Abstract
Bag5 is a member of the BAG family of molecular chaperone regulators and is unusual in that it consists of five BAG domains, which function as modulators of chaperone activity. Bag family proteins play a key role in cellular as well as in cardiac function and their differential expression is reported in heart failure. In this study, we examined the importance of a Bag family member protein, Bag5, in cardiomyocytes during endoplasmic reticulum (ER) stress. We found that expression of Bag5 in cardiomyocytes is significantly increased with the induction of ER stress in a time dependent manner. We have taken gain-in and loss-of functional approaches to characterize Bag5 protein function in cardiomyocytes. Adenoviral mediated expression of Bag5 significantly decreased cell death as well as improved cellular viability in ER stress. Along with this, ER stress-induced CHOP protein expression is significantly decreased in cells that overexpress Bag5. Conversely, we found that siRNA-mediated knockdown of Bag5 caused cell death, increased cytotoxicity and decreased cellular viability in cardiomyocytes. Mechanistically, we found that Bag5 protein expression is significantly increased in the ER during ER stress and that this in turn modulates GRP78 protein stability and reduces ER stress. This study suggests that Bag5 is an important regulator of ER function and so could be exploited as a tool to improve cardiomyocyte function under stress conditions.
Keywords: ER STRESS, BAG5, CARDIOMYOCYTES, APOPTOSIS
The endoplasmic reticulum (ER) is considered an important hub for protein synthesis, post-translational modification and translocation of secretory proteins to different destinations for the maintenance of cellular protein quality and quantity in the cell [Glembotski, 2012]. In addition to this, the ER plays a role in regulation of excitation-contraction coupling by maintaining Ca2+ homeostasis in cardiomyocytes [Stutzmann and Mattson, 2011]. During cardiac stress such as oxidative stress, ischemia, calcium depletion, etc., ER function is disturbed and causes accumulation of unfolded or defective proteins in the ER. Accumulation of defective proteins generates a condition known as ER stress. With induction of ER stress, the cell turns on a protective mechanism known as the unfolded protein response (UPR)[Groenendyk et al., 2010]. The UPR is characterized by inhibition of protein synthesis and enhanced ER chaperone protein production as well as the facilitation of unfolded protein degradation through ER-associated protein degradation (ERAD)[Ruggiano et al., 2014]. Although the UPR is consider as a compensatory mechanism of the cell, prolonged activation of UPR triggers proapoptotic signaling and cell death [Xu et al., 2005].
Recent studies suggest that many cardiovascular diseases such as cardiac hypertrophy, ischemia, diabetic and chemotherapy have increased levels of ER stress [Minamino and Kitakaze, 2010]. In cell culture and animal model studies, it was shown that improving UPR improves ER function as well cardiac performance. The role of some of the ER-associated proteins was validated in ER stress and found to be cardioprotective in the ischemia reperfusion model of heart failure [Fu et al., 2008; Wang et al., 2014b; Brody et al., 2015]. These include: 78 kDa glucose-regulated protein (GRP78), also known as Binding Immunoglobulin Protein (BIP) and heat shock 70 kDa protein 5 (HSPA5); X-box binding protein 1 (XBP1); and thrombospondin.
The Bag family of proteins comprises six isoforms, Bag1-Bag6, which act as co-chaperones in different cellular functions by making a complex with other chaperone proteins [Kabbage and Dickman, 2008; Arakawa et al., 2010]. BAG5 is a member of this family of molecular chaperone regulators and is unique in that it has five BAG domains. The function of Bag domains is thought to be modulation of the activity of chaperones and Bag family proteins play key roles in cellular and cardiac function. Expression of different isoforms has been reported across species suggesting that it is an important class of protein. Bag family proteins are found to be upregulated in stress conditions and they shuttle from the cytosol to other subcellular organelles [Nollen et al., 2000; Bruchmann et al., 2013; Wang et al., 2014a]. Previous studies from our lab as well others have shown that the Bag family member protein, Bag3, is important for cardiomyocyte function and knockdown of Bag3 protein causes reduced cardiomyocyte function, cellular disarray and heart failure [Hishiya et al., 2010]. Bag5 is another member of the Bag family and is known to be upregulated during ER stress and mitochondrial stress [Bruchmann et al., 2013; Wang et al., 2014a]. It has been shown that Bag5 confers cellular protection in different disease models by reducing ER stress and improving cellular viability [Bruchmann et al., 2013]. Although the function of Bag5 has been studied in other cell types, its function in cardiomyocytes has not yet been investigated.
Therefore, in the present study, we explore the role of Bag5 in cardiomyocytes using neonatal rat ventricular cardiomyocytes (NRVCs) as a cell model. We tested the protective role of Bag5 during ER stress-mediated apoptosis. Our data suggest that Bag5 protects cardiomyocytes from ER stress and improves cellular viability.
MATERIALS AND METHODS
Isolation of NRVCs and cell culture
All experiments were performed according to the guidelines and with the permission of the Temple University Institutional Animal Care and Use Committee. NRVCs were isolated from the ventricles of 1-2 day old Harlan Sprague-Dawley rats by enzymatic digestion with trypsin followed by collagenase as described before [Louch et al., 2011]. In brief, animals were decapitated, hearts removed, atria excised, and the ventricles then incubated overnight in 0.05% trypsin/EDTA (Life Technologies) with shaking. The medium was aspirated and the ventricles digested with collagenase/DNase in MEMα for 40 min at 37°C. The cell suspension was homogenized by repeated pipetting, passed through a 45 μm filter and harvested by centrifugation at 100g. Isolated cardiomyocytes were finally resuspended in MEM with 10% FBS and ampicillin/streptomycin and plated for experiments.
Generation of adenovirus and adenoviral infection
The ORF of Bag5 was amplified from human cDNA by PCR. A Flag tag sequence (GACTACAAAGACGATGACGAC AAG) was incorporated at the N-terminus of the gene for easy detection of the protein. Amplified DNA was cloned into the pShuttle vector and adenovirus was generated using AdEasy™ adenoviral Vector System (Agilent Technology, Wilmington DE) according to the Manufacturer’s Instructions. Expression of recombinant adenovirus was checked in the NRVCs by Western blot. At 24h post plating, cells were infected with adenovirus in DMEM (no FBS) for 2h. After infection, supernatants were removed and replaced with DMEM with 2% FBS.
siRNA transfection
Bag5 protein was knockdown by siRNA1 (S-172897,sense 5’ GGCGUACAGAGAUCAGAAAtt 3’, antisense UUUCUGAUCUCUGUACGCCca)) and siRNA2 (S-88750, sense, GGGACAGAACCAUUCCAUUtt3’, antisense AAUGGAAUGGUUCUGUCCCag) in NRVCs. Cells were transfected with siRNAs using Lipofectamine 2000 in Opti-MEM 1reduced serum medium (Thermo Fisher Scientific, Waltham, MA) overnight. The transfection medium was replaced with DMEM with 2% FBS and cells were grown for another 48h in the same medium. Knockdown of Bag5 protein was checked by Western blot.
Isolation of protein and Western blot analysis
Cells were washed with PBS (Sigma-Aldrich, St. Louis MO) twice and lysed in RIPA buffer. Cells were then passed through a 26 gauge needle to break them up completely. Unbroken cells and cell debris were removed by centrifugation and supernatant was used as total cell lysate for Western blots. Protein concentration was determined by the Bradford method (bioWorld, Columbus OH). Equal amounts of proteins were separated on SDS-PAGE and transferred to Odyssey Nitrocellulose membrane (Li-Cor, Lincoln NE) by wet transfer (Bio-Rad, Philadelphia PA). Membranes were blocked with Odyssey blocking buffer (Li-Cor) for 1h at room temperature and probed with primary antibody for overnight at 4°C with rocking. The membranes were further washed thrice with PBST and then probed with respective secondary antibody for 1h at room temperature. Signal was detected by an Odyssey scanner after washing twice with PBST. The following primary antibodies were used for Western blotting: Bag1, Bag2, Bag4 (Novus Biologicals, Littleton CO), Bag3 (Protein Tech, Chicago IL), Bag6 (R&D system, Minneapolis MN), CHOP, GRP78, Bag5 (Abcam, Cambridge MA) PDI, pELFα, ELFα (Cell Signaling, Danvers MA).
Immunolabeling and microscopy
Immunocytochemistry was performed as previously described Gupta et al., 2014]. In brief, primary cells in two well chamber slides were washed with PBS and then fixed with 4% paraformaldehyde (PFA) for 10 min. Cells were then washed twice with PBS and then permeabilized with 0.5% Triton X100 for 10 min. Cells were masked with 0.1M glycine for 30 min at room temperature and then washed twice with PBS. Blocking was done at room temperature with blocking buffer (1% BSA, 0.1% Tween 20 in PBS) for 1h. Cells were incubated with primary antibody in blocking buffer overnight at 4°C. Unbound antibody was removed by washing with PBS and probed with secondary antibody labeled with Alexa Fluor (Thermo Fisher Scientific) in blocking buffer for 1h. For counter staining with a second primary antibody, cells were blocked for 30 min at room temperature before probing with antibody. Cells were mounted with VECTASHIELD hard set mounting medium with DAPI (VECTOR Laboratories,Burlingame,CA). Images were captured under a Zeiss fluorescence microscope.
Measurement of cell cytotoxicity and viability
NRVCs were plated into 96 well plates at a density of 10,000 cells per well. For overexpression of Bag5 protein, cells were transduced with Ad-Bag5 for 48h and as a control, cells were transduced with scramble adenovirus. For knockdown of Bag5, cells were transfected with Bag5 siRNA and control cells were transfected with Ctrl siRNA (scramble) for 48h. All transfections were performed using Lipofectamine 2000 (Life Technologies, Grand Island, NY) according to the manufacturer's protocol. Cells were treated with tunicamycin for 0-24h and cellular cytotoxicity was measured with SYTOX Green cell death assay (Thermo Fisher Scientific). SYTOX Green nucleic acid stain is a green-fluorescent nuclear and chromosome counterstain that is impermeant to live cells, making it an indicator of dead cells within a population. In brief, cells were incubated with SYTOX green for 10 min at room temperature and the OD at 523 nm was read with a spectrophotometer. Viability assays were done with the same cells by incubating with viability assay reagent CellTiter blue (Promega, Madison WI). The CellTiter-Blue cell viability assay provides a homogeneous, fluorescent method for monitoring cell viability based on the ability of living cells to convert a redox dye (resazurin) into a fluorescent end product (resorufin). Nonviable cells lose metabolic capacity and do not generate a fluorescent signal. The homogeneous assay procedure involves adding the resazurin directly to cells cultured in serum-supplemented medium. After an incubation step, the OD at 590 nm was then read with a spectrophotometer.
Apoptosis assay
Apoptotic cells were distinguished from live cells by propidium iodide (PI) and nucleus with Hoechst 33342 labeling (Thermo Fisher Scientific). PI only labels the nuclei of cells undergoing apoptosis while Hoechst 33342 labels the nuclei of all cells regardless of apoptotic status. PI (10 μg/ml) and Hoechst 33342 (100 μg/ml) were added to the culture medium directly and incubated at 37°C for 15 min. Cells were washed once with culture medium and live imaging was done under a fluorescent microscope.
Isolation of endoplasmic reticulum
ER fraction was isolated by differential centrifugation. NRVC cells were washed twice with PBS and then resuspended in hypotonic buffer (250 mM sucrose, 20 mM HEPES, pH 7.5, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA and protease inhibitor) and incubated on ice for 30 min. Cells were passed through a 30 gauge needle to further break them. Unbroken cells and cell debris were removed by centrifugation at 12000g for 10 min at 4°C and the supernatant was kept for ER and cytosol fractionation. For the ER rich fraction, cell lysates were centrifuged at 100000g for 1h at 4°C. The supernatant was used as a cytosolic fraction and the pellet is the ER rich fraction. The pellet was washed once with cell fractionation buffer and then suspended in RIPA buffer.
Immunoprecipitation
Immunoprecipitation was done with NRVC protein using anti-flag M2 affinity magnetic beads (Sigma-Aldrich, St. Louis MO) according to the Manufacturer’s instructions. In brief, protein lysate was prepared in RIPA buffer as described above. Affinity beads were washed with TBS and finally suspended in TBS with help of a magnetic stand. In a 1.5 ml eppendorf tube, 200 μg protein and 20 μl of resin were incubated overnight at 4°C with gentle shaking. To precipitate the bound protein, tubes were placed in the magnetic stand. The precipitate was then washed three times with TBS and finally the bound protein was eluted using 2X Laemmli sample buffer. Samples were boiled and cleared by centrifugation. Flag tagged protein was detected in the supernatant by Western blot using anti-flag antibody (Sigma-Aldrich).
Statistical analysis
Results are shown as mean ± SD. Paired data were compared by Student’s t test. Multiple groups were compared using one-way ANOVA with post hoc Tukey’s Test. A value of P< 0.05 was considered as significance.
RESULTS
The Bag family represents a distinctive class of proteins comprising six isoforms Bag1 - Bag6, and each having at least one conserved Bag domain. Expression of the Bag family proteins has been reported across various species, found to be upregulated during stress conditions and can be detected in different organs. Bag5 protein is one of the Bag family member proteins and is unique in that it has 5 Bag domains (Fig. 1A). Previous studies showed that Bag5 expression is upregulated during stress conditions in different disease models. In this study, we found that expression of Bag family members is upregulated along with other ER signaling proteins during ER stress conditions in NRVCs (Fig. 1B and C).
Figure 1. Expression of Bag5 protein is upregulated during ER stress.
A. Diagrammatic representation of the conserved domains of the Bag protein family. All contain the Bag conserved domain shown in yellow. UBL denotes ubiquitin ligase domains shown in blue. B. Western blot showed that the ER stress marker proteins CHOP, GRP78 and pELF2 alpha expression are upregulated during tunicamycin mediated ER stress. NRVCs were treated with tunicamycin (TUG, 10 μg/ml) for 0-24h. C. Western blot shows that Bag family proteins are upregulated in NRVC cells treated with tunicamycin.
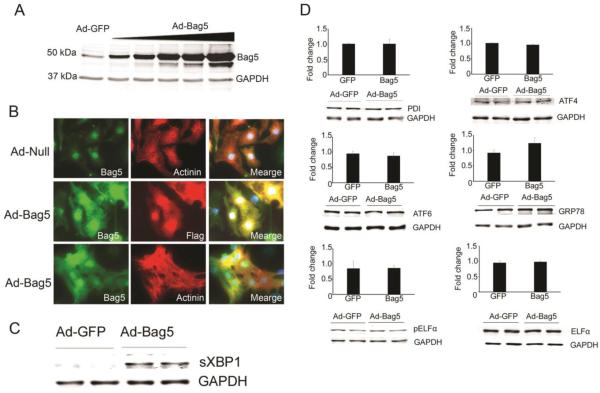
For functional analysis of Bag5 protein, we generated Ad-Bag5 adenovirus to express full length Bag5 protein in cells. The Bag5 ORF was cloned into the pShuttle vector downstream of the CMV promoter. A Flag tag sequence was introduced at the N-terminal of the protein for ease of detection. Expression of Bag5 protein was confirmed in cardiomyocytes infected with Ad-Bag5 for 48h. Western blot showed that Bag5 protein expression goes up in a dose dependent manner (Fig. 2A). Bag5 localization was assessed in cardiomyocytes infected with Ad-Null or Ad-Bag5 for 48h. Consistent with previous studies we found that Bag5 protein is predominantly localized to the perinuclear space with some expression throughout the cytosol (Fig. 2B). Recent studies suggest that Bag family proteins are localized to cytosol as well as different subcellular organelles. Here we have detected the localization of endogenous as well as overexpressed Bag5 protein by immunocytochemistry in NRVCs. Previous studies also suggested that Bag5 acts as a regulator of ER signaling. In this study, we have checked the expression of ER signaling proteins expression in presence of Bag5 and we found that Bag5 protein expression significantly increased the expression of sXBP1 (Fig. 2C). This suggests that Bag5 is a key regulator of the ER stress signaling pathway.
Figure 2. Bag5 expression regulates ER stress signaling.
A. Expression analysis of Bag5 in cardiomyocytes infected with Ad-Bag5 at increasing MOI of adenovirus. B. Immunocytochemistry suggests that Bag5 is predominantly localized to the perinuclear space. NRVCs were transfected with Ad-Null or Ad-Bag5 for 48h and cells were fixed with 4% PFA. Fixed cells were probed with Bag5 (green), Flag (red), and Actinin (red). C. Western blot analysis shows that adenovirus mediated expression of Bag5 upregulates sXBP1. D. Western blot analysis shows that adenovirus mediated expression of Bag5 upregulates the unfolded protein response (UPR) in NRVCs. NRVC cells were infected with Ad-Bag5 for the 72h and then Western blots were done with PDI, ATF4, ATF6, Grp78, ELFα and pELFα antibodies.
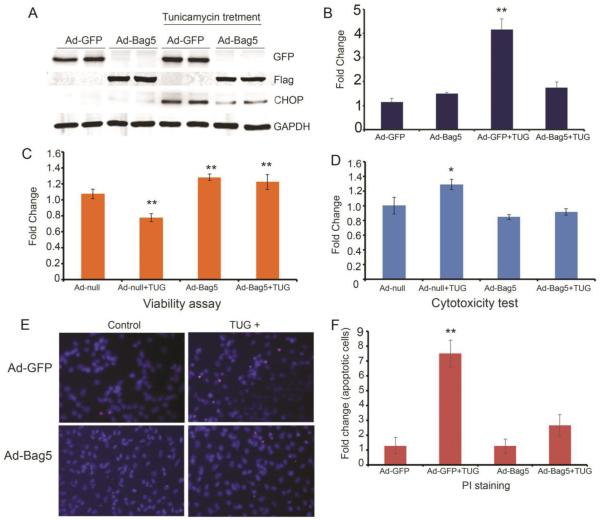
Next, we tested the protective role of Bag5 during ER stress. Tunicamycin is an inhibitor of N-glycosylation of membrane proteins and is known to be potent inducer of ER stress signaling in cardiomyocytes as well as in other cell types. Tunicamycin treatment causes upregulation of ER stress signaling as well as cell death and apoptosis. In this study, we found that tunicamycin treatment of cardiomyocytes caused the upregulation of GRP78, a master regulator of the ER signaling pathway (Fig. 1B). Also, the expression of CHOP protein is upregulated in the presence of tunicamycin (Figs. 1B and 3A). Interestingly, we found that the expression of Bag5 protein caused reduced expression of CHOP protein (Fig. 3A and B) suggesting that Bag5 reduces ER stress signaling in the cardiomyocytes. Bag5 protein expression significantly improved cellular viability and reduced cytotoxicity in NRVC (Fig. 3C and D). Propidium iodide (PI) staining is a widely used method for the detection of apoptotic or dead cells based on membrane permeability. Only dead cells DNA get stained with PI, because live cells are not permeable to this dye. Apoptotic cell-specific labeling with propidium iodide also shows that Bag5 expression significantly reduces cell death during ER stress (Fig. 3E and F).
Figure 3. Bag5 expression reduces ER stress in NRVCs.
A & B. Western blot shows Bag5 expression reduces the ER stress in cardiomyocytes. NRVC cells were infected with Ad-GFP and Ad-Bag5 for 48h and treated with tunicamycin for another 24h. ER stress marker protein expression was detected by probing with CHOP antibody and a graph showing quantification of CHOP protein expression is shown. C & D. Graph showing the viability and cytotoxicity of tunicamycin-treated NRVCs. NRVCs were grown for 48h and treated with tunicamycin (TUG) for 24h. Viability and cytotoxicity were analyzed using cell titer blue and SYTOX green dye. E & F. Apoptotic cells were distinguished from live cells using PI labeling for dead cells and Hoechst 33342 for nucleus in the tunicamycin-treated cardiomyocytes. Images were captured under a fluorescence microscope.* P<0.05, was significant difference between Ad-Null versus Ad-Null+TUG (n=24) . **P<0.001, was significant difference between Ad-Null versus Ad-Null+TUG (n=24). **P<0.001, was significant difference between Ad-GFP versus Ad-GFP+TUG.
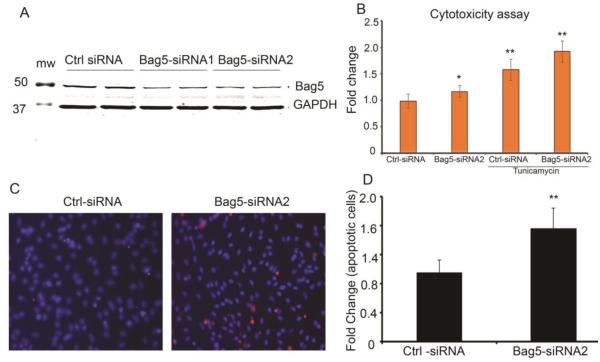
To further investigate the function of Bag5, we used an siRNA knockdown approach. Bag5 expression was knocked down using Bag5 siRNAs 1 and 2. NRVCs cells were treated with Bag5-specific siRNAs or, as a control, scrambled siRNA as described in Materials and Methods. Western blot of siRNA-treated cells showed that Bag5 expression was reduced in cells treated with Bag5 siRNA (Fig. 4A). Cellular cytotoxicity assay suggests that Bag5 siRNA treatment increases cellular toxicity (Fig. 4B). Cellular toxicity of Bag5 siRNA-treated cells is further increased in the presence of tunicamycin (Fig. 4B). This suggests that Bag5 is an important molecule and plays a significant role in cellular function during stress conditions. PI staining showed that apoptotic cell number was increased in Bag5-siRNA treated cells (Fig. 4C and D).
Figure 4. Knockdown of Bag5 causes increased cytotoxicity to NRVC.
A. Western blot showing expression of Bag5 in NRVC cells treated with Bag5-siRNA or Ctrl-siRNA for 48h and probed with Bag5 and GAPDH antibody. B. Cellular cytotoxicity was detected in NRVC cells treated with tunicamycin for 24h using SYTOX green. C & D. Representative images show the apoptotic cells labeled with propidium iodide in the NRVC cells treated with Bag5-siRNA or Ctrl-siRNA. P<0.05, was significant difference between Ctrl-siRNA versus Bag5-siRNA2 (n=24). P<0.001, was significant difference between Ctrl-siRNA versus Ctrl-siRNA and Bag5-siRNA 2 tunicamycin treated (n=24).
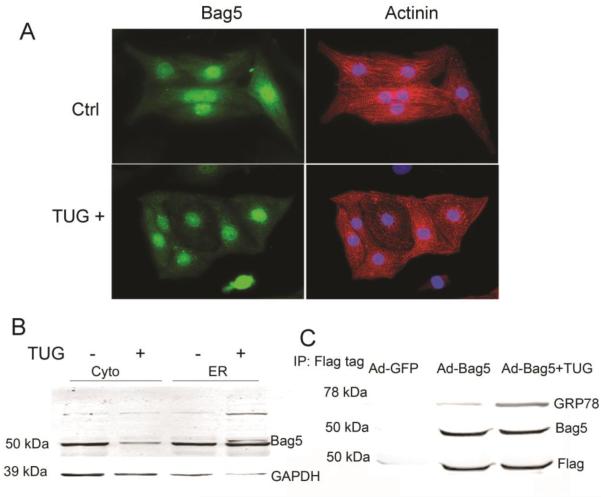
To investigate the Bag5-mediated regulation of ER signaling, we looked at the localization of Bag5 protein during ER stress conditions and found that Bag5 protein expression reduced in the cytosol and became predominantly localized to the perinuclear space in tunicamycin-treated cells (Fig. 5A). Differential localization of Bag5 protein in stressed cells suggests that Bag5 protein plays an important role during cellular stress. Earlier studies suggested that Bag5 protein expression increased in the ER during ER stress [Bruchmann et al., 2013]. In this study, we also found that expression of cytosolic Bag5 decreased during ER stress and that ER resident Bag5 protein expression increased (Fig. 5B). We next studied the interaction of Bag5 protein with GRP78 during ER stress. GRP78 was co-immunoprecipitated with FLAG-tagged Bag5 and we found that Bag5 interaction with GRP78 was increased significantly in tunicamycin-treated cells (Fig. 5C). These results taken together with other studies suggest that Bag5 is an important partner of GRP78 and plays a role in the reduction of ER stress by stabilizing GRP78. The schematic shown in Fig. 6 shows the role of the interaction of Bag5 with GRP78 (BIP) and downstream signaling in the unfolded protein response in cardiomyocytes. Bag5 expression regulates expression of GRP78 and sXBP1. In ER stress, the interaction of Bag5 and GRP78 is increased and may regulate GRP78 function and reduce stress-induced CHOP expression and cell death.
Figure 5. Bag5 interacts with GRP7 and regulates the ER stress pathway.
A. Immunocytochemistry showing Bag5 protein localization during ER stress. NRVC cells were treated with TUG for 24h and then fixed with 4% PFA. Cells were labeled with Bag5 antibody (green) and actinin (red). B. Cellular fractionation shows Bag5 protein expression in the cytosolic and ER fractions during ER stress. C. Bag5-FLAG immunoprecipitation experiment showing co-immunoprecipitation of GRP78 demonstrating interaction of Bag5 with GRP78 during ER stress.
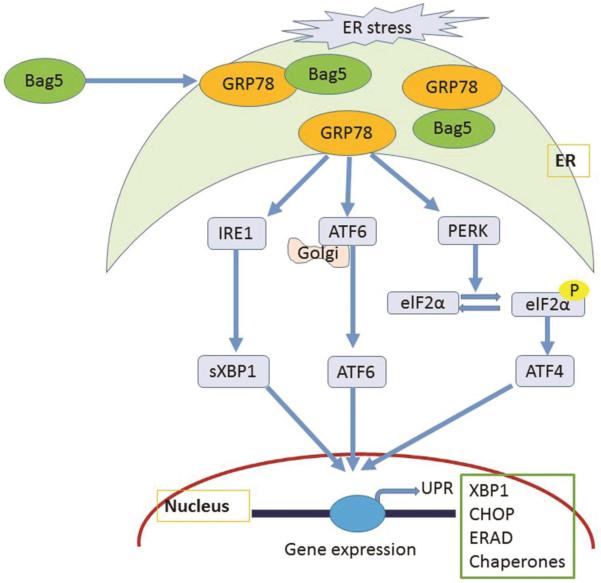
Figure 6. Schematic diagram showing role of Bag5 in unfolded protein response in cardiomyocytes.
Bag5 protein interacts with GRP78 and regulates the downstream signaling of UPR. Expression of Bag5 regulates the expression of GRP78 sXBP1 but other signaling ER signaling ATF4, ATF6, does not change. Interestingly, in ER stress it is found that Bag5 and GRP78 protein interaction increased suggests that Bag5 protein is a one of the regulator of GRP78 function. Importantly, expression of Bag5 protein reduces the ER stress induced CHOP protein expression and attenuated the cell death.
DISCUSSION
Bag family member proteins comprise a special class of proteins that have the conserved Bag domain [Kabbage and Dickman, 2008]. The Bag domain interacts with the ATPase domain of Hsp70 and regulates its function [Liman et al., 2005]. It has been reported that Bag family proteins are upregulated during physiological stress and that they provide cellular protection from stress conditions by increasing the expression of the anti-apoptotic protein Bcl-2 [Doong et al., 2002]. Earlier studies suggested that the Bag member proteins Bag1, Bag2, and Bag3 improve cellular survival by decreasing cell death during stress [Rosati et al., 2011; Kermer et al., 2015; Che et al., 2015]. Previous studies in this laboratory have reported that Bag3 improves cardiac function and survival in the mouse TAC model [Knezevic et al., 2015]. In the case of Bag5, there are 5 Bag domains within the protein and so it is unique among Bag family members. In prostate cancer cells, it has been demonstrated that Bag5 expression is upregulated and in the 22Rv.1 prostate cancer cell line, Bag5 overexpression inhibited ER stress-induced apoptosis in the unfolded protein response by suppressing PERK-eIF2-ATF4 and enhancing the IRE1-Xbp1 activity [Bruchmann et al., 2013]. In a disease model of Parkinson’s disease, it was demonstrated that Bag5 expression protects PC12 cells from 1-methyl-4-phenyl-pyridinnium (MPP+)-induced cell death [Ma et al., 2012]. Another study showed that Bag5 protects cells from oxidative stress by regulation of PTEN-induced kinase 1 (PINK1) degradation and mitochondrial function [Wang et al., 2014a]. Earlier studies also suggested that Bag5 regulates other cellular events such as autophagy by complexing with Rab7 and cyclin associated kinases [Beilina et al., 2014]. While the function of Bag5 is established in several different cell types, its possible role in cardiac cells has not been studied. In this study, we found that Bag5 protein expression is upregulated by ER stress in cardiomyocytes along with other Bag family members. Using a combination of gain-in and loss-of functional approaches, we explored Bag5 function in the cardiomyocytes. Earlier studies suggested that induction of Bag5 protein expression had an anti-apoptotic activity and provided cellular protection. Here, we examined a possible cardioprotective role of Bag5 in ER stress using adenovirus-mediated Bag5 overexpression. We found that Bag5 expression goes up with increasing doses of adenovirus in NRVCs and that the increased expression of Bag5 caused an increase in expression of other ER signaling proteins GRP78 and phospho-ELFα, which suggests that Bag5 is an important regulator of the unfolded protein response in cardiomyocytes. In a gain-in functional approach, we found that Bag5 expressing cells reduced ER stress-induced cytotoxicity and increased cellular viability. Earlier studies suggested that Bag5 reduces cellular apoptosis by increasing expression of Bcl-2 and decreasing the apoptotic proteins factor CHOP and Caspase 3. Consistent with these studies, we found that Bag5 expression reduced the expression of CHOP in tunicamycin-treated cells when compared to control cells expressing GFP only.
It is known from previous studies that the UPR increases expression of ER-associated chaperone proteins so as to improve ER protein folding capacity and reduce ER stress. Thus, overexpression of chaperone proteins such as PDI, GRP78, XBP1 [Fu et al., 2008; Toldo et al., 2011; Wang et al., 2014b] and thrombospondin protects cardiomyocytes from ER stress as well as improving cardiac function. In this study, we found that Bag5 regulates expression of GRP78 and that interaction between Bag5 and GRP78 was significantly increased during tunicamycin treatment. This interaction may be important for the function and stabilization of GRP78 as has been reported by other investigators [Bruchmann et al., 2013]. GRP78 has a dual function: activated GRP78 acts as a chaperone protein to reduce the unfolded protein load in the ER and activated GRP78 also regulates other downstream signaling events to activate the unfolded protein response to counteract stress. Bag proteins are multifunctional and there are increases in the levels of their expression in response to stress suggesting that they have a protective function and survival. In this study, we provide evidence for such a function for Bag5 in cardiomyocytes suggesting that Bag5 is an important protein in these cells and thus may be a suitable therapeutic target to improve heart function.
ACKNOWLEDGEMENTS
We thank past and present members of the Department on Neuroscience and Center for Neurovirology for their insightful discussion and sharing of ideas and reagents. This study utilized services offered by Lewis Katz School of Medicine Comprehensive NeuroAIDS Center at Temple University. This work was supported by grants P30 MH092177 awarded to KK and R01 HL123093 to KK, JYC and AMF.
Contract grant sponsor: NIH
Contract grant number: P30 MH092177 (KK); R01 HL123093 (KK, JYC, AMF).
REFERENCES
- Arakawa A, Handa N, Ohsawa N, Shida M, Kigawa T, Hayashi F, Shirouzu M, Yokoyama S. The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange. Structure. 2010;18:309–319. doi: 10.1016/j.str.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Beilina A, Rudenko IN, Kaganovich A, Civiero L, Chau H, Kalia SK, Kalia LV, Lobbestael E, Chia R, Ndukwe K, Ding J, Nalls MA, International Parkinson’s Disease Genomics Consortium; North American Brain Expression Consortium. Olszewski M, Hauser DN, Kumaran R, Lozano AM, Baekelandt V, Greene LE, Taymans JM, Greggio E, Cookson MR. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc Natl Acad Sci USA. 2014;111:2626–2631. doi: 10.1073/pnas.1318306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody MJ, Schips TG, Vanhoutte D, Kanisicak O, Karch J, Maliken BD, Blair NS, Sargent MA, Prasad V, Molkentin JD. Dissection of thrombospondin-4 domains involved in intracellular adaptive ER Stress responsive signaling. Mol Cell Biol. 2015;36:2–12. doi: 10.1128/MCB.00607-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchmann A, Roller C, Walther TV, Schafer G, Lehmusvaara S, Visakorpi T, Klocker H, Cato AC, Maddalo D. Bcl-2 associated athanogene 5 (Bag5) is overexpressed in prostate cancer and inhibits ER-stress induced apoptosis. BMC Cancer. 2013;13:96. doi: 10.1186/1471-2407-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che XQ, Tang BS, Wang HF, Yan XX, Jiang H, Shen L, Xu Q, Wang GH, Zhang HN, Wang CY, Guo JF. The BAG2 and BAG5 proteins inhibit the ubiquitination of pathogenic ataxin3-80Q. Int J Neurosci. 2015;125:390–394. doi: 10.3109/00207454.2014.940585. [DOI] [PubMed] [Google Scholar]
- Doong H, Vrailas A, Kohn EC. What's in the 'BAG'?--A functional domain analysis of the BAG-family proteins. Cancer Lett. 2002;188:25–32. doi: 10.1016/s0304-3835(02)00456-1. [DOI] [PubMed] [Google Scholar]
- Fu HY, Minamino T, Tsukamoto O, Sawada T, Asai M, Kato H, Asano Y, Fujita M, Takashima S, Hori M, Kitakaze M. Overexpression of endoplasmic reticulum-resident chaperone attenuates cardiomyocyte death induced by proteasome inhibition. Cardiovasc Res. 2008;79:600–610. doi: 10.1093/cvr/cvn128. [DOI] [PubMed] [Google Scholar]
- Glembotski CC. Roles for the Sarco-/Endoplasmic Reticulum in Cardiac Myocyte Contraction, Protein Synthesis, and Protein Quality Control. Physiology. 2012;27:343–350. doi: 10.1152/physiol.00034.2012. [DOI] [PubMed] [Google Scholar]
- Groenendyk J, Sreenivasaiah PK, Kim do H, Agellon LB, Michalak M. Biology of endoplasmic reticulum stress in the heart. Circ Res. 2010;107:1185–1197. doi: 10.1161/CIRCRESAHA.110.227033. [DOI] [PubMed] [Google Scholar]
- Gupta MK, Gulick J, Liu R, Wang X, Molkentin JD, Robbins J. Sumo E2 enzyme UBC9 is required for efficient protein quality control in cardiomyocytes. Circ Res. 2014;115:721–729. doi: 10.1161/CIRCRESAHA.115.304760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiya A, Kitazawa T, Takayama S. BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress. Circ Res. 2010;107:1220–1231. doi: 10.1161/CIRCRESAHA.110.225649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage M, Dickman MB. The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol Life Sci. 2008;65:1390–1402. doi: 10.1007/s00018-008-7535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermer P, Kohn A, Schnieder M, Lingor P, Bahr M, Liman J, Dohm CP. BAG1 is neuroprotective in in vivo and in vitro models of Parkinson's disease. J Mol Neurosci. 2015;55:587–595. doi: 10.1007/s12031-014-0396-2. [DOI] [PubMed] [Google Scholar]
- Knezevic T, Myers VD, Gordon J, Tilley DG, Sharp TE, Wang J, Khalili K, Cheung JY, Feldman AM. BAG3: a new player in the heart failure paradigm. Heart Fail Rev. 2015;20:423–434. doi: 10.1007/s10741-015-9487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman J, Ganesan S, Dohm CP, Krajewski S, Reed JC, Bähr M, Wouters FS, Kermer P. Interaction of BAG1 and Hsp70 mediates neuroprotectivity and increases chaperone activity. Mol Cell Biol. 2005;25:3715–3725. doi: 10.1128/MCB.25.9.3715-3725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louch WE, Sheehan KA, Wolska BM. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol. 2011;51:288–298. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Wang X, Ding X, Jing J, Ma Y, Teng J. Protective effect of BAG5 on MPP+-induced apoptosis in PC12 cells. Neurol Res. 2012;34:977–983. doi: 10.1179/1743132812Y.0000000102. [DOI] [PubMed] [Google Scholar]
- Minamino T, Kitakaze M. ER stress in cardiovascular disease. J Mol Cell Cardiol. 2010;48:1105–1110. doi: 10.1016/j.yjmcc.2009.10.026. [DOI] [PubMed] [Google Scholar]
- Nollen EA, Brunsting JF, Song J, Kampinga HH, Morimoto RI. Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity. Mol Cell Biol. 2000;20:1083–1088. doi: 10.1128/mcb.20.3.1083-1088.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco MC. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2:e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutzmann GE, Mattson MP. Endoplasmic reticulum Ca(2+) handling in excitable cells in health and disease. Pharmacol Rev. 2011;63:700–727. doi: 10.1124/pr.110.003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toldo S, Severino A, Abbate A, Baldi A. The role of PDI as a survival factor in cardiomyocyte ischemia. Methods Enzymol. 2011;489:47–65. doi: 10.1016/B978-0-12-385116-1.00003-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Guo J, Fei E, Mu Y, He S, Che X, Tan J, Xia K, Zhang Z, Wang G, Tang B. BAG5 protects against mitochondrial oxidative damage through regulating PINK1 degradation. PLoS One. 2014a;9:e86276. doi: 10.1371/journal.pone.0086276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZV, Deng Y, Gao N, Pedrozo Z, Li DL, Morales CR, Criollo A, Luo X, Tan W, Jiang N, Lehrman MA, Rothermel BA, Lee AH, Lavandero S, Mammen PP, Ferdous A, Gillette TG, Scherer PE, Hill JA. Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell. 2014b;156:1179–1192. doi: 10.1016/j.cell.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]