Abstract
The genome sequence available for different Plasmodium species is a valuable resource for understanding malaria parasite biology. However, comparative genomics on its own cannot fully explain all the species-specific differences which suggests that other genomic aspects such as regulation of gene expression play an important role in defining species-specific characteristics. Here, we developed a comprehensive approach to measure transcriptional changes of the evolutionary conserved syntenic orthologs during the intraerythrocytic developmental cycle across six Plasmodium species. We show significant transcriptional constraint at the mid-developmental stage of Plasmodium species while the earliest stages of parasite development display the greatest transcriptional variation associated with critical functional processes. Modeling of the evolutionary relationship based on changes in transcriptional profile reveal a phylogeny pattern of the Plasmodium species that strictly follows its mammalian hosts. In addition, the work shows that transcriptional conserved orthologs represent potential future targets for anti-malaria intervention as they would be expected to carry out key essential functions within the parasites. This work provides an integrated analysis of orthologous transcriptome, which aims to provide insights into the Plasmodium evolution thereby establishing a framework to explore complex pathways and drug discovery in Plasmodium species with broad host range.
Abbreviations: IDC, intraerythrocytic developmental cycle; P. falciparum, PF, Plasmodium falciparum; P. vivax, PV, Plasmodium vivax; P. knowlesi, PK, Plasmodium knowlesi; P. berghei, PB, Plasmodium berghei; P. chabaudi, PC, Plasmodium chabaudi; P. yoelii, PY, Plasmodium yoelii; PCC, Pearson correlation coefficient; Ds, Divergent score; Ph, Phase; PBS, Phosphate-buffered saline; Ka, non-synonymous site; Ks, synonymous site; hr, hour; SRCC, Spearman Rank Correlation Coefficient; AU, approximately unbiased; BP, bootstrap probability; MPM, Malaria Parasite Metabolic Pathway
Keywords: Comparative transcriptomics, Plasmodium species, Evolution, Microarray, Transcriptome, Drug targets
Highlights
-
•
Comparison of variations in mRNA abundance across six different Plasmodium species.
-
•
Transcriptional conservation and divergence of Plasmodium syntenic orthologs.
-
•
Pattern of Plasmodium transcriptome evolution are established.
-
•
Transcriptionally conserved orthologs represent attractive intervention targets.
Malaria remains a major public health concern despite global efforts in the fight against this disease. The intraerythrocytic stage of the malaria parasites is currently in the spotlight for anti-malarial intervention and vaccine targets. The primary goal of this study is to generate a comprehensive and directly comparable transcriptome dataset across multiple Plasmodium species originating from different hosts. We establish that specific pathways and intraerythrocytic stages are more transcriptionally diverged than others, reflecting transcriptional evolutionary diversity. We further propose a panel of transcriptionally conserved genes as potential drug targets.
1. Introduction
Protozoan belonging to the Plasmodium species are obligate intracellular parasites that display substantial developmental complexity during their life cycle in the vertebrate hosts and mosquito vectors. The development of single nucleated parasite cells into multi-nucleated schizonts through several rounds of mitosis closely resembles embryonic development of multicellular organisms with a majority of genes changing their expression during this period (Piras et al., 2014, Quint et al., 2012, Bozdech et al., 2003, Irie and Kuratani, 2011). The intraerythrocytic developmental cycle (IDC) exhibits a tightly regulated transcriptional cascade in which essentially every gene in the genome is targeted to a specific stage of the parasite development. This precise control of gene expression ultimately governs critical functional processes for Plasmodium species to thrive within the host erythrocytes (Bozdech et al., 2003, Le Roch et al., 2003). We have previously reported that orthologous expression between two human Plasmodium species, Plasmodium falciparum and Plasmodium vivax, showed transcriptional divergence in 30% of the genes, some of which could be linked to known functional differences between these two species (Bozdech et al., 2008). This suggested that while there is high conservation of the overall pattern of genome activity during the IDC of the two Plasmodium species, there is transcriptional variation of a subset of genes that enable the parasites to adapt to the individual host niches presumably defined by variations in nutrients, metabolites, as well as other cellular components (Kafsack and Llinas, 2010, Srivastava et al., 2015). However, no studies so far have addressed the extent of transcriptional diversity of Plasmodium species from distinct host erythrocytes. Here, we developed a comprehensive analysis of the IDC transcriptional profiles; from uniform data processing to extensive orthology annotation, which allow us to directly compare variations in mRNA abundance across six different Plasmodium species and significantly extends previous work using pairwise comparison (Bozdech et al., 2008). The Plasmodium conserved syntenic orthologs show significant transcriptional divergence at the earliest stages of parasite development, with transcriptional phylogeny pattern that strictly follows the Plasmodium species mammalian hosts. Furthermore, changes in the expression of key putative transcriptional regulators are implicated in the transcriptional diversity. Critically the work also provides the tools for the identification of new and so far uncharacterized drug targets, as the orthologs that show a conserved transcription pattern across the species are likely to carry out critical conserved functions in all Plasmodium species.
2. Materials and methods
2.1. Sample collection for microarray analysis
2.1.1. Rodent malaria parasites
All studies involving mice were approved by the institutional animal care and use committee (IACUC) of the Nanyang Technological University, Singapore. Male BALB/c mice 6–7 weeks old, bred specific pathogen free (SPF) at the Nanyang Technological University Animal Resource Facility, were infected with either cryopreserved stocks of parasites or by syringe passage from a pre-existing infected mouse. Mice were infected by intraperitoneal injections of Plasmodium berghei ANKA, Plasmodium chabaudi AS or Plasmodium yoelii 17 × parasitized erythrocytes and parasitaemia and parasite stages were monitored by thin blood smears stained with Giemsa. For P. berghei and P. yoelii infection, mice were terminal bleed and the stage-specific parasitized erythrocytes were separated via Nycodenz density gradient. The ring stage interface was isolated, washed and subjected to ex vivo culture, which was then collected every 2 hr over the course of 24 hr over a complete IDC life-cycle. Mice infected with P. chabaudi were terminal bled every 2 hr under anesthesia over the course of 24 hr. Blood was collected and filtered through Plasmodipur filters (Eurodiagnostica, Netherlands) to remove white blood cells and then washed with PBS. The washed blood was flash-frozen in liquid nitrogen and stored at − 80 °C until further use.
2.2. RNA extraction, cDNA preparation and DNA microarray hybridization
Total RNA was isolated using a standard protocol using trizol/chloroform extraction as described by (Bozdech et al., 2003). For preparation of the target DNA for microarray hybridization, Switch Mechanism at the 5′ end of Reverse Transcription (SMART) PCR approach was employed (Petalidis et al., 2003). Thereafter, cDNA was synthesized by using the reverse transcriptase (PowerScript, Clontech BD) for 2 hr at 42 °C. This was followed by PCR amplification with Taq Polymerase (NEB) and the resulting PCR product was purified using MiniElute DNA purification kit (Qiagen). The purified DNA was labeled with fluorescent dye Cy5 (Amersham). A reference pool comprising of equal mass of total RNA samples representing all developmental stages of the parasite was prepared and labeled with Cy3 (Amersham). The microarray hybridization was carried at 65 °C in the automated hybridization station (MAUI, USA). In these two channels competitive hybridizations, RNA from each time point was labeled by Cy5 and hybridized against a reference RNA pool labeled with Cy3. Data acquired were analyzed by GenePix Pro software (Axon Instruments USA).
2.3. Microarray data processing and analysis
2.3.1. Reannotation of oligos
All the oligonucleotides used in this study for the rodent malaria parasites microarray were designed by OligoRankPick as previously described (Hu et al., 2007). The rodent malaria parasites microarray contained 13,224 60-mer oligos. The unprocessed microarray hybridization spots for P. falciparum, P. vivax and P. knowlesi were derived from previously published data; P. falciparum Dd2 strain (Foth et al., 2011), P. vivax smru1 strain (Bozdech et al., 2008) and P. knowlesi PkHa strain (Lapp et al., 2015). The oligonucleotides used in the current and previous microarray studies were blasted against the genomes of the respective Plasmodium species from PlasmoDB release 8.2. As a result, a total of 5276 P. falciparum genes, 4700 P. knowlesi genes, 5017 P. vivax genes, 6251 P. yoelii genes, 4486 P. chabaudi genes and 4401 P. berghei genes were uniquely represented on the corresponding microarray datasets.
2.3.2. Normalize and data filtering
All microarray hybridization spots obtained from all six Plasmodium species were subjected to “normexp” background correction followed by LOWESS (locally weighted scatterplot smoothing) normalization within each array and quantile normalization between arrays using Limma package of R. Log2 ratios of Cy5 over Cy3 intensities were calculated for each spot to represent expression value of a particular probe except those with signal intensity < 1.5 times the background intensity for both Cy5 and Cy3 fluorescence. For each gene, the expression value was estimated as the average of all probes representing it. Overall, 4750 (90% of genes designed on the microarray) P. falciparum genes, 4670 (99%) P. knowlesi genes, 4884 (97%) P. vivax genes, 5486 (88%) P. yoelii genes, 3990 (89%) P. chabaudi genes and 3787 (86%) P. berghei genes display expression profiles with one or zero missing value across each IDC life cycle. These “processed” microarray expression dataset was used for subsequent analysis. The raw and processed microarray data for P. yoelii, P. berghei and P. chabaudi have been deposited into Gene Expression Omnibus (GEO accession number: GSE80015).
2.3.3. Phaseogram
The expression profile of each gene is modeled using sine function which has been described in detail (Lapp et al., 2015). Briefly, the formula is E(t) = A × sin (ωt - α) + C.
where E(t) is the log2 ratio sample/reference control at the t time point of sample collection, A is the amplitude of expression profile across life cycle, C is the vertical offset of profile from zero, ω is the angular frequency given by 2π/h and h is the length of a complete IDC duration expressed in hours (details see below), and α is the horizontal offset of profile from zero. α was projected into an interval ranging from 0 to 2π with, π/2 of α representing the peak expression at early ring stage matching to 0. The converted α was subsequently used as phaseogram (Ph) to indicate the IDC timing where gene expression peaks. Ph for each gene was sorted from early to late IDC for complete visualization of genes expressed in the complete IDC.
2.3.4. Estimation of IDC duration and adjustment of phaseogram
The timing for complete IDC varies between the six Plasmodium species and the time points collected for each sample were different. Therefore, h, which is the length of complete IDC duration was optimized and projected within 44 hr to 54 hr for P. falciparum, P. vivax and 20 hr to 36 hr for P. knowlesi and the other three rodent strains to determine the best fit sine function model (Lapp et al., 2015). As a result, the optimized h is 48 hr for P. falciparum, 49 hr for P. vivax, 29 hr for P. knowlesi, 27 hr for P. yoelii, 28 hr for P. chabaudi genes and 29 hr P. berghei. To minimize the effect of asynchronous parasites between species samples, the phaseogram was adjusted for P. vivax, P. knowlesi, P. yoelii, P. chabaudi and P. berghei to P. falciparum reference. For example, for P. vivax phaseogram adjustment, the best matching time points with P. falciparum for each P. vivax time point was estimated by the highest Spearman Rank Correlation Coefficient (SRCC) values between global transcription profiles of syntenic orthologs. Next, the average shift of time using those best matching time point pairs with SRCC < 0.2 was calculated, which resulted in 25 time points for each gene per species on a sine wave model.
2.3.5. Delta phase, ΔPh
The dissimilarity of transcription profiles between two genes is given by:
Where ∆ Ph is the distance of Ph expression timing between gene a and gene b, Pha and Phb is the expression timing of gene a and b respectively.
2.3.6. Medoid gene and Ds value
The metric of transcription divergence among multiple genes, given by Ds, is the average ∆ Ph of each gene to the medoid gene. K-medoids clustering methods was applied, where K equal to 1, and took the genes in question as a cluster. The medoid gene of this cluster was determined based on the dissimilarity matrix, which contained ∆ Ph of all gene pairs. Ds value is the average dissimilarity of this cluster based on its medoid. Calculation was conducted using the package ‘cluster’ of R.
2.3.7. Detection of outlier gene and species
For a syntenic orthologs, outlier of one species is defined as the ortholog gene with the most divergent expression timing or Ph compare to the other species member within an ortholog group. To detect the outlier for each ortholog group, we construct the dissimilarity matrix of species based on ∆ Ph. Outlier is define as the ortholog gene of a particular species which maximally contributes to the sum of ∆ Ph of that dissimilarity matrix.
2.3.8. Dendrogram of transcriptome relationship
Dendrogram was constructed by applying Ward hierarchical clustering method based on the dissimilarity matrix containing the distance of each pair of species, ∆ Ph. The distance of two species is defined as;
where D(A,B) is the distance of species A and B, n is the total number of syntenic orthologous gene which is equal to 2312 and ∆ Phi (A,B) if the distance of expression timing between the ith gene of species A and its orthologous gene in species B. The dendrogram cluster was subjected to 100 times bootstrapping to estimate the percentage of approximately unbiased (AU) p-value and bootstrap probability (BP).
2.3.9. Ka/Ks ratio
Ka/Ks ratio is the ratio of the number of nonsynonymous substitutions per non-synonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks). Selective pressure of protein-coding genes between six Plasmodium species was estimated by calculating Ka/Ks ratio for each pair of syntenic orthologs for a total of 2312 orthologs. The syntenic orthologs were aligned using ClustalW (Larkin et al., 2007) and Ka/Ks were calculated using package ‘seqinr’ of R (Charif and Lobry, 2007). Dendrogram indicating evolution relationship between species was constructed based on the dissimilarity matrix in which the distance of two species was defined by the mode of Ka values.
2.3.10. Functional enrichment analysis
Functional enrichment analysis was carried out by identifying syntenic orthologs sets that are significantly over-represented in studied genes list by comparing to annotated gene sets from P. falciparum MPM database. Hypergeometric test was applied to calculate the level of significance of indicated orthologous gene sets from the MPM database.
3. Results
3.1. Lack of conservation of specific ortholog expression in Plasmodium species' life cycle
Despite the long evolutionary time scale of about 100 million years (Escalante and Ayala, 1995, Escalante and Ayala, 1994), the overall genome organization and content across the sequenced Plasmodium species is highly conserved, with about 4000 conserved syntenic genes located within the central core regions of the 14 chromosomes (Hall et al., 2002, Carlton et al., 2008a, Pain et al., 2008, Gardner et al., 2002, Hall et al., 2005). On the other hand, species-specific differences are attributed to the variant multigene families that reside predominantly at the subtelomeric regions. In this study, we generated the IDC transcriptomes for the rodent malaria parasites P. berghei, P. yoelii and P. chabaudi and compared these with the previously generated data for the human malaria species P. falciparum (Foth et al., 2011), P. vivax (Bozdech et al., 2008), and the simian malaria species P. knowlesi (Lapp et al., 2015) (Supplementary Table 1, Supplementary Fig. 1a). The IDC transcriptional profiles of each gene were first best fitted onto a sine-wave function as described in the experimental procedure section (Fig. 1a, Supplementary Fig. 1b). This allowed us to define several parameters that reflect the temporal aspects of gene expression and thus make direct species-to-species comparisons. The quality of the newly generated data was further verified by comparisons with the recently published P. berghei RNA-seq data showing high levels of correlation (Supplementary Fig. 1c) (Otto et al., 2014). Overall, 3787 to 5486 genes (including species-specific genes) exhibited specific IDC transcriptional profiles in the six Plasmodium species (Table 1). In contrast, between 29 and 1436 genes are unique to individual species with specific transcriptional profiles (Table 1, Supplementary Fig. 1d), where the majority of them are genes restricted to subtelomeric ends and are involved in antigenic variation, host-parasite interactions, cytoadherence and erythrocyte aggregation for all six species, similar to previous genomic studies (Kooij et al., 2005, Kyes et al., 2001, Dzikowski et al., 2006, Galinski and Corredor, 2004). Interestingly, while the distribution of peak time expression remains relatively constant for the syntenic orthologs, the species-specific genes show remarkable variability (Supplementary Fig. 1d).
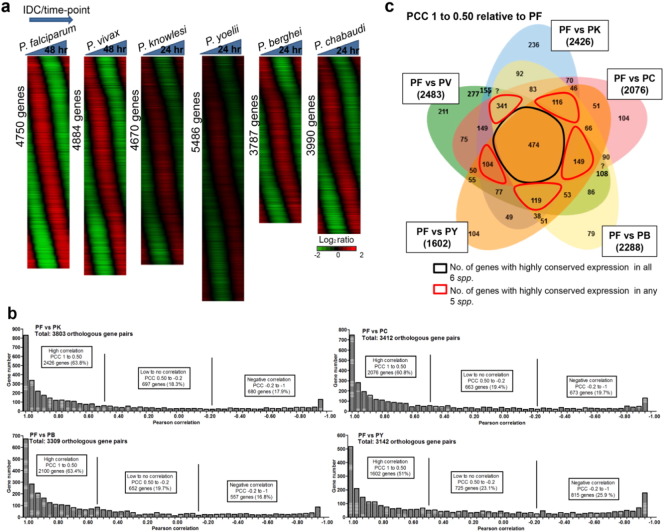
Fig. 1.
Smoothed transcriptome and comparative correlation transcriptomic analyses of P. falciparum, P. vivax, P. knowlesi, P. berghei, P. yoelii and P. chabaudi.
(a) Overall intraerythrocytic developmental cycle (IDC) transcriptome profiling for P. falciparum (4750 genes), P. vivax (4884 genes), P. knowlesi (4670 genes), P. yoelii (5486 genes), P. berghei (3787 genes), and P. chabaudi (3990 genes). The phaseograms were generated from the log2 expression ratio and each profile was median centered. The phaseograms also include the expression of 2312 syntenic orthologous genes present in all 6 Plasmodium species. (b) Histograms showed overall distribution of Pearson correlation coefficients (PCCs) calculated from the smoothed transcriptome dataset between P. knowlesi, P. chabaudi, P. berghei and P. yoelii using P. falciparum as reference species. (c) Venn diagram analysis of co-expressed genes with PCC scores of ≥ 0.5–1.00 in different Plasmodium species. The numbers in brackets beside each species pairs (Pf vs Pv, Pf vs Pk, Pf vs Pb, Pf vs Pc, Pf vs Py) represent total number of genes with PCCs of ≥ 0.50. The numbers inside the Venn diagram represent total number of overlapped orthologous genes between the species pair.
Table 1.
Summary table of number of genes that are syntenic orthologs and with specific transcriptional profile across all six Plasmodium species.
| Microarray data analysis | ||||||
|---|---|---|---|---|---|---|
| Pf | Pk | Pv | Py | Pb | Pc | |
| Reannotation of oligos for microarray data analysis | 5276 | 4700 | 5017 | 6251 | 4401 | 4486 |
| After data normalization and filtering (related to Fig. 1) | 4750 | 4670 | 4884 | 5486 | 3787 | 3990 |
| Syntenic orthologs and species-species specific genes | ||||||
| Syntenic orthologs present in at least one or more species (OrthoMCL DB) | 4148 | 4286 | 4275 | 3577 | 3708 | 3783 |
| Syntenic orthologs present in all six species (OrthoMCL DB) | 3374 | 3374 | 3374 | 3374 | 3374 | 3374 |
| Syntenic orthologs present in at least one or more species with specific transcriptional profile | 3175 | 3182 | 3190 | 3161 | 2951 | 3025 |
| Syntenic orthologs present in all six species with specific transcriptional profile | 2312 | 2312 | 2312 | 2312 | 2312 | 2312 |
| Species-specific genes with specific transcriptional profile (non-orthologs, non-syntenic) | 84 | 245 | 457 | 1436 | 29 | 136 |
Previous studies using Pearson correlation coefficient (PCC) to compare P. falciparum to P. vivax had indicated that PCC between 1 and 0.5 represent strongly correlated pattern, 0.5 to − 0.2 represent a moderate timing shift in gene expression, while PCC < − 0.2 indicate dramatic changes of gene expression along the IDC (Bozdech et al., 2008) (Supplementary Fig. 2). Using the same approach, we compared all orthologous gene pairs with P. falciparum as the reference (Pf-Pk, Pf-Pc, Pf-Pb and Pf-Py) and showed high correlations between 50 and 65%, intermediate to low correlations for 18% to 23% and anti-correlation for 18% to 26% (Fig. 1b). P. yoelii appears most divergent from P. falciparum with 725 (23.1%) and 815 (25.9%) gene pairs with low and anti-correlations, respectively (Fig. 1b). The overall distribution of highly correlated orthologous genes (PCC 1–0.50) showed only 474 genes conserved among the six species (Fig. 1c). Even when using any group of five out of the six species the total number of conserved genes would increase by only 104 to 341 genes (Fig. 1c). This suggests that only a relatively small subset of genes remained transcriptionally conserved throughout Plasmodium evolution while the majority underwent (some level of) transcriptional diversification. The results also indicate a relatively limited constraint on transcriptional timing across the syntenic orthologs providing a considerable flexibility for species-specific adaptation among Plasmodium species.
3.2. Pattern of evolution in Plasmodium transcriptional regulation
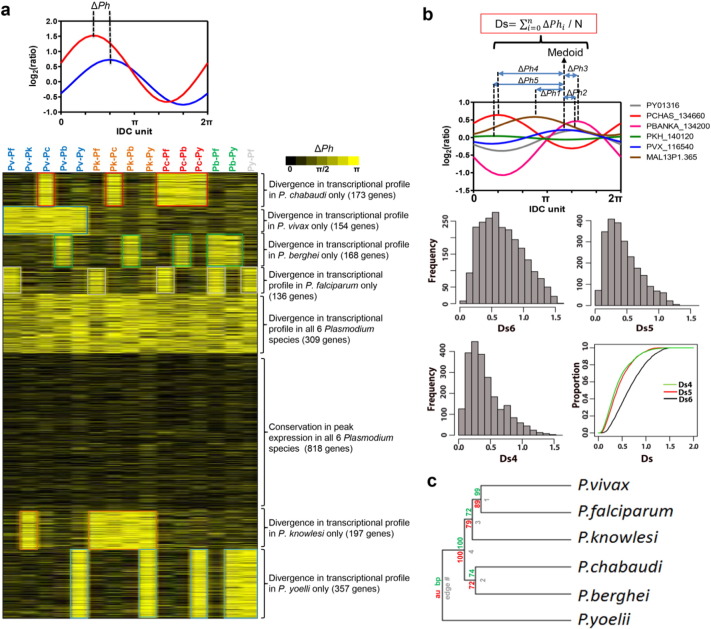
To compare the temporal character of expression between orthologous genes, we define the time-shift (delta phase, ∆ Ph) relationship between the ortholog pairs derived from their IDC gene expression profiles. ∆ Ph between the species' orthologs for each gene expression profile was first projected onto a polar coordinate system with timing of gene expression being depicted by values ranging from 0 to 2π (Supplementary Fig. 3). Phase adjustment, Ph was performed by offsetting the peak of gene expression timing to the peak gene expression time point at the early ring-stage of development (Bozdech et al., 2008) (Supplementary Table 2). This method takes advantage of a uniform periodic character of the gene expression profiles that allows much more efficient and representative normalization of gene expression profiles compared to PCC-based similarity or Euclidean distance tests generally used to study other organisms (Pereira et al., 2009, Glazko and Mushegian, 2010). In total, we established ∆ Ph for the 2312 syntenic ortholog groups with IDC expression datasets for all six Plasmodium species. K-means clustering for the ∆ Ph values revealed eight distinct clusters that represent the overall evolutionary pattern of transcriptional regulation in Plasmodium (Fig. 2a). The eight clusters consist of gene groups that are (i.) fully conserved in all six species; (ii.) show no similarity in gene expression in any species pair; (iii.) genes with diverse expression in one of the species while similar in the other five. In particular, (i) there are 818 genes whose transcriptional timing have been maintained throughout evolution (Fig. 2a), representing 35.4% of the 2312 syntenic orthologs. This transcriptionally conservation suggests a crucial role of these genes in Plasmodium biology. On the other hand, (ii) there are 309 genes that are completely devoid of any transcriptional conservation among the Plasmodium species. Although the role(s) of these genes is generally unknown, this large diversity suggests their involvement in highly dynamic evolutionary processes in Plasmodium. Finally, (iii) there are six clusters ranging from 136 to 357 genes, in which one Plasmodium species shows a diversion from the other five (Fig. 2a). To further clarify these findings we calculated the overall transcriptional divergence (Ds score) among orthologs as the average ∆ Ph between each species gene to the medoid species gene (Fig. 2b). When including all 6 species, about 50% of the ortholog groups have a Ds6 value of < 0.75 (conserved transcription). This can be significantly increased to approximately 80% if one (Ds5) or two (Ds4) outlier gene(s) with the most divergent Ph are removed (Fig. 2b, Supplementary Table 2). This supports the model that the majority of transcriptional diversion occurred in individual single species diverting gene expression from a putative ancestral transcriptional profile. Finally, the ∆ Ph distance measure separates the Plasmodium species into two distinct clades that delineates precisely along the mammalian host species (Fig. 2c). This suggests that transcriptome-based phylogeny is shaped by the adaptation of the different Plasmodium species in their respective mammalian host erythrocytes; the human P. falciparum and P. vivax, the rhesus macaque P. knowlesi and the rodent Plasmodium species.
Fig. 2.
Delta phase (ΔPh) and Ds as the measurement of transcriptional divergence.
(a) ΔPh is calculated based on the absolute difference of Ph or peak in gene expression timing between any two orthologs in two different species. Heatmap shows the syntenic orthologs with K-means clustering (100 runs) of ΔPh of species pair P. vivax; Pv, P. knowlesi; Pk, P. chabaudi; Pc, P. berghei; Pb and P. yoelii; Py with reference to P. falciparum; Pf. Yellow represents highly divergent genes with large delta phase, while black represents highly conserved genes with small delta phase compare to P. falciparum. (b) Ds score is the average ΔPh between each orthologs to the medoid ortholog measuring the transcription divergence of a gene across multiple species (see method). Overall frequency distribution and cumulative proportion of Ds measured from 2312 syntenic orthologous genes present in all six Plasmodium species (Ds6), any five Plasmodium species (Ds5) or any four Plasmodium species (Ds4). (c) Hierarchical clustering of the six Plasmodium species based on the IDC phaseogram using Wards algorithm of clustering and dissimilarity matrix defined by ∆ Ph (see methods). Numbers adjacent to the branch points are percentage of approximately unbiased (AU), P-value (in red) and bootstrap probability (BP) (in green).
3.3. Sequence and transcription-based evolution in Plasmodium species
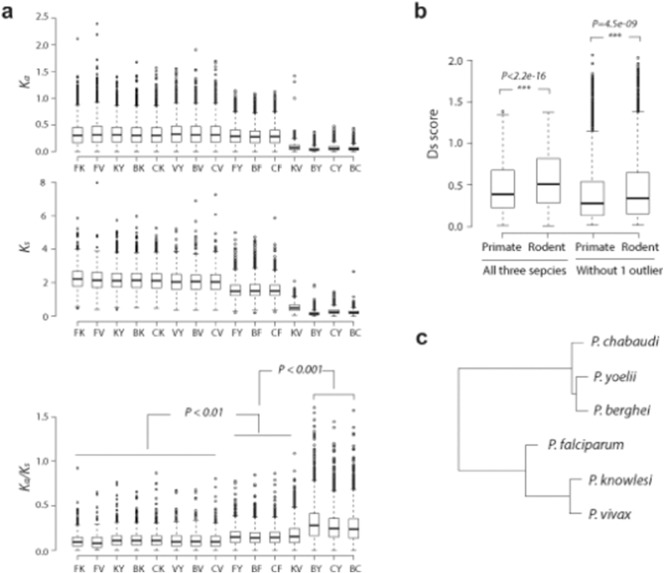
In Plasmodium, diversity of genomic sequence is considered to be a major factor of speciation, in particular involving species-specific gene families encoding of factors of host-parasite interaction (Frech and Chen, 2011). In the next step we wished to evaluate the evolutionary role of transcription in the context of genome sequence diversion. For that, we first estimated the number of non-synonymous substitutions per non-synonymous site (Ka) and the number of synonymous substitutions per synonymous site (Ks) for the syntenic orthologous gene groups for each species pair (Fig. 3a). The distribution of Ka/Ks ratio reflects the level of selection pressure on protein-coding genes throughout the evolution of Plasmodium parasites species (Fig. 3a), with the rodent malaria parasites showing a greater difference in the change of protein-coding genes within the species group. One of the speculated factors of the rodent malaria parasites evolution is the geographical aspect of adaptation to their hosts that subsequently led to significant changes in the coding regions within the rodent species compared to their primate counterparts. Accordingly, the overall transcriptome in the rodent Plasmodium species exhibits a greater divergence compared to the primate-infecting species (Fig. 3b). This suggests broader diversification of the rodent-infecting species driven by both sequence diversity and transcriptional divergence. We next assembled a dendrogram based on non-synonymous Ka ratio, which reflects the phylogenetic relationships among the Plasmodium species based on the sequence homology. Here, P. vivax and P. knowlesi are more closely related than P. falciparum and P. berghei, which forms a separate cluster together with P. yoelii and slightly more distant P. chabaudi (Fig. 3c). This is consistent with the previously constructed phylogenetic topology based on partial mitochondrial genomes (Escalante et al., 1998, Carlton et al., 2008b) as well as genes encoding surface antigens (Weedall et al., 2008). The sequence-based phylogenetic tree(s) are different from the transcriptional differences that is reflective of its mammalian host lineage (as mentioned above) (Fig. 2c). The sequence-based relationships seem to be driven by other (than host) factors such as AT content of the genome (with P. falciparum having a much higher AT content than P. knowlesi and P. vivax) and possibly reticulocyte preferences (Craig et al., 2012).
Fig. 3.
Conservation and diversification of syntenic orthologs at the coding sequence and transcription levels.
(a) Non-synonymous (Ka) and synonymous (Ks) rates and their ratios (Ka/Ks) were calculated for 2312 syntenic orthologous gene, between cross-species gene pairs. F represents for P. falciparum, V for P. vivax, K for P. knowlesi, Y for P. yoelii, C for P. chabaudi and B for P. berghei. (b) Distribution of Ds scores of primate Plasmodium species (PF, PK and PV) and rodent Plasmodium species (PY, PB and PC) in one category of all three species and one category of two species without one outlier. (c) Unrooted tree constructed using mode values of Ka as the distance metric (details see Materials and methods). Statistical significance of differences was measured using Mann-Whitney test.
3.4. Evolutionary divergence and conservation of the Plasmodium transcriptome occurs at specific IDC timing and within selected functional pathways
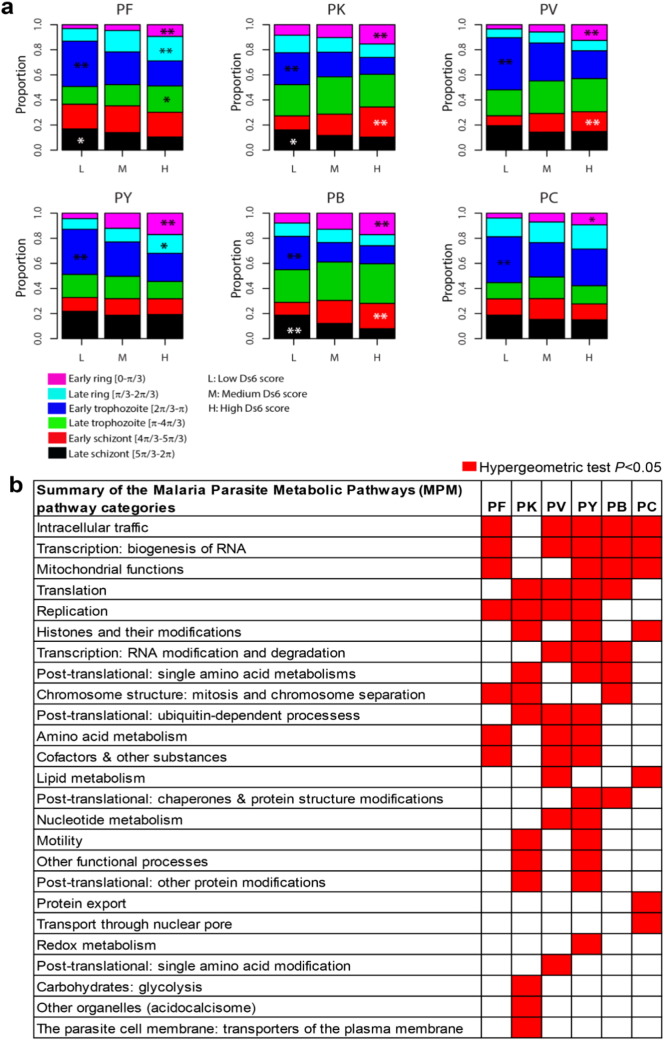
We examined IDC progression to evaluate whether particular stages are more prone to transcriptional conservation/diversity. We analyzed the Ds6 values as a function of gene expression timing, where Ds6 were equally divided into low [0- π/6, medium [π/6- π/3) and high values [π/3- π/2) and segregated equally into six IDC unit time points from 0 to 2π (in a range of π/3) according to the peak of gene expression corresponding to the individual IDC stages (Fig. 4a). Genes whose expression peaks at the early ring stage for any of the Plasmodium species showed significantly higher Ds (P < 0.001). This implies that corresponding orthologs are likely to be expressed at different IDC stages in other species (Fig. 4a). In contrast, the late ring and early trophozoite stages express genes with overall low Ds (P < 0.001), which suggest that in other species these genes are expressed at a similar IDC stage. Genes whose expression peaks at the late trophozoite, early schizont and late schizont stages showed variable enrichment in conservation and divergence across the species (Fig. 4a). For instance, genes expressed at the late schizont stage are only significantly enriched with low Ds in P. falciparum (P < 0.01), P. knowlesi (P < 0.01), P. vivax (P < 0.05), and P. berghei (P < 0.001). The early developmental stage (rings) exhibit the highest diversity, the mid stage (late ring trophozoite) appear to be the most constrained and the late stage (schizonts) are characterized by intermediate levels of diversion, demonstrating that transcriptional conservation of Plasmodium genes is not uniform throughout the IDC. To investigate the biological significance of transcriptional conservation in Plasmodium, we carried out pathway enrichment analyses of the orthologs with an “outlier” in one Plasmodium species; where the gene is expressed at a different time compared to the others (Fig. 2b, Supplementary Table 3, hypergeometric test P < 0.05). Here we find that basic cellular and biochemical pathways such as intracellular trafficking, transcription, translation, mitochondrial functions and DNA replication tend to have genes with an outlier-like expression (Fig. 4b). Interestingly, while each overrepresentation includes the outlier genes within an individual species, these cellular pathways are overrepresented across multiple species. This suggests that each species transcriptionally diversified a different set of genes in the otherwise overlapping pathways. It is feasible to speculate that these genes represent rate limiting steps of these biological processes that facilitate temporal shifts of their overall function. On the other hand there are several pathways with significantly diversified orthologs that are exclusive to one species including protein export (P. chabaudi), redox metabolism (P. yoelii), glycolysis and protein transport (P. knowlesi) and post-translational modifications (P. vivax) (Fig. 4b). This exclusivity may indicate the importance of these pathways for the evolutionary adaptation.
Fig. 4.
Variability of the Plasmodium transcriptome during IDC and enrichment of functional pathways in outlier species.
(a) Temporal expression divergence of the Plasmodium transcriptome during IDC. Proportion of genes with low (L), medium (M) and high (H) Ds value in P. falciparum; PV, P. knowlesi; PK, P. vivax; PV, P. yoelii; PY, P. berghei; PB and P. chabaudi; PC. Peak in gene expression timing, Ph for each gene was bin in range of π/3 from 0 (early ring) to 2π (late schizont). Proportion was calculated based on the number of genes within each Ph range over the total number of genes with H, M, or L Ds value. Significance of association between Ds and Ph proportion for each IDC range were analyzed using Chi-square test (*P < 0.01, **P < 0.001). (b) Summary of functionally significant MPM pathways in outlier genes/species. Orthologs of species with the most divergent expression timing, Ph or outlier species (see Materials and methods) are subjected to pathway enrichment analysis using hypergeometric distribution function. Red-colored panel indicates significant MPM pathway with altered transcriptome profile within each species group with hypergeometric test P < 0.05.
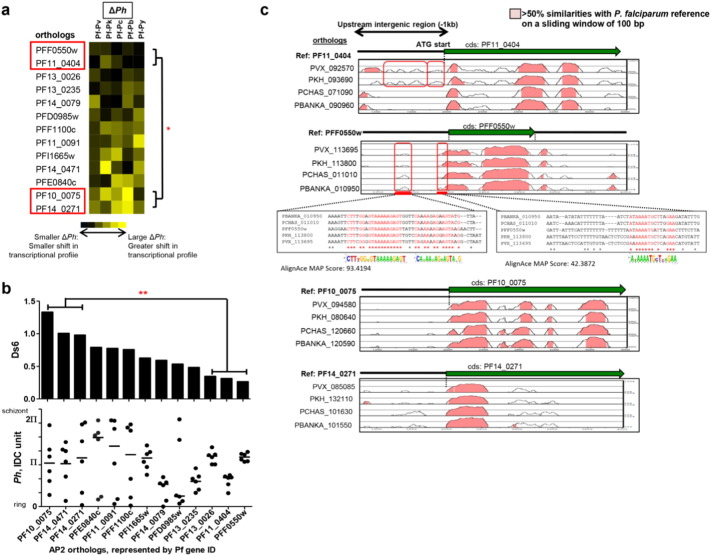
3.5. Transcription conservation and divergence of the AP2 transcriptional regulators and its cis-regulatory region
To better understand the possible mechanism(s) that drive transcriptional divergence, we analyzed the transcriptional profile of the 26 putative and known AP2 transcriptional regulators in all six species (Painter et al., 2011). Of these 26 AP2 genes, only 19 are present as syntenic orthologs in all six species, whereas the remaining 7 AP2 genes have orthologs in between 3 and 5 species. Of the 19 syntenic AP2 orthologs, 13 have transcriptional profiles that were used for subsequent Ds6 analysis (Fig. 5). ΔPh measurement and matched Ds revealed that the 13 AP2 transcriptional profiles differed across the species (Fig. 5a and b). Represented by P. falciparum IDs, the expression of PFF0500w, PF11_0404 and PF13_0026 AP2 orthologs are significantly more conserved in contrast to PF10_0075, PF14_0471 and PF14_0271 (Fig. 5b). Furthermore, AP2 orthologs with high Ds values (PF10_0075, PF14_0471 and PF14_0271) showed more variable stage-specific expression than those with low Ds values (PFF0500w, PF11_0404 and PF13_0026) that were predominantly expressed at the trophozoite stage (Fig. 5b). The conserved temporal expression pattern for a subset of AP2 implies the possibility of similar transcriptional control (either activation or repression) on downstream regulatory events among the Plasmodium species. Four AP2 orthologs were subjected to preliminary bioinformatics analysis on the conservation of cis-regulatory regions within the promoters of AP2 orthologs in different Plasmodium species. Phylogenetic footprint analysis on the AP2 promoters was performed to estimate the percentage of best-matching regions within the 1000 bp nucleotide frame (Janky and van Helden, 2008) (Fig. 5c). Using P. falciparum as the reference, the alignment analysis revealed variable patterns and percentages of sequence conservation upstream of the ATG start sites (Fig. 5c). Genes with greater transcriptional conservation across species (PFF0550w and PF11_0404) tend to have greater conservation at the upstream non-coding sequence, while the transcriptionally diverged genes (PF10_0075 and PF14_0271) showed a low level of conservation (Fig. 5c). In particular, PFF0550w orthologs displayed a clear conserved, interspecies “footprint” at the promoter intergenic region, about − 500 and − 100 bp upstream of the ATG start site. This region may contain putative conserved regulatory elements for transcriptional regulation (Chen and Jiang, 2006). In contrast limited conservation of regulatory sequences indicates transcriptional divergence across species.
Fig. 5.
Comparative analyses of AP2 expression profiles and sequence alignment across six Plasmodium species.
(a) Heatmap shows ΔPh profile for 13 AP2 syntenic orthologs present in all 6 Plasmodium species. Statistical significance of differences was measured between the ΔPh of the highly divergent and conserve AP2 orthologs expression, highlighted with red box, using one-way ANOVA analysis (*P < 0.05). (b) Barplot and scatterplot (median value indicated by black line) show Ds6 and Ph respectively for the 13 AP2 syntenic orthologs. Statistical significance of differences was measured between Ds6 of the highly divergent and conserve AP2 orthologs expression using nonparametric t-test (**P < 0.01). (c) Alignment plot of coding region and -1000 bp upstream region from the ATG start site of four AP2 with the most conserved and diverged gene expression using mLAGAN and wAlignAce. Cutoff of > 50% similarities are highlighted in pink. AP2 orthologs are represented by P. falciparum gene ID on the top-left of each alignment plot.
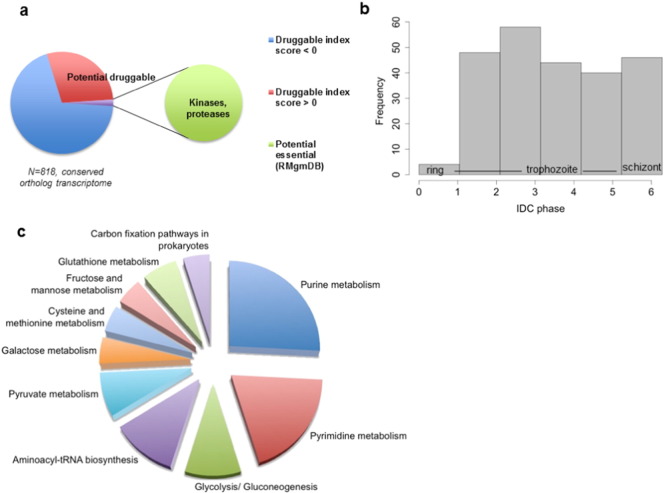
3.6. Druggability prediction of the conserved ortholog transcriptome
Our analysis thus far implies that the different Plasmodium species have evolved to conserve a subset of transcriptional responses with analogous cellular functions and pathways, suggesting a possible selective advantage required for the parasites' development and survival within the host cell. Considering the relatively low number of genes that are transcriptionally conserved across Plasmodium species where about 818 orthologs are transcriptionally conserved (Fig. 2), those that show a conserved transcription pattern would be expected to carry out critical time dependent functions within the parasite and host cells. Drug target prioritization databases from online resources such as Tropical Disease and Research (TDR) v5 (Magarinos et al., 2012, Aguero et al., 2008) short-listed > 30% (240 genes) of the transcriptionally conserved orthologs are potentially druggable with druggability index score (DIS) > 0, where almost one-third (91 genes) was not assign to any putative function (Fig. 6a, Supplementary Table 4). 17 of the transcriptionally conserved, potentially druggable orthologs including a group of kinases and proteases were found to be essential in P. berghei from the rodent orthologs database (RMgmDB) (Janse et al., 2011) (Fig. 6a, Supplementary Table 4). The majority of these potentially druggable orthologs are expressed from trophozoite to schizont stages in all six species (Fig. 6b). Metabolic pathway enrichment assessments suggest that these orthologs are involved in nucleotide metabolism, energy dependent processes, tRNA biosynthesis and carbohydrate metabolism (Fig. 6c) supporting a conserved expression for selected functional pathways, which may be relevant for core essential processes across species. Furthermore, 13 of the transcriptionally conserved, druggable orthologs have been characterized and pharmacologically validated as antimalarial drug targets in blood stage P. falciparum and P. vivax orthologs according to TDR databases (Table 2). This includes the PfFKBP35 petidyl-propyl isomerase encoded by PFL2275c (Harikishore et al., 2013), PfPK5 or cyclin dependent protein kinase 5 encoded by MAL13P1.279 (Harmse et al., 2001), tubulin beta chain encoded by PF10_0084 (Fennell et al., 2006), and glycogen synthase kinase 3 encoded by PFC0525c (Droucheau et al., 2004), all which inhibition of function affected the cellular division of the parasites. Moreover, the conserved blood stage-specific expression of this particular group of orthologs suggests there is potential cross-species anti-malarial targets, in particular against P. vivax and P. falciparum both, the prominent etiological agent of human malaria.
Fig. 6.
Druggability prediction from the conserved orthologs transcriptome.
(a) Proportion of transcriptionally conserved orthologs with positive druggability index score and essentiality data from RMgmDB database. (b) Overall frequency distribution of peak in IDC expression timing (phase) and (c) metabolic pathway enrichment analysis (PlasmoDB) of 240 genes that are both transcriptionally conserved and with positive druggability evidence index score from TDR database.
Table 2.
List of 13 P. falciparum transcriptionally conserved orthologs obtained from TDR databases with pharmacological validation as antimalarial drug targets. Target orthologs are represented by P. falciparum gene ID and its corresponding gene products, predicted transmembrane (tmd) count, signal peptide and druggability index score.
| Gene ID | Gene product | Gene tmd count | Gene signal peptide | Druggability evidence index |
|---|---|---|---|---|
| PF10_0084 | tubulin beta chain, putative | 0 | N | 1 |
| PFC0525c | glycogen synthase kinase 3 | 0 | N | 0.9 |
| MAL13P1.279 | protein kinase 5 | 0 | N | 0.9 |
| PFE1050w | adenosylhomocysteinase(S-adenosyl-l-homocystein e hydrolase) | 0 | N | 0.8 |
| PFI0380c | formylmethionine deformylase, putative | 1 | Y | 0.8 |
| PFL1370w | NIMA-related protein kinase, Pfnek-1 | 0 | N | 0.7 |
| PFL2275c | FK506-binding protein (FKBP)-type peptidyl-propyl isomerase | 0 | N | 0.6 |
| PF14_0641 | 1-deoxy-d-xylulose 5-phosphate reductoisomerase | 0 | N | 0.6 |
| PF10_0086 | adenylate kinase | 0 | N | 0.6 |
| PF11_0282 | deoxyuridine 5′-triphosphate nucleotidohydrolase, putative | 0 | N | 0.6 |
| PFI0925w | gamma-glutamylcysteine synthetase | 0 | N | 0.6 |
| PF10_0123 | GMP synthetase | 0 | N | 0.6 |
| PF11_0301 | spermidine synthase | 0 | N | 0.6 |
4. Discussion
Comparative analysis of large-scale genome wide expression datasets across multiple species, especially time-series expression datasets, was often complicated by several parameters such as oligonucleotide probe affinity, background noises and heterogeneity of samples from different organisms (Kuo et al., 2006). In this study, microarray analysis was performed on Plasmodium oligonucleotides and pan-rodent oligonucleotides designed by OligoRankPick as previously described (Hu et al., 2007, Liew et al., 2010). Taking into account that majority of the genes in P. falciparum, P. vivax and P. knowlesi are expressed in a periodic manner during the IDC stage (Bozdech et al., 2003, Bozdech et al., 2008, Lapp et al., 2015), including the rodent malaria parasite genes, the raw microarray data for each gene from all six species were best-fitted into a sine-wave phaseogram. Phaseogram correction was performed to adjust for the variation in IDC timing among the different Plasmodium species as described in detail elsewhere (Lapp et al., 2015). Nevertheless, the phaseogram correction has its own caveats; not all of the Plasmodium genes will display a distinct periodic expression manner throughout the IDC, particular for very lowly expressed genes, which may exclude their consideration in the analysis of overall transcriptional divergence. Future work using RNA-seq analysis could more effectively assess the contribution that lowly expressed genes make to the overall transcriptional variation. It is interesting to note that for two of the species the IDC microarray data (P. berghei and P. vivax) is highly correlated with the periodic life-cycle transcriptional profiles from RNA-seq studies of the corresponding species (Otto et al., 2014, Zhu et al., 2016), suggesting that a large proportion of the overall gene expression detected by microarray are credible estimates and that only a small fraction of the transcripts are of such low abundance as not to be accurately detected in the microarray. Second, variation in synchrony of the parasites cultures, specifically for the rodent malaria parasites, could potentially contribute to the overall transcriptional variation observed in this study. To minimize such variation, we sampled the Nycodenz ring stage parasitized rodent erythrocytes from different mice infected with either P. berghei or P. yoelii from ex vivo culture, which were then collected for every 2 hr until late schizont stage. A similar ex vivo study was performed for the transcriptome study of P. knowlesi ex vivo culture (Lapp et al., 2015). Furthermore, the complete IDC timing for each species was also adjusted to minimize synchrony issues and sampling time variation, which resulted in 25 time points for each gene. The rigorous effort to minimize these potential limitations, makes us confident that our analysis provides a framework for further hypothesis testing across different malaria parasites.
Multi-gene comparisons provided a robust evolutionary topology of the malaria parasite species by dividing them into several distinct monophyletic clades; the human parasite P. falciparum, a separate clade containing both P. vivax and P. knowlesi, and the rodent parasites clade (Martinsen et al., 2008). In this study, we show that Plasmodium species' phylogeny based on transcriptional divergence contradicts the established sequence based evolutionary model (Perkins and Schall, 2002, Martinsen et al., 2008), suggesting that transcriptional and sequence evolution in Plasmodium species are under different selection pressures. Phenotypic evolution across species can occur at many different levels; from the changes in coding sequences to protein structure and to alteration in the levels and timing of gene and protein expression (Harrison et al., 2012, Carroll, 2008). Moreover, orthologous expression studies from other higher eukaryotes such as humans and mice have highlighted minor selective constraints in the evolution of both species (Su et al., 2002, Yanai et al., 2004). One crucial finding of this study was the observation that particularly the early ring stage, appears to be under the greatest transcriptional divergence. This suggests that host specific variations that the parasite encounters within a newly infected erythrocyte rather than variation of host cell receptors is a key driver of evolution. The high level of transcriptional divergence in the early ring stage, as opposed to other stages of the parasite's development, is intriguing and similar to several other ontogeny studies based on other model organisms such as Drosophila, zebrafish and Arabidopsis, where transcriptional conservation is the highest during the mid-embryonic stage of development (Kalinka et al., 2010, Domazet-Loso and Tautz, 2010, Quint et al., 2012, Irie and Kuratani, 2011). The Plasmodium parasites, like higher multicellular organisms, may undergo initial adaptation responses to the cellular environment through genetic control, in this case within the host erythrocytes at the very early stage of their development.
Our data suggests that rather than coding sequence differences other fundamental features such as transcription factors, upstream promoter regions, or post-transcriptional control such as epigenetic changes, chromatin remodeling events or noncoding RNAs may play a larger part in the modulation of transcription across the Plasmodium species (Coulson et al., 2004, Militello et al., 2004, Van Noort and Huynen, 2006, Templeton et al., 2004, Gupta et al., 2013, Vembar et al., 2014, Ay et al., 2015). The ApiAP2 family of proteins is the largest putative transcriptional regulators identified in Plasmodium species (Balaji et al., 2005) and has been described as the master regulator in both the sexual and asexual stages (Sinha et al., 2014, Kafsack et al., 2014). Our findings here are consistent with changes in AP2 expression driving broader transcriptional changes in the different species. Although the evolutionary role(s) of cis-regulatory elements among the Plasmodium species is relatively unknown as they are not homologous to any known eukaryotic regulatory elements, the significance of variation in cis-regulatory sequences on transcription factor binding and divergent gene expression towards species phenotypic evolution has been largely described in higher eukaryotes (Gompel et al., 2005, Shirangi et al., 2009, Deplancke et al., 2006). The variation in putative cis-regulatory elements in the AP2 promoter regions of different plasmodium species with transcriptional divergence is consistent with recent findings in P. falciparum supporting the role of cis-trans regulatory control events (Russell et al., 2015).
With mounting reports of anti-malarial drug resistance, there is an urgent need to identify new drug targets in the malaria parasite that will lead to the development of new therapeutic compounds to complement the existing repertoire of available drugs. The utility of cross-species transcriptomes for assessment of potential drug targets against common biological processes is recently gaining attention (Okyere et al., 2014, Foth et al., 2014). The conserved orthologous transcriptome of Plasmodium species provides abundant molecular targets, including both annotated and hypothetical gene targets, for further functional studies and target-based approaches to drug discovery. We propose that the transcriptionally conserved orthologs carry out crucial time-dependent functions within parasite development that have yet to be studied and at the same time also represent possible anti-malarial targets for parasite intervention. The common biological processes with conserved IDC transcriptomes are likely to be most critically functional for the parasites, as the different species have strictly maintained these gene expression patterns over the course of evolution. By combining the comparative transcriptome data with web-based resources, we have short-listed a consensus group of potential anti-malarial targets that increases the likelihood of identifying critical functions in parasite biology, and eventually potential anti-malarial drug targets. Ultimately, the comparative transcriptome analysis across multiple species provides important tools and resources for other post-genomic studies and continued assessments of the evolution of Plasmodium parasites.
Author contributions
R.H. and L.Z. analyzed and interpreted the data. L.Z. developed computational algorithms. A.A., O.N., S.M., S.A.L., G.H., and K.L. performed the microarray experiments. M.R.G. and Z.B. reviewed the manuscript critically and provided intellectual content. R.H. and P.R.P. wrote the manuscript with contributions from all authors.
Disclosure
The author(s) declare(s) that there is no conflict of interests.
Acknowledgements and funding sources
This work was supported by the National Medical Research Council (Singapore) grant number NMRC/1292/2011, the US National Institute of Health/National Institute of Allergy and Infectious Diseases, Grant numbers 1R01AI24710 and R01-AI065961, and the Yerkes National Primate Research Center Base Grant (ORIP/OD P51OD011132) awarded by the National Center for Research Resources of the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2016.04.011.
Contributor Information
Zbynek Bozdech, Email: ZBOZDECH@NTU.EDU.SG.
Peter R. Preiser, Email: PRPREISER@NTU.EDU.SG.
Appendix A. Supplementary data
Supplementary material.
References
- Aguero F., Al-Lazikani B., Aslett M., Berriman M., Buckner F.S., Campbell R.K., Carmona S., Carruthers I.M., Chan A.W., Chen F., Crowther G.J., Doyle M.A., Hertz-Fowler C., Hopkins A.L., Mcallister G., Nwaka S., Overington J.P., Pain A., Paolini G.V., Pieper U., Ralph S.A., Riechers A., Roos D.S., Sali A., Shanmugam D., Suzuki T., Van Voorhis W.C., Verlinde C.L. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat. Rev. Drug Discov. 2008;7:900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay F., Bunnik E.M., Varoquaux N., Vert J.P., Noble W.S., Le Roch K.G. Multiple dimensions of epigenetic gene regulation in the malaria parasite Plasmodium falciparum: gene regulation via histone modifications, nucleosome positioning and nuclear architecture in P. falciparum. BioEssays: News Rev. Mol. Cell. Dev. Biol. 2015;37:182–194. doi: 10.1002/bies.201400145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S., Babu M.M., Iyer L.M., Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z., Llinas M., Pulliam B.L., Wong E.D., Zhu J., Derisi J.L. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1 doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z., Mok S., Hu G., Imwong M., Jaidee A., Russell B., Ginsburg H., Nosten F., Day N.P., White N.J., Carlton J.M., Preiser P.R. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.M., Adams J.H., Silva J.C., Bidwell S.L., Lorenzi H., Caler E., Crabtree J., Angiuoli S.V., Merino E.F., Amedeo P., Cheng Q., Coulson R.M., Crabb B.S., Del Portillo H.A., Essien K., Feldblyum T.V., Fernandez-Becerra C., Gilson P.R., Gueye A.H., Guo X., Kang'a S., Kooij T.W., Korsinczky M., Meyer E.V., Nene V., Paulsen I., White O., Ralph S.A., Ren Q., Sargeant T.J., Salzberg S.L., Stoeckert C.J., Sullivan S.A., Yamamoto M.M., Hoffman S.L., Wortman J.R., Gardner M.J., Galinski M.R., Barnwell J.W., Fraser-Liggett C.M. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.M., Escalante A.A., Neafsey D., Volkman S.K. Comparative evolutionary genomics of human malaria parasites. Trends Parasitol. 2008;24:545–550. doi: 10.1016/j.pt.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Charif D., Lobry J.R. Strucutral Approaches to Sequence Evolution. Springer; Berlin Heidelberg: 2007. SeqinR 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. [Google Scholar]
- Chen X., Jiang T. An improved Gibbs sampling method for motif discovery via sequence weighting. Comput. Syst. Bioinf./Life Sci. Soc. Comput. Syst. Bioinf. Conf. 2006:239–247. [PubMed] [Google Scholar]
- Coulson R.M., Hall N., Ouzounis C.A. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A.G., Grau G.E., Janse C., Kazura J.W., Milner D., Barnwell J.W., Turner G., Langhorne J. The role of animal models for research on severe malaria. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Mukhopadhyay A., AO W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Tissenbaum H.A., Mango S.E., Walhout A.J. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Domazet-Loso T., Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- Droucheau E., Primot A., Thomas V., Mattei D., Knockaert M., Richardson C., Sallicandro P., Alano P., Jafarshad A., Baratte B., Kunick C., Parzy D., Pearl L., Doerig C., Meijer L. Plasmodium falciparum glycogen synthase kinase-3: molecular model, expression, intracellular localisation and selective inhibitors. Biochim. Biophys. Acta. 2004;1697:181–196. doi: 10.1016/j.bbapap.2003.11.023. [DOI] [PubMed] [Google Scholar]
- Dzikowski R., Templeton T.J., Deitsch K. Variant antigen gene expression in malaria. Cell. Microbiol. 2006;8:1371–1381. doi: 10.1111/j.1462-5822.2006.00760.x. [DOI] [PubMed] [Google Scholar]
- Escalante A.A., Ayala F.J. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante A.A., Ayala F.J. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5793–5797. doi: 10.1073/pnas.92.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante A.A., Freeland D.E., Collins W.E., Lal A.A. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell B.J., Naughton J.A., Dempsey E., Bell A. Cellular and molecular actions of dinitroaniline and phosphorothioamidate herbicides on Plasmodium falciparum: tubulin as a specific antimalarial target. Mol. Biochem. Parasitol. 2006;145:226–238. doi: 10.1016/j.molbiopara.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Foth B.J., Tsai I.J., Reid A.J., Bancroft A.J., Nichol S., Tracey A., Holroyd N., Cotton J.A., Stanley E.J., Zarowiecki M., Liu J.Z., Huckvale T., Cooper P.J., Grencis R.K., Berriman M. Whipworm genome and dual-species transcriptome analyses provide molecular insights into an intimate host-parasite interaction. Nat. Genet. 2014;46:693–700. doi: 10.1038/ng.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth B.J., Zhang N., Chaal B.K., Sze S.K., Preiser P.R., Bozdech Z. Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum. Mol. Cell Proteomics: MCP. 2011;10(M110):006411. doi: 10.1074/mcp.M110.006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech C., Chen N. Genome comparison of human and non-human malaria parasites reveals species subset-specific genes potentially linked to human disease. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski M.R., Corredor V. Variant antigen expression in malaria infections: posttranscriptional gene silencing, virulence and severe pathology. Mol. Biochem. Parasitol. 2004;134:17–25. doi: 10.1016/j.molbiopara.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Paulsen I.T., James K., Eisen J.A., Rutherford K., Salzberg S.L., Craig A., Kyes S., Chan M.S., Nene V., Shallom S.J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M.W., Vaidya A.B., Martin D.M., Fairlamb A.H., Fraunholz M.J., Roos D.S., Ralph S.A., Mcfadden G.I., Cummings L.M., Subramanian G.M., Mungall C., Venter J.C., Carucci D.J., Hoffman S.L., Newbold C., Davis R.W., Fraser C.M., Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G., Mushegian A. Measuring gene expression divergence: the distance to keep. Biol. Direct. 2010;5:51. doi: 10.1186/1745-6150-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N., Prud'Homme B., Wittkopp P.J., Kassner V.A., Carroll S.B. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Gupta A.P., Chin W.H., Zhu L., Mok S., Luah Y.H., Lim E.H., Bozdech Z. Dynamic epigenetic regulation of gene expression during the life cycle of malaria parasite Plasmodium falciparum. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall N., Karras M., Raine J.D., Carlton J.M., Kooij T.W., Berriman M., Florens L., Janssen C.S., Pain A., Christophides G.K., James K., Rutherford K., Harris B., Harris D., Churcher C., Quail M.A., Ormond D., Doggett J., Trueman H.E., Mendoza J., Bidwell S.L., Rajandream M.A., Carucci D.J., Yates J.R., III, Kafatos F.C., Janse C.J., Barrell B., Turner C.M., Waters A.P., Sinden R.E. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- Hall N., Pain A., Berriman M., Churcher C., Harris B., Harris D., Mungall K., Bowman S., Atkin R., Baker S., Barron A., Brooks K., Buckee C.O., Burrows C., Cherevach I., Chillingworth C., Chillingworth T., Christodoulou Z., Clark L., Clark R., Corton C., Cronin A., Davies R., Davis P., Dear P., Dearden F., Doggett J., Feltwell T., Goble A., Goodhead I., Gwilliam R., Hamlin N., Hance Z., Harper D., Hauser h., Hornsby t., Holroyd S., Horrocks P., Humphray S., Jagels K., James K.D., Johnson D., Kerhornou A., Knights A., Konfortov B., Kyes S., Larke N., Lawson D., Lennard N., Line A., Maddison M., Mclean J., Mooney P., Moule S., Murphy L., Oliver K., Ormond D., Price C., Quail M.A., Rabbinowitsch E., Rajandream M.A., Rutter S., Rutherford K.M., Sanders M., Simmonds M., Seeger K., Sharp S., Smith R., Squares R., Squares S., Stevens K., Taylor K., Tivey A., Unwin L., Whitehead S., Woodward J., Sulston J.E., Craig A., Newbold C., Barrell B.G. Sequence of Plasmodium falciparum chromosomes 1, 3-9 and 13. Nature. 2002;419:527–531. doi: 10.1038/nature01095. [DOI] [PubMed] [Google Scholar]
- Harikishore A., Niang M., Rajan S., Preiser P.R., Yoon H.S. Small molecule Plasmodium FKBP35 inhibitor as a potential antimalaria agent. Sci. Report. 2013;3:2501. doi: 10.1038/srep02501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmse L., Van Zyl R., Gray N., Schultz P., Leclerc S., Meijer L., Doerig C., Havlik I. Structure-activity relationships and inhibitory effects of various purine derivatives on the in vitro growth of Plasmodium falciparum. Biochem. Pharmacol. 2001;62:341–348. doi: 10.1016/s0006-2952(01)00644-x. [DOI] [PubMed] [Google Scholar]
- Harrison P.W., Wright A.E., Mank J.E. The evolution of gene expression and the transcriptome-phenotype relationship. Semin. Cell Dev. Biol. 2012;23:222–229. doi: 10.1016/j.semcdb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Llinas M., Li J., Preiser P.R., Bozdech Z. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinf. 2007;8:350. doi: 10.1186/1471-2105-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N., Kuratani S. Comparative transcriptome analysis reveals vertebrate phylotypic period during organogenesis. Nat. Commun. 2011;2:248. doi: 10.1038/ncomms1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janky R., Van Helden J. Evaluation of phylogenetic footprint discovery for predicting bacterial cis-regulatory elements and revealing their evolution. BMC Bioinf. 2008;9:37. doi: 10.1186/1471-2105-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse C.J., Kroeze H., Van Wigcheren A., Mededovic S., Fonager J., Franke-Fayard B., Waters A.P., Khan S.M. A genotype and phenotype database of genetically modified malaria-parasites. Trends Parasitol. 2011;27:31–39. doi: 10.1016/j.pt.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Kafsack B.F., Llinas M. Eating at the table of another: metabolomics of host-parasite interactions. Cell Host Microbe. 2010;7:90–99. doi: 10.1016/j.chom.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafsack B.F., Rovira-Graells N., Clark T.G., Bancells C., Crowley V.M., Campino S.G., Williams A.E., Drought L.G., Kwiatkowski D.P., Baker D.A., Cortes A., Llinas M. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507:248–252. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka A.T., Varga K.M., Gerrard D.T., Preibisch S., Corcoran D.L., Jarrells J., Ohler U., Bergman C.M., Tomancak P. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–814. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- Kooij T.W., Carlton J.M., Bidwell S.L., Hall N., Ramesar J., Janse C.J., Waters A.P. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 2005;1 doi: 10.1371/journal.ppat.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo W.P., Liu F., Trimarchi J., Punzo C., Lombardi M., Sarang J., Whipple M.E., Maysuria M., Serikawa K., Lee S.Y., Mccrann D., Kang J., Shearstone J.R., Burke J., Park D.J., Wang X., Rector T.L., Ricciardi-Castagnoli P., Perrin S., Choi S., Bumgarner R., Kim J.H., Short G.F., III, Freeman M.W., Seed B., Jensen R., Church G.M., Hovig E., Cepko C.L., Park P., Ohno-Machado L., Jenssen T.K. A sequence-oriented comparison of gene expression measurements across different hybridization-based technologies. Nat. Biotechnol. 2006;24:832–840. doi: 10.1038/nbt1217. [DOI] [PubMed] [Google Scholar]
- Kyes S., Horrocks P., Newbold C. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 2001;55:673–707. doi: 10.1146/annurev.micro.55.1.673. [DOI] [PubMed] [Google Scholar]
- Lapp S.A., Mok S., Zhu L., Wu H., Preiser P., Bozdech Z., Galinski M.R. Plasmodium knowlesi gene expression differs in ex vivo compared to in vitro blood-stage cultures. Malar. J. 2015;14:110. doi: 10.1186/s12936-015-0612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., Mcgettigan P.A., Mcwilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Le Roch K.G., Zhou Y., Blair P.L., Grainger M., Moch J.K., Haynes J.D., De La Vega P., Holder A.A., Batalov S., Carucci D.J., Winzeler E.A. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Liew K.J., Hu G., Bozdech Z., Peter P.R. Defining species specific genome differences in malaria parasites. BMC Genomics. 2010;11:128. doi: 10.1186/1471-2164-11-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magarinos M.P., Carmona S.J., Crowther G.J., Ralph S.A., Roos D.S., Shanmugam D., Van Voorhis W.C., Aguero F. TDR Targets: a chemogenomics resource for neglected diseases. Nucleic Acids Res. 2012;40:D1118–D1127. doi: 10.1093/nar/gkr1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen E.S., Perkins S.L., Schall J.J. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol. Phylogenet. Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Militello K.T., Dodge M., Bethke L., Wirth D.F. Identification of regulatory elements in the Plasmodium falciparum genome. Mol. Biochem. Parasitol. 2004;134:75–88. doi: 10.1016/j.molbiopara.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Okyere J., Oppon E., Dzidzienyo D., Sharma L., Ball G. Cross-species gene expression analysis of species specific differences in the preclinical assessment of pharmaceutical compounds. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto T.D., Bohme U., Jackson A.P., Hunt M., Franke-Fayard B., Hoeijmakers W.A., Religa A.A., Robertson L., Sanders M., Ogun S.A., Cunningham D., Erhart A., Billker O., Khan S.M., Stunnenberg H.G., Langhorne J., holder A.A., Waters A.P., Newbold C.I., Pain A., Berriman M., Janse C.J. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014;12:86. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain A., Bohme U., Berry A.E., Mungall K., Finn R.D., Jackson A.P., Mourier T., Mistry J., Pasini E.M., Aslett M.A., Balasubrammaniam S., Borgwardt K., Brooks K., Carret C., Carver T.J., Cherevach I., Chillingworth T., Clark T.G., Galinski M.R., Hall N., Harper D., Harris D., Hauser H., Ivens A., Janssen C.S., Keane T., Larke N., LAPP S., Marti M., Moule S., Meyer I.M., Ormond D., Peters N., Sanders M., Sanders S., Sargeant T.J., Simmonds M., Smith F., Squares R., Thurston S., Tivey A.R., Walker D., White B., Zuiderwijk E., Churcher C., Quail M.A., Cowman A.F., Turner C.M., Rajandream M.A., Kocken C.H., Thomas A.W., Newbold C.I., Barrell B.G., Berriman M. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter H.J., Campbell T.L., Llinas M. The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol. Biochem. Parasitol. 2011;176:1–7. doi: 10.1016/j.molbiopara.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira V., Waxman D., Eyre-Walker A. A problem with the correlation coefficient as a measure of gene expression divergence. Genetics. 2009;183:1597–1600. doi: 10.1534/genetics.109.110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S.L., Schall J.J. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J. Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Petalidis L., Bhattacharyya S., Morris G.A., Collins V.P., Freeman T.C., Lyons P.A. Global amplification of mRNA by template-switching PCR: linearity and application to microarray analysis. Nucleic Acids Res. 2003;31 doi: 10.1093/nar/gng142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras V., Tomita M., Selvarajoo K. Transcriptome-wide variability in single embryonic development cells. Sci. Rep. 2014;4:7137. doi: 10.1038/srep07137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M., Drost H.G., Gabel A., Ullrich K.K., Bonn M., Grosse I. A transcriptomic hourglass in plant embryogenesis. Nature. 2012;490:98–101. doi: 10.1038/nature11394. [DOI] [PubMed] [Google Scholar]
- Russell K., Emes R., Horrocks P. Triaging informative cis-regulatory elements for the combinatorial control of temporal gene expression during Plasmodium falciparum intraerythrocytic development. Parasit. Vectors. 2015;8:81. doi: 10.1186/s13071-015-0701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi T.R., Dufour H.D., Williams T.M., Carroll S.B. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A., Hughes K.R., Modrzynska K.K., Otto T.D., Pfander C., Dickens N.J., Religa A.A., Bushell E., Graham A.L., Cameron R., Kafsack B.F., Williams A.E., Llinas M., Berriman M., Billker O., Waters A.P. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014;507:253–257. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Creek D.J., Evans K.J., De Souza D., Schofield L., Muller S., Barrett M.P., Mcconville M.J., Waters A.P. Host reticulocytes provide metabolic reservoirs that can be exploited by malaria parasites. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Patapoutian A., Hampton G.M., Schultz P.G., Hogenesch J.B. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton T.J., Iyer L.M., Anantharaman V., ENOMOTO S., Abrahante J.E., Subramanian G.M., Hoffman S.L., Abrahamsen M.S., Aravind L. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Noort V., Huynen M.A. Combinatorial gene regulation in Plasmodium falciparum. Trends Genet.: TIG. 2006;22:73–78. doi: 10.1016/j.tig.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Vembar S.S., Scherf A., Siegel T.N. Noncoding RNAs as emerging regulators of Plasmodium falciparum virulence gene expression. Curr. Opin. Microbiol. 2014;20:153–161. doi: 10.1016/j.mib.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedall G.D., Polley S.D., Conway D.J. Gene-specific signatures of elevated non-synonymous substitution rates correlate poorly across the Plasmodium genus. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I., Graur D., Ophir R. Incongruent expression profiles between human and mouse orthologous genes suggest widespread neutral evolution of transcription control. Omics-J. Integr. Biol. 2004;8:15–24. doi: 10.1089/153623104773547462. [DOI] [PubMed] [Google Scholar]
- Zhu L., Mok S., Imwong M., Jaidee A., Russell B., Nosten F., Day N.P., White N.J., Preiser P.R., Bozdech Z. New insights into the Plasmodium vivax transcriptome using RNA-Seq. Sci. Report. 2016;6 doi: 10.1038/srep20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.