Abstract
A strategy for the production of side-chain-to-tail cyclic peptides from ribosomally derived polypeptide precursors is reported. Two genetically encodable unnatural amino acids, bearing either an aryl or alkyl amino group, were investigated for their efficiency toward promoting the formation of medium to large-sized peptide macrocycles via intein-mediated side-chain-to-C-terminus cyclization. While only partial cyclization was observed with precursor proteins containing para-amino-phenylalanine, efficient peptide macrocyclization could be achieved using O-2-aminoethyl-tyrosine as the reactive moiety. Conveniently, the latter was generated upon quantitative, post-translational reduction of the azido-containing counterpart, O-2-azidoethyl-tyrosine, directly in E. coli cells. This methodology could be successfully applied for the production of a 12mer cyclic peptide with enhanced binding affinity for the model target protein streptavidin as compared to the acyclic counterpart (KD: 5.1 μM vs. 22.4 μM), thus demonstrating its utility toward the creation and investigation of novel, functional macrocyclic peptides.
Introduction
Owing to their peculiar structural and conformational properties, macrocyclic peptides provide attractive scaffolds for targeting complex biomolecular interfaces.1–5 The occurrence of macrocyclic backbones amongst biologically active peptides6 found in nature further motivates the increasing interest toward this structural class.1–5 As illustrated by a growing body of studies, macrocyclization can help overcome important limitations of linear peptides such as limited proteolytic stability7–9 and membrane permeability.10–14 The conformational rigidification imparted by macrocyclization on the peptide structure can also aid in pre-organizing the molecule into a bioactive conformation, resulting in enhanced selectivity and binding affinity for the target biomolecule.15–18
While a variety of synthetic methods have been investigated for the synthesis of cyclic peptides,19 the ability to cyclize ribosomally derived polypeptides can offer key advantages toward the synthesis, combinatorial diversification, and functional evaluation of collections of peptide macrocycles.20,21 Indeed, the application of these strategies has enabled the successful identification of cyclopeptide inhibitors for a variety of target proteins and enzymes.22–29
Our group has reported methods to rapidly generate structurally diverse Macrocyclic Organo Peptide Hybrids (MOrPHs), via a dual ligation strategy involving arbitrary non-peptidic linkers and ribosomally produced intein-fusion proteins bearing an unnatural amino acid equipped with a bioorthogonal functional group.9,30–33 More recently, complementary methodologies has been developed to enable the biosynthesis of cyclic and bicyclic peptides of arbitrary sequence in living bacterial cells.34–36 As illustrated in these and other studies,18,37 the type of backbone connectivity and intramolecular linkage can play a significant role in affecting the functional properties (e.g., protein binding affinity) of the resulting macrocyclic peptide. Accordingly, we have been interested in exploring alternative approaches for creating peptide macrocycles through the post-translational processing of ribosomally derived polypeptides.
Here, we report the development of a new strategy to achieve this goal, which involves the side-chain-to-tail cyclization of intein-fused polypeptides by means of amino-functionalized unnatural amino acids. We also demonstrate how this cyclization strategy led to the development of a cyclic peptide with improved binding affinity toward a model target protein.
Results and Discussion
Peptide macrocyclization strategy
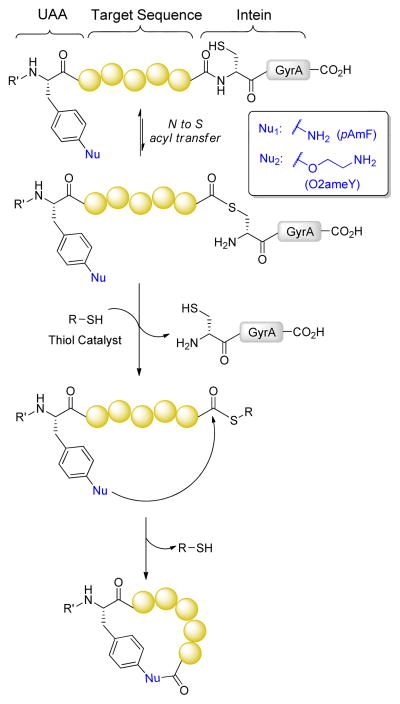
Scheme 1 illustrates the general strategy investigated in the present study. This approach involves the use of a recombinant biosynthetic precursor in which an arbitrary target sequence is framed between an amino-functionalized unnatural amino acid and a C-terminal intein. In the latter, the conserved C-terminal asparagine residue (Asn198 in GyrA intein) and cysteine residue at the intein+1 position is mutated (N198A) and removed, respectively, to allow for the formation of a transient thioester linkage at the junction between the peptide target sequence and intein while preventing splicing of the intein itself. We envisioned that thiol-induced cleavage of the intein moiety on this precursor polypeptide would form a reactive C-terminal thioester intermediate, which would then undergo aminolysis by action of the side-chain amino group from the unnatural amino acid upstream of the target sequence to yield the desired side-chain-to-tail cyclopeptide product (Scheme 1). Extending upon our previous studies on the cyclization of intein-fused polypeptides with amino-thiol containing amino acids,35 this strategy would enable the formation of macrocyclic peptides featuring alternative side-chain-to-tail connectivities, thus broadening the repertoire of peptide macrocycles accessible via posttranslational elaboration of ribosomally produced polypeptides.
Scheme 1.
Synthesis of macrocyclic peptides via thiol-catalyzed cyclization of biosynthetic precursor proteins containing amino-functionalized unnatural amino acids. UAA: Unnatural amino acid, GyrA: GyrA mini-intein from Mycobacterium xenopi. Nu1 corresponds to p-amino-phenylalanine (pAmF) and Nu2 corresponds to O-2-aminoethyl-tyrosine (O2ameY).
Para-amino-phenylalanine-mediated cyclization
The realization of the reaction scheme outlined above was anticipated to require the choice of an amino-functionalized unnatural amino acid with suitable structural and reactivity properties. First, it should exhibit the desired nucleophilic reactivity in aqueous media and under conditions in which the cyclization through other nucleophilic amino acids (e.g., lysine) is not favorable. Secondly, it should be amenable to the ribosomal incorporation into the precursor polypeptide through convenient methods such as amber stop codon suppression38 with orthogonal aminoacyl-tRNA synthetases. In light of these considerations, we selected p-amino-phenylalanine (pAmF) (Scheme 1, Nu1) as a promising candidate for mediating the peptide cyclization process. Because of its significantly lower pKa compared to lysine (4.5 vs. 10.5 for Lys ε-NH2), the aryl amino group in pAmF is expected to remain unprotonated and to possess higher nucleophilic reactivity in aqueous buffer at near-neutral pH. Indeed, aniline and derivatives thereof have been successfully applied as nucleophilic catalysts for promoting condensation reactions under physiological conditions.39,40 In addition, an orthogonal tRNA/tRNA amino acyl synthetase (AARS) pair was previously made available to enable the incorporation of pAmF into recombinant proteins in response to an amber stop codon.41
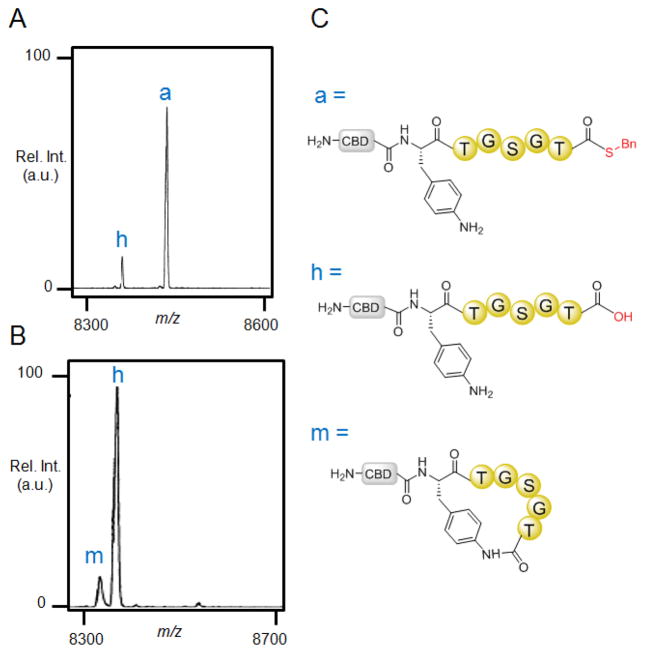
To investigate the feasibility of pAmF-mediated peptide cyclization, a model precursor polypeptide was initially tested, which comprises a N-terminal chitin-binding domain (CBD) followed by pAmF, a 5mer target sequence (TGSGT), and the GyrA mini-intein from Myobacterium xenopi42 (Table 1, Entry 1). This protein construct was expressed in BL21(DE3) E. coli cells and purified by nickel-affinity chromatography using a polyhistidine tag fused to the C-terminus of the GyrA intein. The purified protein was then incubated with either benzylmercaptan or thiophenol in phosphate buffer at pH 7.0. Tris(2-carboxyethyl)phosphine (TCEP) was also added to the reaction mixture to maintain the thiols in the protein and reagents in the reduced form. As shown in Figure 1, MALDI-TOF mass spectrometry (MS) analysis of the reaction in the presence of benzyl mercaptan showed the formation of only the acyclic thioester intermediate (‘a’) together with a smaller amount of the hydrolysis byproduct (‘h’). In contrast, the reaction in the presence of thiophenol was found to result in formation of the desired macrocyclic peptide product (‘m’, Figure 1), along with the linear byproduct resulting from hydrolysis of the thiophenol thioester (‘h’). Due to the presence of a large N-terminal tag (CBD protein) and identical target sequence, it is reasonable to assume that the cyclic and acyclic species generated in these experiments have similar ionization properties. Accordingly, the ratio of macrocycle : hydrolysis product in the thiophenol reaction was estimated to be approximately 1:5. Furthermore, no significant changes in product distribution were observed over a 24 hour period.
Table 1.
Biosynthetic precursors investigated in this study. CBD: Chitin Binding Domain, OpgY: O-propargyl-tyrosine. MeaF: 3-(2-mercapto-ethyl)amino-phenylalanine.
| Entry | Construct Name | UAA | Target Sequence |
|---|---|---|---|
| 1 | CBD-5mer(pAmF) | pAmF | TGSGT |
| 2 | CBD-6mer(pAmF) | pAmF | TGSYGT |
| 3 | CBD-7mer(pAmF) | pAmF | TGSEYGT |
| 4 | CBD-8mer(pAmF) | pAmF | TGSAEYGT |
| 5 | CBD-10mer(pAmF) | pAmF | TGSKLAEYGT |
| 6 | CBD-10mer(OpgY) | OpgY | TGSKLAEYGT |
| 7 | 5mer(O2ameY) | OameY | TGSGT |
| 8 | 6mer(O2ameY) | OameY | TGSYGT |
| 9 | 8mer(O2ameY) | OameY | TGSAEYGT |
| 10 | Strep-11mer(O2ameY) | OameY | FTNVHPQFANA |
| 11 | Strep-11mer(OpgY) | OpgY | FTNVHPQFANA |
| 12 | Strep-11mer(MeaF) | MeaF | FTNVHPQFANA |
Figure 1.
pAmF-mediated cyclization of 5mer peptide sequence. A–B) MALDI-TOF MS spectra of small MW products from reaction of CBD-5mer(pAmF) (100 μM) with 10 mM thiol benzylmercaptan (A) or thiophenol (B), and TCEP (10 mM) after 24 hours. ‘m’ : macrocycle, ‘h’: hydrolysis product. [m+H]+ calc: 8345.2, obs: 8345.4; [h+H]+ calc: 8363.2, obs: 8363.6; [a+H]+ calc: 8469.4, obs: 8469.0 C) Schematic structures of the macrocyclic product (m), thioester intermediate (a), and hydrolysis (h) byproduct.
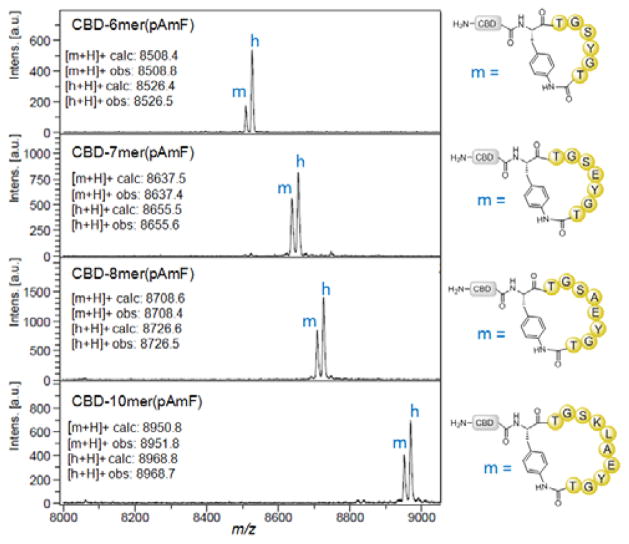
To gain insight into the effect of the length of the target peptide sequence on the efficiency of the macrocyclization reaction, four additional biosynthetic precursors with 6 to 10 amino acid-long target sequences were generated (Table 1, entries 2–5). As for the 5mer peptide, these sequences of arbitrary composition were previously found to have no particular propensity either toward cyclization or against it,31,32 thus providing ideal model structures for testing the functionality of the present method. After purification, the corresponding proteins were incubated with thiophenol and TCEP as described above. Importantly, MALDI-TOF MS analysis showed the formation of the desired macrocyclic peptide for all the reactions, thus demonstrating that pAmF-mediated macrocyclization can be achieved across target peptide sequences of variable length (5–10mer) and composition (Figure 2). As observed for the 5mer target sequence, however, these reactions also led to the accumulation of the hydrolysis byproduct in approximately 3:1 (6mer) to 2:1 (7mer to 10mer) excess over the macrocycle. Since the 10mer sequence also contains a lysine residue, this construct also provided an opportunity to probe the chemoselectivity of the cyclization reaction. Accordingly, a control protein construct (Table 1, Entry 6) was prepared by incorporating O-propargyl-tyrosine (OpgY)43 instead of pAmF upstream of the 10mer target sequence. OpgY lacks a nucleophilic side-chain and is thus not expected to react with the C-terminal thioester. Incubation of the CBD-10mer(OpgY) construct with thiophenol resulted in the formation of hydrolysis product (Figure S1), thus confirming that lysine does not mediate the cyclization process under the applied conditions.
Figure 2.
pAmF-mediated cyclization of 6mer to 10mer peptide sequences. MALDI-TOF MS spectra for the reactions with the indicated precursor proteins after incubation with 10 mM thiophenol and 10 mM TCEP (24 hours). The calculated and observed m/z values for the macrocyclic (m) and hydrolyzed (h) products are shown.
Although the results above supported the feasibility of the general strategy outlined in Scheme 1, they also showed that the desired thioester aminolysis reaction mediated by pAmF does not effectively outcompete hydrolysis of the thioester intermediate. This phenomenon can be attributed to the limited nucleophilicity of aryl amino group of pAmF in the context of the present reaction. The noticeable increase in the pAmF-mediated macrocyclization efficiency observed with the longer, 7mer to 10mer target sequences (Figures 1–2) also suggests that unfavourable conformational constraints could be another factor contributing to the limited cyclization efficiency of the pAmF-containing proteins in the context of smaller rings (i.e., for 5mer to 6mer target sequences).
Identification of O-2-aminoethyl-tyrosine-tRNA synthetase
The considerations above prompted the design of an alternative amino-functionalized unnatural amino acid capable of mediating macrocyclization with higher efficiency. To this end, we identified O-2-aminoethyl-tyrosine (O2ameY, Scheme 1) as a promising candidate. Compared to pAmF, the side-chain alkyl amino functionality in O2ameY was expected to be more sterically accessible to nucleophilic attack to the C-terminal thioester group (Scheme 1), thus favouring cyclization over hydrolysis of the thioester intermediate. This hypothesis was also supported by our previous observations in the context of peptide cyclization promoted by amino-thiol unnatural amino acids.35 Furthermore, the estimated pKa of the amino group in O2ameY (~9) was deemed suitable for mediating the desired cyclization process under near-neutral conditions.
Although a wide variety of tyrosine- and phenylalanine-derivatives have been successfully incorporated into proteins using engineered variants of the tyrosyl-tRNA synthetase from Methanococcus jannaschii or the pyrrolysyl-tRNA synthetase from Methanosarcina sp.,38,44,45 the incorporation of unnatural amino acids containing polar side-chains have been very challenging due to the hydrophobic nature of the active sites in these enzymes.46 Accordingly, finding a AARS variant for the efficient recognition and incorporation of O2ameY into the precursor protein was expected to be very difficult. To overcome this problem, we envisioned an alternative approach that entails the incorporation of the unnatural amino acid O-2-azidoethyl-tyrosine (O2azeY), followed by unmasking of the desired alkyl amino functionality via a Staudinger reduction of the azide group by means of the phosphine-based reducing agent (i.e., TCEP) added during the cyclization reaction.
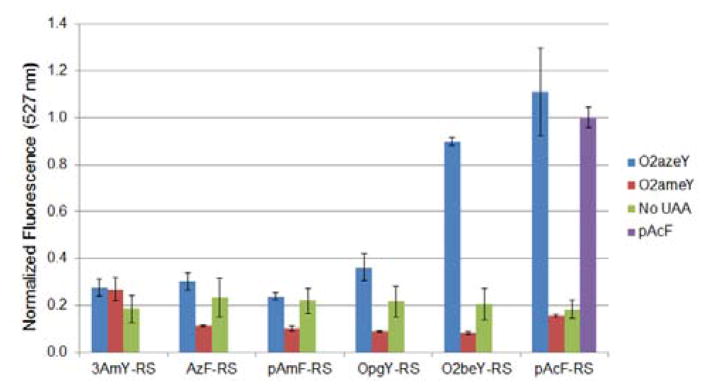
To identify a suitable AARS for incorporation of O2azeY, we screened a panel of M. jannaschii tyrosyl-tRNA synthetase variants previously engineered for the incorporation of para-substituted tyrosine and phenylalanine derivatives such as 3-aminotyrosine (3AmY)47, p-azidophenylalanine (AzF)48, p-aminophenylalanine (pAmF)49, O-propargyltyrosine (OpgY)43, O-(2-bromoethyl)-tyrosine (O2beY)36, and p-acetylphenylalanine (pAcF)50 (Figure 3). The ability of these AARSs to incorporate O2azeY in response to an amber stop codon was measured by means of a Yellow Fluorescent Protein (YFP) reporter protein containing a TAG codon within the N-terminal region of the protein sequence.35 In this assay, the suppression efficiency and fidelity of each synthetase is measured based on the relative expression levels of the reporter YFP protein in the presence and in the absence of O2azeY, respectively. As shown in Figure 3, both O2beY-RS and pAcF-RS were found to efficiently incorporate O2azeY into the reporter YFP protein. Notably, the protein yield obtained with pAcF-RS in the presence of O2azeY is comparable to that observed in the presence of pAcF, for which this enzyme variant was selected. Despite the structural differences between pAcF and O2azeY, this result is in line with the ‘polyspecificity’ often exhibited by engineered AARS enzymes toward unnatural amino acids other than those involved in the functional selection process.51 As expected, none of the tested AARS variants were able to efficiently incorporate O2ameY (Figure 3), likely reflecting the incompatibility of its polar side-chain with the hydrophobic environment surrounding the para position of the tyrosine substrate in the M. jannaschii tyrosyl-tRNA synthetase structure.52 Because of its superior performance in the YFP assay, pAcF-RS was selected for preparation of the O2azeY-containing proteins.
Figure 3.
Screening of engineered aminoacyl-tRNA synthetases for ribosomal incorporation of O2azeY. The graph reports the relative fluorescence measured in the YFP reporter assay for the panel of AARSs in the presence of O2azeY, O2ameY, and no unnatural amino acid. Data are normalized to the fluorescence value obtained with pAcF-RS in the presence of pAcF.
To our surprise, the purified O2azeY-containing YFP protein did not show the expected molecular weight but rather a mass that is consistent with the quantitative reduction of the side-chain azido group of O2azeY to the corresponding amine derivative, to effectively yield a O2ameY-containing protein. Since pAcF-RS is unable to incorporate O2ameY directly (Figure 3), it can be derived that the ‘in vivo’ conversion of O2azeY to O2ameY must occur at the post-translational level, that is after O2azeY is charged onto the suppressor tRNA and incorporated into the protein by ribosomal synthesis. Partial reduction of azido-containing groups in living cells has been previously reported53 but not in the context of compounds like OazeY.54 Importantly, our serendipitous discovery that O2azeY is efficiently converted to O2ameY during protein expression in E. coli cells provides a streamlined approach to obtain O2ameY-containing recombinant proteins for the present and other applications.
O2ameY-mediated macrocyclization
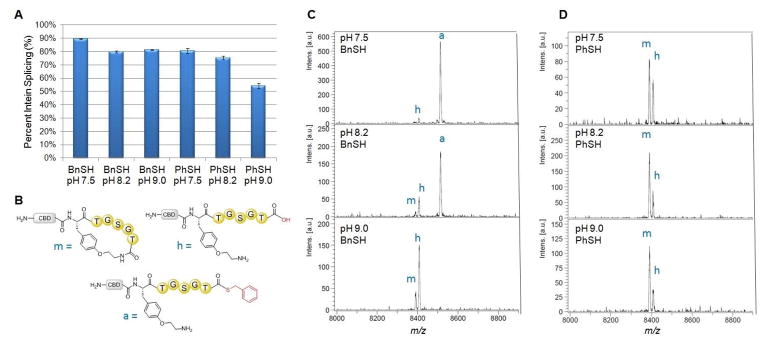
Having acquired the capability of producing O2ameY-containing proteins in an efficient manner, we proceeded to investigate the feasibility and functionality of the envisioned O2ameY-mediated macrocyclization strategy (Scheme 1). To this end, we expressed and isolated the model 5mer construct CBD-5mer(O2ameY), in which O2ameY is placed upstream of the target sequence TGSGT. To identify optimal conditions for macrocyclization, this precursor protein was incubated under reducing (TCEP) conditions in the presence of either benzylmercaptan or thiophenol and at varying pH (7.5, 8.2, and 9.0), (Figure 4). The products of these reactions were analysed by MALDI-TOF MS and the extent of intein cleavage was measured by SDS-PAGE followed by densitometric analysis of the gel bands corresponding to full-length protein and cleaved GyrA intein (Figures S2 and S3).35
Figure 4.
O2ameY-mediated peptide macrocyclization. A) Extent of intein splicing as measured by SDS-PAGE gel densitometry for CBD-5mer(O2ameY) after 24 h with benzylmercaptan (BnSH) or thiophenol (PhSH) at varying pH. b) Representative structures of ‘m’ , ‘h’ , and ‘a’. MALDI-TOF mass spectra of CBD-5mer(O2ameY) after 24 h with c) benzyl mercaptan and d) thiophenol at pH 7.5, 8.2, and 9.0. ‘m’ : macrocycle, ‘h’ : acyclic hydrolysis product, ‘a’ : acyclic thioester product.
As summarized in Figure 4A, a high degree of intein splicing (75–90%) was observed across most of the conditions tested, consistent with the ability of both thiol nucleophiles to induce intein cleavage via transthioesterification. Despite the generally higher levels of intein cleavage in the presence of benzyl mercaptan, in particular at pH 7.5, these reactions mainly resulted in the formation of the acyclic thioester intermediate (‘a’) along with small quantities of the hydrolysis byproduct (‘h’). At higher pH (9.0) however, the acyclic intermediate is fully converted to desired macrocyclic product (‘m’) along with the hydrolysis byproduct (Figure 4C) in an approximately 1:4 ratio.
In stark contrast, the thiophenol-catalyzed reactions were found to yield the desired macrocyclic peptide as the major product across all the tested pH values (Figure 4D). As the pH is raised from pH 7.5 to 9.0, macrocyclization becomes more favourable over hydrolysis, as reflected by an increase of the macrocycle (‘m’) : hydrolysis product (‘h’) ratio from 3:2 to 7:3. Interestingly, the thiophenol thioester intermediate is not observed in these reactions, even at earlier time points (data not shown), indicating that, once formed, this species is rapidly converted to the macrocyclic product via O2ameY-mediated aminolysis or is hydrolyzed. The higher efficiency of the macrocyclization process at higher pH can be rationalized considering that a larger fraction of O2ameY side-chain amine is deprotonated at more alkaline conditions and thus become available for nucleophilic attack to the thiophenol thioester group. On the other hand, the better results obtained with PhSH vs. BnSH is consistent with the higher reactivity of thiophenol thioesters as compared to the benzyl mercaptan thioester counterparts.55 On the basis of these these experiments, we established that macrocyclization of the OameY-containing constructs occurs most efficiently at pH 8.2 with the addition of thiophenol as the catalyst, as these conditions provide an optimal combination of high intein cleavage (75%) with high conversion to the desired macrocyclic product (70–80%). These results also clearly demonstrated the superiority of the O2ameY-based cyclization strategy over that based on pAmF-containing constructs described earlier.
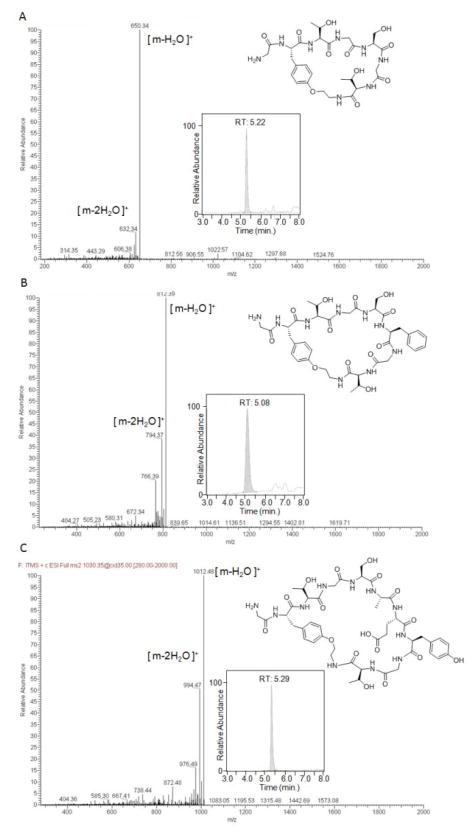
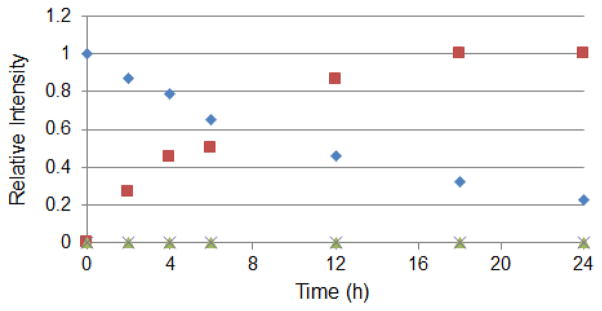
To assess the value of the O2ameY-based approach toward generating cyclic peptides of low to medium MW (600–1,000 Da), 5mer to 8mer target peptide sequences lacking a N-terminal tail were investigated (Table 1, entries 7–9). Following expression and purification, the corresponding precursor proteins were subjected to the optimized reaction conditions (10 mM PhSH, 10 mM TCEP, pH 8.2), followed by LC-MS analysis. Notably, all of these reactions led to the desired small-MW peptide macrocycle as the only product, whose cyclic structure was further corroborated by the characteristic MS/MS fragmentation profile (Figure 5). Based on the measured extent of intein cleavage and the lack of observation of the thioester intermediate or hydrolysis byproduct, the macrocycle conversion yield of these reactions was estimated to be around 65–80%. To gain further insight into the kinetics of these reactions, the cyclization of the 5mer(O2ameY) construct was monitored over time (Figure 6). These studies releaved that the precursor protein had undergone 50% intein cleavage after 6 hours and reached a maximum of 80% cleavage after 18–24 hours (Figure S4). The incomplete cleavage even at extended reaction times suggests that a fraction of the precursor protein carries a non-functional GyrA intein, which is unable to undergo the thiol-induced transthioesterification, possibly due to partial misfolding during the expression and/or purification step. In concomitance with the cleavage of the precursor protein, the macrocyclic product was found to progressively accumulate over time. Interestingly, no accumulation of the thioester intermediate was observed at any of the time points. Altogether, these results indicate that the rate-limiting step of the overall reaction is the thiophenol-induced transthioesterification step and that all of the precursor protein capable of reacting with the thiol catalyst undergoes O2ameY-mediated cyclization. As shown by the plot of Figure 6, about 50% and 80% of the total macrocyclic product is formed after 4.5 and 12 hours, respectively.
Figure 5.
Chemical structures, LC-MS ion extract chromatograms (inserts), and MS/MS spectra for macrocyclic peptide products obtained via O2ameY-mediated cyclization of 5mer(OameY), 6mer(OameY), and 8mer(OameY) precursor proteins.
Figure 6.
Time course experiment for cyclization of 5mer(OameY) construct. The graph plots the % of intein cleavage (
 ) and the relative amount of macrocyclic product (
) and the relative amount of macrocyclic product (
 ), thioester intermediate (
), thioester intermediate (
 ), and hydrolysis byproduct (
), and hydrolysis byproduct (
 ) over time as measured by LC-MS (Figure S4).
) over time as measured by LC-MS (Figure S4).
Synthesis of streptavidin-binding cyclic peptide
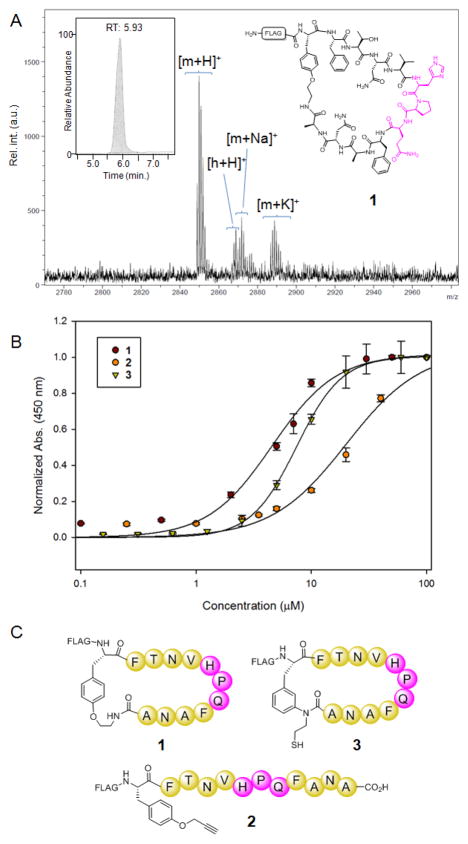
To demonstrate the utility of this approach toward generating functional peptide macrocycles, a biosynthetic precursor encompassing a histidine-proline-glutamine (HPQ) motif known to bind streptavidin56–58 was generated (Table 1, entry 10). Specifically, this protein construct contained an N-terminal FLAG affinity tag (MDYKDDDYK), followed by a linker sequence (GSSG), and a 11mer target sequence (FTNVHPQFANA) flanked by O2ameY and GyrA. Subjecting the precursor protein Strep-11mer(O2ameY) to the standard reaction conditions led to the production of the desired macrocyclic peptide 1, as confirmed by LC-MS and MALDI-TOF MS (Figure 7A). In the reaction only a minimal fraction (<5%) of the hydrolysis byproduct was observed (‘h’, Figure 7A), demonstrating the efficiency of the macrocyclization reaction also in context of a 11mer target sequence and in the presence of alternative residues (i.e., Ala instead of Thr) at the ‘I-1’ position preceding the intein. Based on the observed extent of intein cleavage (63%), the macrocycle yield for this transformation was estimated to be ~60%. In contrast, sufficient amounts of the pAmF-cyclized peptide for streptavidin binding studies could not be isolated, primarily due to inefficient cyclization in the presence of this unnatural amino acid as observed in the context of the model peptide sequences (Figure 2).
Figure 7.
Streptavidin-binding peptides. A) Chemical structure, LCMS ion extract chromatogram (insert), and MALDI-TOF MS spectrum of macrocycle 1 obtained via cyclization of Strep-11mer(O2ameY). [m+H]+ calc: 2849.1; obs: 2849.8. B–C) Streptavidin binding curves and schematic structure for O2ameY-cyclized peptide 1 (KD: 5.1 μM ± 0.4), MeaF-cyclized peptide 3 (KD = 7.7 μM ± 0.6), and the linear peptide 2 (KD = 22.4 μM ± 1.8).
In order to evaluate the effect of cyclization on the streptavidin binding affinity of the peptide, the control linear peptide FLAG-(OpgY)-FTNVHPQFANA-CO2H (2) was also prepared. The latter carries the non-nucleophilic O-propargyl-tyrosine (OpgY) in place of O2ameY and was produced by thiophenol-induced cleavage of the corresponding precursor protein (Table 1, entry 11) followed by thioester hydrolysis under alkaline conditions (pH 8.0). The successful isolation of this peptide in the linear form further demonstrated the excellent chemoselectivity of the O2ameY-mediated cyclization process, as none of the several nucleophilic residues present in the target sequence and FLAG tag caused cyclization of the peptide under the applied reaction conditions.
The dissociation constants (KD) for the cyclic and linear peptide were determined by means of an immunoassay, in which the FLAG-tagged peptides were incubated on streptavidin-coated plates at varying concentrations between 0.05–40 μM. The amount of the streptavidin-bound peptide was then measured based on the corresponding colorimetric signal (450 nm), after incubation with a horseradish peroxidase (HRP) conjugated anti-FLAG antibody followed by addition of the HRP substrate o-phenylinediamine. In this assay, the O2ameY-containing macrocycle exhibited a KD of 5.1 μM, whereas the linear peptide exhibited a KD of 22.4 μM (Figure 7B). The 4-fold lower KD for the cyclic peptide compared to its linear counterpart clearly evidenced the beneficial effect of the macrocyclic backbone toward improving the binding affinity for the target protein. A direct comparison was also made with the cyclopeptide 3 (Figure 7C), which comprises an identical target sequence as 1 (FTNVHPQFANA) but features a different side-chain-to-tail linkage as obtained via cyclization by means of 3-(2-mercapto-ethyl)amino-phenylalanine (MeaF)35. In this assay, the MeaF-cyclized peptide 3 exhibited an approximately 1.5-fold higher KD (7.7 μM) than the O2ameY-cyclized peptide 1 (Figure 7B). Since the peptide-streptavidin interaction is primarily driven by the HPQ motif35 and is not expected to directly involve the intramolecular linkage, the differential binding affinity of these compounds nicely illustrates the subtle effects of the side-chain-to-tail linkage in modulating the protein binding properties of these macrocyclic peptides.
Conclusion
We have developed a new methodology to achieve the formation of macrocyclic peptides via an intramolecular ligation of amino-functionalized unnatural amino acids to the C-terminus of a genetically encoded peptide. Using this approach, 5- to 11-amino acid-long peptides could be efficiently cyclized to generate medium- to large-membered rings (23–44 atoms). In these compounds, the N-terminal tail can be readily altered to include either a single residue or various affinity tags to facilitate the immobilization, isolation, and/or detection of these macrocyles for characterization purposes. Importantly, this strategy could be applied to generate a cyclic peptide with improved streptavidin-binding affinity, highlighting the effect of macrocyclization and the type of side-chain-to-tail connectivity toward modulating this functional property.
Another important result of this study concerns the discovery that the azido-containing unnatural amino acid O2azeY is consistently and quantitatively reduced to the amine-containing residue O2ameY in living E. coli cells. While the exact mechanism of this process was not investigated, it is most likely dependent upon the reducing intracellular environment provided by the bacterial host. As exemplified by the present study, this in vivo azide reduction process could be exploited to produce proteins containing O2ameY as well as other amino-functionalized unnatural amino acids, a currently challenging task using commonly adopted aminoacyl-tRNA synthetases for amber stop codon suppression. These results are also insightful in that they provide a cautionary note about the often assumed but clearly not universal, bioorthogonal nature of azido-based functionalities in biological systems.
Experimental Procedures
General
Chemical reagents and solvents were purchased from Sigma–Aldrich, Acros Organics, and Fluka. Silica gel chromatography purifications were carried out by using AMD Silica Gel 60 230–400 mesh. 1H and 13C NMR spectra were recorded on Bruker Avance spectrometers by using solvent peaks as reference. LC-MS analyses were performed on a Thermo Scientific LTQ Velos ESI/ion-trap mass spectrometer coupled to an Accela U-HPLC. MALDI-TOF spectra were acquired on a Bruker Autoflex III MALDI-TOF spectrometer by using a stainless steel MALDI plate and sinapinic acid as matrix.
Synthesis of unnatural amino acids
Synthetic procedures and spectral data relative to the preparation and characterization of O2azeY and O2ameY are reported in the Supplementary Information. OpgY was synthesized as described previously.32
Plasmid constructs
The biosynthetic precursors described in Table 1 were expressed using pET22-based plasmids and the cloning procedures for preparation of these constructs were reported previously.32,36 The plasmids for expression of the amber stop codon suppression system were derived from pEVOL59 and their preparation was described previously.35,36
Protein Expression and Purification
Proteins were expressed in E. coli BL21(DE3) cells cotransformed with the pET22-based plasmid for expression of the protein precursor and the pEVOL-based vector for expression of the appropriate AARS (pAmF-RS, pAzF-RS, 3AmY-RS, pAcF-RS, OpgY-RS, O2beY-RS). After overnight growth, cells were used to inoculate M9 media supplemented with ampicillin (50 mg L−1), chloramphenicol (34 mg L−1) and glycerol (1%). At an OD600 of 0.6, the culture was supplemented with the appropriate unnatural amino acid (pAmF, O2azeY, or OpgY) at a final concentration of 2 mM and expression of the AARS was induced by addition of L-arabinose (0.06%). After one hour of shaking at 30ºC, protein expression was induced by addition of 0.25mM isopropyl-β-D-thiogalactopyranoside (IPTG). Cultures were grown for an additional 12 hours at 27ºC and harvested by centrifugation at 3400 g. Cells were resuspended in Tris buffer (50 mM; pH 7.4) containing NaCl (150 mM) and imidazole (20 mM) and lysed by sonication. Protein was purified from the cell lysate by Ni-NTA affinity chromatography using Tris buffer (50 mM; pH 7.4; NaCl 150 mM) with 20 mM imidazole for washing and 300 mM imidazole for elution. The recovered protein was exchanged into potassium phosphate buffer (50 mM, NaCl 150 mM, pH 7.4), concentrated, and stored at −80C.
Macrocyclization reactions
Reactions were carried out incubating the precursor protein (100 μM) in potassium phosphate buffer (50 mM, NaCl 150 mM) in the presence of TCEP (10 mM) and thiophenol or benzyl mercaptan (10 mM). The pH of the reaction was adjusted to the specified value, and the reaction was gently mixed for up to 24 hours at room temperature. Protein splicing of the CBD-fused substrates was monitored and quantified by SDS-PAGE and densitometric analysis of the gel bands corresponding to the full-length protein (24–31 kDa) and spliced GyrA intein (~22 kDa) using ImageJ software. The low MW products (2–9 kDa) were analyzed by MALDI-TOF MS. The products of the reactions with the biosynthetic precursors without N-terminal CBD were analysed by LC-MS. The extent of intein cleavage was determined based on the deconvoluted MS spectra for the full-length protein and spliced GyrA intein, whereas the relative amount of the macrocyclic product, hydrolysis product, and thiophenol thioester intermediate were estimated based on the corresponding extracted-ion chromatograms. Each reaction was carried out at least in duplicate.
Fluorescence assay for AARS screening
E. coli BL21(DE3) cells were co-transformed with a pET22-based plasmid encoding for MetGly(amber stop)YFP-His6 and a pEVOL-based plasmid encoding for an appropriate aminoacyl-tRNA synthetase and cognate amber suppressor tRNA. Cells were then grown in LB media containing ampicillin (50 mg/L) and chloramphenicol (26 mg/L) at 37°C overnight. The overnight cultures were used to inoculate 96-deep well plates containing M9 minimal media. At an OD600 of 0.6, cell cultures were induced by adding arabinose (0.06%), IPTG (0.2 mM), and the appropriate unnatural amino acid (final concentration of 1 mM for L-isomer). After overnight growth at 27°C, the cell cultures were diluted (1:1) with phosphate buffer (50 mM, 150 mM NaCl, pH 7.5) and fluorescence intensity (λex = 514 nm; λem = 527 nm) was determined using a Tecan Infinite 1000 plate reader. Cell cultures containing no unnatural amino acid were included as negative controls. Each sample was measured in triplicate. The fluorescence values were normalized to that obtained for cells expressing pAcF-RS in the presence of pAcF.
Streptavidin binding studies
Cyclic peptide 1 was prepared according to the macrocyclization conditions described above. Linear peptide 2 was produced by thiophenol-induced cleavage of the corresponding precursor protein (Table 1, entry 11) followed by thioester hydrolysis under alkaline conditions (pH 9.0). Following completion of the reactions (24 hours), the pH was adjusted to 7.0 and the solution was dialyzed against water overnight with a 500 Da cut-off membrane. The reaction was then filtered through a 10 kDa cut-off centrifugal membrane (Millipore) at 3600 g. The filtrate was lyophilized and the peptide was resuspended in Tris buffer (50 mM, 150 mM NaCl, pH 7.4). A solution of each peptide (100 uL), was added to NeutrAvidin-coated 96-well plates (Pierce) at various concentrations (0.1 – 40 μM) in triplicate. After incubation at room temperature for 2 hours, the wells were then washed three times with 200 μL of Tris buffer (50 mM, 150 mM NaCl, pH 7.4) containing 0.5% Tween 20. A solution of anti-FLAG-antibody-HRP conjugate (100 uL, 1:2500 dilution in Tris buffer) was added to each well and incubated for 1 hour. The wells were washed three times with 200 μL Tris buffer containing 0.5% Tween 20. Finally, 100 μL of SigmaFast OPD solution was added to each well, and the absorbance at 450 nm was measured after 20 minutes. KD values were calculated with SigmaPlot via fitting of the dose-response curves using a 1:1 binding model.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institutes of Health (grant R21 CA187502). MS instrumentation was supported by the U.S. National Science Foundation (grants CHE-0840410 and CHE-0946653).
Footnotes
Electronic Supplementary Information (ESI) available: Synthetic procedures and spectral data for O2azeY and O2ameY. See DOI: 10.1039/b000000x/
References
- 1.Marsault E, Peterson ML. J Med Chem. 2011;54:1961. doi: 10.1021/jm1012374. [DOI] [PubMed] [Google Scholar]
- 2.Cardote TA, Ciulli A. ChemMedChem. 2015 doi: 10.1002/cmdc.201500450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villar EA, Beglov D, Chennamadhavuni S, Porco JA, Jr, Kozakov D, Vajda S, Whitty A. Nat Chem Biol. 2014;10:723. doi: 10.1038/nchembio.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bionda N, Fasan R. 2015;16:2011. doi: 10.1002/cbic.201500179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yudin AK. Chem Sci. 2015;6:30. doi: 10.1039/c4sc03089c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan EM, Walsh CT. Chembiochem. 2009;10:34. doi: 10.1002/cbic.200800438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fairlie DP, Tyndall JD, Reid RC, Wong AK, Abbenante G, Scanlon MJ, March DR, Bergman DA, Chai CL, Burkett BA. J Med Chem. 2000;43:1271. doi: 10.1021/jm990315t. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Liao W, Arora PS. Angew Chem Int Ed Engl. 2005;44:6525. doi: 10.1002/anie.200501603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JM, Frost JR, Fasan R. Chem Commun (Camb) 2014;50:5027. doi: 10.1039/c4cc01199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezai T, Bock JE, Zhou MV, Kalyanaraman C, Lokey RS, Jacobson MP. J Am Chem Soc. 2006;128:14073. doi: 10.1021/ja063076p. [DOI] [PubMed] [Google Scholar]
- 12.Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. J Am Chem Soc. 2006;128:2510. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- 13.Bock JE, Gavenonis J, Kritzer JA. ACS Chem Biol. 2013;8:488. doi: 10.1021/cb300515u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian Z, Liu T, Liu YY, Briesewitz R, Barrios AM, Jhiang SM, Pei D. ACS Chem Biol. 2013;8:423. doi: 10.1021/cb3005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizo J, Gierasch LM. Annu Rev Biochem. 1992;61:387. doi: 10.1146/annurev.bi.61.070192.002131. [DOI] [PubMed] [Google Scholar]
- 16.Dias RL, Fasan R, Moehle K, Renard A, Obrecht D, Robinson JA. J Am Chem Soc. 2006;128:2726. doi: 10.1021/ja057513w. [DOI] [PubMed] [Google Scholar]
- 17.Diana D, Basile A, De Rosa L, Di Stasi R, Auriemma S, Arra C, Pedone C, Turco MC, Fattorusso R, D’Andrea LD. J Biol Chem. 2011;286:41680. doi: 10.1074/jbc.M111.257402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quartararo JS, Wu P, Kritzer JA. Chembiochem. 2012;13:1490. doi: 10.1002/cbic.201200175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White CJ, Yudin AK. Nat Chem. 2011;3:509. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- 20.Smith JM, Frost JR, Fasan R. J Org Chem. 2013;78:3525. doi: 10.1021/jo400119s. [DOI] [PubMed] [Google Scholar]
- 21.Frost JR, Smith JM, Fasan R. Curr Opin Struct Biol. 2013;23:571. doi: 10.1016/j.sbi.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 22.O’Neil KT, Hoess RH, Jackson SA, Ramachandran NS, Mousa SA, DeGrado WF. Proteins. 1992;14:509. doi: 10.1002/prot.340140411. [DOI] [PubMed] [Google Scholar]
- 23.Wrighton NC, Farrell FX, Chang R, Kashyap AK, Barbone FP, Mulcahy LS, Johnson DL, Barrett RW, Jolliffe LK, Dower WJ. Science. 1996;458 doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 24.Tavassoli A, Benkovic SJ. Angew Chem Int Ed Engl. 2005;44:2760. doi: 10.1002/anie.200500417. [DOI] [PubMed] [Google Scholar]
- 25.Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. ACS Chem Biol. 2008;3:757. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 26.Morimoto J, Hayashi Y, Suga H. Angew Chem Int Ed Engl. 2012;51:3423. doi: 10.1002/anie.201108118. [DOI] [PubMed] [Google Scholar]
- 27.Heinis C, Rutherford T, Freund S, Winter G. Nat Chem Biol. 2009;5:502. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 28.Kawakami T, Ishizawa T, Fujino T, Reid PC, Suga H, Murakami H. ACS Chem Biol. 2013 doi: 10.1021/cb300697h. [DOI] [PubMed] [Google Scholar]
- 29.Baeriswyl V, Rapley H, Pollaro L, Stace C, Teufel D, Walker E, Chen S, Winter G, Tite J, Heinis C. ChemMedChem. 2012;7:1173. doi: 10.1002/cmdc.201200071. [DOI] [PubMed] [Google Scholar]
- 30.Smith JM, Vitali F, Archer SA, Fasan R. Angew Chem Int Ed Engl. 2011;50:5075. doi: 10.1002/anie.201101331. [DOI] [PubMed] [Google Scholar]
- 31.Satyanarayana M, Vitali F, Frost JR, Fasan R. Chem Commun (Camb) 2012;48:1461. doi: 10.1039/c1cc13533c. [DOI] [PubMed] [Google Scholar]
- 32.Frost JR, Vitali F, Jacob NT, Brown MD, Fasan R. Chembiochem. 2013;14:147. doi: 10.1002/cbic.201200579. [DOI] [PubMed] [Google Scholar]
- 33.Smith JM, Hill NC, Krasniak PJ, Fasan R. Org Biomol Chem. 2014;12:1135. doi: 10.1039/c3ob42222d. [DOI] [PubMed] [Google Scholar]
- 34.Bionda N, Fasan R. Chembiochem. 2015;16:2011. doi: 10.1002/cbic.201500179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frost JR, Jacob NT, Papa LJ, Owens AE, Fasan R. ACS Chem Biol. 2015;10:1805. doi: 10.1021/acschembio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bionda N, Cryan AL, Fasan R. ACS Chem Biol. 2014;9:2008. doi: 10.1021/cb500311k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Morales-Sanfrutos J, Angelini A, Cutting B, Heinis C. Chembiochem. 2012;13:1032. doi: 10.1002/cbic.201200049. [DOI] [PubMed] [Google Scholar]
- 38.Liu CC, Schultz PG. Annu Rev Biochem. 2010;79:413. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 39.Dirksen A, Hackeng TM, Dawson PE. Angew Chem Int Ed Engl. 2006;45:7581. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 40.Crisalli P, Kool ET. J Org Chem. 2013;78:1184. doi: 10.1021/jo302746p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. J Am Chem Soc. 2003;125:935. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]
- 42.Telenti A, Southworth M, Alcaide F, Daugelat S, Jacobs WR, Perler FB. J Bacteriol. 1997;179:6378. doi: 10.1128/jb.179.20.6378-6382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deiters A, Schultz PG. Bioorg Med Chem Lett. 2005;15:1521. doi: 10.1016/j.bmcl.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 44.Lang K, Chin JW. Chem Rev. 2014;114:4764. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 45.Dumas A, Lercher L, Spicer CD, Davis BG. Chem Sci. 2015;6:50. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen DP, Elliott T, Holt M, Muir TW, Chin JW. J Am Chem Soc. 2011;133:11418. doi: 10.1021/ja203111c. [DOI] [PubMed] [Google Scholar]
- 47.Seyedsayamdost MR, Xie J, Chan CT, Schultz PG, Stubbe J. J Am Chem Soc. 2007;129:15060. doi: 10.1021/ja076043y. [DOI] [PubMed] [Google Scholar]
- 48.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. J Am Chem Soc. 2002;124:9026. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 49.Santoro SW, Wang L, Herberich B, King DS, Schultz PG. Nat Biotechnol. 2002;20:1044. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- 50.Wang L, Zhang Z, Brock A, Schultz PG. Proc Natl Acad Sci USA. 2003;100:56. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young DD, Young TS, Jahnz M, Ahmad I, Spraggon G, Schultz PG. Biochemistry. 2011;50:1894. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Nat Struct Biol. 2003;10:425. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 53.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. Biochemistry. 2013;52:1828. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuley A, Wang YS, Fang X, Kurra Y, Rezenom YH, Liu WR. Chem Commun (Camb) 2014;50:2673. doi: 10.1039/c3cc49068h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dawson PE, Muir TW, Clarklewis I, Kent SBH. Science. 1994;266:776. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 56.Bayer EA, Ben-Hur H, Wilchek M. Methods Enzymol. 1990;184:80. doi: 10.1016/0076-6879(90)84262-f. [DOI] [PubMed] [Google Scholar]
- 57.Katz BA. Biochemistry. 1995;34:15421. doi: 10.1021/bi00047a005. [DOI] [PubMed] [Google Scholar]
- 58.Giebel LB, Cass RT, Milligan DL, Young DC, Arze R, Johnson CR. Biochemistry. 1995;34:15430. doi: 10.1021/bi00047a006. [DOI] [PubMed] [Google Scholar]
- 59.Young TS, Ahmad I, Yin JA, Schultz PG. J Mol Biol. 2010;395:361. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.