid amide hydrolase. J Neurosci. 2003;23(9):3916–3923. doi: 10.1523/JNEUROSCI.23-09-03916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cravatt BF, et al. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci U S A. 2001;98(16):9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuehl FA, et al. The Identification of N-(2-Hydroxyethyl)-Palmitamide as a Naturally Occurring Anti-Inflammatory Agent. Journal of the American Chemical Society. 1957;79(20):5577–5578. [Google Scholar]
- 32.Astarita G, Piomelli D. Lipidomic analysis of endocannabinoid metabolism in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2755–2767. doi: 10.1016/j.jchromb.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ezzili C, Otrubova K, Boger DL. Fatty acid amide signaling molecules. Bioorg Med Chem Lett. 2010;20(20):5959–5968. doi: 10.1016/j.bmcl.2010.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen KB, et al. 2-Oleoyl glycerol is a GPR119 agonist and signals GLP-1 release in humans. J Clin Endocrinol Metab. 2011;96(9):E1409–E1417. doi: 10.1210/jc.2011-0647. [DOI] [PubMed] [Google Scholar]
- 35.Saghatelian A, et al. A FAAH-regulated class of N-acyl taurines that activates TRP ion channels. Biochemistry. 2006;45(30):9007–9015. doi: 10.1021/bi0608008. [DOI] [PubMed] [Google Scholar]
- 36.Zygmunt PM, et al. Vanilloid receptors on sensory nerves mediate the vasodilator action of anandamide. Nature. 1999;400(6743):452–457. doi: 10.1038/22761. [DOI] [PubMed] [Google Scholar]
- 37.Ahern GP. Activation of TRPV1 by the satiety factor oleoylethanolamide. J Biol Chem. 2003;278(33):30429–30434. doi: 10.1074/jbc.M305051200. [DOI] [PubMed] [Google Scholar]
- 38.McHugh D, et al. N-arachidonoyl glycine, an abundant endogenous lipid, potently drives directed cellular migration through GPR18, the putative abnormal cannabidiol receptor. BMC Neurosci. 2010;11:44. doi: 10.1186/1471-2202-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McHugh D, et al. Delta(9) -Tetrahydrocannabinol and N-arachidonyl glycine are full agonists at GPR18 receptors and induce migration in human endometrial HEC-1B cells. Br J Pharmacol. 2012;165(8):2414–2424. doi: 10.1111/j.1476-5381.2011.01497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradshaw HB, et al. The endocannabinoid anandamide is a precursor for the signaling lipid N-arachidonoyl glycine by two distinct pathways. BMC Biochem. 2009;10:14. doi: 10.1186/1471-2091-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradshaw HB, et al. Novel endogenous N-acyl glycines identification and characterization. Vitam Horm. 2009;81:191–205. doi: 10.1016/S0083-6729(09)81008-X. [DOI] [PubMed] [Google Scholar]
- 42.Rouzer CA, Marnett LJ. Endocannabinoid oxygenation by cyclooxygenases, lipoxygenases, and cytochromes P450: cross-talk between the eicosanoid and endocannabinoid signaling pathways. Chem Rev. 2011;111(10):5899–5921. doi: 10.1021/cr2002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Console-Bram L, Marcu J, Abood ME. Cannabinoid receptors: nomenclature and pharmacological principles. Prog Neuropsychopharmacol Biol Psychiatry. 2012;38(1):4–15. doi: 10.1016/j.pnpbp.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294(5548):1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 45.Zhu C, et al. Proinflammatory stimuli control N-acylphosphatidylethanolamine-specific phospholipase D expression in macrophages. Mol Pharmacol. 2011;79(4):786–792. doi: 10.1124/mol.110.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geurts L, et al. Adipose tissue NAPE-PLD controls fat mass development by altering the browning process and gut microbiota. Nat Commun. 2015;6:6495. doi: 10.1038/ncomms7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marnett LJ. The COXIB experience: a look in the rearview mirror. Annu Rev Pharmacol Toxicol. 2009;49:265–290. doi: 10.1146/annurev.pharmtox.011008.145638. [DOI] [PubMed] [Google Scholar]
- 48.Chen R, et al. Delta9-THC-caused synaptic and memory impairments are mediated through COX-2 signaling. Cell. 2013;155(5):1154–1165. doi: 10.1016/j.cell.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amateau SK, McCarthy MM. Induction of PGE2 by estradiol mediates developmental masculinization of sex behavior. Nat Neurosci. 2004;7(6):643–650. doi: 10.1038/nn1254. [DOI] [PubMed] [Google Scholar]
- 50.Liang X, et al. Deletion of the prostaglandin E2 EP2 receptor reduces oxidative damage and amyloid burden in a model of Alzheimer's disease. J Neurosci. 2005;25(44):10180–10187. doi: 10.1523/JNEUROSCI.3591-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao JS, et al. Increased neuroinflammatory and arachidonic acid cascade markers, and reduced synaptic proteins, in the postmortem frontal cortex from schizophrenia patients. Schizophr Res. 2013;147(1):24–31. doi: 10.1016/j.schres.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim HW, Rapoport SI, Rao JS. Altered arachidonic acid cascade enzymes in postmortem brain from bipolar disorder patients. Mol Psychiatry. 2011;16(4):419–428. doi: 10.1038/mp.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez-Gras I, et al. The anti-inflammatory prostaglandin 15d-PGJ2 and its nuclear receptor PPARgamma are decreased in schizophrenia. Schizophr Res. 2011;128(1–3):15–22. doi: 10.1016/j.schres.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 54.Cheon Y, et al. Chronic olanzapine treatment decreases arachidonic acid turnover and prostaglandin E(2) concentration in rat brain. J Neurochem. 2011;119(2):364–376. doi: 10.1111/j.1471-4159.2011.07410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rao JS, Rapoport SI, Kim HW. Altered neuroinflammatory, arachidonic acid cascade and synaptic markers in postmortem Alzheimer's disease brain. Transl Psychiatry. 2011;1:e31. doi: 10.1038/tp.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Keleshian VL, et al. Aging is associated with altered inflammatory, arachidonic acid cascade, and synaptic markers, influenced by epigenetic modifications, in the human frontal cortex. J Neurochem. 2013;125(1):63–73. doi: 10.1111/jnc.12153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Szekely CA, et al. Nonsteroidal anti-inflammatory drugs for the prevention of Alzheimer's disease: a systematic review. Neuroepidemiology. 2004;23(4):159–169. doi: 10.1159/000078501. [DOI] [PubMed] [Google Scholar]
- 58.Nomura DK, et al. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol. 2011;18(7):846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taha AY, et al. Altered fatty acid concentrations in prefrontal cortex of schizophrenic patients. J Psychiatr Res. 2013;47(5):636–643. doi: 10.1016/j.jpsychires.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozak KR, et al. Metabolism of the endocannabinoids, 2-arachidonylglycerol and anandamide, into prostaglandin, thromboxane, and prostacyclin glycerol esters and ethanolamides. J Biol Chem. 2002;277(47):44877–44885. doi: 10.1074/jbc.M206788200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.