Abstract
Toxin–antitoxin systems are encoded by bacteria and archaea to enable an immediate response to environmental stresses, including antibiotics and the host immune response. During normal conditions, the antitoxin components prevent toxins from interfering with metabolism and arresting growth; however, toxin activation enables microbes to remain dormant through unfavorable conditions that might continue over millions of years. Intense investigations have revealed a multitude of mechanisms for both regulation and activation of toxin–antitoxin systems, which are abundant in pathogenic microorganisms. This minireview provides an overview of the current knowledge regarding type II toxin–antitoxin systems along with their clinical and environmental implications.
Keywords: Bacteria, biomedical, microbiology, protein–protein interactions, proteolysis, enzymes
Introduction
Toxin–antitoxin (TA) modules are bacterial operons that are part of the mobilome, moving from one organism to another via horizontal gene transfer,1,2 and these loci are important for facilitating a state of dormancy in bacteria under stressful conditions. Multiple TA loci have been found in hundreds of sequenced bacterial genomes to date, including clinical isolates of Mycobacterium tuberculosis, Pseudomonas spp., methicillin-resistant Staphylococcus aureus, uropathogenic Escherichia coli, Vibrio cholerae, and Yersinia pestis.3–6 These systems consist of a co-transcribed gene pair which encodes both a toxin that acts within the bacterium itself to arrest growth, and an antitoxin that interferes with the toxin activity. Five families of TA modules have been identified to date and are characterized by the nature of the associated antitoxins. Type I systems consist of an antisense RNA antitoxin that binds to the toxin mRNA, preventing ribosome binding and likely targeting the resulting RNA duplex for degradation by RNase III.7 Type II TA modules encode a protein antitoxin and toxin that form a tight non-toxic complex upon translation.8,9 Type III systems consist of a protein toxin bound by an RNA pseudoknot antitoxin formed from a tandem array of repeats.10–12 Type IV TA systems are characterized by a protein antitoxin that does not bind the toxin, but instead interferes with the binding between the toxin and its target.13,14 One representative of a type V toxin–antitoxin system has been described, in which an antitoxin protein specifically cleaves the mRNA of the toxin.15 In this minireview, we will focus on the type II TA systems, which have been the most extensively studied among the five families.
Toxin–antitoxin loci were first identified on plasmids and thought to be merely addiction systems that ensured plasmid stability.16 However, with advances in DNA sequencing came the ability to sequence and analyze numerous bacterial chromosomes, and investigators discovered that these gene pairs were nearly ubiquitous and found in the genomes of archaea, as well as Gram-negative and Gram-positive bacteria, often in multiple copies. Because bacterial chromosomes had no need for an addiction system, the principle of parsimony suggested that maintaining these operons must confer some sort of benefit to the organism. Although researchers were not certain why these modules were so highly conserved, the evidence strongly favored the notion that the maintenance of TA loci in bacterial genomes supported important biological functions. It was an exciting time for scientists, as the number of reports of new and heretofore unknown types of TA systems (and TA-like genes) started to increase rapidly.
The first goal was to determine the mechanisms of these modules, which would suggest how the systems might benefit the bacterial host. These early investigations were initially performed in vitro, because it was much easier to control conditions and there were numerous validated protocols and reagents available. However, it was imperative to eventually employ in vivo models to determine the biological relevance of the in vitro findings to clinical isolates. This led to the discovery that TA loci were important for bacterial survival in a mammalian host, including Salmonella enterica serovar Typhimurium, E. coli, M. tuberculosis, and Haemophilus influenzae infections.5,17–20 In addition, some loci have since been shown to be active in bacteria during infections of plant hosts.21,22
Clinical importance of toxin–antitoxin modules
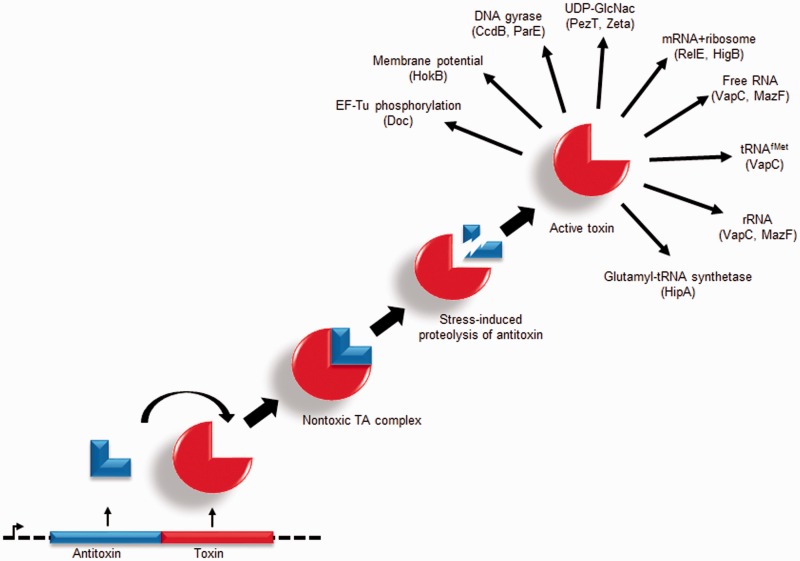
One of the roles of TA systems is to increase bacterial survival under stressful environmental conditions, which includes antibiotic, oxidative, nutrient, pH or heat stress, and attack by bacteriophages or the host immune response.8,17,19,23–26 For type II TA modules, the microbial general stress response induces bacterial cellular proteases, such as Lon and ClpXP, that degrade the labile antitoxin portion of the TA protein complex, freeing the more stable toxin protein to exert its effects (Figure 1).27 Many type II TA toxins are ribonucleases that degrade bacterial mRNA in a sequence-specific or non-specific fashion, causing the organism to enter a state of bacteriostasis.3,28–34 This is beneficial to a pathogenic bacterium, particularly during an infection, as growth arrest significantly decreases its metabolic burden, accumulation of DNA damage from reactive oxygen species, as well as minimizing the production of molecular patterns that signal receptors of the host’s immune response. Further, because TA complexes are pre-formed, activation of this mechanism does not require transcription and translation of effectors, allowing the organism to mount an immediate response to its microenvironment.35 Also, because most antimicrobials disrupt an essential function for replicating cells (i.e. DNA, RNA, and protein synthesis), treatment with these compounds is not effective when the bacterium is in a non-replicative state.36 However, if stress is removed from the microenvironment, surviving cells can resume growth and again display their natural antibiotic susceptibilities.37 These findings highlight the importance of TA modules in facilitating this rapid, effective and highly conserved survival mechanism.
Figure 1.
Canonical type II TA module expression and stress-induced degradation of the antitoxin, leading to an active toxin. A number of targets are shown (arrows), along with the names of representative toxins displaying similar activities. See text for details and references
Bigger38 was the first to provide empirical evidence that bacterial pathogens (in this case, S. aureus) formed tolerant cells which he termed “persisters” that survived treatment with penicillin. Following this ground-breaking study, McDermott39 reported that persister cells could also be formed upon starvation, and that at least some species displayed differences in morphology upon entering a persister state. Further, he hypothesized that the ability of a bacterium to “play dead” was important for its survival in the host, and that dormant infections resulted from alterations in the host environment, whereas latent infections required a change in the pathogen itself. Finally, he coined the term “drug indifference” to describe the ability of persisters to survive antibiotic treatment in vivo, but display susceptibility to the same drug when grown in vitro.39
Most clinically relevant bacteria cannot form endospores, which are tough seed-like structures that allow the organism to exist in a suspended animation state for many years, decades, or even longer. These constructs are resistant to environmental conditions that are normally lethal, such as nutrient deprivation, desiccation, and the presence of noxious chemicals and ionizing radiation. A few medically important Gram-positive human pathogens can form spores, including Bacillus anthracis (anthrax), Clostridium tetani (tetanus), Clostridium botulinum (botulism), and Clostridium difficile (antibiotic treatment-induced colitis).40,41 In non-spore-forming bacteria, TA loci facilitate entry into a bacteriostatic state upon stressful conditions.37 However, a recent study has implicated TA systems in the possible facilitation of recurrent infections with C. difficile, a spore-forming pathogen,40 so these modules might be functional and affect virulence in both groups. Holberger et al.42 provided evidence that the PF04740 proteins found in both pathogenic and non-pathogenic Bacillus species represented a new class of type II TA toxins, in which the N-terminal regions defined the protein family, whereas the C-terminal portion (CT) of each toxin assayed was responsible for its ribonuclease activity. The organization of these modules is reversed compared to canonical TA modules, such that the toxin preceded its antitoxin, reminiscent of the higBA locus. Although the CT portions were highly divergent, the N-terminal regions were conserved and contained an ESAT-6/WXG100 motif, a putative secretion signal in Bacillus. Two purposes were proposed for these modules: that the RNase toxins could be released into the environment to scavenge nucleosides prior to sporulation, or that release of the toxins could enhance competition for environmental niches.
Structural and mechanistic characterizations of type II TA systems
Type II toxins have been shown to target ribonucleic acid by: (a) cleaving free RNA (VapC from Haemophilus strains, MazF, Kid/PemK, HicA, and MqsR),5,28,43–45 (b) cleaving RNA in the context of a ribosome during translation (YafO, RelE, HigB, and YoeB),34,46–48 and (c) cleaving tRNAfMet (VapC from S. enterica serovar Typhimurium).49 Other targets include DNA gyrase (CcdB and ParE);16 uridine diphosphate-N-acetylglucosamine, a peptidoglycan precursor (PezT and Zeta toxins);25 decreasing membrane potential (HokB);50 and phosphorylating glutamyl-tRNA synthetase (HipA)51,52 or EF-Tu (Doc).53 The obg gene, which encodes a conserved P-loop GTPase, has been implicated in persistence using antisense RNA (asRNA), because the gene is essential for viability in E. coli. The Obg protein was shown to increase persistence in both E. coli and P. aeruginosa via the mechanism of inducing expression of the HokB toxin, which results in membrane depolarization leading to both dormancy and antibiotic tolerance.50 While a substantial number of type II TA systems have been characterized by structural and mechanistic studies,54–56 several selected examples are highlighted below and organized according to the mechanism of toxicity.
Ribosome-independent ribonuclease TA systems
Toxins of the VapBC (virulence associated proteins) TA family contain a PIN (PilT N-terminus) domain, which is associated with Mg2+ or Mn2+-dependent ribonuclease activity.30 Indeed, purified VapC from Mycobacterium smegmatis was shown to cleave RNA preferentially at AUAU and AUAA sites in vitro and in vivo.57 The VapC-1 toxin of non-typeable H. influenzae (NTHi) was also shown to function as a ribonuclease with no activity against DNA.28 Similarly, in vitro studies with VapC-5 from M. tuberculosis suggested that the toxin acts on free RNA as both an endo- and exo-ribonuclease.58 While VapC toxin homologues from Shigella flexneri and S. enterica serovar Typhimurium were found to cleave initiator tRNA between the anticodon stem and loop,49 VapC20 of M. tuberculosis cleaves the sarcin-ricin loop of 23S ribosomal RNA, based on secondary structural recognition rather than sequence recognition.59 Although VapC-mt4 from M. tuberculosis demonstrated sequence-specific endo-ribonuclease activity against ACGC or AC(A/U)GC motifs, the activity was relatively weak and translational inhibition and growth arrest preceded RNA cleavage, suggesting that toxin activity might be mediated primarily by RNA binding rather than cleavage.60 Recent crystallographic studies of the VapC30 toxin from M. tuberculosis reveal a homodimer with a canonical α/β/α sandwich fold containing four parallel β-strands flanked on both sides by six α-helices.61 Overall, the structure shared similar architecture with VapC from S. flexneri, VapC2 from Rickettsia felis, VapC15 from M. tuberculosis, and VapC3 from P. aerophilum (Table 1). The structure of the VapB30 antitoxin is distinct from other VapB family members, aside from the N-terminal α-helix. Interestingly, the C-terminal region of VapB30 blocks the activity of the distal dimeric toxin through a swapped inactivation process,61 whereas the structures of M. tuberculosis VapBC362 and VapBC558 show individual antitoxins bound with a cognate toxin to directly block the active site.63
Table 1.
Three-dimensional structures of the type II TA systems described in this minireview
| TA Family | Toxin | Antitoxin | Mechanism of Toxicity | PDB IDs | Method | Organisms |
|---|---|---|---|---|---|---|
| vapBC | VapC | VapB | Ribosome-independent mRNA cleavage | 1W8I, 2FE1, 3TND, 4CHG, 4XGQ, 4XGR, 3ZVK, 3H87, 3DBO, 1Y82 | X-ray diffraction | Archaeoglobus fulgidus, Pyrobaculum aerophilum, Shigella flexneri, Mycobacterium tuberculosis, Rickettsia felis, Pyrococcus furiosus |
| mazEF | MazF | MazE | Ribosome-independent mRNA cleavage | 3NFC, 1UB4, 4MDX, 4ME7, 4MZP, 4MZT, 2MF2, 4MZM, 4OF1 | X-ray diffraction and solution NMR | Escherichia coli, Bacillus subtilis, Staphylococcus aureus |
| relBE | RelE | RelB | Ribosome-dependent mRNA cleavage | 1WMI, 2K29, 3BPQ, 2KC9, 2KC8, 3G5O, 2KHE, 3OEI, 4FXI, 4FXH, 4FXE, 4V7J, 4V7K | X-ray diffraction and solution NMR | Escherichia coli, Pyrococcus horikoshii, Methanocaldococcus jannaschii, Mycobacterium tuberculosis, Thermus thermophilus |
| higBA | HigB | HigA | Ribosome-dependent mRNA cleavage | 4YPB, 4YZV, 4MCX, 4MCT, 2ICT, 4PX8 | X-ray diffraction | Escherichia coli, Proteus vulgaris |
| ccd | CcdB | CcdA | Inhibits DNA gyrase | 2H3A, 2H3C, 3TCJ, 2ADL, 2H3C, 2ADN, 3G7Z, 3HPW, 3VUB, 2VUB, 2KMT, 3JRZ, 3JSC, 1X75, 4VUB, 1VUB, 4ELZ, 4ELY | X-ray diffraction and solution NMR | Escherichia coli, Aliivibrio fischeri, Shigella flexneri |
| parDE | ParE | ParD | Inhibits DNA gyrase | 3KXE, 2AN7 | X-ray diffraction and solution NMR | Escherichia coli, Caulobacter crescentus |
| hipBA | HipA | HipB | Phosphorylates glutamyl-transfer RNA synthase | 4YG7, 4YG4, 4YG1, 3DNV, 3HZI, 3FBR, 3DNU, 3DNT, 2WIU, 3TPB, 3TPD, 3TPT, 3TPV, 4PU3, 4PU4, 4PU5, 4PU7, 4PU8 | X-ray diffraction | Escherichia coli, Shewanella oneidensis |
| phd-doc | Doc | PhD | Inhibits the 30S ribosome and Phosphorylates EF-Tu | 3K33, 3KH2, 3HS2, 3HRY, 3DD7, 3DD9 | X-ray diffraction | Escherichia coli, Enterobacteria phage P1, Enterobacteria phage P2 |
Note: The information was collected from the Protein Data Bank (www.rcsb.org).63
The mazEF TA family, encoding the MazE antitoxin and MazF toxin, has been studied extensively and MazF homologues have been identified in archaea and bacteria, with the majority of studies focusing on the E. coli system.64 The first crystallographic structures reported were of the E. coli chromosomal MazEF complex, which form a linear heterohexamer of alternating toxin and antitoxin homodimers (MazF2-MazE2-MazF2).65 The MazF toxin is an endo-ribonuclease that cleaves single-stranded mRNA, with specificity for ACA sequences, to reduce overall protein synthesis through rapid degradation of bulk mRNA.65 MazF also targets the 5′ untranslated region among a distinct subset of transcripts, sometimes removing the Shine-Dalgarno sequence to generate leaderless mRNA. Additionally, MazF cleaves 16S rRNA to remove the anti-Shine-Dalgarno sequence, thereby generating specific ribosomes that allow translation of MazF-processed mRNAs.66,67 Operons for mazEF in M. tuberculosis were shown to encode endo-ribonucleases that target sequence-specific regions of the era mRNA, with MazF-mt1 cleaving U*AC triplet sequences (* indicates the cleavage site) and MazF-mt6 cleaving U-rich regions.68 Similarly, MazF-mt3 demonstrates specific cleavage of RNA at UU*CCU or CU*CCU and MazF-mt7 cleaves at U*CGCU. It was proposed that MazF toxins might alter protein expression via differential cleavage of mRNA.69 Recently, MazF-mt6 was reported to cleave 23S rRNA at a single UU*CCU sequence in the ribosomal aminoacyl (A) site which contacts tRNA and ribosome recycling factor. MazF-mt6-mediated cleavage of rRNA was demonstrated to inhibit protein synthesis even in the absence of mRNA cleavage. Additionally, MazF-mt6 was found to destabilize the 50S-30S ribosomal subunit association.70 Similarly, another mechanism of toxicity was discovered for MazF-mt7, based on interactions with topoisomerase I that inhibit the activity of both enzymes.71 Collectively, these results suggest a variety of ways that MazF proteins might mediate toxicity. Studies with MazF from Bacillus subtilis demonstrated sequence-specific cleavage at U*ACAU.72 Crystal structures were determined of B. subtilis MazF, including complexes of MazF with MazE (Figure 2(a)) or mRNA containing the uncleavable dUACAU sequence (Figure 2(b)).73 The MazF-mRNA complex showed dUACAU bound to MazF with the bases projecting towards the dimer interface of the toxin and the phosphate backbone moieties projecting away from the surface. Residues R25 and T48, which are highly conserved among MazF homologues, form hydrogen bonds with the oxygen atoms of the scissile phosphate between dU3 and A4 of the mRNA and either R25A or T48A mutations resulted in a loss of toxicity.73 The mRNA binding and cleavage by MazF are blocked through the positioning of the MazE C-terminal helical region within the RNA binding channel of the MazF dimer (Figure 2(a)).73
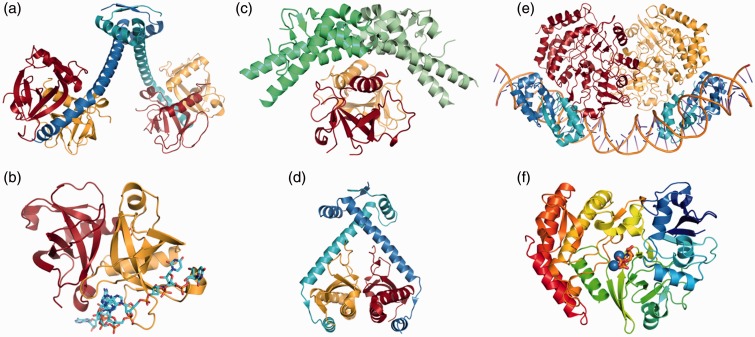
Figure 2.
Cartoon representations of selected toxin and antitoxin structures deposited in the PDB database.63 The selection includes toxins with different mechanisms, including ribonuclease, DNA gyrase inhibitor and kinase (from left). (a) MazF toxin homodimers from B. subtilis (red and gold) are inhibited when bound to the C-terminal region of MazE antitoxin proteins (blue and cyan) (PDB ID: 4ME7).73 (b) A dimeric structure of MazF from B. subtilis (red and gold) is in complex with RNA (cyan sticks) (PDB ID: 4MDX).73 (c) A homodimer of the DNA gyrase subunit A dimerization domain (green and light green) is bound to a CcdB homodimer (red and gold) from E. coli (PDB ID: 1X75).74 (d) A ParE toxin homodimer from Caulobacter vibrioides (red and gold) is inhibited by a ParD antitoxin homodimer (blue and cyan) (PDB ID: 3KXE).75 (e) A multi-molecular complex of E. coli HipA toxin homodimers (red and gold) and HipB antitoxin homodimers (blue and cyan) bound to DNA operator sites O1 and O2 (orange) (PDB ID: 4YG7).76 (f) The ribbon diagram of a HipA toxin structure is colored as a rainbow from blue (N-terminus) to red (C-terminus) and shows two bound Mg2+ ions (blue spheres) and ATP (orange sticks) (PDB ID: 3DNT).77 All cartoon diagrams were generated with the program PYMOL.78
Ribosome-dependent ribonuclease TA systems
The relBE TA family toxin, RelE, has been extensively studied both in vitro and in vivo and has been shown to function as a ribosome-dependent endonuclease.79 Under normal conditions, RelE is inhibited by forming a complex with the antitoxin, RelB, which has been revealed at the atomic level by structural studies of RelBE from E. coli, M. tuberculosis, Methanocaldococcus jannaschii, and Pyrococcus horikoshii (Table 1). Both in vitro and in vivo studies showed that the overall RelB:RelE ratio regulates transcriptional repression. Binding to the relO operator is enhanced by RelE up to a RelB:RelE ratio of 2:1, beyond which the affinity for DNA is reduced, which has been described as “conditional cooperativity.”80–82 Following degradation of RelB by Lon protease, one of the bacterial stress-induced proteases, RelE binds to the ribosome and cleaves mRNA in the A site.79,83 Crystal structures were determined of E. coli RelE bound to Thermus thermophilus 70S ribosomes in complex with mRNA and tRNAfMet before and after mRNA cleavage.84 The structures show that binding of RelE to the A site on the 30S subunit significantly reorganizes the mRNA, leading to 2′-OH-induced hydrolysis between the second and third codon positions.84 Although RelE was shown to cleave codons preferentially in vitro,79 recent studies suggest that RelE specificity might be broader than originally thought.85,86
Another well-studied ribosome-dependent ribonuclease TA system is the higBA (host inhibition of growth) family, which is related to the relBE family.34 In contrast to many other TA families, including relBE, the higBA operon has an inverted gene structure with the toxin gene preceding the antitoxin gene.87 Crystallographic structures of a tetrameric HigB-(HigA)2-HigB TA complex from Proteus vulgaris have been reported.88 The overall tertiary fold of the HigB toxin is similar to the RelE family; however, the structures revealed some unusual molecular details about the HigBA TA system. Interestingly, the HigA antitoxin makes minimal interactions with HigB and does not block the active site, which is solvent-exposed.88 Additionally, HigA monomers contain a DNA-binding helix-turn-helix motif that binds an individual operator sequence, which is unlike antitoxins of other superfamilies that require dimerization to form DNA-binding motifs.88 The tetrameric complex was critical for binding the DNA operator, as HigBA heterodimers were unable to bind.88 Recently, crystal structures were determined of P. vulgaris HigB bound to the T. thermophilus 70S ribosome containing either an AAA or ACA codon in the A site.89 The structures revealed that a microbial RNase-like nucleotide loop of HigB is able to recognize either cytosine or adenosine at the second A-site position. The residue N71 of HigB along with nucleotide C1054 of 16S rRNA contributes to a pocket at the third A-site nucleotide that is specific for adenosine at the +6 position.89 Toxin recognition of mRNA substrates through the third nucleotide of the codon is in contrast to the way tRNAs utilize an anticodon stem loop to recognize A site codons.89 Together, these studies provide a mechanistic explanation for the way ribosomes enable HigB and possibly other ribosome-dependent ribonucleases to achieve substrate specificity.
TA systems that target DNA gyrase
DNA gyrase is an essential bacterial topoisomerase that hydrolyzes ATP to introduce negative supercoils into DNA and is also a target of the quinolone family of antibiotics. The active enzyme is a heterotetramer of two subunits, GyrA, which cleaves DNA, and GyrB that hydrolyzes ATP.74 The ccd (coupled cell division) system of the E. coli F plasmid, comprises a CcdB toxin and a CcdA antitoxin.16 The CcdB toxin is a 101 amino acid residue protein that binds to GyrA with strong affinity90 and inhibits DNA gyrase by either poisoning a covalent DNA-gyrase complex or binding free gyrase, both of which involve identical interactions between CcdB and GyrA (Figure 2(c)).91 Inhibition of gyrase by CcdB is reversible, as the CcdA antitoxin can compete with gyrase to bind and sequester CcdB, thereby allowing DNA gyrase to regain function.92 The CcdA antitoxin is a protein of 72 amino acid residues and two independent domains. The N-terminal domain contains a ribbon-helix-helix fold that facilitates DNA binding and dimerization, whereas the C-terminal domain, which is intrinsically unstructured, binds and inhibits the CcdB toxin.93 The high flexibility of the C-terminal region is thought to increase the susceptibility of CcdA to cleavage by the Lon protease.93 Structural studies of the CcdB toxin and the CcdA antitoxin proteins by both nuclear magnetic resonance (NMR) and X-ray crystallography have been reported (Table 1).
Similar to CcdB, the ParE toxin also blocks DNA replication through inhibition of DNA gyrase.94 A crystal structure of the ParD-ParE complex from Caulobacter crescentus was reported and reveals an α2β2 heterotetrameric complex (Figure 2(d)), which is supported by solution studies.75 The ParD antitoxin contains an N-terminal ribbon-helix-helix DNA binding motif, which facilitates dimerization, as well as an extended α2 helix and an α3 helix towards the C-terminus that both interact with ParE.75 Interestingly, structural comparisons between toxins revealed hydrophobic grooves for antitoxin recognition and binding that are conserved between the parDE and relBE families.75 Although ParE shares both primary and tertiary structural homology with the RelE toxin, ParE lacks the three catalytic residues required for mRNA cleavage on the ribosome.75,84
TA systems that encode kinases
Within the hipBA (high persistence) family of E. coli, hipA encodes the 440-residue toxin protein, HipA, which is co-transcribed with a smaller upstream gene, hipB, that encodes an 88-residue antitoxin protein, HipB. The HipA toxin is a member of the phosphatidylinositol 3/4-kinase superfamily95 and structural studies by multiple groups have confirmed that HipA has a eukaryotic serine/threonine kinase-like fold that is most similar to human CDK2/cyclin A kinase.77,96 Initially, HipA was reported to phosphorylate the transcription factor, EF-Tu;77,97 however, more recent results indicate that the target is S239 of glutamyl-tRNA synthetase, which is located near the enzyme active site.51,52 The kinase activity of HipA is regulated by phosphorylation of the residue S150, which forms part of the ATP-binding P loop motif and inactivates the kinase by disrupting the ATP binding pocket.97 Previously, mutations in the E. coli HipA protein had been associated with a substantial increase in persister cells.98 Recent structural studies showed that these mutations localize to the HipA N-subdomain-1, which is distal to the toxin active site and HipB binding site; however, this region facilitates HipA dimerization within higher order complexes of HipA-HipB and multiple operators in hipBA promoters (Figure 2(e)).76 The HipA–HipA interface blocks the kinase active sites (Figure 2(f)), so persister-associated mutations in the N-subdomain-1 are thought to release HipA from the inactive state.76
The phd-doc TA module encodes the 126-residue toxin, Doc (death on curing), and 73-residue antitoxin, Phd (prevent host death), to maintain the plasmid-prophage P1 in E. coli.99 Regulation of the phd/doc operon involves conditional cooperativity that is regulated by complexes of Phd and Doc.100 Doc is a member of the Fic (filamentation induced by cyclic AMP) family of AMPylation enzymes, which are found in all kingdoms of life.101 The toxicity of Doc was initially reported to be inhibition of translation elongation through interactions with the 30S ribosomal subunit, similar to the aminoglycoside antibiotic hygromycin B.53 More recently, it was discovered that Doc is a new type of kinase that phosphorylates the translation elongation factor EF-Tu on the conserved threonine, T382, thereby preventing EF-Tu from binding aminoacylated tRNAs.102
Microbial dormancy
Dormancy is the state of most of the prokaryotes on Earth, whose total numbers are estimated to be in the range of 4.0–6.0 × 1030 cells in three enormous habitats: soil, seawater, and the marine sediment or terrestrial subsurface.103 Microbes in these environments display extremely low metabolic activity, but are not completely inactive due to the need to maintain their DNA integrity.104,105 Price and Sowers105 estimated that the energy demands of a cell in a dormant state were three orders of magnitude lower than that of a replicating organism. However, a bacterium in a dormant state must maintain not only an intact genome, but also an energized membrane to allow ATP synthesis, both of which are requirements for successful regrowth upon improved conditions. Resuscitation triggers, such as nutrient upshift or specific biomolecules,106,107 as well as stochastic processes of transcriptional regulation and the “microbial scout” hypothesis,108–110 have been implicated in the ability of these dormant bacteria to awaken. For example, this might entail a mechanism of the spontaneous resuscitation of random cells in a population, which subsequently produce a protein known as “resuscitation promoting factor” (Rpf) if conditions are conducive for growth. This molecule then binds to and signals the remaining dormant population to resuscitate.107
Geomicrobiologists have found that conditions in Siberian permafrost soil, which are hundreds of thousands of years old,111 and in deep sub-seafloor sediments, which are millions of years old,112–114 harbor a diverse community of microorganisms at 103 to 108 cells/cm3.115–117 D’Hondt et al.115 found that the sediments in the South Pacific Gyre were characterized by extremely low biomass and metabolic activity. Sub-seafloor sediments harbor between 55% and 85% of all the microbes on the planet,103,112 and Schippers et al.118 provided direct evidence that many of these microbes are alive and capable of metabolism (i.e. not in endospores) by determining the presence of ribosomes using catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH). Given the amount of carbon availability and usage, the estimated average turnover rate of organisms in such sediments is on the order of 1000 years, and might be as long as several thousand years.103,119
These surprising findings have been shown to be characteristic of microbial life on Earth. Inagaki et al.120 hypothesized the existence of a “paleome,” microbial fossil biomolecules encased in black shale identified in deep subsurface sediments. In an intriguing study, these investigators utilized molecular genetic culture-independent analyses to demonstrate that the preserved DNA from deep sections of terrestrial core sediment (250 to 300 cm below the surface) corresponding to the Cretaceous lower Albian period (∼100 million years old) contained phylotypes that were consistent with those found in extant marine environments.120 Interestingly, another careful study found that the bacterial community in deep marine sediment east of Japan (>2 km below the seafloor) harbored a group of microorganisms that resembled a terrestrial community, while those in the shallow sediment at the same sample site reflected a marine population.121 This suggests that at least some microbes have survived for tens of millions of years after burial in the sediment, which challenges our notions of the longevity of prokaryotes and the stability of nucleic acid in such an environment.
The dormant state induced by TA systems can be achieved by spontaneous switching under stable environmental conditions, creating a subpopulation of persister cells that can respond quickly to a changing environment.122,123 Lennon and Jones113 have described the benefit of these persisters as being microbial “seed banks” that ensure the long-term viability of the bacterial population by providing for genetic diversity. Indeed, Ayrapetyan et al.37 suggested that the viable but non-culturable (VBNC) state identified in non-sporulating bacteria and the drug-tolerant bacterial persister cell are both part of a shared “dormancy continuum.” Their hypothesis was that the persister state is a gateway into the VBNC state, and this is why persisters both (a) occur in lower numbers, and (b) resume growth more rapidly upon antibiotic removal, because the so-called VBNC cells can resuscitate, but require more time to do so than persister cells.24 These studies have therefore raised concerns about the validity of measuring viability via CFU counts. Type II TA loci have been linked to both states in various organisms.
Future outlook
It is likely that new types of TA modules will continue to be discovered as the genomes of novel organisms are sequenced. It is also possible that coordinate regulation of various loci and groups of modules might be identified. Currently, unanswered questions of importance to the field include (but are not limited to) the following: are there specific triggers for various TA loci, and can groups be identified? How do the different TA types interact with one another (e.g. type II ribonucleases are activated by protein antitoxin degradation, which might then degrade the RNA antitoxins of type III toxins), and does the threshold level of response to various stresses differ when the organism is infecting a host? Does the specific stressor correlate with the activation of certain TA loci? Further, are all the loci activated simultaneously, or is there a hierarchy in the cascade of toxin activation across types in response to stress? When are these systems induced during the formation of a biofilm? Helaine and Kugelberg124 noted a need for new methods that will allow the study of the multifactorial processes that result in dormant persister cells. However, a related issue is whether advanced techniques, such as the examination of the behavior of single cells in a microfluidics model, can be reflective of actual conditions within an infecting population.
The number of TA systems in the genome has been shown to correlate with the virulence capacity of the organism.125 For example, M. tuberculosis strains encode at least 79 TA modules, whereas the non-pathogenic species M. smegmatis maintains only four.126 Targeting bacterial TA systems might be an effective strategy for the development of novel antibiotics. Indeed, the deletion of a VapBC module in NTHi significantly decreased its virulence in primary human tissues and in an animal model of otitis media.19 Wen et al.127 recently highlighted the remarkable overlap between the targets of TA toxins and antibiotics. Likewise, many investigators have proposed that TA modules have therapeutic potential as novel antimicrobials.55,128–130 In a comprehensive recent review, Chan et al.130 discussed the “druggability” of various type II systems, with a view toward disrupting the protein–protein interactions of the toxin–antitoxin complex in order to free the toxin. However, this tactic also has the potential to induce persister cells that are difficult to treat.131 Furthermore, the redundancy of TA systems and other pathways to persistence presents significant challenges that must be addressed.132,133 In the final analysis, it is possible that targeting multiple TA modules using a range of approaches might result in the most successful antimicrobial strategy.
Concluding remarks
Given the ubiquity of toxin–antitoxin systems among bacteria and their critical roles in adaptation to adverse conditions, we anticipate that future studies will reveal additional complexities in the way these systems operate and function together to sustain life and optimize survival within a range of environments (Figure 3). Knowledge obtained from the study of TA systems has already enabled the development of novel antiviral strategies and biotechnology applications, such as positive selection plasmids and biosensors.130 Therefore, further studies to elucidate the molecular details underlying these important systems will likely continue to drive advancements in biotechnology, synthetic biology and medicine.
Figure 3.
Toxin–antitoxin systems significantly impact a wide range of areas, from human health and enhancing microbial survival in the environment to enabling discoveries in molecular biology and medicine
Acknowledgements
Work in our laboratories was supported by NIH grants R01DC010187 and U01DC014756, along with the NCATS Division of Pre-Clinical Innovation Intramural Program. We thank Dr. D.E. Sonenshine for valuable comments about our manuscript and L.J. Coussens for her contributions to Figure 3. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Space limitations of this minireview prevented citation of all relevant literature.
Authors’ contributions
All authors participated in the writing of this minireview.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 2008; 36: 6688–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 2005; 3: 722–32. [DOI] [PubMed] [Google Scholar]
- 3.Frampton R, Aggio RB, Villas-Boas SG, Arcus VL, Cook GM. Toxin-antitoxin systems of Mycobacterium smegmatis are essential for cell survival. J Biol Chem 2012; 287: 5340–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams JJ, Halvorsen EM, Dwyer EM, DiFazio RM, Hergenrother PJ. Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 2011; 322: 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norton JP, Mulvey MA. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog 2012; 8: e1002954.–e1002954.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goulard C, Langrand S, Carniel E, Chauvaux S. The Yersinia pestis chromosome encodes active addiction toxins. J Bacteriol 2010; 192: 3669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brantl S. Bacterial type I toxin-antitoxin systems. RNA Biology 2012; 9: 1488–90. [DOI] [PubMed] [Google Scholar]
- 8.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 2003; 301: 1496–9. [DOI] [PubMed] [Google Scholar]
- 9.Chukwudi CU, Good L. The role of the hok/sok locus in bacterial response to stressful growth conditions. Microb Pathog 2015; 79: 70–9. [DOI] [PubMed] [Google Scholar]
- 10.Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GP. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol 2009; 191: 6029–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A 2009; 106: 894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao F, Short FL, Voss JE, Blower TR, Orme AL, Whittaker TE, Luisi BF, Salmond GP. Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein-RNA toxin-antitoxin system. Nucleic Acids Res 2015; 43: 9529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda H, Tan Q, Awano N, Wu KP, Inouye M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol 2012; 84: 979–89. [DOI] [PubMed] [Google Scholar]
- 14.Dy RL, Przybilski R, Semeijn K, Salmond GP, Fineran PC. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res 2014; 42: 4590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol 2012; 8: 855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogura T, Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A 1983; 80: 4784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De la Cruz MA, Zhao W, Farenc C, Gimenez G, Raoult D, Cambillau C, Gorvel JP, Meresse S. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog 2013; 9: e1003827–e1003827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Lobato-Marquez D, Moreno-Cordoba I, Figueroa V, Diaz-Orejas R, Garcia-del Portillo F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci Rep 2015; 5: 9374–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren D, Walker AN, Daines DA. Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbiol 2012; 12: 263–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 2009; 5: e1000767–e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shavit R, Lebendiker M, Pasternak Z, Burdman S, Helman Y. The vapB-vapC operon of Acidovorax citrulli functions as a bona-fide toxin-antitoxin module. Fronti Microbiol 2015; 6: 1499–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodogai M, Ferenczi S, Bashtovyy D, Miclea P, Papp P, Dusha I. The ntrPR operon of Sinorhizobium meliloti is organized and functions as a toxin-antitoxin module. Mol Plant Microbe Interact 2006; 19: 811–22. [DOI] [PubMed] [Google Scholar]
- 23.Short FL, Blower TR, Salmond GP. A promiscuous antitoxin of bacteriophage T4 ensures successful viral replication. Mol Microbiology 2012; 83: 665–8. [DOI] [PubMed] [Google Scholar]
- 24.Ayrapetyan M, Williams TC, Baxter R, Oliver JD. Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infect Immun 2015; 83: 4194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mutschler H, Meinhart A. epsilon/zeta systems: their role in resistance, virulence, and their potential for antibiotic development. J Mol Med (Berl) 2011; 89: 1183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian D, Natarajan J. Network analysis of S. aureus response to ramoplanin reveals modules for virulence factors and resistance mechanisms and characteristic novel genes. Gene 2015; 574: 149–62. [DOI] [PubMed] [Google Scholar]
- 27.Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol 2004; 51: 1705–17. [DOI] [PubMed] [Google Scholar]
- 28.Daines DA, Wu MH, Yuan SY. VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol 2007; 189: 5041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahidjo BA, Kuhnert D, McKenzie JL, Machowski EE, Gordhan BG, Arcus V, Abrahams GL, Mizrahi V. VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PloS One 2011; 6: e21738–e21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arcus VL, McKenzie JL, Robson J, Cook GM. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng Des Sel 2011; 24: 33–40. [DOI] [PubMed] [Google Scholar]
- 31.Arcus VL, Rainey PB, Turner SJ. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol 2005; 13: 360–5. [DOI] [PubMed] [Google Scholar]
- 32.Cook GM, Robson JR, Frampton RA, McKenzie J, Przybilski R, Fineran PC, Arcus VL. Ribonucleases in bacterial toxin-antitoxin systems. Biochim Biophys Acta 2013; 1829: 523–31. [DOI] [PubMed] [Google Scholar]
- 33.Boss L, Labudda L, Wegrzyn G, Hayes F, Kedzierska B. The axe-txe complex of Enterococcus faecium presents a multilayered mode of toxin-antitoxin gene expression regulation. PloS One 2013; 8: e73569.–e73569.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurley JM, Woychik NA. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J Biol Chem 2009; 284: 18605–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes F, Kedzierska B. Regulating toxin-antitoxin expression: controlled detonation of intracellular molecular timebombs. Toxins 2014; 6: 337–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 2004; 230: 13–8. [DOI] [PubMed] [Google Scholar]
- 37.Ayrapetyan M, Williams TC, Oliver JD. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol 2015; 23: 7–13. [DOI] [PubMed] [Google Scholar]
- 38.Bigger JW. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 1944; 244: 497–500. [Google Scholar]
- 39.Mc Dermott W. Microbial persistence. Yale J Biol Med 1958; 30: 257–91. [PMC free article] [PubMed] [Google Scholar]
- 40.Gil F, Pizarro-Guajardo M, Alvarez R, Garavaglia M, Paredes-Sabja D. Clostridium difficile recurrent infection: possible implication of TA systems. Future Microbiol 2015; 10: 1649–57. [DOI] [PubMed] [Google Scholar]
- 41.Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol 2012; 14: 2870–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holberger LE, Garza-Sanchez F, Lamoureux J, Low DA, Hayes CS. A novel family of toxin/antitoxin proteins in Bacillus species. FEBS Lett 2012; 586: 132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripathi A, Dewan PC, Siddique SA, Varadarajan R. MazF-induced growth inhibition and persister generation in Escherichia coli. J Biol Chem 2014; 289: 4191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butt A, Higman VA, Williams C, Crump MP, Hemsley CM, Harmer N, Titball RW. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochem J 2014; 459: 333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figueira R, Brown DR, Ferreira D, Eldridge MJ, Burchell L, Pan Z, Helaine S, Wigneshweraraj S. Adaptation to sustained nitrogen starvation by Escherichia coli requires the eukaryote-like serine/threonine kinase YeaG. Sci Rep 2015; 5: 17524–17524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Yamaguchi Y, Inouye M. Characterization of YafO, an Escherichia coli toxin. J Biol Chem 2009; 284: 25522–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korch SB, Malhotra V, Contreras H, Clark-Curtiss JE. The Mycobacterium tuberculosis relBE toxin:antitoxin genes are stress-responsive modules that regulate growth through translation inhibition. J Microbiol 2015; 53: 783–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janssen BD, Garza-Sanchez F, Hayes CS. YoeB toxin is activated during thermal stress. Microbiologyopen 2015; 4: 682–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A 2011; 108: 7403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versees W, Hofkens J, Jansen M, Fauvart M, Michiels J. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 2015; 59: 9–21. [DOI] [PubMed] [Google Scholar]
- 51.Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular mechanism of bacterial persistence by HipA. Mol Cell 2013; 52: 248–54. [DOI] [PubMed] [Google Scholar]
- 52.Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, Glaser G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun 2013; 4: 3001–3001. [DOI] [PubMed] [Google Scholar]
- 53.Liu M, Zhang Y, Inouye M, Woychik NA. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc Natl Acad Sci U S A 2008; 105: 5885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blower TR, Salmond GP, Luisi BF. Balancing at survival's edge: the structure and adaptive benefits of prokaryotic toxin-antitoxin partners. Curr Opin Struct Biol 2011; 21: 109–18. [DOI] [PubMed] [Google Scholar]
- 55.Park SJ, Son WS, Lee BJ. Structural overview of toxin-antitoxin systems in infectious bacteria: a target for developing antimicrobial agents. Biochim Biophys Acta 2013; 1834: 1155–67. [DOI] [PubMed] [Google Scholar]
- 56.Loris R, Garcia-Pino A. Disorder- and dynamics-based regulatory mechanisms in toxin-antitoxin modules. Chem Rev 2014; 114: 6933–47. [DOI] [PubMed] [Google Scholar]
- 57.McKenzie JL, Robson J, Berney M, Smith TC, Ruthe A, Gardner PP, Arcus VL, Cook GM. A VapBC toxin-antitoxin module is a posttranscriptional regulator of metabolic flux in mycobacteria. J Bacteriol 2012; 194: 2189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miallau L, Faller M, Chiang J, Arbing M, Guo F, Cascio D, Eisenberg D. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J Biol Chem 2009; 284: 276–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winther KS, Brodersen DE, Brown AK, Gerdes K. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat Commun 2013; 4: 2796–2796. [DOI] [PubMed] [Google Scholar]
- 60.Sharp JD, Cruz JW, Raman S, Inouye M, Husson RN, Woychik NA. Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J Biol Chem 2012; 287: 12835–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee IG, Lee SJ, Chae S, Lee KY, Kim JH, Lee BJ. Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides. Nucleic Acids Res 2015; 43: 7624–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min AB, Miallau L, Sawaya MR, Habel J, Cascio D, Eisenberg D. The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. tuberculosis reveals a Mg(2)(+) ion in the active site and a putative RNA-binding site. Protein Sci 2012; 21: 1754–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res 2000; 28: 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertram R, Schuster CF. Post-transcriptional regulation of gene expression in bacterial pathogens by toxin-antitoxin systems. Front Cell Infect Microbiol 2014; 4: 6–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamada K, Hanaoka F, Burley SK. Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell 2003; 11: 875–84. [DOI] [PubMed] [Google Scholar]
- 66.Sauert M, Wolfinger MT, Vesper O, Muller C, Byrgazov K, Moll I. The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res. Epub ahead of print 22 February 2016. DOI: 10.1093/nar/gkw115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 2011; 147: 147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, Inouye M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem 2006; 281: 18638–43. [DOI] [PubMed] [Google Scholar]
- 69.Zhu L, Phadtare S, Nariya H, Ouyang M, Husson RN, Inouye M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol Microbiol 2008; 69: 559–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schifano JM, Edifor R, Sharp JD, Ouyang M, Konkimalla A, Husson RN, Woychik NA. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc Natl Acad Sci U S A 2013; 110: 8501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang F, He ZG. Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic Acids Res 2010; 38: 8219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park JH, Yamaguchi Y, Inouye M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett 2011; 585: 2526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simanshu DK, Yamaguchi Y, Park JH, Inouye M, Patel DJ. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis Mol Cell 2013;52:447–58. [DOI] [PMC free article] [PubMed]
- 74.Dao-Thi MH, Van Melderen L, De Genst E, Afif H, Buts L, Wyns L, Loris R. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol 2005; 348: 1091–102. [DOI] [PubMed] [Google Scholar]
- 75.Dalton KM, Crosson S. A conserved mode of protein recognition and binding in a ParD-ParE toxin-antitoxin complex. Biochemistry 2010; 49: 2205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schumacher MA, Balani P, Min J, Chinnam NB, Hansen S, Vulic M, Lewis K, Brennan RG. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 2015; 524: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 2009; 323: 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeLano WL. PyMOL molecular viewer: updates and refinements. Abstract Paper Am Chem Soc 2009; 238: Abstract 1-1–Abstract 1-1. [Google Scholar]
- 79.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 2003; 112: 131–40. [DOI] [PubMed] [Google Scholar]
- 80.Overgaard M, Borch J, Jorgensen MG, Gerdes K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol Microbiol 2008; 69: 841–57. [DOI] [PubMed] [Google Scholar]
- 81.Overgaard M, Borch J, Gerdes K. RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB. J Mol Biol 2009; 394: 183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cataudella I, Trusina A, Sneppen K, Gerdes K, Mitarai N. Conditional cooperativity in toxin-antitoxin regulation prevents random toxin activation and promotes fast translational recovery. Nucleic Acids Res 2012; 40: 6424–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol 2003; 48: 1389–400. [DOI] [PubMed] [Google Scholar]
- 84.Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 2009; 139: 1084–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goeders N, Dreze PL, Van Melderen L. Relaxed cleavage specificity within the RelE toxin family. J Bacteriol 2013; 195: 2541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hurley JM, Cruz JW, Ouyang M, Woychik NA. Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from the 5' end of coding regions in vivo. J Biol Chem 2011; 286: 14770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tian QB, Ohnishi M, Tabuchi A, Terawaki Y. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem Biophys Res Commun 1996; 220: 280–4. [DOI] [PubMed] [Google Scholar]
- 88.Schureck MA, Maehigashi T, Miles SJ, Marquez J, Cho SE, Erdman R, Dunham CM. Structure of the Proteus vulgaris HigB-(HigA)2-HigB toxin-antitoxin complex. J Biol Chem 2014; 289: 1060–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schureck MA, Dunkle JA, Maehigashi T, Miles SJ, Dunham CM. Defining the mRNA recognition signature of a bacterial toxin protein. Proc Natl Acad Sci U S A 2015; 112: 13862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Jonge N, Simic M, Buts L, Haesaerts S, Roelants K, Garcia-Pino A, Sterckx Y, De Greve H, Lah J, Loris R. Alternative interactions define gyrase specificity in the CcdB family. Mol Microbiol 2012; 84: 965–78. [DOI] [PubMed] [Google Scholar]
- 91.Bahassi EM, O'Dea MH, Allali N, Messens J, Gellert M, Couturier M. Interactions of CcdB with DNA gyrase. Inactivation of Gyra, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J Biol Chem 1999; 274: 10936–44. [DOI] [PubMed] [Google Scholar]
- 92.De Jonge N, Garcia-Pino A, Buts L, Haesaerts S, Charlier D, Zangger K, Wyns L, De Greve H, Loris R. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol Cell 2009; 35: 154–63. [DOI] [PubMed] [Google Scholar]
- 93.Madl T, Van Melderen L, Mine N, Respondek M, Oberer M, Keller W, Khatai L, Zangger K. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J Mol Biol 2006; 364: 170–85. [DOI] [PubMed] [Google Scholar]
- 94.Jiang Y, Pogliano J, Helinski DR, Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol 2002; 44: 971–9. [DOI] [PubMed] [Google Scholar]
- 95.Correia FF, D'Onofrio A, Rejtar T, Li L, Karger BL, Makarova K, Koonin EV, Lewis K. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J Bacteriol 2006; 188: 8360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Evdokimov A, Voznesensky I, Fennell K, Anderson M, Smith JF, Fisher DA. New kinase regulation mechanism found in HipBA: a bacterial persistence switch. Acta Crystallogr D Biol Crystallogr 2009; 65: 875–9. [DOI] [PubMed] [Google Scholar]
- 97.Schumacher MA, Min J, Link TM, Guan Z, Xu W, Ahn YH, Soderblom EJ, Kurie JM, Evdokimov A, Moseley MA, Lewis K, Brennan RG. Role of unusual P loop ejection and autophosphorylation in HipA-mediated persistence and multidrug tolerance. Cell Rep 2012; 2: 518–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol 2003; 50: 1199–213. [DOI] [PubMed] [Google Scholar]
- 99.Lehnherr H, Maguin E, Jafri S, Yarmolinsky MB. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol 1993; 233: 414–28. [DOI] [PubMed] [Google Scholar]
- 100.Garcia-Pino A, Balasubramanian S, Wyns L, Gazit E, De Greve H, Magnuson RD, Charlier D, van Nuland NA, Loris R. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 2010; 142: 101–11. [DOI] [PubMed] [Google Scholar]
- 101.Engel P, Goepfert A, Stanger FV, Harms A, Schmidt A, Schirmer T, Dehio C. Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 2012; 482: 107–10. [DOI] [PubMed] [Google Scholar]
- 102.Castro-Roa D, Garcia-Pino A, De Gieter S, van Nuland NA, Loris R, Zenkin N. The Fic protein doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol 2013; 9: 811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 1998; 95: 6578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morono Y, Terada T, Nishizawa M, Ito M, Hillion F, Takahata N, Sano Y, Inagaki F. Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc Natl Acad Sci USA 2011; 108: 18295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Price PB, Sowers T. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci USA 2004; 101: 4631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morishige Y, Fujimori K, Amano F. Differential resuscitative effect of pyruvate and its analogues on VBNC (viable but non-culturable) Salmonella. Microbes Environ 2013; 28: 180–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Puspita ID, Kamagata Y, Tanaka M, Asano K, Nakatsu CH. Are uncultivated bacteria really uncultivable? Microbes Environ 2012; 27: 356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Epstein SS. Microbial awakenings. Nature 2009; 457: 1083–1083. [DOI] [PubMed] [Google Scholar]
- 109.Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl Environ Microbiol 2012; 78: 3221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. Microbial scout hypothesis and microbial discovery. Appl Environ Microbiol 2012; 78: 3229–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johnson SS, Hebsgaard MB, Christensen TR, Mastepanov M, Nielsen R, Munch K, Brand T, Gilbert MT, Zuber MT, Bunce M, Ronn R, Gilichinsky D, Froese D, Willerslev E. Ancient bacteria show evidence of DNA repair. Proc Natl Acad Sci USA 2007; 104: 14401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jorgensen BB, D'Hondt S. Ecology. A starving majority deep beneath the seafloor. Science 2006; 314: 932–4. [DOI] [PubMed] [Google Scholar]
- 113.Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 2011; 9: 119–30. [DOI] [PubMed] [Google Scholar]
- 114.Haruta S, Kanno N. Survivability of microbes in natural environments and their ecological impacts. Microbes Environ 2015; 30: 123–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.D'Hondt S, Spivack AJ, Pockalny R, Ferdelman TG, Fischer JP, Kallmeyer J, Abrams LJ, Smith DC, Graham D, Hasiuk F, Schrum H, Stancin AM. Subseafloor sedimentary life in the South Pacific Gyre. Proc Natl Acad Sci USA 2009; 106: 11651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vreeland RH, Rosenzweig WD, Powers DW. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature 2000; 407: 897–900. [DOI] [PubMed] [Google Scholar]
- 117.Parkes RJ. A case of bacterial immortality? Nature 2000; 407: 844–5. [DOI] [PubMed] [Google Scholar]
- 118.Schippers A, Neretin LN, Kallmeyer J, Ferdelman TG, Cragg BA, Parkes RJ, Jorgensen BB. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 2005; 433: 861–4. [DOI] [PubMed] [Google Scholar]
- 119.Jorgensen BB. Deep subseafloor microbial cells on physiological standby. Proc Natl Acad Sci USA 2011; 108: 18193–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Inagaki F, Okada H, Tsapin AI, Nealson KH. Microbial survival: the paleome: a sedimentary genetic record of past microbial communities. Astrobiology 2005; 5: 141–53. [DOI] [PubMed] [Google Scholar]
- 121.Inagaki F, Hinrichs KU, Kubo Y, Bowles MW, Heuer VB, Hong WL, Hoshino T, Ijiri A, Imachi H, Ito M, Kaneko M, Lever MA, Lin YS, Methe BA, Morita S, Morono Y, Tanikawa W, Bihan M, Bowden SA, Elvert M, Glombitza C, Gross D, Harrington GJ, Hori T, Li K, Limmer D, Liu CH, Murayama M, Ohkouchi N, Ono S, Park YS, Phillips SC, Prieto-Mollar X, Purkey M, Riedinger N, Sanada Y, Sauvage J, Snyder G, Susilawati R, Takano Y, Tasumi E, Terada T, Tomaru H, Trembath-Reichert E, Wang DT, Yamada Y. Deep biosphere. Exploring deep microbial life in coal-bearing sediment down to ∼2.5 km below the ocean floor. Science 2015; 349: 420–4. [DOI] [PubMed] [Google Scholar]
- 122.Kussell E, Kishony R, Balaban NQ, Leibler S. Bacterial persistence: a model of survival in changing environments. Genetics 2005; 169: 1807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science 2005; 309: 2075–8. [DOI] [PubMed] [Google Scholar]
- 124.Helaine S, Kugelberg E. Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol 2014; 22: 417–24. [DOI] [PubMed] [Google Scholar]
- 125.Georgiades K, Raoult D. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PloS One 2011; 6: e17962–e17962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sala A, Bordes P, Genevaux P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins 2014; 6: 1002–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wen Y, Behiels E, Devreese B. Toxin-antitoxin systems: their role in persistence, biofilm formation, and pathogenicity. Pathog Dis 2014; 70: 240–9. [DOI] [PubMed] [Google Scholar]
- 128.Williams JJ, Hergenrother PJ. Exposing plasmids as the Achilles' heel of drug-resistant bacteria. Curr Opin Chem Biol 2008; 12: 389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Williams JJ, Hergenrother PJ. Artificial activation of toxin-antitoxin systems as an antibacterial strategy. Trends Microbiol 2012; 20: 291–8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chan WT, Balsa D, Espinosa M. One cannot rule them all: are bacterial toxins-antitoxins druggable? FEMS Microbiol Rev 2015; 39: 522–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wood TK. Combatting bacterial persister cells. Biotechnol Bioeng 2016; 113: 476–83. [DOI] [PubMed] [Google Scholar]
- 132.Lewis K. Antibiotics: recover the lost art of drug discovery. Nature 2012; 485: 439–40. [DOI] [PubMed] [Google Scholar]
- 133.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov 2013; 12: 371–87. [DOI] [PubMed] [Google Scholar]