Abstract
Macromolecular crystallography at synchrotron sources has proven to be the most influential method within structural biology, producing thousands of structures since its inception. While its utility has been instrumental in progressing our knowledge of structures of molecules, it suffers from limitations such as the need for large, well-diffracting crystals, and radiation damage that can hamper native structural determination. The recent advent of X-ray free electron lasers (XFELs) and their implementation in the emerging field of serial femtosecond crystallography (SFX) has given rise to a remarkable expansion upon existing crystallographic constraints, allowing structural biologists access to previously restricted scientific territory. SFX relies on exceptionally brilliant, micro-focused X-ray pulses, which are femtoseconds in duration, to probe nano/micrometer sized crystals in a serial fashion. This results in data sets comprised of individual snapshots, each capturing Bragg diffraction of single crystals in random orientations prior to their subsequent destruction. Thus structural elucidation while avoiding radiation damage, even at room temperature, can now be achieved. This emerging field has cultivated new methods for nanocrystallogenesis, sample delivery, and data processing. Opportunities and challenges within SFX are reviewed herein.
Introduction
Since the invention of the first light bulb by Thomas Alva Edison in 1879, light sources have been the primary tools for the investigation of matter. But it was the discovery of X-rays in 1895 by Wilhelm Conrad Röentgen which catalyzed a revolutionary change in our understanding of the physical world. The ability of X-rays to reveal the invisible has made them one of the most important research and diagnostic tools in medicine, chemistry, and physics in the last century. One of the most powerful sources of X-rays in modern research are synchrotrons, in which electrons are accelerated by radiofrequency cavities to extremely high energies (typically ≥ 3 GeV) and transferred to a storage ring in which they can be maintained at high current in stable ‘bunches’ for hours. The electrons are then subsequently perturbed in an undulating electromagnetic field to induce X-ray emission. The first synchrotron facility was built in the early 1970's and due to the increasing demand from both the scientific community and private research sector; the number of synchrotron facilities has grown quickly since. In the last three decades, synchrotrons have been built or are in the construction phase in 24 countries. APS at the Argonne National Laboratory (Chicago, USA), the ESRF (Grenoble, France), PETRA III (Hamburg, Germany), Spring-8 (Harima Science Park City, Japan), and DIAMOND (Oxfordshire, England) are currently the largest and most powerful synchrotron light sources. X-ray methods have also played an increasingly more important role in the life sciences as evidenced by medical X-ray imaging, X-ray scattering, and highlighted here, X-ray crystallography. The latter typically makes use of X-rays from 3.5 keV to 20 keV (3.5 Å to 0.6 Å) to enable the determination of the atomic structure of matter. The most difficult and challenging biomolecules, such as large complexes and membrane proteins, are almost exclusively solved with synchrotron radiation. However, in spite of the usefulness and power of these facilities, there are two important areas in which synchrotron facilities are limited in addressing the full range of current scientific challenges. The main limitations are primary and secondary X-ray damage which cannot be outrun by pulse durations currently available at synchrotron sources (tens of ps). Secondary damage can be minimized by freezing but primary damage is unavoidable.
To overcome these limitations a new light source has emerged in the form of the X-ray free electron laser (XFEL) which has improved upon many of the properties of synchrotron radiation sources, in some cases by orders of magnitude. Free electron lasers (FELs) have been used over many years since their conception by John Madey in the early 70's7, initially operating at infrared wavelengths. More recently, visible and near ultraviolet wavelengths were achieved9. Since its discovery, the idea of extending FELs to shorter wavelengths, in particular to the X-ray regime, have been considered and explored by many scientists (see 9 for a review on the development of FELs). After many years of theoretical and experimental work along with new technological advances, three XFELs are currently operational. Namely, they are the Deutsches Elektronen-Synchrotron's (DESY) Free-electron LASer in Hamburg (FLASH), SLAC's Linac Coherent Light Source (LCLS), and RIKEN's Spring-8 Angstrom Compact free electron LAser (SACLA). At its core, an XFEL consists of an electron source, a linear accelerator, and undulator magnets spaced to produce X-ray wavelengths (typically ranging from 0.01 to 10 nm) 7,12,13. Briefly, a relativistic electron beam is accelerated to almost the speed of light in a linear accelerator prior to interaction with the undulator. Upon interaction with the undulator, the electrons move in curved paths via the magnets. The long length of an XFEL undulator allows the relativistic electrons to interact with their emitted radiation which causes bunching of the electrons with spacing equal to the wavelength of the emitted X-rays. As the electrons bunch, their emission becomes more coherent and allows for a stronger interaction between the two. This results in beam with highly coherent pulses with femtosecond duration. The coherence causes radiative emission proportional to the number of electrons squared in contrast to a synchrotron (incoherent) in which the radiative emission scales proportional to the number of electrons due to cancelation effects from the out of phase generated electromagnetic fields16. Because the number of electrons in a bunch is on the order of a billion, this causes a massive increase in flux compared to synchrotron sources. Furthermore, the high longitudinal coherence (and thus, ultrashort duration) of the pulses pushes the peak brilliance orders of magnitude higher than that achievable at a synchrotron. The output wavelength can by tuned by modulating the electron energy and magnetic field strength. Figure 1 illustrates this principle. XFELs generate high gain, ultrashort pulsed X-rays with only a single undulator pass18, accomplished by the interaction between oscillating electrons, moving at relativistic speeds, with their emitted electromagnetic waves (for a more detailed overview of XFEL physics please refer to Ackermann, et al. 19 and McNeil, et al. 21). The interaction between the electrons and their radiation causes spatial redistribution of the electrons into compact bunches perpendicular to their direction of motion. This generates coherent X-ray pulses with durations of tens of femtoseconds. The first hard XFEL, LCLS, was built at the SLAC National Accelerator Laboratory in California and has been in operation since April 200923. It produces X-ray pulses of approximately 3 mJ energy at 120 Hz. Each pulse has a duration that ranges from around 300 fs down to a few femtoseconds with up to 1013 coherent photons per pulse. Thus, the LCLS instrument has set a new standard, with a peak X-ray brilliance over ten orders of magnitude higher than that of the most powerful synchrotron radiation sources26. XFELs are unique light sources that can be used to explore matter at atomic length and femtosecond time scales. The increase in brightness along with ultra-short pulses has facilitated the appearance of a new application of XFEL technology in the field of structural biology via serial femtosecond crystallography (SFX) 2,28. A typical setup for SFX data collection is illustrated in Figure 2.
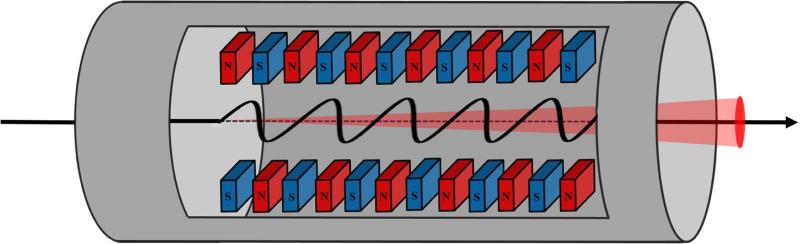
Figure 1.
Schematic representation of an undulator segment at an XFEL instrument. A relativistic electron beam (solid line) is brought to high energy in a linear accelerator (not pictured) prior to interaction with the undulator. The electrons then travel on a sinusoidal path, induced by a special arrangement of magnets called an undulator, a periodic array of magnetic dipoles shown as red and blue boxes. Because the electrons move in curved paths via the magnets, the change in momentum causes the emission of monochromatic radiation the same way a synchrotron does (depicted as a red cone).
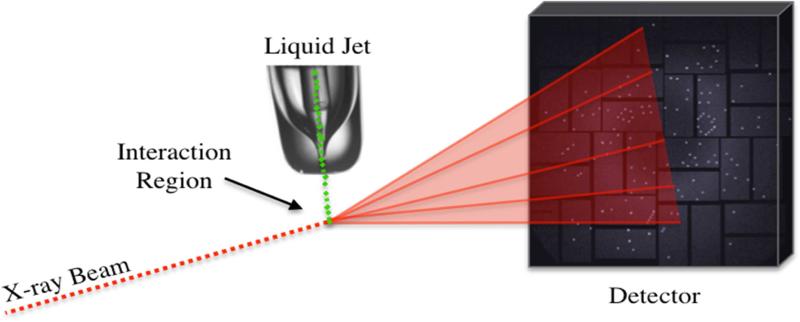
Figure 2.
Schematic representation of the experimental setup of a typical SFX experiment at LCLS. Randomly oriented nanocrystals (green) in their mother liquor are delivered into the focus of X-ray beam by a gas-focused liquid injector. The X-ray beam, which is transverse to the jet, hits the crystals in the interaction region. Diffraction snapshots of single crystals are recorded using a Cornell-SLAC pixel array detector (CSPAD) located in the forward-scattering region.
The technique of macromolecular X-ray crystallography and its historical success in determining the structure of biological macromolecules has always suffered from a major bottleneck, namely the production of well diffracting crystals. This problem is especially prevalent in the structure determination of membrane proteins, which are notorious for their difficulty in forming high diffraction quality crystals. In some cases, years have been devoted to determining crystallization conditions for a membrane protein and it is not uncommon that only showers of nanocrystals (crystals between 200 nm to 10 μm) are observed while attempts to grow larger crystals remain unsuccessful. These nanocrystals were once seen as only a possible intermediate towards achieving useable crystals31. Modern microfocus beamlines have expanded the usefulness of these nanocrystals but even they experience constraints on what is achievable mainly due to severe X-ray damage by long exposure times, often requiring cryo-cooling and necessitating crystals larger than 10 μm, which must be even larger in the case of large unit cells (common in membrane proteins and large complexes)32. With the advent of XFELs, crystals which would have previously been too small for use have been shown to be suitable for structure determination2. Generally, these small crystals are easier to produce and possess significantly less long range disorder. Furthermore, nanocrystals are ideal for time-resolved studies as a greater percentage of molecules in the crystal can be activated homogeneously by light or rapid mixing. For light activated reactions, time domains ranging from the femtosecond to microsecond regimes can be probed and temporal resolution of reactions induced by mixing can range from seconds down to microseconds 1,20,33-35.
In addition to the challenges of sample preparation, the problem of X-ray induced radiation damage has hindered progress in structure determination even in well-diffracting crystals. Traditionally, cryogenic cooling of crystals has been the most successful way to minimize damage, increasing the radiation dose tolerance by a factor between 30 and 5036-38. Cryo-cooling works by slowing the dispersion of radiation-induced reactive products that occur during the X-ray exposure. Upon exposure to X-rays, photoionization and processes related to Auger decay take place, leading to heat as well as the production of radicals and photo-ions. These diffuse within the crystal at rates dependent on available kinetic energy. This can lead to subsequent chemical reactions that result in the breaking of chemical bonds and thus a reduction in the diffraction quality of the crystal. While cryo-cooling does not influence the primary X-ray damage (i.e. photoionization and Auger decay), it slows down diffusion of the secondary processes of X-ray induced damage, radicals and heat progression. This method thus allows crystals to tolerate longer exposure times, improving the signal to noise ratio. Nonetheless, even at cryogenic temperatures site-specific radiation damage remains a problem, in which rapid photo-damage by the X-ray beam in areas with high X-ray cross-sections can occur, decreasing diffraction until it ultimately terminates. In such cases, partial data sets from many crystals must be merged in order to obtain a structural model. Furthermore, these models can exhibit significant artifacts from damage which can be substantially different from the native structure39. Examples of radiation damage include the breakage of disulfide bonds and salt bridges, tyrosine residues becoming hydrolyzed, decarboxylation of glutamate and aspartate residues, B-factor increase, and loss of diffraction progressing from high to low resolution8,40-42. Radiation damage is particularly troublesome in limiting resolution of high-Z catalytic centers, causing metalloproteins to be the most prone to local radiation damage. For example, spectroscopy has shown that X-ray radiation exposure to cryo-cooled, photosystem II crystals leads to the metal center becoming photo-reduced below the native oxidation state. This effect was still seen at 4% of average doses used for crystallography43.
In 2000 Neutze et al.44 conducted molecular dynamic simulations on the time course of the Coulomb explosion of T4 lysozyme at a total flux of 3.8 × 106 12 keV photons per Å2 over pulse durations of 2, 10, and 50 fs (the presumed theoretical flux achievable by an XFEL) in vacuum. At this time, XFELs were still in the planning phases. They predicted that the onset of the Coulomb explosion would occur at 5-10 fs and predicted that if the pulses are short, diffraction of the molecule could be collected before it is destroyed referred to as “diffraction before destruction”. This principle relies on an XFELs ability to deliver a sufficient number of photons (~1013 photons/pulse, orders of magnitude greater than that required to form a plasma) for structure determination on time scales competing with primary radiation damage events. The first experimental demonstration of this principle was carried out using the soft XFEL, FLASH, located at DESY45. In this experiment an intense 25 fs, 4 × 103 W/cm2 pulse, containing 1012 photons, produced a coherent diffraction pattern from a non-periodic object before destruction occurred46. Thus, XFELs have the potential to minimize the effects of radiation damage and reduce the size restrictions on crystals suitable for X-ray structure determination. SFX requires new data processing and handling as the determination of crystallographic structures is based on thousands of snapshot diffraction patterns at room temperature by continuous delivery of orientationally unrelated nanocrystals in their mother liquor2,5,11,47-49.
In December 2009 the first SFX experiment was carried out at LCLS, using nanocrystals of photosystem I (PSI) as the first sample. PSI mediates the conversion of light energy from the sun to chemical energy in plants, green algae, and cyanobacteria, is one of the most complex membrane proteins crystalized so far, the entire complex consisting of 36 proteins and 381 cofactors50. Tens of thousands of diffraction patterns were collected, which allowed for the determination of the PSI structure, providing a proof of concept for SFX2. Furthermore, this study resolved interference fringes between the Bragg peaks which were originally suggested by Sayre in 195251. He proposed that diffraction from crystals with a countable number of unit cells, would show the Fourier transforms of the crystals in the diffraction pattern leading to fringes between the Bragg peaks directly related to the number of unit cells (n-1)52. This theory came to fruition in the first SFX experiments when these fringes were detected in the diffraction patterns. In the future, with higher spatial resolution detectors, these shape transforms can be used for direct phasing53.
Six years after the first SFX experiments were carried out 2, evidence has continued to mount that XFELs make overcoming radiation damage in protein crystallography an attainable realization4-6,8,10,14,30,47,54-60. In addition, SFX not only mitigates radiation damage but is suitable for crystals with as little as a few hundred unit cells. Thereby, data can be collected from nanocrystals eliminating the need to grow large crystals. Experimental evidence is emerging that nanocrystals may show significantly less long-range disorder than their larger counterparts, making them ideal candidates for the structure determination of challenging proteins.
Nanocrystallization and Characterization
Because nanocrystals were previously seen as merely a stepping stone for the desired growth of large crystals, nanocrystal growth methods have remained largely unexplored. Due to the serial nature of SFX, a few unique characteristics must be considered for the development of nanocrystallization techniques. First, a different crystal in a random orientation is used for each diffraction pattern, with patterns being captured in a serial fashion2. Thus, to constantly replenish the sample between X-ray pulses, crystals are delivered to the X-ray interaction region by a liquid jet (in a typical SFX experiment) composed of crystals in their mother liquor at room temperature2. The delivery of the crystals to the X-ray region is much more rapid than the X-ray repetition rate. Consequently, most crystals do not interact with the X-rays11. Additionally, since diffraction patterns represent “still frame” slices through the Ewald sphere, only partial reflections can be recorded (i.e. there is no goniometer and therefore no rotation to record full profiles of reflections). In order to measure accurate structure factors of the Miller indices (h, k, l), high redundancy of the data sets (> 50) are required via Monte Carlo data analysis methods61. Due to approximately one in every ten thousand crystals resulting in a diffraction pattern and the necessity for high multiplicity of the data, tens to hundreds of milligrams of sample may be needed for successful SFX data sets, where samples are delivered in a liquid jet. The hit rate (percentage of pulses that result in crystal diffraction) is largely a function of crystal density and as such, must be considered in growth methods and characterization of the crystals.
Since data from many crystals must be merged in analysis of SFX data, it is also important to consider crystal homogeneity. For example, if there is a large size distribution in the sample, then the data set will be composed of diffraction patterns with varying relative Bragg peak intensities (directly related to the number of unit cells and hence, the size). This can lead to scaling issues which become problematic in subsequent analysis. Furthermore, the presence of some outlying (e.g. larger) crystals may necessitate attenuation of the beam to prevent damage to the detector. This in turn would limit resolution on images obtained from the more plentiful smaller crystals. Size inhomogeneity can also lead to instabilities or clogging in the liquid jet62. Therefore, it is imperative to consider and monitor crystal size distribution while screening conditions for nanocrystallization.
These considerations become even more pronounced in time-resolved experiments where the crystal size is the primary parameter in reaction initiation homogeneity20,63. To meet the need for large sample volumes, optimized crystal density and crystal size homogeneity, in addition to the traditional optimization of crystal diffraction quality, nanocrystallization techniques are rapidly emerging to address the unique challenges present in SFX.
Nanocrystallogenesis
As is the case in the growth of macroscopic crystals, knowledge of a proteins phase space is highly beneficial for the optimization of crystallization conditions. Driven by thermodynamics, the formation of protein crystals requires a controlled decrease in solubility of the protein that ideally avoids the formation of non-ordered precipitation. Traditionally, the supersaturated area of phase space is typically reached by gradually increasing the precipitant concentration, allowing for few nucleation sites to form that grow by addition of the free protein to the nuclei over time, resulting in a few large crystals64. In contrast to this approach, nanocrystallization is best achieved by inducing a high number of nucleation sites, whereby the free protein concentration is rapidly decreased by the formation of a plethora of nuclei followed by their growth into a multitude of small crystals62,65. Typically this is achieved by increase of the concentration of protein, precipitating agents, or both as compared to optimized conditions for large crystal growth for the same protein62. While the approaches for each size regime differ, they both rely fundamentally on the knowledge of the proteins phase space and may be even more important for growth of well-ordered nanocrystals as formation of amorphous precipitate is best avoided.
Many of the existing methods for macro-crystallogenesis are also applicable for nanocrystallization with modification necessary to occupy a different area of phase space3,22,27,57,62,66. Batch methods, in which the protein and precipitating conditions are mixed to homogeneity initially, provide a good example of well-established methodology that needs little modification for suitability in SFX (outside of parameter values). This is illustrated in Figure 3 where a batch approach for macro- and nanocrystals differs only by starting point within the phase space67. It has been reported in some cases that large crystals grown by traditional methods can also be mechanically crushed to obtain the smaller crystals needed for SFX6,68. This is, however, not generally applicable and may lead to loss of quality or destruction of fragile crystals, such as those common amongst membrane proteins. Nanocrystalline showers with traditional methods such as vapor diffusion have been obtained, using a brute force approach with many individual setups to scale for SFX experiments69. While this poses no theoretical issue, the concern for this type of nanocrystal preparation is due to the tedious nature of sample preparation involved to obtain enough material for full data set collection. While this can in part be alleviated with sample conserving delivery systems3,25,29 (assuming they are compatible), sample density and homogeneity remain challenging and necessitate specific consideration.
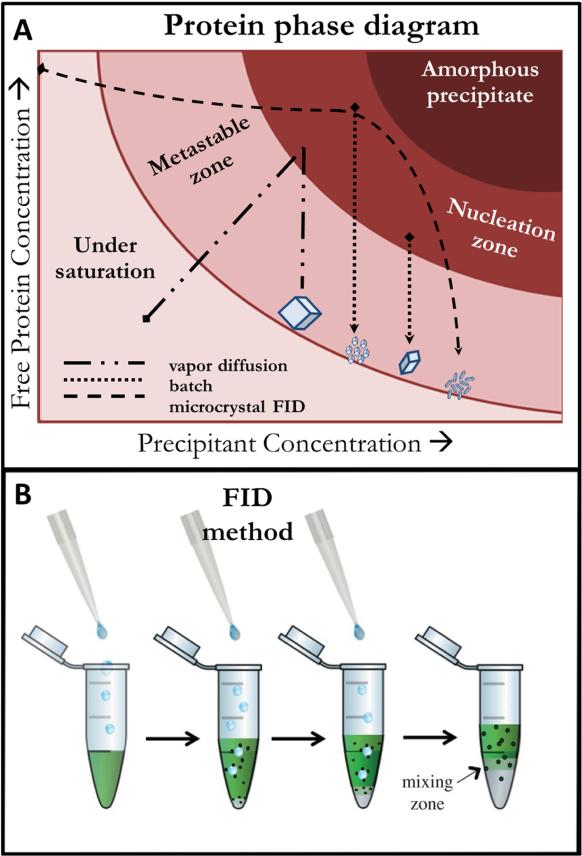
Figure 3.
A) 2-dimensional slice of a typical free protein phase diagram with selected crystallogenesis methods exemplifying the general relationship between phase space occupation and resultant crystalline protein. B) Depiction of the microcrystalline FID method in the case of a denser precipitant being dropped through a protein solution and the subsequent interface resulting in microcrystal pelleting (B reproduced with permission from 1).
The nanocrystalline free interface diffusion (FID) method is an example of a technique that has been developed from an existing method for SFX applications62. In this method, one solution (either the protein or precipitant solution) is added drop wise to the other. The less dense of the two (typically the protein/buffer solution) is first placed in a vessel, typically a microcentrifuge tube. The denser solution (typically precipitants such as high concentration salts, polyethylene glycol, etc.) is then added drop wise, thereby allowing for a high nucleation rate at the protein/precipitant interface of the two solutions but still inducing some mixing immediately. Nanocrystals will form at the interface, owing to the access of areas of phase space that typically results in high nucleation rates (i.e. high concentrations of both protein and precipitants). Additional benefits arise in the common scenario where the denser solution is the precipitant since this allows crystals that form at the interface to settle due to gravity into the precipitant rich layer. This provides a way to effectively quench crystal growth as well as accumulate a high density of nanocrystals, allowing practical optimization of crystal density by resuspension to a desired concentration. Gentle centrifugation of the setup has also been shown to expedite crystal formation (e.g. photosystem II forming nanocrystals in as little as 30 minutes with centrifugation in contrast to 1 day without centrifugation62) and has led to improvement of crystal size and homogeneity due to the tendency for crystals to spend less time in relatively high concentrations of free protein after nuclei formation, resulting in more uniform growth throughout the sample. It is important to note that over time, full mixing of the two layers will occur (rate proportional to miscibility). This could lead to the dissolution of the crystals if the mixture represents an undersaturated portion of phase space. Oswald ripening and decreased size homogeneity can also occur if there is high mobility or an appreciable amount of free protein left over after complete mixing has occurred. This can be avoided by harvesting the crystal pellet or removal of the free protein layer before considerable mixing occurs.
In-vivo crystallization
Crystallization in vivo using insect and mammalian cells is a highly innovative approach towards nanocrystallography that has been recently discovered6,15,59,70. This technique was first applied in SFX in 2012 after nanocrystals were identified by electron microscopy inside insect cells when cathepsin B from Trypanosoma brucei was overexpressed using the recombinant baculovirus system59. Redecke and co-workers observed needle shaped nanocrystals protruding out of the cell after 70 hours but were still surrounded by the cell membrane6. The crystals were 10-15 μm in length and about 0.5-1 μm in width. These crystals were isolated and used for SFX analysis6. It has been proposed that this spontaneous crystallization may occur due to accumulation of the protein in a specific organelle such as the endoplasmic reticulum, peroxisomes, or secretory granules59,71. In vivo crystallization is not limited to the Sf9 insect cells and has been demonstrated for multiple proteins in cockroaches, seeds, and bacterial cells14,70,71. However, little is understood about the mechanism of in-vivo crystallization as it was initially hypothesized to be a rare occurrence. Additionally, the crystals were not probed when they were first found as they were too small for macro-crystallography59,71. With the advent of XFELs, in vivo crystallization could potentially become a major method for crystallization, thus removing the bottleneck of determining crystallization conditions for applicable proteins. Before in vivo crystallization comes to fruition as a more general method, further research needs to be done towards understanding the necessary mechanistic components involved.
Growing crystals using LCP
Lipidic cubic phase (LCP), a bicontinuous mesophase that acts as a membrane-mimetic, has been used to crystallize a variety of membrane proteins, notably G coupled-protein receptors, ion channels, and transporters72,73. Unlike membrane protein crystals grown in surfo (in partial or full detergent micelles), which usually exhibit type II micelle crystal packing and often have a high solvent content, crystals grown in LCP feature type I crystal packing74. This packing allows for hydrophilic protein-protein and hydrophobic protein-lipid-protein interactions and thus, often leads to tighter and more rigid packing, in turn possibly leading to a lower solvent content and better diffraction75. To crystallize membrane proteins in LCP, purified protein at high concentration (usually > 20 mg/mL) is mixed with molten monoolein in Hamilton gas-tight syringes using a syringe-mixer76. For standard crystallography, special robots (e.g. Flexus Crystal IMP, Gryphon LCP, NT8-LCP, Mosquito LCP, ProCrys Meso) can be used to dispense the LCP and precipitant and crystals are often grown in micro-batch assays of <1 nL to 100 nL. Cherezov and colleagues have had notable success in adapting the LCP crystallization method for SFX8,10,24,75,77. In this method, protein laden LCP is injected in another syringe containing precipitate solution and incubated for 24 hours to two weeks to permit crystallization8,77. Excess precipitate solution is removed and the crystals embedded in the LCP are delivered using a high-viscosity-injector11.
Crystal Detection and Characterization
Some SFX experiments have been carried out with crystals that are > 5 μm and can therefore be identified by established methods such as polarized light microscopy in combination with UV-fluorescence microscopy. Nanocrystals are very difficult to identify and differentiate from amorphous precipitate, particularly if the crystal size is on the order or smaller than 1 μm, nearing the limit of resolution for optical microscopy methods. Techniques such as tryptophan fluorescence and birefringence may also be limited since the signal for each is proportional to crystal size. Alternative methods for detecting small crystals have proven to overcome the aforementioned difficulties. One of the most useful methods for rapid feedback during initial and optimization stages of crystallogenesis is the use of second-order harmonic generation (SHG) spectroscopy, in particular the SONICC (second order non-linear imaging of chiral crystals) instrument invented by G. Simpson78. SONICC can identify nanocrystals of chiral molecules as small as 100 nm78. When a chiral crystal is exposed to two 1024 nm photons in a strong field, frequency doubling occurs due to inherent polarization anisotropy, allowing a detector to measure the 512 nm photon output. Constructive interference arising from crystalline translational symmetry increases the probability of this occurring, allowing for practical measurements to be taken. It should be cautioned that the signal strength depends on the space group (with higher symmetry leading to decreased signal in general) and also the specific molecular polarization susceptibility (higher SHG is typical in molecules with a chromophore)79. While truly centrosymmetric space groups are impossible in natural protein crystals, many of the high symmetry space groups can still lead to attenuation of signal below detection, resulting in false negatives. Furthermore, some precipitants are chiral (e.g. sugars, chiral salts) and can crystallize in space groups that can be active in SHG (i.e. false positives) so care must be taken when interpreting results.
The most trustworthy method to verify the existence of diffracting crystals is X-ray powder diffraction, which can be carried out at either cryogenic or ambient temperatures. For room temperature powder diffraction measurements, a high density pellet of crystals is transferred to an X-ray transparent capillary80-82. In contrast with cryogenic powder diffraction, room temperature powder diffraction measurements are ideal for optimizing relative resolution for an XFEL since this ensures that there are not artifacts arising from the freezing process. It is also important to include buffer in the capillary to avoid drying out of the small crystals which can affect diffraction quality. While powder diffraction data can be collected from nanocrystals at low flux X-ray home sources, it requires several microliters of dense crystal sample which is more than is produced from commercial screens68. It also depends highly on the sample quality and density so establishing conditions for powder diffraction typically requires synchrotron radiation to produce measurable diffraction for un-optimized samples83.
Transmission electron microscopy (TEM) is another reliable method to monitor nanocrystal quality and requires low sample volumes. Strong correlations exist between crystals that have highly ordered lattices visible through TEM and crystals that feature better X-ray diffraction quality83. While this technique can provide insightful characterization, it requires elaborate sample preparation and often involves negative staining which can affect electron diffraction and is only suitable for very thin nanocrystals (< 200 nm)83,84. Large crystals can be mechanically crushed in order to achieve dimensions suitable for TEM68.
In protein crystallography, dynamic light scattering (DLS) is commonly used to analyze protein homogeneity since a monodisperse (i.e. homogenous and non-aggregated) sample is ideal for crystallization85. In addition, DLS is also used in SFX experiments to determine nanocrystal size distribution and homogeneity across multiple conditions86. DLS measures the light scattered by particles in solution. As molecules in solution undergo Brownian motion, the change in intermolecular distance leads to constructive and destructive interference of the scattered light. Fluctuations in intensity over time are indicative of the particle size and can be derived from the Stokes-Einstein relation. DLS only requires a few microliters of sample for analysis at broad range of concentrations (about 108–1012 crystals/mL)87. However, large particles such as dust or aggregates can affect the accuracy of size determination88. Another important parameter to take into account in SFX experiments is the crystal density. Crystal density can be optimized by using nanoparticle tracking analysis (NTA), which, like DLS, infers the Brownian motion of particles in solution and relates this movement to an equivalent hydrodynamic radius. Unlike DLS, NTA uses video to track the Brownian motion and thus, captures the light scattering signal from individual particles89. This not only allows for particle size to be determined on a particle-by-particle basis but also allows for particle concentration to be estimated which is extremely helpful for optimizing crystal density for SFX experiments87. However only particles < 1 μm can be detected and 1 mL of particle concentrations of 107 –109 crystals/mL87 is required. Therefore, DLS and NTA provide a means to monitor nanocrystal size. Furthermore, microfluidic devices using dielectrophoresis have been invented that can sort nanocrystals according to size90.
Sample delivery methods
The majority of SFX data has been collected from a jet of small crystals (typically 200 nm - 10 μm) in their mother liquor. This scheme allows the sample to be constantly replenished for each XFEL pulse as the crystals are destroyed with each shot. While this has shown great success and breadth, limitations of this method have trigged multiple strategies to deliver crystals to the X-ray interaction region to be born over the last 6 years. In this section the current crystal delivery methods used at XFELs are reviewed (summarized in Table 1).
Table 1.
Summary of sample delivery methods currently being deployed for SFX.
| Sample Delivery Method | Average Flow Rate | Advantages | Limitations | Experimental Environment | Examples of Systems/Proteins Studied |
|---|---|---|---|---|---|
| Gas Dynamic Virtual Nozzle91 | 10-25 μL/min67 | amenable for TR-SFX1,54, compatible with most systems | high sample consumption3,11,48, freezing around nozzle tip48, fragile crystals might be damaged during jetting83,92, requires crystals to be filtered107 | vacuum | photosystem I & II1,99, photosystem I-ferredoxin54, lysozyme5, cathepsin B6, photosynthetic reaction center4, Cry3A14, photoactive yellow protein20, P-type ATPase22, myoglobin34, phycocyanin29, B subunit of cholera toxin-membrane-proximal region of gp41100, CPV17 polyhedrin15, trans-acting acyl transferase68, RNA polymerase B II68, parathyroid hormone receptor l83 |
| Electrospinning47 | 140-3100 nL/min47 | low sample consumption47, compatible with membrane proteins 101 | requires cryoprotectant to prevent the jet from freezing47, unknown if high electric fields affect crystals25 | vacuum | photosystem II101, 30S ribosomal subunit92, thermolysin102 |
| Lipidic Cubic Phase11 | 50-200 nL/min8 | low sample consumption8, crystals grown in LCP do not need to be harvested8, some soluble crystals post-crystallization can be mixed to conserve sample29 | Debye Sherrer rings25, membrane protein crystals cannot be mixed post crystallization25, crystals grown in LCP cannot be filtered before injection | both | 5-HT2B receptor8, smoothened receptor11, human delta opioid receptor10, human angiotensin receptor24, rhodopsin arrestin complex30, diacylglycerol kinase17, lysozyme29, phycocyanin29 |
| Grease3 | 460-480 nL/min3 | low sample consumption3 | Debye-Sherrer rings25, not shown to be compatible with membrane proteins25 | atmosphere | lysozyme3, glucose isomerase3, thaumatin3, fatty acid–binding protein type 33, luciferin-regenerating enzyme27 |
| Agarose25 | 100-200 nL/min25 | low sample consumption25, compatible with membrane and soluble proteins25, low background25 | requires cryoprotectant to prevent the jet from freezing when operated in vacuum25 | both | phycocyanin25, photosystem I & II25, sindbis virus69 |
| fixed target: sample supports (wafers57,103,104 and chips105,106) | NA | low sample consumption105, compatible with 2D crystals103, target can be rotated about a fixed angle105 | commercial wafers/chips material is not X-ray transparent104,105, low data collection rates (<10 hz, limited by velocity of sample holder)57, crystal traps can produce crystal orientation biases105 and fill rate is limited to ~50% to prevent crystal stacking106, in vacuum experiments requires emerging crystals in oil57,104, unless crystals are grown on a chip they must be transferred105,106 | both | rapid encystment protein 24 kDa57, bacteriorhodopsin103, anthrax toxin protective antigen104, lysozyme105, sperm whale myoglobin106 |
| Fixed Target: Goniometer107(Grids/Mesh/Loops) | NA | amenable to micro- and to macro- crystals107, compatible with standard goniometers107 | requires cryopreservation and freezing107, not amenable for TR-SFX, requires hitting the crystal in a different position each shot to avoid damage | atmosphere | RNA polymerase II107, general transcription factor IIB107, large nucleic acid scaffold107, monomeric iron-containing hydrogenase 107, sperm whale myoglobin107, cytochrome c oxidase108, synaptotagmin-1 SNARE complex109, β2-adrenoreceptor/nanobody complex107, cyclophilin A110 |
Gas-focused liquid injectors
In preparation for the first SFX experiments, the gas dynamic virtual nozzle (GDVN) was invented to continuously deliver crystals to the X-ray pulses49,91. Based on the principle that flowing gas encompassing a liquid can focus the liquid jet to a smaller diameter (1/10 the original capillary size) the GDVN produces a liquid jet that is only a few microns in diameter49,91. This is achieved by mounting a smaller capillary inside a larger capillary. Crystals in their mother liquor are delivered through the smaller, inner capillary and high-pressured gas flows through the interstitial space between the larger, outer capillary and the inner capillary (see figure 4). The GDVN has been used successfully for many SFX experiments (see Table 1) including time-resolved studies1,6,54 and has thus far been the workhorse for the majority of SFX experiments. While the GDVN has experienced much success, several limitations exist. Namely, clogging of the smaller, inner capillary can occur due to blockage from aggregating crystals, the shear forces experienced during jetting have been suspected to damage some fragile crystals83,92, and ice can form on the nozzle by back-spraying of debris from the explosion at the X-ray interaction region91,92. Clogging events can be decreased by filtering the sample prior to sample loading and using in-line filters upstream from the nozzle. However, filtering could result in loss of sample and damage to crystals, thus highlighting the need for knowledge and control over the size distribution92. One challenge when using the GDVN is that the X-ray pulse repetition rates currently available (≤120 Hz) are low compared to the minimal flow rate of the GDVN at 10 μL/min, thus the majority of crystals never interact with the X-ray beam since they flow through the interaction region between pulses48. Therefore, large sample volumes are needed to produce a complete SFX dataset using this method. Data sets using the GDVN can require 10 mL of crystal suspensions, typically containing on the order of 109-1011 crystals/mL2,62,93,94.
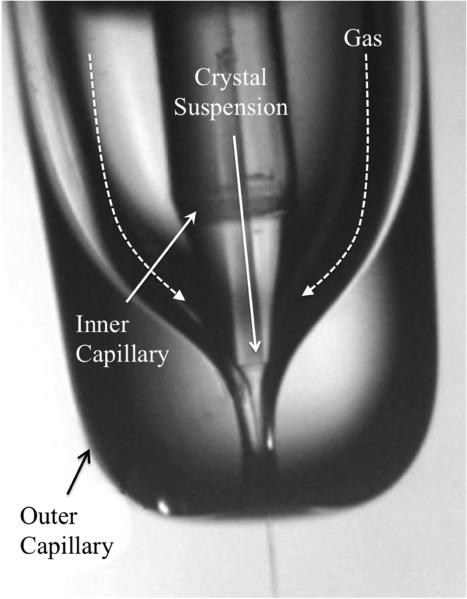
Figure 4.
Schematic of GDVN. The GDVN is assembled by placing a smaller, hollow-core fused silica optical fiber (inner capillary) inside a larger, borosilicate glass capillary (outer capillary). Crystals are passed through the inner capillary while a focusing gas is passed through the outer capillary and thus occupies the space in between the two capillaries. A thin, micrometer jet is produced when the co-flowing gas meets the crystals as they exit the inner capillary. The scale bar is 0.1 mm.
Several strategies have been proposed to reduce the flow rate of the GDVN. First, the GDVN can be operated in a pulsed mode whereby the jet is switched on and off, using a high performance liquid chromatography (HPLC) valve, as to minimize sample loss between pulses. It has been reported that flow times of 300 s separated by times of 2 ms without flow could reduce the GDVN sample consumption by a factor of four48. Similar to a pulsed method, droplets can be used to deliver crystals to the X-ray beam. In acoustic droplet ejection, droplets can be produced by a piezoelectric transducer95. To optimally synchronize the crystal filled drops, the droplets would be trigged by the XFEL. However, droplet speeds produced tend to be unstable on the necessary timescales, making synchronization difficult. Additional challenges faced by this method include large droplet size and thus high background96,97, crystal settling without the addition of a viscous solution48, and compatibility with high vacuum48. To date, neither the pulsed liquid jet nor acoustic droplet ejection has been employed at an XFEL successfully.
Electrospinning injectors
In one method of counteracting the fast consumption of sample by the GDVN, an electrospun liquid microjet, which uses high electric fields instead of gas to focus the liquid, has been implemented47. Electrospinning allows for a slower moving jet and thus, fewer sample flows between XFEL pulses and the amount of sample needed for a dataset can be reduced11. The slow speed is also suitable for experiments with long pump-probe delay times as it allows for longer incubation times compared to those achievable with the GDVN47. However, to form a continuous jet and prevent crystal settling, the crystals must be suspended in a suitable viscous media (e.g. glycerol, PEG, or sucrose); otherwise the electrospun jet breaks up into highly charged droplets once surface tension is overcome. The embedding media also serves as a cryo-protectant, necessary to prevent jet dehydration due to the length of time spent in vacuum, making this method not universally suitable. Caution of the impact of the electric field and the high charge of the jet and droplets on the sample must also be considered as they are unknown at present47.
High-viscosity media injectors
LCP has a viscosity similar to vacuum grease, which makes harvesting tens of thousands of small crystals from their sticky, viscous environment impractical. Thus, an alternative method was devised so that the crystal laden LCP could be delivered directly to an X-ray beam. To extrude such a viscous material, the high-viscosity injector (commonly referred to as the “LCP injector”) amplifies the pressure of an HPLC using a hydraulic stage to extrude the viscous media out of a capillary. A co-flowing gas stream ensures that the highly viscous fluid does not curl back onto the nozzle and forms a stable jet11. Unlike the GDVN, the jet is not focused to a smaller size and maintains the inner diameter of the capillary it is extruded from (10-100 μm). In addition, the high viscosity injector has been shown to work both in vacuum and in air98. To decrease sample consumption for soluble nanocrystals, crystals have been successfully manually mixed post-crystallization with LCP and grease3,29. Recently, agarose-based gels have been shown to be compatible for the sample delivery of both soluble and membrane proteins which are embedded into pre-gelled agarose using a coupled syringe setup as described in Conrad et al25. The agarose jet also offers an improved background over grease3 or LCP, leading to higher quality data25.
Fixed Targets
For most experiments that use a flowing jet, either liquid, viscous, or electrospun, the optimal crystal hit rate is usually around 10-30%, as higher crystals densities tend to lead to clogging. Flowing jets remain the ideal method for sample delivery for most SFX experiments as they do not require freezing and are the most practical technique for conducting time-resolved experiments. But for a small subset of experiments, fixed targets may be beneficial since this approach has the potential to decrease the sample quantities necessary for a data set to be collected. Fixed targets vary from traditional goniometers, to windowed sample support wafers/grids (analogous to electron microscopy holders), and to microfluidic chips45,104,106. The main advantage of sample support wafers is their compatibility with 2D nanocrystals, which require a scaffolding and thus, cannot be delivered in a flowing jet102. In order to prevent dehydration, crystals must be immersed in oil before being painted onto the support grid. Microfluidic devices have also been used for fixed target experiments at XFELs and have been designed to trap single crystals104,112. These traps can theoretically ensure that the X-ray beam interacts with only one crystal at a time but with current designs, crystal stacking will occur before all the traps are filled. Fixed targets can result in crystal orientation bias113. Currently, fixed target approaches are severely limited by slow data acquisition. Data acquisition rates are primarily limited by the velocity of the fixed target stage and the time it takes to replace the sample grid or chip. For example, if the stage motor could operate at 120 Hz and automated scripts aligned the X-rays to each window/trap, a fixed target containing 800 window/traps, the sample holder would need to be replaced every 6.67 minutes. In addition, some fixed targets produce high background due to the support material103-105.
Recently, data has also been collected at XFELs with a conventional goniometer approach on very large crystals (up to millimeters in size) under cryogenic conditions. This method has been used to determine a dark structure of photosystem II with minimal X-ray damage at 1.95 Å resolution114. However, this approach is low throughput and tedious as it can require days or even weeks of very precious data collection time at FELs on hundreds of individually mounted crystals111. The experiment required freezing, attenuation, and translation of the beam focus by 50 μm at an X-ray spot size of 1 μm to avoid X-ray damage. Furthermore, freezing and the large crystal size prohibits time-resolved experiments from being feasible. Except for cases like photosystem II, where X-ray damage significantly alters the structure of the catalytic metal complex, it is questionable if this is advantageous compared to standard macromolecular crystallography at synchrotron sources.
Structural dynamics and molecular movies: challenges and opportunities
In addition to broadening the array of macromolecules available to crystallography, SFX also expands upon the information that can be obtained regarding functional dynamics. Macromolecular crystals typically exhibit extremely high solvent content and weak electrostatic contacts in comparison to their smaller molecular counterparts. While this has always presented a challenge to crystallogenesis of well-diffracting crystals, the porous nature of macromolecular crystals lend them to intact catalytic or functional activity in the crystalline phase112-115. This is because large solvent channels that are present in the crystals can allow for conformational movement of the proteins or even the diffusion of a substrate to active sites in crystals. This property indicates an exciting opportunity to see intermediate structures over the course of a functional reaction. By achieving time-resolved structure determination (i.e. multiple structures along a reaction pathway), one can begin to directly understand the relationship between structure and function of macromolecules. This has numerous applications such as rational drug design and renewable energy by unraveling the mechanism of biochemical processes. However, some important considerations must be taken into account in order to realize reliable time-resolved structures.
Time-resolved crystallography has historically been performed on large single crystals by Laue multi-wavelength crystallography (for a review see 116) and has exhibited considerable gains over the past twenty years, moving from the millisecond117 to the picosecond118 temporal resolution regimes. This has opened the door to viewing conformationally dynamic proteins in action on an atomic scale. While this technique continues to improve, both within its experimental parameters and subsequent data analysis, it faces some hard limitations due to the nature of large crystals and current synchrotron sources, namely: 1) temporal resolution is limited by pulse length achievable using a synchrotron, 2) irreversible reactions cannot be studied as the induction of a reaction would cause a permanent modification of the molecules in the crystal, 3) only light activated, pump-probe type experiments are generally feasible at present, with limited light penetration being one of the major obstacles, and 4) homogeneity of reaction initiation must be considered and presents a challenge.
SFX presents an opportunity to complement the Laue method by providing access to experiments previously impossible by overcoming the above challenges via its unique experimental characteristics. XFELs have pulse durations on the order of tens of femtoseconds, which, within the framework of time-resolved crystallography, allow access to detect and resolve fast time points in catalytic reaction processes. Thereby, more temporally constrained intermediates can be detected along a reaction pathway. Recent work has pushed temporal resolution to the sub-picosecond regime34,119, which is utterly out of reach at existing synchrotron sources. This allows insight into ultrafast intermediates and therefore, the promise of a much finer understanding of catalytic mechanisms.
With irreversible reactions one must consider the way in which data is collected. In traditional crystallography a crystal is rotated during data collection and each diffraction pattern corresponds to a rotational increment of the reciprocal space. If an irreversible reaction is to be probed fully, data collection would require thousands of large crystals for each time point as the crystal would be permanently altered after the induction of the reaction. This would require an oppressively large number of crystals to collect a complete data set from individual crystals. The serial “diffraction before destruction” nature of SFX experiments completely bypasses this constraint, as only one diffraction pattern is collected from each crystal. Since the sample is constantly replenished, there is no constraint on the reversibility of a reaction. The utilization of Monte Carlo merging of an immense number of individual crystals in random orientations leads to accurate structure factors for time-resolved experiments due to implications from the central limit theorem.
Considerations of structural homogeneity and the constraint of Laue to predominantly pump-probe style experiments are highly intertwined due to one parameter: diffusion. Thus far, Laue crystallography has been predominantly limited to photo-activated reactions since chemically activated reactions would necessitate diffusion of a substrate throughout the crystal, limiting reaction timescales available. While the large solvent channels in crystalline macromolecules often allow for diffusion to take place via soaking with a diffusing substrate120-122 (assuming the active site is unobstructed and the substrate is sufficiently small relative to the solvent channels), the size of the crystal limits diffusion times, thereby constraining the time regime of reaction intermediates accessible in an experiment. There have been chemically activated time-resolved experiments performed successfully by incorporating photo-activated caged substrates into the crystal123-125, thus reducing diffusion times to that of the photo-penetration of the pump laser. However, the incorporation of caged substrates requires extensive knowledge about a given system and is not generally compatible126. The small size of the crystals used in SFX theoretically allow for diffusion times on microsecond time scales, allowing access to many reactions on the short millisecond and even microsecond regimes in the absence of caged substrates. For example, a crystal with dimensions of 0.5×0.5×0.5 μm3 has been modeled to exhibit a diffusion time of 17 μs, while a 3×4×5 μm3 crystal is estimated to take 1 ms, and a large 300×400×500 μm3 crystal would take 9.5 s126. Many biological reactions occur with intermediates observed in the time range of ms and faster but significantly less occur in the longer regime of seconds. One must also consider reaction homogeneity since Bragg diffraction relies upon translational symmetry of the molecules in the crystal. This means that in order to observe an intermediate structure, a sufficient proportion of the molecules must be in a single conformation during probing in order for a structure to be elucidated.
Photoactivated reactions have one major advantage over substrate or other diffusion based reactions as initiation is homogeneous and rapid. However, the degree of reaction initiation by photoinduction is also limited by the size of the crystal since molecules absorb the light as it travels through the crystal causing a decrease in transmission with increasing path length. When considering faster time points, the reaction homogeneity of the molecules in the crystal is sensitive to the lifetime of the intermediate state as compared to the time difference of reaction initiation from the front and back surfaces of the crystal relative to the pump beam. The distance traveled with respect to the front and back of photo-activated crystals is not the main cause of this temporal offset (though this is on the order of picoseconds for large crystals); instead, it is the attenuation of the pump intensity as it proceeds through the crystal, undergoing absorption and scattering. Depending on the particular robustness of the sample, one can attempt to address this by increasing the power of the laser but this approach is constrained by photo-damage and heating effects that can occur. Thus, the size of nanocrystals provides an advantage, allowing a much smaller difference of reaction initiation throughout the crystal and requiring decreased pump intensities for maximal reaction initiation. Furthermore, the volume of the crystal itself is so small that in practice, the whole crystal can be illuminated by the pump, minimizing considerations of the intensity profile of the pump laser. Because of these attributes, SFX provides the basis for a much higher reaction initiation yield, a point which as has already been shown in practice20.
Time-Resolved Serial Femtosecond Crystallography
The structure-mechanistic relationship found in biological reactions is rarely explained through resolution of a single, static structure. Instead, the dynamics between initial and final states embody the mechanism. TR-SFX permits access to transient intermediates that occur as a reaction is proceeding, providing still snapshots of these states. Snapshots of the initial, intermediate, and final state(s) can be viewed in a quick succession from start to finish to reveal motion of the enzyme and thus a “molecular movie” can be obtained to unravel how these biological molecules proceed in nature.
The first time-resolved SFX (TR-SFX) experiment was carried out on PSI-ferredoxin co-crystals which undergo electron transfer reactions that lead to the subsequent undocking of ferredoxin and dissolution of the crystals upon light excitation, leading to a rapid loss of diffracting quality upon pumping. Large differences have been obtained in the diffraction patterns between the excited and uninitiated reactions as revealed by the Wilson plot. The timing of these differences agrees with the time range previously found by spectroscopic methods for electron transfer127,128. These experiments provided the proof of concept for TR-SFX, (though an electron density map was not obtained due to data limitations)54.
The first successful TR-SFX experiment was performed using photosystem II, the membrane protein responsible for splitting water into its constituent protons, electrons and oxygen. This process provides the electrons for the photosynthetic electron transfer chain in oxygenic photosynthesis. The catalytic oxygen evolving cluster, structurally the most interesting domain in photosystem II, is particularly susceptible to radiation damage due to the presence of 5 Mn atoms. The X-ray photo-damage processes are virtually eliminated by the “diffraction before destruction” nature of SFX. A further concern of time-resolved studies with X-rays is that local photo-damage can be induced by repeated illumination of the crystal by the pump laser in Laue time-resolved experiments. Again, this problem is mitigated by TR-SFX since relatively low pump energy and flux can be applied per crystal due to smaller, more permeable crystals. Indeed, TR-SFX has succeeded in ‘shining the first light’ on the undamaged ground state of PSII using single and multiple laser excitations prior to diffraction, allowing multiple states to be studied along the multiple-excitation reaction pathway. This revealed large conformational changes occurring in the photo-excited “double flash state” of PSII, which includes movement of the protein via its coordination of the oxygen evolving cluster1. Though higher resolution is needed to provide a deeper understanding of the water splitting process, TR-SFX in general provides the most feasible path forward toward obtaining a series of initial, final and intermediate structures during a reaction, i.e. molecular movies of biomolecules at work.
More recently, atomic resolution has been achieved for TR-SFX using PYP20. This experiment also pushed the limits on the time-resolution available at an XFEL, capturing reaction time points down to the sub-picosecond regime119. These studies improve upon the temporal resolution that has been achieved in Laue crystallography118 by orders of magnitude and is only significantly limited by beam characteristics (i.e. time-jitter)119. As developments continue with future generation sources, temporal resolution will be pushed even further. In addition to ultrafast time resolution, it is notable that the TR-SFX studies on PYP have also shown huge improvement on reaction initiation (40% at XFELs with nanocrystals compared to 10-15% achieved at a synchrotron with larger crystals20,33), allowing for stronger signal from transient intermediates to be detected, leading to data that can be easily interpretable. Even more recently, research in this time domain has led to novel observations of global structural changes in myoglobin upon lysis of an Fe-CO bond that occur within a few picoseconds34.
Though no mixing TR-SFX work has been published to date, the stage is set for ‘on the fly’ mixing experiments that take advantage of the nature of SFX. Many enzymes have biologically interesting intermediates that occur on time scales faster than those accessible to soaking experiments of large crystals. Although one could scale down the size of the crystals and thereby, narrow the reaction initiation temporal profile, the tradeoff with decreased diffraction severely limits the range of this method. In theory, the sizes of crystals available for SFX alleviates this concern, decreasing necessary mixing times such that a new regime of reactions is accessible. Delivery methods based on the GDVN have already been developed for time-resolved mixing experiments e.g. a double focused mixing jet35 where a jet containing crystals is mixed within a stream containing the desired substrate prior to discharging from the nozzle . For a more in depth look at substrate mixing techniques, the reader is referred to 126.
Indeed, the most promising areas for XFELs to push the boundaries within structural biology is the ability to study temporal dynamics with femtosecond pulses, allowing access to ultrafast timescales and thereby, short lived intermediates. Without sacrificing resolution, TRSFX paves the way for dynamic structural elucidation of biological processes that feature fast (<100 ps) conformational changes, irreversible reactions, non-photo-activated inductions, and to those which are limited by crystal size. TR-SFX can also avoid the local radiation damage often experienced in the active sites of macromolecules and generate a higher fraction of intermediate states20. This has far reaching implications for many fields, notably alternative energy and drug design, which both rely heavily on understanding the relationship between structure and mechanism. The reactions accessible to TR-SFX studies will continue to broaden as ‘mix and inject’ methods for chemo-activated reactions continue development. With TR-SFX and other time-resolved XFEL methods (such as time-resolved wide angle X-ray scattering, e.g.129), it is evident that XFELs provide new ways to explore a novel regime of time-resolved structural biology, leading towards true movies of dynamic macromolecules in action.
Data acquisition and data processing in SFX
The short duration pulses delivered by XFELs have necessitated the development of new detector technologies capable of integrating all of the photons that arrive within the time duration of a few femtoseconds, while sustaining full-frame readout at the XFEL pulse repetition rate. Although X-ray charge coupled device (CCD) detectors are very common at synchrotrons, few CCD detectors have readout speeds that match the LCLS repetition rate. Two CCD's were used at LCLS for the first experiments (at 2 keV), the pnCCD and the fCCD130,131. Due to their very low noise and high quantum efficiency over a large range of energies, they have been used for imaging and spectroscopy experiments. However, a larger dynamic range then the CCDs can currently cover (a few hundred thousands of photons) is necessary for SFX experiments at atomic resolution (energies >6 keV). The first detector specifically designed for higher energies presently used for SFX experiments at LCLS was the Cornell-SLAC pixel array detector (CSPAD)132-135. Today the CSPAD is the principal detector used for SFX experiments at LCLS. It consists of 64 separate modules (194×185 pixels each), allowing cost-effective replacement and experimental flexibility. The CSPAD is tiled to produce a 2.3 megapixel detector, with readout speeds matching the repetition rate of 120 Hz133. The panel distribution leaves an adjustable sized hole in the middle to allow for adjustable incident beam focuses, preventing the beam from damaging the detector (the beam is powerful enough to melt through a conventional beam stop)136.
The use of detectors composed of multiple modules introduces unique concerns, namely the exact location of each module must be known in order to correctly assess the data. While the experimental geometry may be known to low precision prior to an experiment, it can be subsequently refined using the collected data. In this case, assembling a physically correct image during initial processing is futile and a known calibration sample is used to refine the experimental geometry. The detector geometry is specified in a pixel location map containing the coordinates of each detector pixel in a suitably defined coordinate system. All constraints for an experimental geometry description are saved in a single text file which can be implemented within any of the available SFX software102,136,137. The success of indexing, predicting spot locations using a crystal orientation matrix and integrating reflection intensities depends upon the precise knowledge of the location of these sensors in three-dimensional space. This means that an accurate calibration and refinement of the tile metrology is critical.
Data analysis in SFX has unique challenges with respect to data sets collected at synchrotrons. This is due to the serial nature from snapshot diffraction patterns of randomly oriented crystals with unknown partiality and shot to shot variation in the beam characteristics. Furthermore, the images collected during an experiment consist not only of single crystal hits (one crystal in the beam) but also blank patterns (no crystal in the beam) and multi-hits (multiple crystals in the beam). The XFELs at LCLS and SACLA operate up to 120 Hz and 60 Hz, respectively, resulting in hundreds of thousands of patterns collected per hour, thus creating terabytes of data. Due to these challenges, conventional crystallographic data processing methods cannot be efficiently used for SFX data collection. Thus, new data analysis tools for SFX have been developed. In order to optimize efficiency during data collection it is important to have rapid feedback during data collection. First, data is reduced by eliminating blank and multi-hit patterns which is implemented within the programs Cheetah137 and CASS138. They perform the data pre-processing steps, evaluate the quality of each data frame, and reject all those frames that are not suitable for further analysis. Once the data size is reduced, detector artifacts are removed and background subtraction is performed. Each frame is then subjected to Bragg peak location analysis (so called ‘peak finding’) in which Bragg peaks are identified by searching for clusters of connected pixels based on a series of parameters including minimum numbers of pixels per peak, the number of peaks in a frame, intensity thresholds, and signal to noise ratio. A minimum number of Bragg peaks must be identified in the diffraction pattern as a further constraint, so that patterns with too few peaks (i.e. impossible to index) are rejected.
The background corrected and sorted diffraction patterns identified during the hit finding process are subjected to indexing and integration by programs such as CrysFEL136, cctbx.xfel102, and nXDS139. The purpose of these programs is to identify Bragg peaks in the hits and then to perform indexing. Determining the unit cell parameters and the orientation of a crystal is carried out by the widely used algorithms such as MOSFLM140-142, DIRAX143, LABELIT144, and XDS139, based on the Bragg peak locations in a diffraction pattern. Once each pattern has been successfully indexed, the intensities are merged and integrated using the Monte Carlo method61,145. One of the largest problems in SFX data analysis is the indexing ambiguity which occurs when the Bravais symmetry is higher than the space group symmetry. An indexing ambiguity arises in some polar space groups (like P63) where each pattern has multiple ways it can be indexed. In standard crystallography this indexing ambiguity is solved by exploration of the data set with both indexing options, where only one option leads to a correct X-ray structure. However, since many individual patterns are merged in SFX, the ambiguity must be solved before any additional processing to avoid artificial twinning. In order to overcome the indexing ambiguity problem, CrystFEL has implemented an algorithm based on the expectation maximization approach146 which has been successfully applied and validated using both simulated and experimental diffraction146.
As with conventional crystallography, the “phase problem” has to be solved in order to reconstruct a real-space electron density map from the measured SFX intensities. Indeed, this presents itself as a primary challenge in serial crystallography data analysis. Until recently, all crystallographic structures so far determined by XFEL have been phased by molecular replacement, using phases from known or related structures2,5,6,47,58. This method limits the target protein molecules to be investigated by SFX since the protein of interest must have a known homologous structure. Even if it does, there is risk that phase bias can be introduced. However, some conventional phasing methods have also been suggested to be applied to SFX data such as multiple isomorphous replacement or multi/single-wavelength anomalous dispersion (MIR, MAD, SAD)34,55,147,148. Barends55 and co-workers have demonstrated for the first time that the conventional phasing method of SAD can also be successfully used for SFX experiments55. In this experiment they collected and solved the structure to 2.1 Å resolution of a lysozyme heavy atom derivative that gives a strong anomalous signal from two gadolinium atoms per asymmetric unit. The MAD phasing method has also been recently adapted to SFX by using modified Karle-Hendrickson equations147,148. This proposed generalized version of MAD phasing method offers another potential for experimental phasing for structural determination in SFX.
In addition to the classical methods, new phasing techniques have been proposed for SFX data analysis. Methods such as ab initio phasing by the evaluation of the shape transforms (or the oversampling method)2,53,149,150 and the “high-intensity radiation induced phasing” (HI-RIP)93 have been proposed. The phase transform method exploits the intensity scattered between neighboring Bragg peaks (or fringes) from crystals that contain less than 20 unit cells in each crystal direction. This phenomenon has historically been obscured by noise in large crystals due to inverse scaling of inter-Bragg intensity with the number of unit cells. Each recorded Bragg peak represents a particular portion of the Ewald sphere and phasing is achieved by oversampling between the Bragg peaks. The HI-RIP method takes advantage of the ionization process of the atoms that occur within the femtosecond timescale of the pulse in SFX experiments151. The change in the electronic configuration of atoms leads to the modification of the atomic scattering factors during diffraction148. In the case of SFX this would introduce the possibility of determining phases by varying the scattering factors of the heavy atoms. This new approach, which has been already tested with Trypanosoma brucei cathepsin B protein, would represent a powerful method of experimental phasing without the need to modify the protein crystals assuming sulfur atoms are present in the native structure93.
Challenges and outlook
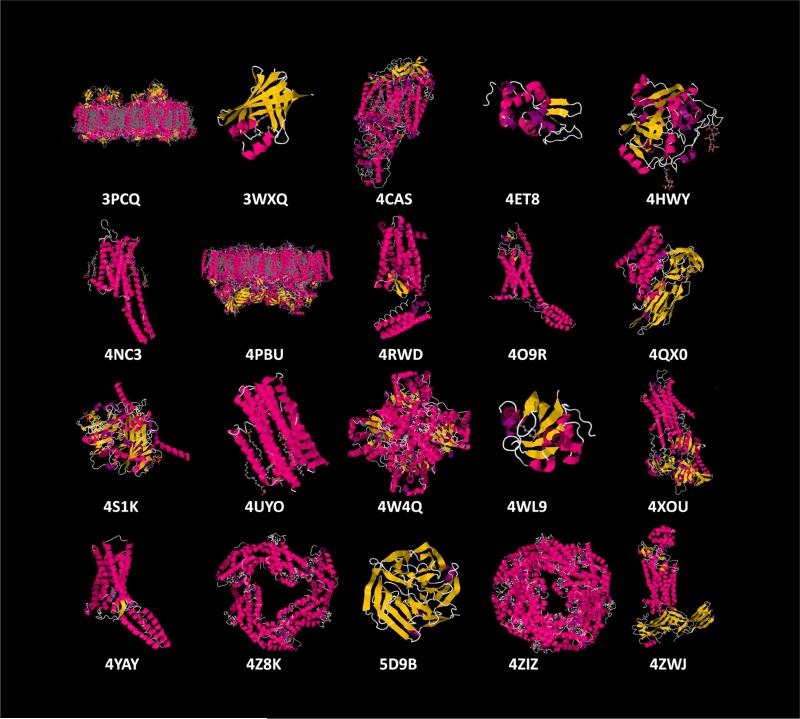
Nearly six years after the first SFX data were collected using an XFEL at LCLS and four years after the proof of principle of SFX was published2, a new era in structural biology has emerged. Figure 5 shows a gallery illustrating the breadth of protein structures successfully solved with XFELs to date. The unique properties of XFELs (ultrashort, extremely intense pulses with high frequency and coherence) have attracted considerable attention of a wide community of scientists in fields ranging from material science, to chemistry, to structural biology, and to high energy physics. This has led to ground breaking discoveries but new science brings new challenges and access to experimental time at XFELs is presently one of the major limitations. Currently, there are only two high energy XFELs in operation in the world (i.e. only two experiments can take place in the world at the same time). It is exceedingly unfortunate that due to this, beamtime is so scarce, thus becoming a major limiting factor as the field progresses out of its initial stages. Fortunately, three new XFELs are currently under construction, in South Korea (PAL), in Switzerland (SwissFEL), and in Germany (European XFEL), which are expected to enter the commissioning phase in 2016 (PAL) and 2017 (European XFEL and SwissFEL). Furthermore, XFEL facilities are planned or are under initial stages of construction in Italy and China.
Figure 5.
Select structures solved using XFELs by PDB code. From top left: photosystem I (3PCQ2), FABP3 (3WXQ3), purple bacterial reaction center (4CAS4), lysozyme (4ET85), cathepsin B (4HWY6), 5-HT2B (4NC38), photosystem II1), δ-opiod receptor (4RWD10), smoothened receptor (11), Cry3A (4QX014), CPV17 polyhedron (4S1K15), diacylglycerol kinase (4UYO17), xylose isomerase (4W4Q3), photoactive yellow protein (4WL920), SR Ca2+-ATPase (4XOU22), angiotensin II receptor (4YAY24), phycocyanin (4Z8K25), luciferin-regenerating enzyme (5D9B27), C-phycocyanin (4ZIZ29), rhodopsin-arrestin complex (4ZWJ30)
Along with the unprecedented new scientific opportunities, the successes of LCLS and SACLA have opened the eyes of the community for the need for further novel instrumentation developments in the field of XFELs. One challenge is the production of coherent photons of higher energies (50 keV or more). Another is the production of single spike pulses that are shorter than 1 fs. In addition, optics, diagnostics, detectors, sample delivery, and data acquisition must continue to be developed to keep up with the new development in data acquisition speed and X-ray pulse duration and intensity. Lower noise and higher dynamic range detectors are needed to take full advantage of these scientific opportunities. In this sense, a second generation of XFEL-capable detectors, the ePix family, is being developed at SLAC for this purpose. In parallel with the development of second generation XFELs, a new generation of detectors are being developed to meet the technical specifications required (such as the AGIPD152 at European XFEL or the Jungfrou at SwissFEL)153.
Research in the field of structural biology can now collect nearly damage-free X-ray data on biomacromolecular nanocrystals and continues to greatly impact the field by addressing many limitations faced by traditional crystallography. Going beyond SFX, computational simulations using rubisco have shown that a serial femtosecond imaging technique on individual molecules, in combination with the oversampling phasing method, could open a new horizon of structural elucidation of macromolecules without the need to first crystallize them154. However, with the photon flux of current XFELs, the ultimate goal of collecting atomic resolution data on a solution of individual, non-crystallized molecules remains currently out of reach. As the technique progresses, single particle imaging to atomic resolution may become feasible with future development of XFELs and data processing. In order to obtain high resolution 3D structural information of single large protein complexes, there are several challenges that need to be overcome. First, the pulse fluence of XFELs is not yet high enough to allow the measurement of high resolution diffraction signal from a single large protein complex. This can, in principle, be overcome by improving the XFEL peak intensity and using better focusing optics. Second, the dynamic range of the detectors presently used for single-particle imaging with XFELs is ~103. This has to be increased by at least 1 – 2 orders of magnitude. Finally, background free sample delivery is critical for single particle imaging with XFELs.
New future XFEL facilities such as the European XFEL will increase the repetition rate from 120 Hz to 27 kHz due to its superconducting linear accelerator155. This will represent a significant increase of the pulse repetition rate available today. However, the pulses will not be evenly distributed over the time of 1 second but are delivered in form of ten 600 μs pulse trains152. This poses further challenges for sample injection as jets have to run very fast during the duration of the pulse train, whereas sample would run without interception with the X-rays in the time between the arrivals of the pulse trains. Sample delivery systems that could match these increased repetition rates are challenging. Neither current fixed target options nor viscous jets can match the increased data collection rates. At SLAC a new XFEL, LCLS II, is under development, which will operate in the low to medium energy regime (current planning includes a maximum energy of 5 keV), which will allow for data to be collected at repetition rates up to 1 M Hz. Faster sample delivery injectors are presently under development to meet the needs of second-generation XFEL sources. Additionally, efficient diffraction-pattern screening algorithms and parallelized execution will be necessary to reduce the raw data stream into a more manageable set of data frames containing only diffraction patterns which have a high likelihood of being usable for indexing and intensity integration. This means that saving each and every frame for post analysis will no longer be practical and data reduction will have to be performed in real time. Developing programs further like Cheetah137 and CASS138 to do both faster on-the-fly analysis would allow researchers to pass this information directly to indexing programs for auto-indexing on-the-fly without the need to save any intermediate data. Due to their effectiveness and high speed, Cheetah137 and CASS138, have been demonstrated to be essential in the first stages of SFX data treatment.
To support next generation XFEL facilities, new features are continually under development to meet the constantly evolving needs of new experiments. In fact, while originally designed for implementation at LCLS, Cheetah has undergone several updates in the last year and a half and has now been implemented in serial millisecond crystallography experiments at synchrotron sources (https://github.com/antonbarty/cheetah98) and for SFX data collected at SACLA (https://github.com/biochem-fan/cheetah/commits/online). In addition to Cheetah and CASS software, a new software system called Karabo is currently under development at the European XFEL. European users will have access to CrystFEL through Karabo to enable fast data analysis required by the immense data acquisition challenge.
With proof of principle experiments already displaying the breadth of the technique, it is clear that the future is not just bright but brilliant. As SFX emerges from its infancy, it is apparent that molecular movies will provide a bedrock for new advances and discoveries from structural biology, to medicine, to energy conversion.
Table 2.
Details of the structures depicted in Figure 5.
| Protein Name | Organism | PDB Code | Protein Classification | Resolution | Space Group | Unit cell | Rwork, Rfree | Year | Reference |
|---|---|---|---|---|---|---|---|---|---|
| photosystem I | Thermosynechococcus elongatus | 3PCQ | membrane | 8.98 Å | P63 | 281.0, 281.0, 165.2 90, 90, 120 |
0.25, 0.23 | 20101 | (Chapman et al., 2011) |
| FABP3 | Homo sapiens | 3WXQ | soluble | 1.6 Å | P212121 | 133.25, 226.26, 307.09 90, 90, 90 |
0.178, 0.222 | 20142 | (Sugahara et al., 2015) |
| purple bacterial reaction center | Blastochloris viridis | 4CAS | membrane | 3.5 Å | P212121 | 104.8, 104.8, 104.8 90, 90, 90 |
0.295, 0.329 | 20131 | (Johansson et al., 2013) |
| lysozyme | Gallus gallus | 4ET8 | soluble | 1.9 Å | P43212 | 33.71, 54.85, 70,66 90, 90, 90 |
0.196, 0.229 | 20121 | (Boutet et al., 2012) |
| cathepsin B | Trypanosoma brucei | 4HWY | soluble | 2.1 Å | P42212 | 57.90, 84.80, 384.30 90, 90, 90 |
0.182, 0.213 | 20121 | (Redecke et al., 2013) |
| serotonin receptor 5-HT2B | Homo sapiens | 4NC3 | membrane | 2.8 Å | C2221 | 79.0, 79.0, 38.0 90, 90, 90 |
0.227, 0.270 | 20131 | (Liu et al., 2013) |
| photosystem II | Thermosynechococcus elongatus | 4PBU | membrane | 5.0 Å | P212121 | 125.40, 125.40, 54.56 90, 90, 90 |
0.262, 0.261 | 20141 | (Kupitz et al., 2014) |
| δ-opioid receptor | Homo sapiens | 4RWD | membrane | 2.7 Å | C121 | 61.50, 122.20, 168.50 90, 90, 90 |
0.208, 0.238 | 20141 | (Fenalti et al., 2015) |
| smoothened receptor | Homo sapiens | 4O9R | membrane | 3.2 Å | P1211 | 40.50, 157.30, 52.40 90, 97, 90 |
0.232, 0.278 | 20141 | (Weierstall et al., 2014) |
| Cry3A | Bacillus thuringiensis | 4QX0 | soluble | 2.8 Å | C2221 | 116.88, 134.81, 105.15 90, 90, 90 |
0.165, 0.192 | 20141 | (Sawaya et al., 2014) |
| CPV17 polyhedrin | Uranotaenia sapphirina cypovirus | 4S1K | soluble | 2.2 Å | I23 | 156.23, 89.29, 96.42 90, 92.30, 90 |
0.14, 0.19 | 20151 | (Ginn et al., 2015) |
| diacylglycerol kinase | Escherichia coli | 4UYO | membrane | 2.18 Å | P212121 | 75.30, 91.80, 141.70 90, 90, 90 |
0.208, 0.236 | 20141 | (Li et al., 2015) |
| xylose isomerase | Streptomyces rubiginosus | 4W4Q | soluble | 2.0 Å | I222 | 94.00, 100.00, 103.00 90, 90, 90 |
0.159, 0.196 | 20142 | (Sugahara et al., 2015) |
| photoactive yellow protein | Halorhodospira halophila | 4WL9 | soluble | 1.6 Å | P63 | 66.90, 66.90, 40.80 90, 90, 90 |
0.198, 0.231 | 20141 | (Tenboer et al., 2014) |
| SR Ca2+-ATPase | Oryctolagus cuniculus | 4XOU | membrane | 2.8 Å | C121 | 162.00, 76.30, 151.10 90, 109, 90 |
0.304, 0.343 | 20151 | (Bublitz et al., 2015) |
| angiotensin II receptor | Homo sapiens | 4YAY | membrane | 2.9 Å | C121 | 72.80, 41.00, 167.70 90, 99.40, 90 |
0.228, 0.274 | 20151 | (Zhang et al., 2015) |
| phycocyanin | Thermosynechococcus elongatus | 4Z8K | soluble | 2.5 Å | P63 | 153.40, 153.40, 39.64 90, 90, 120 |
0.187, 0.255 | 20151 | (Conrad et al., 2015) |
| luciferin regenerating enzyme | Photinus pyralis | 5D9B | soluble | 1.5 Å | P212121 | 186.38, 186.38, 60.34 90, 90, 120 |
0.184, 0.232 | 20152 | (Yamashita et al., 2015) |
| C-phycocyanin | Thermosynechococcus elongatus | 4ZIZ | soluble | 1.75 Å | H32 | 109.24, 109.24, 452.64 90, 90, 90 |
0.204, 0.254 | 20151 | (Fromme et al., 2015) |
| rhodopsin-arrestin complex | Homo sapiens | 4ZWJ | membrane, soluble | 3.3 Å | P212121 | 48.21, 77.59, 84.76 90, 90, 90 |
0.252, 0.293 | 20151 | (Kang et al., 2015) |
denotes experiments done at LCLS
denotes experiments done at SACLA.
Acknowledgements
This work was supported by the STC Program of the National Science Foundation through BioXFEL under Agreement No. 1231306, the National Institutes of Health Femtosecond Nano-crystallography of Membrane Proteins Award 617095583, the PSI:Biology Center MPID U54GM094625, and the Center for Applied Structure Discovery.
Abbreviations
- XFEL
X-ray free electron laser
- FEL
free electron laser
- SFX
serial femtosecond crystallography
- DESY
Deutsches Elektronen-Synchrotron
- LCLS
Linac Coherent Light Source
- SACLA
Spring-8 Angstrom Compact free electron LAser
- PSI
photosystem I
- DLS
dynamic light scattering
- SONICC
second order of non-linear imaging of chiral crystals
- SHG
second harmonic generation
- CSPAD
Cornell SLAC pixel array detector
- FID
free interface diffusion
- LCP
lipidic cubic phase
- TEM
transmission electron microscopy
- NTA
nanoparticle tracking analysis
- GDVN
gas dynamic virtual nozzle
- HPLC
high performance liquid chromatography
- TR-SFX
time-resolved serial femtosecond crystallography
- CCD
charged coupled device
- HI-RIP
high-intensity radiation induced phasing
- MIR
multiple isomorphous replacement
- MAD
multi-wavelength anomalous dispersion
- SAD
single-wavelength anomalous dispersion
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kupitz C, et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513:261–265. doi: 10.1038/nature13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman HN, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470:73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugahara M, et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nature methods. 2015;12:61–63. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- 4.Johansson LC, et al. Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography. Nature communications. 2013;4 doi: 10.1038/ncomms3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutet S, et al. High-resolution protein structure determination by serial femtosecond crystallography. Science. 2012;337:362–364. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redecke L, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2013;339:227–230. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madey JM. Stimulated emission of bremsstrahlung in a periodic magnetic field. Journal of Applied Physics. 1971;42:1906–1913. [Google Scholar]
- 8.Liu W, et al. Serial femtosecond crystallography of G protein–Coupled receptors. Science. 2013;342:1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellegrini C. The history of X-ray free-electron lasers. EPJ H. 2012;37:659–708. [Google Scholar]
- 10.Fenalti G, et al. Structural basis for bifunctional peptide recognition at human δ-opioid receptor. Nature structural & molecular biology. 2015 doi: 10.1038/nsmb.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weierstall U, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nature communications. 2014;5 doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dattoli G, Renieri A. Laser handbook. 4(1985) [Google Scholar]
- 13.Saldin E, Schneidmiller E, Yurkov MV. The physics of free electron lasers. Springer Science & Business Media; 2013. [Google Scholar]
- 14.Sawaya MR, et al. Protein crystal structure obtained at 2.9 Å resolution from injecting bacterial cells into an X-ray free-electron laser beam. Proceedings of the National Academy of Sciences. 2014;111:12769–12774. doi: 10.1073/pnas.1413456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginn HM, et al. Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data. Nature communications. 2015;6 doi: 10.1038/ncomms7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrini C, Stöhr J. X-ray free-electron lasers—principles, properties and applications. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment. 2003;500:33–40. [Google Scholar]
- 17.Li D, et al. Ternary structure reveals mechanism of a membrane diacylglycerol kinase. Nature communications. 2015;6 doi: 10.1038/ncomms10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrini C, Reiche S. The development of X-ray free-electron lasers. Selected Topics in Quantum Electronics, IEEE Journal of. 2004;10:1393–1404. [Google Scholar]
- 19.Ackermann W, et al. Operation of a free-electron laser from the extreme ultraviolet to the water window. Nature photonics. 2007;1:336–342. [Google Scholar]
- 20.Tenboer J, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346:1242–1246. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNeil BW, Thompson NR. X-ray free-electron lasers. Nature photonics. 2010;4:814–821. [Google Scholar]
- 22.Bublitz M, et al. Structural studies of P-type ATPase–ligand complexes using an X-ray free-electron laser. IUCrJ. 2015;2:409–420. doi: 10.1107/S2052252515008969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emma P, et al. First lasing and operation of an ångstrom-wavelength free-electron laser. nature photonics. 2010;4:641–647. [Google Scholar]
- 24.Zhang H, et al. Structure of the angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015;161:833–844. doi: 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conrad CE, et al. A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ. 2015;2:421–430. doi: 10.1107/S2052252515009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altarelli M. From 3rd-to 4th-generation light sources: Free-electron lasers in the X-ray range. Crystallography Reports. 2010;55:1145–1151. [Google Scholar]
- 27.Yamashita K, et al. An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography. Scientific reports. 2015;5 doi: 10.1038/srep14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spence J, Weierstall U, Chapman H. X-ray lasers for structural and dynamic biology. Reports on Progress in Physics. 2012;75:102601. doi: 10.1088/0034-4885/75/10/102601. [DOI] [PubMed] [Google Scholar]
- 29.Fromme R, et al. Serial femtosecond crystallography of soluble proteins in lipidic cubic phase. IUCrJ. 2015;2:545–551. doi: 10.1107/S2052252515013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cusack S, et al. Small is beautiful: protein micro-crystallography. Nature Structural & Molecular Biology. 1998;5:634–637. doi: 10.1038/1325. [DOI] [PubMed] [Google Scholar]
- 32.Smith JL, Fischetti RF, Yamamoto M. Micro-crystallography comes of age. Current opinion in structural biology. 2012;22:602–612. doi: 10.1016/j.sbi.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt M, et al. Protein energy landscapes determined by five-dimensional crystallography. Acta Crystallographica Section D: Biological Crystallography. 2013;69:2534–2542. doi: 10.1107/S0907444913025997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barends TR, et al. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science. 2015;350:445–450. doi: 10.1126/science.aac5492. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Weierstall U, Pollack L, Spence J. Double-focusing mixing jet for XFEL study of chemical kinetics. Journal of synchrotron radiation. 2014;21:1364–1366. doi: 10.1107/S160057751401858X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holton JM. A beginner's guide to radiation damage. Journal of synchrotron radiation. 2009;16:133–142. doi: 10.1107/S0909049509004361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howells MR, et al. An assessment of the resolution limitation due to radiation-damage in x-ray diffraction microscopy. Journal of electron spectroscopy and related phenomena. 2009;170:4–12. doi: 10.1016/j.elspec.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owen RL, Rudiño-Piñera E, Garman EF. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carugo O, Carugo KD. When X-rays modify the protein structure: radiation damage at work. Trends in biochemical sciences. 2005;30:213–219. doi: 10.1016/j.tibs.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Ravelli RB, McSweeney SM. The ‘fingerprint’that X-rays can leave on structures. Structure (London, England : 1993) 2000;8:315–328. doi: 10.1016/s0969-2126(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 41.Burmeister WP. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallographica Section D: Biological Crystallography. 2000;56:328–341. doi: 10.1107/s0907444999016261. [DOI] [PubMed] [Google Scholar]
- 42.Garman EF, Owen RL. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography. 2006;62:32–47. doi: 10.1107/S0907444905034207. [DOI] [PubMed] [Google Scholar]
- 43.Yano J, et al. X-ray damage to the Mn4Ca complex in single crystals of photosystem II: a case study for metalloprotein crystallography. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12047–12052. doi: 10.1073/pnas.0505207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–757. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 45.Ayvazyan V, et al. First operation of a free-electron laser generating GW power radiation at 32 nm wavelength. The European Physical Journal D-Atomic, Molecular, Optical and Plasma Physics. 2006;37:297–303. [Google Scholar]
- 46.Chapman HN, et al. High-resolution ab initio three-dimensional x-ray diffraction microscopy. JOSA A. 2006;23:1179–1200. doi: 10.1364/josaa.23.001179. [DOI] [PubMed] [Google Scholar]
- 47.Sierra RG, et al. Nanoflow electrospinning serial femtosecond crystallography. Acta Crystallographica Section D: Biological Crystallography. 2012;68:1584–1587. doi: 10.1107/S0907444912038152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weierstall U. Liquid sample delivery techniques for serial femtosecond crystallography. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130337. doi: 10.1098/rstb.2013.0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weierstall U, Spence J, Doak R. Injector for scattering measurements on fully solvated biospecies. Review of Scientific Instruments. 2012;83:035108. doi: 10.1063/1.3693040. [DOI] [PubMed] [Google Scholar]
- 50.Jordan P, et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 51.Sayre D. Some implications of a theorem due to Shannon. Acta Crystallographica. 1952;5:843–843. [Google Scholar]
- 52.Sayre D. The squaring method: a new method for phase determination. Acta Crystallographica. 1952;5:60–65. [Google Scholar]
- 53.Spence JC, et al. Phasing of coherent femtosecond X-ray diffraction from size-varying nanocrystals. Optics express. 2011;19:2866–2873. doi: 10.1364/OE.19.002866. [DOI] [PubMed] [Google Scholar]
- 54.Aquila A, et al. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Optics express. 2012;20:2706–2716. doi: 10.1364/OE.20.002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barends TR, et al. De novo protein crystal structure determination from X-ray free-electron laser data. Nature. 2014;505:244–247. doi: 10.1038/nature12773. [DOI] [PubMed] [Google Scholar]
- 56.Barty A, et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nature Photonics. 2012;6:35–40. doi: 10.1038/nphoton.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunter MS, et al. Fixed-target protein serial microcrystallography with an x-ray free electron laser. Scientific reports. 2014;4 doi: 10.1038/srep06026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johansson LC, et al. Lipidic phase membrane protein serial femtosecond crystallography. Nature Methods. 2012;9:263–265. doi: 10.1038/nmeth.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koopmann R, et al. In vivo protein crystallization opens new routes in structural biology. Nature methods. 2012;9:259–262. doi: 10.1038/nmeth.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lomb L, et al. Radiation damage in protein serial femtosecond crystallography using an x-ray free-electron laser. Physical Review B. 2011;84:214111. doi: 10.1103/PhysRevB.84.214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirian RA, et al. Femtosecond protein nanocrystallography—data analysis methods. Optics express. 2010;18:5713–5723. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kupitz C, et al. Microcrystallization techniques for serial femtosecond crystallography using photosystem II from Thermosynechococcus elongatus as a model system. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130316. doi: 10.1098/rstb.2013.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ibrahim M, et al. Improvements in serial femtosecond crystallography of photosystem II by optimizing crystal uniformity using microseeding procedures. Structural Dynamics. 2015;2:041705. doi: 10.1063/1.4919741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McPherson A. Crystallization of biological macromolecules. Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 65.McPherson A, Cudney B. Optimization of crystallization conditions for biological macromolecules. Acta Crystallographica Section F: Structural Biology Communications. 2014;70:1445–1467. doi: 10.1107/S2053230X14019670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu W, et al. Batch crystallization of rhodopsin for structural dynamics using an X-ray free-electron laser. Acta Crystallographica Section F: Structural Biology Communications. 2015;71:856–860. doi: 10.1107/S2053230X15009966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schlichting I. Serial femtosecond crystallography: the first five years. IUCrJ. 2015;2:246–255. doi: 10.1107/S205225251402702X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stevenson HP, et al. Use of transmission electron microscopy to identify nanocrystals of challenging protein targets. Proceedings of the National Academy of Sciences. 2014;111:8470–8475. doi: 10.1073/pnas.1400240111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lawrence RM, et al. Serial femtosecond X-ray diffraction of enveloped virus microcrystals. Structural Dynamics. 2015;2:041720. doi: 10.1063/1.4929410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gallat F-X, et al. In vivo crystallography at X-ray free-electron lasers: the next generation of structural biology? Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130497. doi: 10.1098/rstb.2013.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doye JP, Poon WC. Protein crystallization in vivo. Current opinion in colloid & interface science. 2006;11:40–46. [Google Scholar]
- 72.Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallographica Section F: Structural Biology Communications. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao J, et al. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335:686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 74.Katona G, Andreasson U, Landau EM, Andreasson L-E, Neutze R. Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.35 Å resolution. Journal of molecular biology. 2003;331:681–692. doi: 10.1016/s0022-2836(03)00751-4. [DOI] [PubMed] [Google Scholar]
- 75.Liu W, Ishchenko A, Cherezov V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nature protocols. 2014;9:2123–2134. doi: 10.1038/nprot.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chemistry and Physics of Lipids. 1998;95:11–21. doi: 10.1016/s0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 77.Liu W, Wacker D, Wang C, Abola E, Cherezov V. Femtosecond crystallography of membrane proteins in the lipidic cubic phase. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130314. doi: 10.1098/rstb.2013.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wampler RD, et al. Selective detection of protein crystals by second harmonic microscopy. Journal of the American Chemical Society. 2008;130:14076–14077. doi: 10.1021/ja805983b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Newman JA, et al. Intercalating dyes for enhanced contrast in second-harmonic generation imaging of protein crystals. Acta Crystallographica Section D: Biological Crystallography. 2015;71:1471–1477. doi: 10.1107/S1399004715008287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated β-sheet structure for amyloid. Proceedings of the National Academy of Sciences. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Steitz TA, Richmond TJ, Wise D, Engelman D. The lac repressor protein: molecular shape, subunit structure, and proposed model for operator interaction based on structural studies of microcrystals. Proceedings of the National Academy of Sciences. 1974;71:593–597. doi: 10.1073/pnas.71.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Von Dreele RB. Multipattern Rietveld refinement of protein powder data: an approach to higher resolution. Journal of Applied Crystallography. 2007;40:133–143. [Google Scholar]
- 83.Stevenson H, et al. Transmission electron microscopy as a tool for nanocrystal characterization pre-and post-injector. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130322. doi: 10.1098/rstb.2013.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wen JG. Practical Materials Characterization. Springer; 2014. pp. 189–229. [Google Scholar]
- 85.Proteau A, Shi R, Cygler M. Application of dynamic light scattering in protein crystallization. Current protocols in protein science. 2010 doi: 10.1002/0471140864.ps1710s61. 17.10. 11-17.10. 19. [DOI] [PubMed] [Google Scholar]
- 86.Schubert R, et al. Reliably distinguishing protein nanocrystals from amorphous precipitate by means of depolarized dynamic light scattering. Journal of Applied Crystallography. 2015;48:1476–1484. [Google Scholar]
- 87.Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharmaceutical research. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berne BJ, Pecora R. Dynamic light scattering: with applications to chemistry, biology, and physics. Courier Corporation; 1976. [Google Scholar]
- 89.Carr B, Malloy A. 2006 [Google Scholar]
- 90.Abdallah BG, Roy-Chowdhury S, Coe J, Fromme P, Ros A. High Throughput Protein Nanocrystal Fractionation in a Microfluidic Sorter. Analytical chemistry. 2015;87:4159–4167. doi: 10.1021/acs.analchem.5b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.DePonte D, et al. Gas dynamic virtual nozzle for generation of microscopic droplet streams. Journal of Physics D: Applied Physics. 2008;41:195505. [Google Scholar]
- 92.Demirci H, et al. Serial femtosecond X-ray diffraction of 30S ribosomal subunit microcrystals in liquid suspension at ambient temperature using an X-ray free-electron laser. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 2013;69:1066–1069. doi: 10.1107/S174430911302099X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galli L, et al. Towards RIP using free-electron laser SFX data. Journal of synchrotron radiation. 2015;22:249–255. doi: 10.1107/S1600577514027854. [DOI] [PubMed] [Google Scholar]
- 94.Schlichting I, Miao J. Emerging opportunities in structural biology with X-ray free-electron lasers. Current opinion in structural biology. 2012;22:613–626. doi: 10.1016/j.sbi.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen AU, Basaran OA. A new method for significantly reducing drop radius without reducing nozzle radius in drop-on-demand drop production. Physics of fluids. 2002;14:L1–L4. [Google Scholar]
- 96.Roessler CG, et al. Acoustic methods for high-throughput protein crystal mounting at next-generation macromolecular crystallographic beamlines. Journal of synchrotron radiation. 2013;20:805–808. doi: 10.1107/S0909049513020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Soares AS, et al. Acoustically mounted microcrystals yield high-resolution X-ray structures. Biochemistry. 2011;50:4399–4401. doi: 10.1021/bi200549x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nogly P, et al. Lipidic cubic phase serial millisecond crystallography using synchrotron radiation. IUCrJ. 2015;2:168–176. doi: 10.1107/S2052252514026487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kern J, et al. Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature. Science. 2013;340:491–495. doi: 10.1126/science.1234273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee H-H, et al. Expression, purification and crystallization of CTB-MPR, a candidate mucosal vaccine component against HIV-1. IUCrJ. 2014;1:305–317. doi: 10.1107/S2052252514014900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kern J, et al. Room temperature femtosecond X-ray diffraction of photosystem II microcrystals. Proceedings of the National Academy of Sciences. 2012;109:9721–9726. doi: 10.1073/pnas.1204598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hattne J, et al. Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Nature methods. 2014;11:545. doi: 10.1038/nmeth.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pedrini B, et al. 7 Å resolution in protein two-dimensional-crystal X-ray diffraction at Linac Coherent Light Source. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130500. doi: 10.1098/rstb.2013.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feld GK, et al. Low-Z polymer sample supports for fixed-target serial femtosecond X-ray crystallography. Journal of Applied Crystallography. 2015;48:1072–1079. [Google Scholar]
- 105.Lyubimov AY, et al. Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array. Acta Crystallographica Section D: Biological Crystallography. 2015;71:928–940. doi: 10.1107/S1399004715002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mueller C, et al. Fixed target matrix for femtosecond time-resolved and in situ serial micro-crystallography. Structural Dynamics. 2015;2:054302. doi: 10.1063/1.4928706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cohen AE, et al. Goniometer-based femtosecond crystallography with X-ray free electron lasers. Proceedings of the National Academy of Sciences. 2014;111:17122–17127. doi: 10.1073/pnas.1418733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hirata K, et al. Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL. Nature methods. 2014;11:734–736. doi: 10.1038/nmeth.2962. [DOI] [PubMed] [Google Scholar]
- 109.Zhou Q, et al. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature. 2015;525:62–67. doi: 10.1038/nature14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Keedy DA, et al. Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography. eLife. 2015;4:e07574. doi: 10.7554/eLife.07574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Suga M, et al. Native structure of photosystem II at 1.95 A resolution viewed by femtosecond X-ray pulses. Nature. 2015;517:99–103. doi: 10.1038/nature13991. [DOI] [PubMed] [Google Scholar]
- 112.Moffat K. Time-resolved biochemical crystallography: a mechanistic perspective. Chemical reviews. 2001;101:1569–1582. doi: 10.1021/cr990039q. [DOI] [PubMed] [Google Scholar]
- 113.Fromme P, Bottin H, Krauss N, Sétif P. Crystallization and electron paramagnetic resonance characterization of the complex of photosystem I with its natural electron acceptor ferredoxin. Biophysical journal. 2002;83:1760–1773. doi: 10.1016/S0006-3495(02)73942-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li D, et al. Crystal structure of the integral membrane diacylglycerol kinase. Nature. 2013;497:521–524. doi: 10.1038/nature12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hofbauer W, et al. Photosystem II single crystals studied by EPR spectroscopy at 94 GHz: The tyrosine radical Y. Proceedings of the National Academy of Sciences. 2001;98:6623–6628. doi: 10.1073/pnas.101127598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hajdu J, Johnson LN. Progress with Laue diffraction studies on protein and virus crystals. Biochemistry. 1990;29:1669–1678. doi: 10.1021/bi00459a001. [DOI] [PubMed] [Google Scholar]
- 117.Genick UK, et al. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- 118.Schotte F, et al. Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography. Proceedings of the National Academy of Sciences. 2012;109:19256–19261. doi: 10.1073/pnas.1210938109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pande K, et al. Femtosecond Structural Dynamics Drives the Trans/Cis Isomerization in Photoactive Yellow Protein. 2015. in submission. [DOI] [PMC free article] [PubMed]
- 120.Bolduc JM, et al. Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase. Science. 1995;268:1312–1318. doi: 10.1126/science.7761851. [DOI] [PubMed] [Google Scholar]
- 121.Stoddard BL, Farber GK. Direct measurement of reactivity in the protein crystal by steady-state kinetic studies. Structure (London, England : 1993) 1995;3:991–996. doi: 10.1016/s0969-2126(01)00235-0. [DOI] [PubMed] [Google Scholar]
- 122.Helliwell JR, et al. Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase. Faraday discussions. 2003;122:131–144. doi: 10.1039/b201331b. [DOI] [PubMed] [Google Scholar]
- 123.Ursby T, et al. Cryophotolysis of caged compounds: a technique for trapping intermediate states in protein crystals. Acta Crystallographica Section D: Biological Crystallography. 2002;58:607–614. doi: 10.1107/s0907444902002135. [DOI] [PubMed] [Google Scholar]
- 124.Stoddard BL, Cohen BE, Brubaker M, Mesecar AD, Koshland DE. Millisecond Laue structures of an enzyme–product complex using photocaged substrate analogs. Nature Structural & Molecular Biology. 1998;5:891–897. doi: 10.1038/2331. [DOI] [PubMed] [Google Scholar]
- 125.Schlichting I, et al. Biochemical and crystallographic characterization of a complex of c-Ha-ras p21 and caged GTP with flash photolysis. Proceedings of the National Academy of Sciences. 1989;86:7687–7690. doi: 10.1073/pnas.86.20.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schmidt M. Mix and inject: reaction initiation by diffusion for time-resolved macromolecular crystallography. Advances in Condensed Matter Physics 2013. 2013 [Google Scholar]
- 127.Setif PQ, Bottin H. Laser flash absorption spectroscopy study of ferredoxin reduction by photosystem I: spectral and kinetic evidence for the existence of several photosystem I-ferredoxin complexes. Biochemistry. 1995;34:9059–9070. doi: 10.1021/bi00028a015. [DOI] [PubMed] [Google Scholar]
- 128.Diaz-Quintana A, Leibl W, Bottin H, Sétif P. Electron transfer in photosystem I reaction centers follows a linear pathway in which iron-sulfur cluster FB is the immediate electron donor to soluble ferredoxin. Biochemistry. 1998;37:3429–3439. doi: 10.1021/bi972469l. [DOI] [PubMed] [Google Scholar]
- 129.Arnlund D, et al. Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nature methods. 2014;11:923–926. doi: 10.1038/nmeth.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doering D, et al. Development of a compact fast CCD camera and resonant soft x-ray scattering endstation for time-resolved pump-probe experiments. Review of Scientific Instruments. 2011;82:073303. doi: 10.1063/1.3609862. [DOI] [PubMed] [Google Scholar]
- 131.Strüder L, et al. Large-format, high-speed, X-ray pnCCDs combined with electron and ion imaging spectrometers in a multipurpose chamber for experiments at 4th generation light sources. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment. 2010;614:483–496. [Google Scholar]
- 132.Koerner LJ, Philipp HT, Hromalik MS, Tate MW, Gruner SM. X-ray tests of a Pixel Array Detector for coherent x-ray imaging at the Linac Coherent Light Source. Journal of Instrumentation. 2009;4:P03001. [Google Scholar]
- 133.Hart P, et al. SPIE Optical Engineering+ Applications. International Society for Optics and Photonics; 85040C-85040C-85011. [Google Scholar]
- 134.Philipp HT, Hromalik M, Tate M, Koerner L, Gruner SM. Pixel array detector for X-ray free electron laser experiments. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment. 2011;649:67–69. [Google Scholar]
- 135.Philipp HT, Koerner LJ, Hromalik MS, Tate MW, Gruner SM. Femtosecond radiation experiment detector for x-ray free-electron laser (XFEL) coherent x-ray imaging. Nuclear Science, IEEE Transactions on. 2010;57:3795–3799. [Google Scholar]
- 136.White TA, et al. Crystallographic data processing for free-electron laser sources. Acta Crystallographica Section D: Biological Crystallography. 2013;69:1231–1240. doi: 10.1107/S0907444913013620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barty A, et al. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. Journal of applied crystallography. 2014;47:1118–1131. doi: 10.1107/S1600576714007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Foucar L, et al. CASS—CFEL-ASG software suite. Computer Physics Communications. 2012;183:2207–2213. [Google Scholar]
- 139.Kabsch W. Xds. Acta Crystallographica Section D: Biological Crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Powell HR. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallographica Section D: Biological Crystallography. 1999;55:1690–1695. doi: 10.1107/s0907444999009506. [DOI] [PubMed] [Google Scholar]
- 141.Powell HR, Johnson O, Leslie AG. Autoindexing diffraction images with iMosflm. Acta Crystallographica Section D: Biological Crystallography. 2013;69:1195–1203. doi: 10.1107/S0907444912048524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rossmann MG, van Beek CG. Data processing. Acta Crystallographica Section D: Biological Crystallography. 1999;55:1631–1640. doi: 10.1107/s0907444999008379. [DOI] [PubMed] [Google Scholar]
- 143.Duisenberg AJ. Indexing in single-crystal diffractometry with an obstinate list of reflections. Journal of applied crystallography. 1992;25:92–96. [Google Scholar]
- 144.Sauter NK, Grosse-Kunstleve RW, Adams PD. Robust indexing for automatic data collection. Journal of applied crystallography. 2004;37:399–409. doi: 10.1107/S0021889804005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kirian RA, et al. Structure-factor analysis of femtosecond microdiffraction patterns from protein nanocrystals. Acta Crystallographica Section A: Foundations of Crystallography. 2011;67:131–140. doi: 10.1107/S0108767310050981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu H, Spence JC. The indexing ambiguity in serial femtosecond crystallography (SFX) resolved using an expectation maximization algorithm. IUCrJ. 2014;1:393–401. doi: 10.1107/S2052252514020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Son S-K, Chapman HN, Santra R. Determination of multiwavelength anomalous diffraction coefficients at high x-ray intensity. Journal of Physics B: Atomic, Molecular and Optical Physics. 2013;46:164015. [Google Scholar]
- 148.Son S-K, Chapman HN, Santra R. Multiwavelength anomalous diffraction at high x-ray intensity. Physical review letters. 2011;107:218102. doi: 10.1103/PhysRevLett.107.218102. [DOI] [PubMed] [Google Scholar]
- 149.Kirian RA, et al. Phasing coherently illuminated nanocrystals bounded by partial unit cells. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130331. doi: 10.1098/rstb.2013.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu H, Poon BK, Saldin DK, Spence JC, Zwart PH. Three-dimensional single-particle imaging using angular correlations from X-ray laser data. Acta Crystallographica Section A: Foundations of Crystallography. 2013;69:365–373. doi: 10.1107/S0108767313006016. [DOI] [PubMed] [Google Scholar]
- 151.Hau-Riege SP, London RA, Szoke A. Dynamics of biological molecules irradiated by short X-ray pulses. Physical Review E. 2004;69:051906. doi: 10.1103/PhysRevE.69.051906. [DOI] [PubMed] [Google Scholar]
- 152.Allahgholi A, et al. AGIPD, a high dynamic range fast detector for the European XFEL. Journal of Instrumentation. 2015;10:C01023. [Google Scholar]
- 153.Denes P, Schmitt B. Pixel detectors for diffraction-limited storage rings. Journal of synchrotron radiation. 2014;21:1006–1010. doi: 10.1107/S1600577514017135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Miao J, Chapman HN, Kirz J, Sayre D, Hodgson KO. Taking X-Ray Diffraction to the Limit: Macromolecular Structures from Femtosecond X-Ray Pulses and Diffraction Microscopy of Cells with Synchrotron Radiation*. Annu. Rev. Biophys. Biomol. Struct. 2004;33:157–176. doi: 10.1146/annurev.biophys.33.110502.140405. [DOI] [PubMed] [Google Scholar]
- 155.Altarelli M, Mancuso AP. Structural biology at the European X-ray free-electron laser facility. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369:20130311. doi: 10.1098/rstb.2013.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]