Abstract
Objective:
To review progress in spinocerebellar ataxias (SCAs) and novel approaches to treatment.
Results and conclusions:
Autosomal dominant ataxias are now referred to as SCAs, with polyglutamine expansion mutations constituting the most common cause of SCAs. Phenotypic variation in patients with SCA is remarkable even in patients with identical mutations. In patients with SCA2, cerebellar ataxia is typically associated with slowed saccadic eye movements. In addition to classic cerebellar and brainstem signs, however, SCA2 can also present as a parkinsonian syndrome or as amyotrophic lateral sclerosis. After identifying the SCA2 gene (gene symbol ATXN2) in 1996, we generated several mouse models that recapitulated salient features of the human disease. In these models, behavioral and physiologic changes preceded cell death. Modified antisense oligonucleotides (ASOs) provide a unique tool to target mRNA transcripts in vivo with extended stability of ASOs and better activation of RNAse H. We generated methoxyethyl group–gapmer ASOs that reduced ATXN2 expression >80% in vitro and then progressed the lead ASO to in vivo testing in an SCA2 mouse model. Compared to intraventricular injection of saline, treatment with ASO resulted in significant knockdown of endogenous mouse and human transgenic ATXN2. In addition, progression of the motor phenotype was slowed and Purkinje cell firing in the acute cerebellar slice normalized. ASO-based therapies are underway in humans providing hope that this approach will also be applicable to patients with cerebellar degenerations.
Neurodegenerative diseases were conventionally divided into phenotypically and pathologically distinct categories. Cerebellar degenerative diseases were no different and the full extent of their phenotypic spectrum has only recently been appreciated. This was largely enabled by discovery of disease genes and their analysis in patients with degenerative phenotypes thought to be completely distinct.
Degenerative ataxias in humans comprise a diverse group of diseases that affect the cerebellum and various other regions of the nervous system. Owing to heterogeneity in clinical signs, age at onset, disease, and progression, the classification of these disorders has been complex. Neuronal degeneration in the cerebellum may occur sporadically or follow mendelian segregation.1,2 Autosomal dominantly inherited ataxias are now exclusively referred to as spinocerebellar ataxias (SCAs).
Although good prevalence figures are available for the major neurodegenerative diseases, this is not the case for the cerebellar degenerations. Probably one of the best estimates of a population-wide prevalence comes from a registry-based study in Japan.1 This study estimated prevalence at approximately 1 in 5,000 individuals. This estimate included mendelian forms of ataxia with mutations in single genes as well as sporadic forms including patients with the cerebellar form of multiple system atrophy. Although the precise composition of mendelian forms of ataxia shows significant geographic and ethnic variation, worldwide about one-quarter are due to mendelian alleles.1,2 For example, spinocerebellar ataxia type 2 (SCA2) is common in Eastern Cuba3 and Friedreich ataxia is extremely rare in Asia.4
What is now known as SCA2 was first described in India5 and Cuba.6 Investigators in these places noticed that a subgroup of families with ataxia segregated a phenotype that showed slowed ocular saccades in addition to cerebellar signs. This particular phenotype was especially prevalent in a founder population in the eastern part of Cuba and reached a prevalence of an order of magnitude greater than in the rest of the world.5 The resource of large pedigrees in Cuba combined with other pedigrees around the world enabled mapping of the trait to the long arm of chromosome 12 in 1993.7 Subsequently, we described a family in northern New York State with a similar cerebellar and oculomotor phenotype that was also linked to chromosome 12q24.8
It had been noted that age at onset (AO) of disease in Cuban pedigrees varied greatly even in closely related individuals. We had noticed in our pedigree that AO was not only highly variable, but that it appeared to have a progressively earlier onset in subsequent generations.8 This phenomenon had been described in other neurologic diseases such as myotonic dystrophy, but had often been attributed to biased ascertainment in younger generations. Having tightly linked genetic markers in hand, we were able to show that anticipation was not artefactual and that there was true anticipation of 14.4 ± 7.9 years per generation. The nature of the SCA2 mutation, however, was not clear at that point, nor was the mechanism for anticipation.
Isolating the gene causing SCA2 proved difficult and it took 3 years after establishment of linkage to identify the gene.9–11 The causative mutation was expansion of a DNA CAG repeat located in the coding region of a gene of unknown function. CAG repeat expansion resulted in an extended polyglutamine (polyQ) domain that was located in the N-terminal portion of this novel protein.
The gene mutated in SCA2 was designated ataxin-2 (gene symbol ATXN2). In Caucasian control individuals, the ATXN2 gene most commonly contained 22 CAG repeats; in SCA2 patients, the repeat was expanded to ≥33 units and most commonly contained 37 repeats. As in other polyQ diseases such as Huntington disease, increasing repeat length was associated with decreasing AO (figure 1). Once expanded, the ATXN2 repeat does not remain stable during meiosis, but is a dynamic mutation with a propensity for expansion. This tendency for expansion explains the phenomenon of anticipation at the molecular level. Despite the deterministic nature of this mutation, variability in AO for a given repeat length is significant (figure 1).
Figure 1. Age at onset (AO) and CAG repeat length are inversely related.
Box and whisker plot after logarithmic transformation of AO. Outliers are indicated by asterisks. Only about half of the AO variance (r2 = 0.54) is explained by CAG repeat length. Data are from references 3 and 13.
For all SCAs, the salient symptom, and often the first, is gait ataxia. This is also correct for SCA2, although some individuals complain of muscle cramps at onset. Other cardinal symptoms of cerebellar degeneration follow, often in unpredictable order and severity. These are appendicular ataxia, cerebellar ocular signs such as nystagmus and dysmetria, instability of stance, and dysarthria. In addition to these symptoms and signs that are clearly referable to cerebellar neurons and circuits, SCA2 may impair a number of neuronal groups such as oculomotor brainstem neurons giving rise to slowed or even absent saccades, one of the more typical signs of SCA2. In addition, dystonia, myoclonus, spasticity, and neuropathy are common in patients with SCA2, as is frontal-executive dysfunction.12–14
The genotypic dissection of ataxia phenotypes has permitted us and others to define not only the typical phenotypic spectrum observed in most patients with SCA2, but also to discover outlier phenotypes. Among the SCAs, SCA2 mutations are particularly prone to manifest—at least initially—with symptoms and signs completely outside the cerebellar spectrum. Chief among these are patients who present with either l-DOPA-responsive parkinsonism or with amyotrophic lateral sclerosis (ALS).14,15 These patients were often seen in specialty movement disorder or ALS clinics as garden variety patients in their respective disease category, attesting to the relatively pure phenotype. It is not known what proportion of ATXN2-associated ALS or Parkinson disease (PD) is accompanied by asymptomatic cerebellar atrophy.
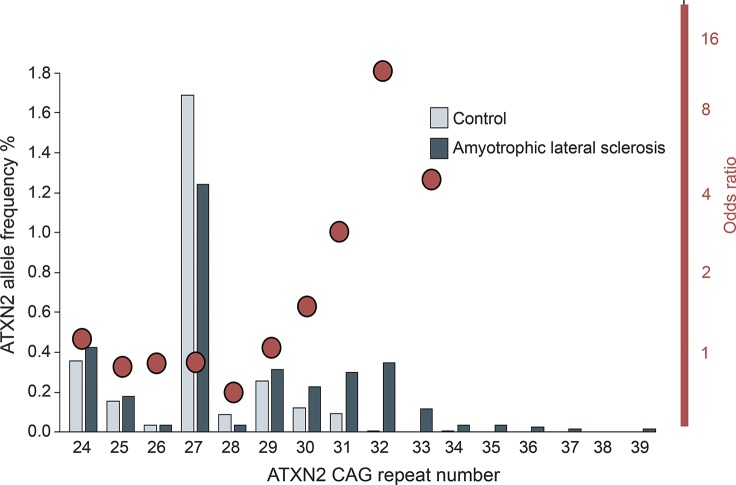
In addition to causative mutations for PD and ALS in patients with ≥33 ATXN2 CAG repeats, repeats at the longer end of normal have recently garnered attention as risk factors for ALS. Based on interaction between ATXN2 and TDP43,15 a protein involved in ALS pathogenesis, Elden et al.15 examined ATXN2 alleles as risk for ALS. They reported that individuals with ATXN2 alleles containing ≥27 repeats carried an increased risk of ALS. We recently performed a meta-analysis of all reported studies of ATXN2 repeats in patients with ALS and age-matched controls and found that ALS risk was actually slightly reduced for individuals with 27 and 28 ATXN2 repeats, but then strongly increased to an 11-fold increased risk for individuals with 32 repeats (figure 2).16 Although ATXN2 alleles with 31 and 32 repeats are rare in the population, they represent one of the most significant risk factor for ALS. There are also initial reports that the ATXN2 ALS phenotype may show a more rapid progression.17
Figure 2. Long normal ATXN2 alleles are risk factors for amyotrophic lateral sclerosis (ALS).
ATXN2 allele frequencies are shown as bars for controls and ALS patients. Odds ratios for a given allele are shown as red circles. Odds ratio for developing ALS is increased more than 10-fold for individuals with 32 CAG repeats. Modified from reference 16 with permission.
For most human neurodegenerative diseases, naturally occurring mouse models do not exist. In the case of SCA2, this is not surprising as the mouse has only one glutamine instead of the most common human 22-repeat allele.18 There are a range of approaches to model human SCAs, all with their own advantages and challenges. As Box and Draper19 said, “Essentially, all models are wrong, but some are useful.” Mouse models for polyQ diseases have ranged from those expressing a truncated cDNA to full-length cDNA constructs, and use of endogenous or strong neuronal promoters.20 These models always express multiple copies of the respective transgene in the presence of 2 copies of the endogenous mouse gene. Advantages of these models are their relative ease of generation, early onset of disease, and rapid progression. By use of a cell-type specific promoter, expression of the transgene can be targeted to one or a few cell types, thus facilitating analysis.
To circumvent nonphysiologic overexpression of the transgene, some groups have used knock-in models, whereby an expanded polyQ repeat replaces the shorter mouse repeat. Although the polyQ expansion occurs in the proper genetic dosage and a physiologic promoter is used, these models lack human control elements for expression and splicing. Often, expression of the mutant allele is low, resulting in late onset and slow progression. A relatively new approach constitutes introduction of the entire human gene into the mouse germline. This approach uses bacterial artificial chromosomes (BACs). Like all transgenic models, BAC copy number may vary, but promoter and intronic sequences are all represented by human DNA, making these models particularly suitable in the investigation of compounds aimed at altering expression or splicing of a disease gene.
To understand SCA2 pathogenesis in vivo, we have generated a number of different mouse models.21–25 A mouse model expressing full-length ATXN2Q58 targeted to Purkinje cells (PCs) displayed a progressive motor phenotype with onset at 6 months.21 PCs contained cytoplasmic but not nuclear aggregates. To accelerate the development of a phenotype for preclinical studies, we generated a mouse model expressing a longer repeat. This Pcp2-ATXN2Q127 model developed a motor phenotype much earlier at 8 weeks.22 Spontaneous firing frequency of PCs mirrored the decline in motor function. Whereas PC firing in wild-type animals remained nearly constant at different ages, transgenic mice showed a progressive decrease in firing. Despite the significant decrease in frequency, the variability in firing (interspike interval) remained unchanged in the presence of mutant ATXN2.22
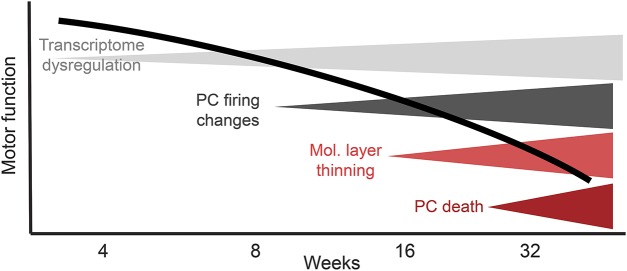
Deep RNA sequencing is a novel tool that allows the simultaneous monitoring of thousands of transcripts in the brain. We purified cerebellar RNAs from wild-type and transgenic animals at 1 day and 3 and 6 weeks of age. Already at day 1, about 200 transcripts were dysregulated, but this number rose steadily with age in transgenic animals.23 This was not due to loss of PC dendrites or cell bodies as morphologic changes occurred much later in this model. The changes are summarized in figure 3 and somewhat arbitrarily separated into 4 stages with dendritic changes and cell loss occurring many weeks after transcriptome, physiologic, and behavioral changes. In other words, dysfunction precedes cell loss and if extrapolated to humans, provides hope that early intervention could ameliorate or prevent neuronal loss.
Figure 3. The 4 stages of disease progression in an SCA2 rodent model22.
Note that physiologic changes in the acute cerebellar slice coincide with the onset of behavioral abnormalities and that both are preceded by changes of steady-state mRNA abundance. Changes in the width of the molecular layer and Purkinje cell (PC) death occur late.
Although transgenic mouse models supported the hypothesis that polyQ-expanded ATXN2 acted in a gain-of-function fashion, we wanted to exclude formally that mutant ATXN2 had a partial loss-of-function component. To test this hypothesis we disrupted the mouse ATXN2 gene in the germline resulting in the complete loss of ATXN2 protein.24,25 Knockout (KO) mice were born without obvious malformations and did not show a neurologic phenotype, abnormal cerebellar morphology, or PC physiology. As we had shown significant transcriptome changes in Pcp2-ATXN2Q127 mice, we performed deep RNA sequencing of KO cerebellar RNAs. In contrast to transcriptomes in transgenic mice, few transcripts were altered in KO mice.
Identifying treatments for neurodegenerative diseases has remained a great challenge in neuroscience. Two different approaches have emerged for mendelian neurodegenerative diseases: one based on targeting pathways in which a mutant protein acts and the other one based on targeting the mutant gene directly. A pathway-based approach has the potential advantage that the targeted pathway may be involved in a number of neurodegenerative diseases, be they genetic or sporadic, and thus benefit a larger group of patients. On the other hand, a mutant protein may act in and disrupt a number of different cellular pathways, making the selection of one crucial pathway difficult. We have employed both strategies either targeting Ca++ release from the endoplasmic reticulum or targeting the ATXN2 gene directly with antisense oligonucleotides (ASOs).
Analysis of the ATXN2 protein by sequence comparison to proteins with known functions revealed relatively little.9,18 ATXN2 was found to contain 2 LSM domains, suggesting that ATXN2 may associate with RNA. This notion was further confirmed by showing that ATXN2 associated with another RNA-binding protein RBFOX1, also called A2BP1.26 As is the case for other disease proteins, a significant number of interacting proteins have been identified. These range from proteins known to be present in stress granules, such as DDX6, to proteins involved in growth factor trafficking.26–29 However, none of these protein–protein interactions showed an altered interaction for mutant ATXN2, making them unlikely to be causative in the pathogenesis of SCA2.
An important interaction of mutant but not wild-type ATXN2 was discovered by the Bezprozvanny group.30 Using a protein interaction screen in yeast cells, they identified the inositol-triphosphate receptor type 1 (ITPR1). This interaction was negligible for wild-type ATXN2, but strong for mutant ATXN2. ITPR1 is highly expressed in PCs and critically involved in calcium handling by regulating calcium release from the endoplasmic reticulum. In PCs isolated from ATXN2 transgenic animals (Pcp2-tg-ATXN2Q58), presence of mutant ATXN2Q58 caused an exaggerated calcium release from the endoplasmic reticulum with increased cell death.21,30 Blocking of the functionally coupled ryanodine receptor with dantrolene reduced abnormal calcium release and cell death in culture.
We then tested whether these observations in vitro would translate to similar effects in vivo. Wild-type and ATXN2Q58 transgenic animals were treated with oral dantrolene or vehicle beginning at 8 weeks of age (prior to onset of the motor phenotype in this mouse model) and treatment continued for 9 months. Chronic dantrolene treatment prevented motor deterioration in rotarod and balance beam assays.30 This was observed despite the acute side effects of dantrolene after each dose consisting of sedation and ataxia seen in both wild-type and transgenic animals. Dantrolene modified disease progression as improved motor behavior persisted after a 1-month washout period. A disease-modifying effect was further supported by improved PC survival in dantrolene-treated animals at the end of the trial.
Although our experience targeting calcium homeostasis showed preclinical success, we wanted to develop methods to target the primary cause of SCA2, the expression of polyQ-expanded ATXN2. One way to target the ATXN2 gene or more precisely its mRNA transcripts is offered by use of ASOs. ASOs can enter a cell and hybridize to the complementary mRNA. The formation of this DNA-RNA duplex is recognized by RNase H, a ubiquitous enzyme that destroys the mRNA, but not the ASO. The ASO is released and can then enter another round of target hybridization and target destruction.
ASOs have had a long history in biomedical science with varying success owing to significant off-target effects and limited stability of the ASO in vivo. In recent years, chemists have modified the bases in oligonucleotides in multiple ways to enhance stability and tissue distribution.31 A common modification is the addition of phosphorothioate groups, in which one of the nonbridging oxygen atoms of the natural phosphodiester linkage is replaced with sulfur. Second-generation ASOs are chimeric single-stranded ASOs with a central region of 8–10 nucleotides flanked on either end by nucleotides with additional modifications such as addition of a 2′-O-methoxyethyl group (MOE). This ASO design is referred to as a MOE-gapmer and increases protection against nuclease-mediated degradation without impairing duplex formation and supports RNase H-mediated cleavage. MOE-gapmer ASOs have been extensively studied in vivo.32
In contrast to other dominant disease genes, it is not technically feasible to target only the mutant allele in the case of ATXN2. There is no single nucleotide polymorphism tightly linked to the mutation and the mutation itself, a long stretch of CAG repeats, is found in many transcripts, especially transcription factors. Therefore, our only option was targeting both wild-type and mutant transcripts at the same time. Given our analysis of the Atxn2 KO mouse, we had reasonable expectations that ASO-mediated knockdown of ATXN2 would not lead to adverse effects.
In collaboration with Ionis Pharmaceuticals (Carlsbad, CA), we set out to identify MOE-gapmer ASOs that targeted ATXN2 mRNAs. We performed a cell-based in vitro screen identifying the most potent ASOs for downregulating human ATXN2. Due to high sequence identity between mouse and human ATXN2, many of these ASOs were predicted to downregulate ATXN2 in both species. We were able to identify a number of different ASOs that reduced ATXN2 expression by 80% or more.
As current ASOs do not cross the blood–brain barrier, we used a stereotactic technique to deliver ASOs into the right lateral ventricle of the mouse. Initial tests assessed the ability of the best in vitro ASOs to reduce mouse Atxn2 expression in vivo. We also monitored induction of a microglial response using both immunocytochemistry to Aif1 or qPCR to Aif1 transcripts. Finally, we assessed whether a given ASO distributed to PCs and brainstem neurons. From these experiments, we chose one lead ASO (ASO7) that showed little or no Iba1 induction and significant downregulation of Atxn2 mRNA and protein for up to 70 days with a single injection.
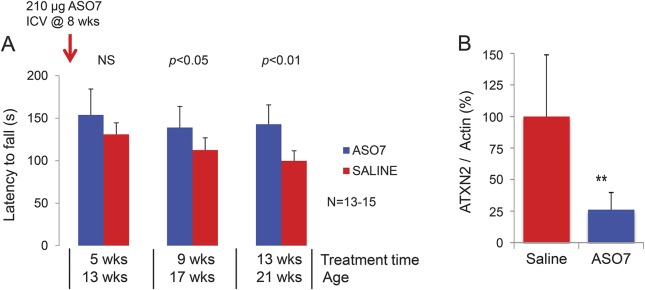
We took ASO7 forward to conduct a blinded preclinical treatment trial in our transgenic mouse model (figure 4). ASO7 or saline were injected in the right lateral ventricle of mice at 8 weeks of age at a time when abnormal motor behavior first appears. Mice were tested at 13, 17, and 21 weeks of age. Saline-treated mice exhibited progressive decline of their motor function, whereas progression was halted by treatment with ASO7. When animals were killed at the end of the experiment, both endogenous mouse and human transgenic ATXN2 mRNAs were significantly downregulated (figure 4B). We also tested PC physiology in a subset of animals. ASO treatment partially reversed abnormally low PC firing frequency in transgenic animals (data not shown). Studies are ongoing to test ASO7 in an additional SCA2 BAC mouse model,23 to test other ASOs and to examine prolonged treatments with repeated intracerebroventricular injections.
Figure 4. Effects of 2′-O-methoxyethyl group–gapmer antisense oligonucleotides (ASOs) on motor behavior and ATXN2 mRNA abundance.
(A) Compared with saline-treated animals, spinocerebellar ataxia type 2 mice (Pcp2-tgATXN2Q127) show significantly reduced progression of motor disability. Mean latency to fall in 3 trials on day 3 of rotarod testing is shown. (B) Cerebellar endogenous mouse and human transgenic ataxin-2 gene (ATXN2) are reduced by ASO treatment 14 weeks after intracerebroventricular (ICV) injection compared with saline. NS = not significant.
The isolation of many genes causing autosomal dominant SCAs has led to a genotypic dissection of degenerative cerebellar phenotypes. Polyglutamine expansion mutations constitute the most common cause of dominant ataxias. Phenotypic variation is remarkable even in patients with identical mutations. In addition to classic cerebellar and brainstem signs, SCA2 can present as a parkinsonian syndrome or as ALS. Animal models of SCA2 have characterized the phenotype at the physiologic and biochemical level. Targeting of ATXN2 in vivo using ASOs results in phenotypic improvements in rodent behavior and physiology. Given that ASO-based therapies in humans are currently underway,33 there is justified hope that this approach will also be applicable to patients with cerebellar degenerations.
ACKNOWLEDGMENT
The author thanks ataxia patients and researchers for their dedication to finding cures. Daniel Scoles, PhD, Karla P. Figueroa, MS, Thomas Otis, PhD, Pratap Meera, PhD, Warunee Dansithong, PhD, Sharan Paul, PhD, Duong Huynh, PhD, Frank Rigo, PhD, Frank Bennett, PhD, and Matthew Schneider made major contributions to the work summarized in this lecture.
GLOSSARY
- ALS
amyotrophic lateral sclerosis
- AO
age at onset
- ASO
antisense oligonucleotide
- ATXN2
ataxin-2 gene
- BAC
bacterial artificial chromosome
- ITPR1
inositol-triphosphate receptor type 1
- KO
knockout
- MOE
2′-O-methoxyethyl group
- PC
Purkinje cell
- PD
Parkinson disease
- polyQ
polyglutamine
- SCA
spinocerebellar ataxia
- SCA2
spinocerebellar ataxia type 2
AUTHOR CONTRIBUTIONS
Stefan Pulst: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, accepts responsibility for conduct of research and final approval.
STUDY FUNDING
Supported by R37NS033123 and R21NS081182 from NINDS and a grant from Target ALS.
DISCLOSURE
S. Pulst has served on the editorial boards of Journal of Cerebellum, NeuroMolecular Medicine, Continuum, Experimental Neurology, Neurogenetics, and Nature Clinical Practice Neurology and as Editor-in-Chief of Current Genomics; conducts research supported by the NIH, Target ALS, and the National Ataxia Foundation; has consulted for Ataxion Therapeutics; has served on a speakers' bureau for Athena Diagnostics, Inc.; is a stockholder of Progenitor Life Sciences; has received license fee payments from Cedars-Sinai Medical Center; has given expert testimony for Hall & Evans, LLC; has received publishing royalties from Churchill Livingston (The Ataxias), AAN Press (Genetics in Neurology and Molecular Genetic Testing in Neurology, 2nd–5th editions), Academic Press (Genetics of Movement Disorders), and Oxford University Press (Neurogenetics); holds patents for Nucleic acids encoding ataxin-2 binding proteins, Nucleic acid encoding Schwannomin-binding proteins and products related thereto, Transgenic mouse expressing a polynucleotide encoding a human ataxin-2 polypeptide, Methods of detecting spinocerebellar ataxia-2 nucleic acids, Nucleic acid encoding spinocerebellar ataxia-2 and products related thereto, Schwannomin-binding proteins, and Compositions and methods for spinocerebellar ataxia; and receives an honorarium from the AAN as the Editor of Neurology: Genetics. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Tsuji S, Onodera O, Goto J, Nishizawa M, Study Group on Ataxic D. Sporadic ataxias in Japan: a population-based epidemiological study. Cerebellum 2008;7:189–197. [DOI] [PubMed] [Google Scholar]
- 2.Abele M, Burk K, Schols L, et al. The aetiology of sporadic adult-onset ataxia. Brain 2002;125:961–968. [DOI] [PubMed] [Google Scholar]
- 3.Pulst SM, Santos N, Wang D, et al. Spinocerebellar ataxia type 2: polyQ repeat variation in the CACNA1A calcium channel modifies age of onset. Brain 2005;128:2297–2303. [DOI] [PubMed] [Google Scholar]
- 4.Pandolfo M. Friedreich ataxia: the clinical picture. J Neurol 2009;256(suppl 1):3–8. [DOI] [PubMed] [Google Scholar]
- 5.Wadia NH, Swami RK. A new form of heredo-familial spinocerebellar degeneration with slow eye movements (nine families). Brain 1971;94:359–374. [DOI] [PubMed] [Google Scholar]
- 6.Orozco G, Estrada R, Perry TL, et al. Dominantly inherited olivopontocerebellar atrophy from eastern Cuba: clinical, neuropathological, and biochemical findings. J Neurol Sci 1989;93:37–50. [DOI] [PubMed] [Google Scholar]
- 7.Gispert S, Twells R, Orozco G, et al. Chromosomal assignment of the second locus for autosomal dominant cerebellar ataxia (SCA2) to chromosome 12q23-24.1. Nat Genet 1993;4:295–299. [DOI] [PubMed] [Google Scholar]
- 8.Pulst SM, Nechiporuk A, Starkman S. Anticipation in spinocerebellar ataxia type 2. Nat Genet 1993;5:8–10. [DOI] [PubMed] [Google Scholar]
- 9.Pulst SM, Nechiporuk A, Nechiporuk T, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet 1996;14:269–276. [DOI] [PubMed] [Google Scholar]
- 10.Sanpei K, Takano H, Igarashi S, et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet 1996;14:277–284. [DOI] [PubMed] [Google Scholar]
- 11.Imbert G, Saudou F, Yvert G, et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet 1996;14:285–291. [DOI] [PubMed] [Google Scholar]
- 12.Geschwind DH, Perlman S, Figueroa CP, Treiman LJ, Pulst SM. The prevalence and wide clinical spectrum of the spinocerebellar ataxia type 2 trinucleotide repeat in patients with autosomal dominant cerebellar ataxia. Am J Hum Genet 1997;60:842–850. [PMC free article] [PubMed] [Google Scholar]
- 13.Ashizawa T, Figueroa KP, Perlman SL, et al. Clinical characteristics of patients with spinocerebellar ataxias 1, 2, 3 and 6 in the US; a prospective observational study. Orphanet J Rare Dis 2013;8:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gwinn-Hardy K, Chen JY, Liu HC, et al. Spinocerebellar ataxia type 2 with parkinsonism in ethnic Chinese. Neurology 2000;55:800–805. [DOI] [PubMed] [Google Scholar]
- 15.Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010;466:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neuenschwander AG, Thai KK, Figueroa KP, Pulst SM. Amyotrophic lateral sclerosis risk for spinocerebellar ataxia type 2 ATXN2 CAG repeat alleles: a meta-analysis. JAMA Neurol 2014;71:1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chio A, Calvo A, Moglia C, et al. ATXN2 polyQ intermediate repeats are a modifier of ALS survival. Neurology 2015;84:251–258. [DOI] [PubMed] [Google Scholar]
- 18.Nechiporuk T, Huynh DP, Figueroa K, Sahba S, Nechiporuk A, Pulst SM. The mouse SCA2 gene: cDNA sequence, alternative splicing and protein expression. Hum Mol Genet 1998;7:1301–1309. [DOI] [PubMed] [Google Scholar]
- 19.Box GEP, Draper NR. Empirical Model-building and Response Surface. New York: Wiley; 1987. [Google Scholar]
- 20.Ingram MA, Orr HT, Clark HB. Genetically engineered mouse models of the trinucleotide-repeat spinocerebellar ataxias. Brain Res Bull 2012;88:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynh DP, Figueroa K, Hoang N, Pulst SM. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat Genet 2000;26:44–50. [DOI] [PubMed] [Google Scholar]
- 22.Hansen ST, Meera P, Otis TS, Pulst SM. Changes in Purkinje cell firing and gene expression precede behavioral pathology in a mouse model of SCA2. Hum Mol Genet 2013;22:271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dansithong W, Paul S, Figueroa KP, et al. Ataxin-2 regulates RGS8 translation in a new BAC-SCA2 transgenic mouse model. PLOS Genet 2015;11:e1005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh DP, Maalouf M, Silva AJ, Schweizer FE, Pulst SM. Dissociated fear and spatial learning in mice with deficiency of ataxin-2. PLoS One 2009;4:e6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiehl TR, Nechiporuk A, Figueroa KP, Keating MT, Huynh DP, Pulst SM. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem Biophys Res Commun 2006;339:17–24. [DOI] [PubMed] [Google Scholar]
- 26.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet 2000;9:1303–1313. [DOI] [PubMed] [Google Scholar]
- 27.Nonhoff U, Ralser M, Welzel F, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell 2007;18:1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet 2006;15:2523–2532. [DOI] [PubMed] [Google Scholar]
- 29.Nonis D, Schmidt MH, van de Loo S, et al. Ataxin-2 associates with the endocytosis complex and affects EGF receptor trafficking. Cell Signal 2008;20:1725–1739. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Tang TS, Tu H, et al. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. J Neurosci 2009;29:9148–9162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigo F, Seth PP, Bennett CF. Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv Exp Med Biol 2014;825:303–352. [DOI] [PubMed] [Google Scholar]
- 32.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 2010;50:259–293. [DOI] [PubMed] [Google Scholar]
- 33.Evers MM, Toonen LJ, van Roon-Mom WM. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv Drug Deliv Rev 2015;87:90–103. [DOI] [PubMed] [Google Scholar]