Abstract
The molecular biology of metazoan eye development is an area of intense investigation. These efforts have led to the surprising recognition that although insect and vertebrate eyes have dramatically different structures, the orthologs or family members of several conserved transcription and signaling regulators such as Pax6, Six3, Prox1 and Bmp4 are commonly required for their development. In contrast, our understanding of post-transcriptional regulation in eye development and disease, particularly regarding the function of RNA binding proteins (RBPs), is limited. We examine the present knowledge of RBPs in eye development in the insect model Drosophila, as well as several vertebrate models such as fish, frog, chicken and mouse. Interestingly, of the 42 RBPs that have been investigated with for their expression or function in vertebrate eye development, 24 (~60%) are recognized in eukaryotic cells as components of RNA granules such as Processing bodies (P-bodies), Stress granules, or other specialized ribonucleoprotein (RNP) complexes. We discuss the distinct developmental and cellular events that may necessitate potential RBP/RNA granule-associated RNA regulon models to facilitate post-transcriptional control of gene expression in eye morphogenesis. In support of these hypotheses, three RBPs and RNP/RNA granule components Tdrd7, Caprin2 and Stau2 are linked to ocular developmental defects such as congenital cataract, Peters anomaly and microphthalmia in human patients or animal models. We conclude by discussing the utility of interdisciplinary approaches such as the bioinformatics tool iSyTE (integrated Systems Tool for Eye gene discovery) to prioritize RBPs for deriving post-transcriptional regulatory networks in eye development and disease.
Keywords: Lens, Retina, Cataract, Post-transcriptional regulation, P-bodies, Stress granules, mRNP interactome, Tdrd7, Caprin2, iSyTE
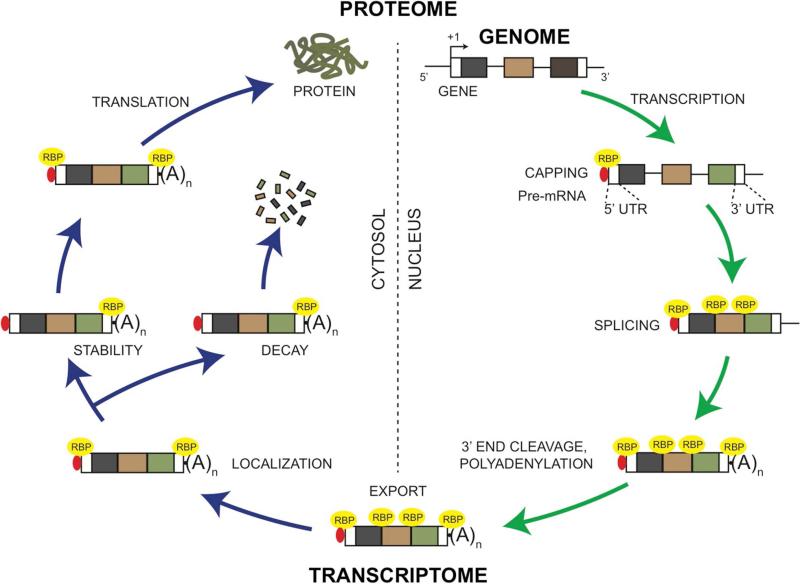
Development of a multicellular organism is orchestrated by the proper expression of its genome, and so far, several families of transcription factors and signaling molecules involved in regulating distinct developmental processes have been identified. Indeed, this rich mechanistic information has now initiated the assembly of detailed gene regulatory networks (GRNs) in specific developmental systems.1,2 In the past two decades however, several revolutionary discoveries in the field of gene expression regulation have revealed that this process is far more complex than was earlier anticipated.3–8 In particular, it is now clear that in addition to transcription and signaling, proper functioning of cells and their decisions to differentiate or remain multipotent requires a myriad of post-transcriptional control mechanisms. For example, these involve regulation of specific events in the life of an mRNA that determine its splicing, export, localization, stability, decay, silencing, and ultimately the extent of its translation into protein (Fig. 1).9 Further, the role of non-coding RNA and RNA binding proteins (RBPs) in mediating post-transcriptional regulation to achieve cell-specific proteomes is now well-appreciated. Yet, compared to the detailed understanding of DNA binding proteins in transcriptional control, function of conserved RBPs in post-transcriptional regulation in vertebrate organogenesis is not well defined. For example, although the mouse genome encodes ~1500 RBPs, few are currently directly associated with organogenesis and developmental disorders in vertebrates.10–14 This reflects a profound gap in our understanding of developmental gene expression, especially because there now is evidence that similar to transcription factors, RBPs participate in combinatorial control, forming RNA operons or higher-order RNA regulons, and unlike transcription factors may even associate in specialized structures such as RNA granules or ribonucleoprotein (RNP) complexes for efficient generation and modulation of the proteome.3,4,15–19 Thus, there lies an urgent need to characterize RBPs that function in morphogenesis of specific tissues, such as the eye.
Figure 1. Control of the eukaryotic mRNA by RNA binding proteins.
RNA binding proteins (RBPs) function in distinct regulatory events in the mRNA life-cycle. During transcription of a gene to pre-mRNA, the nascent transcript is capped with 7-methylguanosine to stabilize the mRNA, a process that is facilitated by RBPs such as RAM. RBPs bind to the 5’-Cap to form the Cap binding complex and mediate further control. Excision of the intronic regions from the pre-mRNA can occur co-transcriptionally, a process in which RBPs can bind to the splicing machinery or the exon-intron junctions to drive tissue-specific splicing reactions. The 3’ end of the pre-mRNA is cleaved at a specific site followed by addition of 150-200 adenosine residues (Poly(A) tail) to form a mature mRNA, a process that is facilitated by RBPs such as Poly(A)-binding protein (Pabp). The mature mRNA is then bound by specialized RBPs and exported to the cytosol. In the cytosol, binding of RBPs (e.g. Stau1 or Zbp1) to either the 3’ UTR or the 5’ UTR facilitate the localization mRNA to specific regions for site-specific translation in cells such as neurons or fibroblasts. The localized mRNA is either stabilized or degraded by RBPs binding to sequence-specific sites such as the ARE (AU-rich element) in its 3’UTR. Within the cytosol, RBPs facilitate translation of mRNA into polypeptide. Alternately, mRNA can be recruited to RNA granules such as Processing bodies (P-bodies), Stress granules or other ribonucleoprotein (RNP) complexes for stability, localized translation, silencing or decay (not shown).
Historically, the eye has served as a valuable model for embryonic and developmental genetics-based studies because of its accessibility, and the ready detection and characterization of associated phenotypic defects. The vertebrate eye lens, for example, has been a focus of scientific inquiry since the early 1900s, when Hans Spemann first demonstrated that the optic vesicle (future retina) could induce formation of the lens, thereby suggesting a fundamental principle in development that tissues coordinately influence their morphogenesis through inductive mechanisms.20 Investigations over the past few decades have now provided fundamental molecular insights into the underlying signaling and transcriptional factors that coordinate eye development.2,21,22 In addition to informing on the basic biology of this complex sensory organ, these studies have expedited ocular disorder gene discovery in human patients. Indeed, mutations in several eye development regulatory genes have been recognized to cause distinct human inherited eye defects such as congenital cataracts (opacity of the lens), retinitis pigmentosa (degeneration of photoreceptor cells of the retina) and various corneal dystrophies (involving loss of corneal stromal or endothelial cells), among many other ocular disorders.23 Although not yet deeply investigated in the eye tissue, there is already some evidence that distinct post-transcriptional regulatory mechanisms have a critical role in eye development.24,25 For example, targeted conditional deletion in the developing mouse lens and corneal tissue of Dicer, which encodes a ribonuclease involved in miRNA processing, leads to severe microphthalmia (small eye) with lens dystrophy and corneal epithelium defects, suggesting that miRNAs have a major function in these developmental processes.26–28 Further, there is evidence that alternative splice forms of regulatory proteins function in eye development. For example, both spliced isoforms of an important transcriptional regulator (Pax6, Pax6(5a)) have overlapping yet distinct functions and are required for the normal development of different eye components such as the cornea, iris, lens and the retina.29 Finally, there also exists evidence that translational-level control is important in eye development. For example, data from Dr. David Beebe's laboratory shows that fiber cell-enriched transcripts are expressed but not translated until later stages in lens epithelial cells.30 These various post-transcriptional regulatory events in the eye likely involve, among other molecules, function of RBPs. In this article, we will comprehensively examine the current understanding of RBPs in the eye from all the major developmental model organisms. Just as Drosophila genetics has impacted the discovery of major transcriptional regulators (e.g. Pax6) in vertebrate eyes, we will investigate if the fly RBP knowledge gained so far can influence RBP gene discovery in vertebrate eye development. Moreover, focusing on the lens tissue, we will highlight some of the specific cellular events that may involve RNA regulons along with specialized RNP complexes such as RNA granules to orchestrate eye development. And finally, we will discuss how interdisciplinary approaches can be applied to further expedite RBP gene discovery in the eye, eventually leading to the derivation of comprehensive post-transcriptional regulatory networks in eye development.
Eye development in vertebrates and insects
The eye in insects such as Drosophila has a dramatically different structure compared to that in vertebrates. Fly eyes are classified as a compound eye and contain several hundred individual units called ommatidia, each with their refractive and photosensitive structures. In contrast, vertebrate eyes resemble a single camera-like unit, with refractive cornea and lens tissues located in the front transmitting and focusing light on the photosensitive retina located in the back. Development of both types of eyes is well understood on the molecular level, and is discussed in detail elsewhere.2,21,22,31,32 Due to space constraints, only a brief introduction to these topics – to enable appreciation of later discussions – is provided here.
Development of the eye in vertebrates
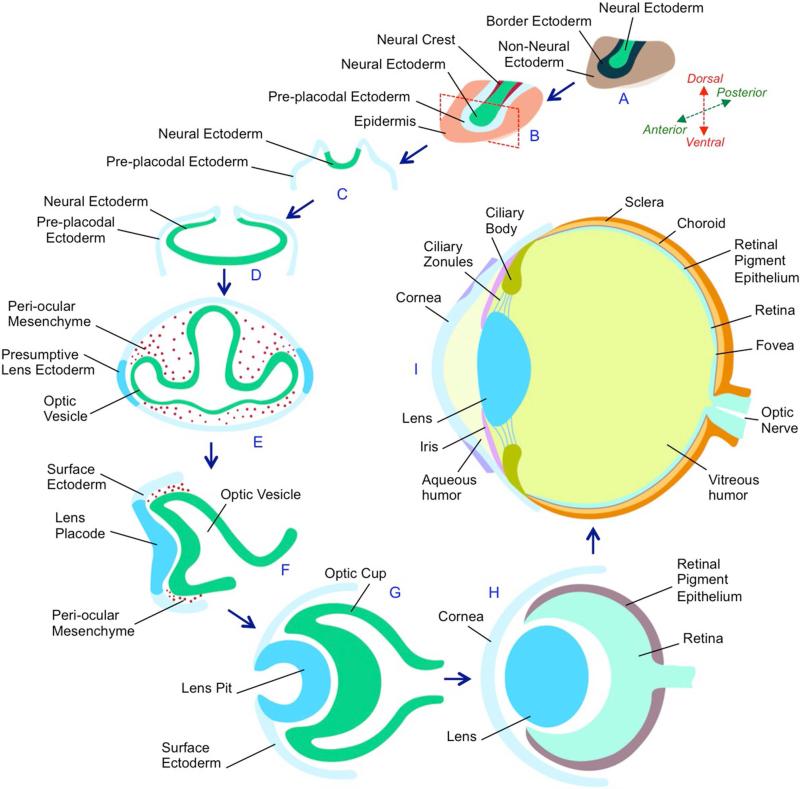
Eye development in vertebrates is a complex process that begins early in embryogenesis during the late-gastrulation stage. The eye-field (future retina) within the neural ectoderm in the anterior neural plate responds to sonic hedgehog signaling and bifurcates into two regions (Fig. 2). After neural tube formation these regions outpouch from each side leading to the formation of bilateral optic sulci that later develop into optic vesicles. Factors such as Bmp4 and activities of other transcription factors such as Hes1, Rx, Lhx2, Mab21l2 in the optic vesicle cause the overlying pre-placodal lens ectoderm to form a thickening called the lens placode. Activities of transcription factors Six3 and Pax6 within the lens placode are important for its subsequent development. The lens placode coordinately invaginates with the optic vesicle, resulting in the formation of the lens pit and a double-layered optic cup, respectively. The inner cell layer of the optic cup will form the neural retina while the outer cell layer will differentiate into the retinal pigment epithelium (RPE). The lens pit pinches off from the surface ectoderm to form the lens vesicle, and the overlying surface ectoderm reassembles and contributes toward forming the cornea. Activities of Pax6 and Foxe3 are necessary for this separation event (Fig. 3), without which an abnormal tissue connection between the lens and cornea remains, which is a feature of a human eye disorder called Peters anomaly. The posterior cells of the lens vesicle terminally differentiate into primary fiber cells that elongate to fill the lumen of the vesicle, a process that requires activity of transcription factors such as Prox1. The epithelium in the anterior part of the lens contains cells that divide in a proliferative zone, and near the lens equator in the transition zone, exit the cell cycle and differentiate into posteriorly localized secondary fiber cells that make up the bulk of the tissue (Fig. 3). The secondary fiber cells lose their organelles and migrate towards the center of the lens. This process of epithelial to fiber cell differentiation occurs throughout the life of the animal. Meanwhile, subsequent differentiation of cells within the optic cup results in mature retinal tissue that is composed of eleven distinct layers of cells (Fig. 3). The retinal tissue contains the rod and cone photoreceptors that sense light and convert it into electrical signals, which are then transferred by ganglion cells through the optic nerve to the brain for interpretation of vision. Other cell types such as RPE function to absorb light and recycle photo-oxidized components of the photoreceptor cells. The adult eye has multiple distinct components such as the outer cornea, iris, lens, ciliary body and zonules, retina, sclera and choroid, among others (Fig. 2, 3).
Figure 2. Eye development in vertebrates.
(A) During gastrulation, the ectoderm is divided into three distinct regions - Neural ectoderm, non-neural ectoderm and the border ectoderm region between these tissues. (B) The border ectoderm gives rise to pre-placodal ectoderm neural crest cells. Red dotted rectangle indicates a section through the embryo that is represented in (C). (C) The neural ectoderm cells comprising the neural plate fold inwards to form the neural tube. (D) A region of cells within the neural ectoderm (anterior neural plate) is specified by eye field transcription factors to form a single eye field, which by Sonic Hedgehog signaling, is partitioned into bilateral optic sulci. (E) Each of the optic sulci develops into an optic vesicle that migrates towards the non-neural ectoderm, which is specified as the surface ectoderm or pre-placodal ectoderm. (F) Interactions between the optic vesicle and the pre-placodal ectoderm results into the latter forming the lens placode. The surrounding peri-ocular mesenchyme inhibits the surface ectoderm that does not appose the optic vesicle from acquiring lens fate. (G) Subsequently, the lens placode and the optic vesicle coordinately invaginate to form the lens pit and optic cup, respectively. (H) The lens pit continues to invaginate the optic cup until it pinches off to form the lens, while the overlying surface ectoderm contributes toward the cornea. The optic cup forms the neuro-retina and the retinal pigment epithelium. (I) Subsequent development and differentiation results in the formation of a multicomponent eye. In the anterior region, the adult eye contains the outer cornea, the iris, the lens, the ciliary body and ciliary zonules, while in the posterior region, it contains the retina. The space between the cornea and the lens is occupied by aqueous humor, while that between the lens and the retina is occupied by the vitreous humor. Light is focused by the cornea and the lens on the retina. The iris responds to the intensity of the light and changes its pinhole similar to the aperture of a camera. The focusing power of the lens is mediated by the ciliary zonules, arising from the ciliary body. Photoreceptor cells within the retina convert the photon energy in light into electrical signals that are transmitted by the optic nerve to the brain where it is interpreted as an image. The retinal pigment epithelium has several functions such as light absorption, nutrient transport, and reduction of photo-oxidative stress by photoreceptor membrane renewal. The fovea is a location in the retina where there is a high concentration of cone photoreceptor cells and where visual sharpness is high.
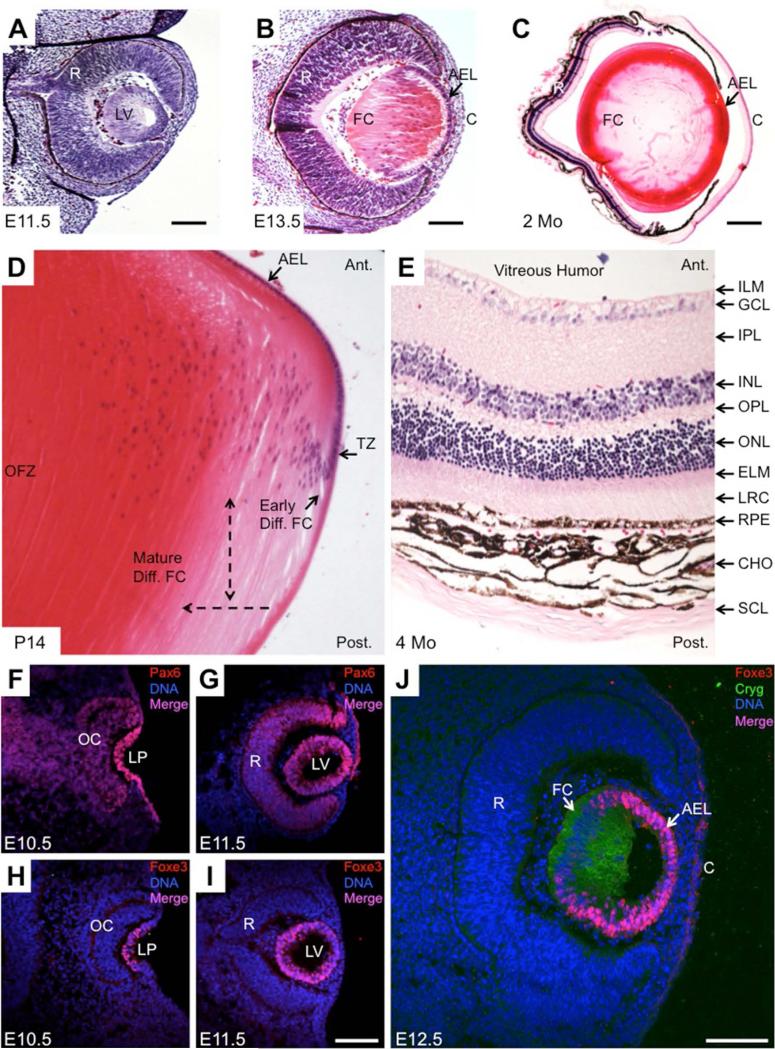
Figure 3. Phenotypic characteristics of the developing mouse eye.
Representative embryonic and postnatal mouse eye tissue stained with hematoxylin and eosin stains that bind to nucleic acid and protein rich regions in the cell, respectively, are shown in A through E. Immunofluorescence maker analyses of key genes in mouse lens development are shown in F through J. (A) At E11.5, a hollow lens vesicle is observed, in which posteriorly located cells have initiated differentiation into primary fiber cells. Also observed is the retina that is primarily composed of the retinal progenitor and ganglion cells. (B) At E13.5, the lumen of the lens vesicle is filled with elongated primary fiber cells and the epithelial cells can be seen in the anterior of the lens. The overlying surface ectoderm will form the cornea. (C) At age 2 months, an adult lens exhibits an epithelial cell layer at the anterior region and fiber cells in the posterior region. The cornea has several layers of cells and the retina is developed with a laminated structure. (D) High magnification image of a post-natal day (P) 14 mouse lens. In the transition zone, cells of the anterior epithelium exit the cell cycle and initiate differentiation of fiber cells. Single head broken arrow indicates direction of early to mature differentiating fiber cells, while double head arrow indicates direction of fiber cell elongation. Terminally differentiated mature fiber cells form a central nuclear-free region called the organelle free zone in the lens. (E) Adult mouse retina is a laminated structure comprising of eleven distinct layers of cells. The sclera originates from the neural ectoderm and protects the eye globe. (F) At E10.5, a critical regulator of eye development Pax6 exhibits expression and nuclear localization in cells of the lens pit and the optic cup. (G) At E11.5, Pax6 continues to be expressed in the lens vesicle and in the retina. (H) A lens-enriched transcription factor Foxe3 is expressed in the lens pit at E10.5. (I) At E11.5, Foxe3 is expressed in all the cells of the lens vesicle. (J) In the following stages, Foxe3 protein is restricted to the cells of the anterior epithelium of the lens. Gamma-crystallin staining is a marker for differentiated lens fiber cells. Abbreviations: LV, Lens Vesicle; R, Retina; AEL, Anterior Epithelium of the Lens; C, Cornea; FC, Fiber Cells; TZ, Transition Zone; OFZ, Organelle Free Zone; Ant., Anterior; Post., Posterior; ILM, Inner Limiting Membrane; GCL, Ganglion Cell Layer; IPL, Inner Plexiform Layer; INL, Inner Nuclear Layer; OPL, Outer Plexiform Layer; ONL, Outer Nuclear Layer; ELM, External Limiting Membrane; LRC, Layer of Rods and Cones; RPE, Retinal Pigment Epithelium; CHO, Choroid; SC, Sclera; OC, Optic Cup; LP, Lens Pit. Scale bar in A, B, I, J, is 100 μm; C is 400 μm.
Development of the eye in Drosophila
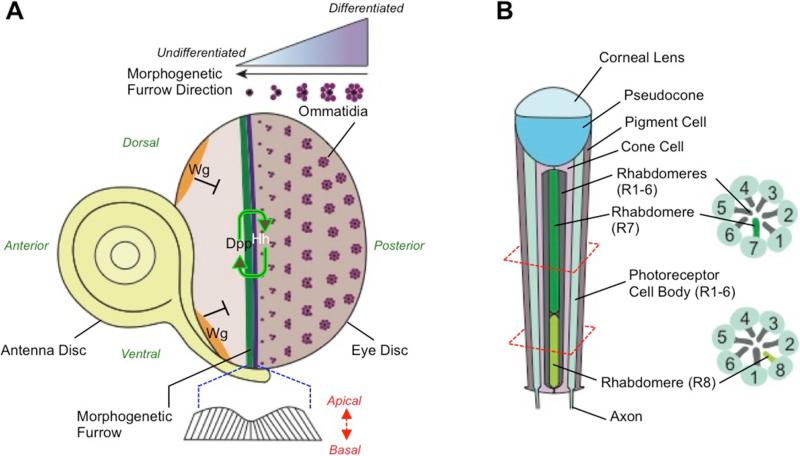
Drosophila eye development begins during the second larval stage when the eye field is specified in the eye imaginal disc by expression of the transcription factors, Eyeless (Ey) and Twin of eyeless (Toy). These transcription factors initiate the expression of other transcription regulators such as Eyes absent (Eya), mutations in which results in no eye tissue formation.33 By the third larval stage, Hedgehog (Hh) and Decapentaplegic (Dpp) signaling initiates retinal fate specification that results in a wave of cell differentiation, termed morphogenetic furrow (MF), which begins at the posterior end of the eye imaginal disc (Fig. 4).34 The MF travels to the anterior region of the eye imaginal disc, and as it does so, behind it immature ommatidia undergo differentiation and photoreceptor recruitment to form mature ommatidia (Fig. 4). The MF can be observed as a shortening of cells, which results from apical constriction.35 The movement of MF is regulated by Dachshund (Dac), which is a transcription factor regulated by Ey, Toy and Eya.36 Wnt signaling through expression of Wingless (Wg) is restricted to the anterior margins where it represses the expression of Eya and Dac, thus inhibiting eye formation.37 Hh in the MF regulates the expression of atonal that functions to differentiate the first photoreceptor cell, R8, which recruits seven other photoreceptors (R1-R8).38 Seven in absentia (Sina) is implicated in the recruitment of the last photoreceptor R7.39 In wildtype flies, the photoreceptors in ommatidia are arranged as mirror images in the ventral and the dorsal eye imaginal discs. During development, each ommatidia rotates 90° towards the dorso-ventral midline. Photoreceptors R3 and R4 then move into asymmetrical positions resulting in ommatidia with opposite chirality on either side of the midline. This is followed by the recruitment of four non-neural cone cells. Each adult ommatidia consists of a corneal lens and a pseudocone, four cone cells and eight photoreceptors with their cellular processes called rhabdomeres that receives light.
Figure 4. Eye development in Drosophila.
(A) In the third larval stage of Drosophila development, Hedgehog (Hh) and Decapentaplegic (Dpp) signaling initiates eye development. The signaling results in the formation of a morphogenetic furrow in the epithelium that moves towards the anterior end. New ommatidia are formed posterior to the morphogenetic furrow. The movement of the furrow is inversely related to development of the ommatidia. (B) Each adult ommatidium consists of a biconvex corneal lens, a pseudocone, 8 photoreceptors (R1-R8) and 4 non-neuronal cone cells.
RNA binding proteins in vertebrate eye development and disease
Presently, over 40 RBPs have been implicated in developing eye tissues across multiple vertebrate species, either by their suggestive expression or by the characterization of their direct function in this process (Table 1). Fish (Zebrafish, Danio rareo), amphibians (Frog, Xenopus laevis, X. tropicalis; Newt, Cynops pyrrhogaster), birds (Chicken, Gallus gallus), and rodents (Mouse, Mus musculus; Rat, Rattus norvegicus) have been used as model organisms in these studies. Mutations in human or targeted gene knockout or knockdown experiments in model organisms indicate that RBPs have important functions in ocular morphogenesis and homeostasis. In the following sections, these RBPs are discussed in detail with regards to their expression and function in the lens, retina and other ocular tissues.
Table 1.
RNA binding proteins in vertebrates eye tissues
| RNA binding protein | Expression in Eye Tissue | Linked to Eye Phenotype | Animal Mutant Model Tested/Available | References |
|---|---|---|---|---|
| Caprin1 | Lens, Retina (Mouse) | Eye examination not reported | Knockout mouse mutants available | 52 |
| Caprin2 | Lens (Chicken, Mouse) | Compaction of lens central fiber region, Lenti-corneal stalk feature of Peter's anomaly | Conditional knockout mouse mutants examined | 51,52 |
| Celf1 (Cugbp1) | Lens (Fish, Frog, Chicken, Mouse) | Detailed eye examination not reported | Knockout mouse mutants available | 69 – 72 |
| Celf3-a (Brul-1) | GCL, ONL (Frog) | - | - | 77 |
| Cerkl | RPE (Rat) | - | - | 89–91, 180 |
| Cpeb1 (Cpeb) | GCL (Frog) | Eye examination not reported | Knockout mouse mutants available Knockdown in Xenopus |
78 |
| Ddx6 | Lens (Mouse) | - | - | 15 |
| Eavl1 (HuR) | Cornea (Human), Lens (Mouse) | Down-regulation in human patients with Keratoconus | Knockdown in Human primary tissue culture | 15,66,104 |
| Elavl2 (HuB, Elrb) | Lens (Mouse), GCL (Frog) | - | - | 66,77 |
| Elavl3 (HuC, Elrc) | Lens (Mouse), GCL, INL (Frog) | - | - | 66,77 |
| Elavl4 (HuD, Elrd) | Lens (Mouse), GCL, INL (Frog) | HuD-target splice forms in transgenic mouse lens | HuD overexpression in transgenic mouse | 66,77 |
| Esrp2 (Rbm35B) | Lacrimal Gland (Human) | - | - | 110 |
| Fmr1 (Fmrp) | IPL, Inner retina (Chicken, Mouse) | - | - | 96 |
| Fxr1 (Xfxr1p) | Lens, Retina (Frog) | - | - | 75 |
| hnRNPK | GCL (Frog) | Inhibition of optic nerve axon regeneration | Knockdown in Xenopus | 79,87 |
| Igf2bp1 | GCL, IPL, INL, Photoreceptors (Fish) | Microphthalmia (Zebrafish) | Knockdown in Zebrafish | 80 |
| Igf2bp3-a (Vg1-RBP) | GCL (Frog) | Microphthalmia (Frog) | Knockdown in Xenopus | 81 |
| Mbnl1 | Cornea (Human) | Fuch's endothelial corneal dystrophy (Human) | Abnormal nuclear RNA foci due to CUG repeats | 107 |
| Msi1 (Musashi-1) | Lens (Mouse), RPE (Newt, Frog, Mouse), Photoreceptors (Newt, Frog, Mouse), Regenerating retina (Newt), CMZ (Newt), Iris (Human) | Photoreceptor degeneration, RPE65 protein down-regulation in RPE cells | Knockout mouse mutants available, Newt retinal regeneration | 73,74,98,100,181 |
| Nono (Xp54nrb) | Eye Vesicles, Developing Neural Retina (Frog) | Knockdown in Xenopus | 101 | |
| Nrp1 | Optic vesicle, CMZ, Photoreceptors, RPE (Frog) | - | - | 77 |
| Pabp | Retina, Lens (Frog) | - | - | 76 |
| Pcbp3 | INL (Mouse) | - | - | 177 |
| Pnn | Cornea (Mouse, Human) | Eye examination not reported | Conditional knockout mouse available | 109 |
| Puf60 | Eye (Fish, Human) | - | - | 8,102 |
| Ptbp1 | Lens (Mouse) | Early embryonic lethality in mouse mutants | Knockout mouse mutants available | 66,182 |
| Ptbp2 | Lens (Mouse) | Eye examination not reported | Knockout mouse mutants available | 66,183,184 |
| Rbfox1 | Lens (Mouse), GCL (Mouse) | - | - | 68,177 |
| Rbm15 | ONL (Mouse) | Eye examination not reported | Knockout mouse mutants available | 177 |
| Rbm24 | Lens (Fish, Frog, Chicken, Mouse) | Microphthalmia (Fish), Lens alpha-crystallin down-regulation (Frog) | Knockdown in Zebrafish, Knockdown in Xenopus | 40,59–63 |
| Rbm38 (Xseb4r) | Developing Neural Retina CMZ (Frog) | Delayed retinal cell differentiation (Frog) | Knockdown in Xenopus | 97 |
| Rbm4 | Eye (Frog) | - | - | 65 |
| Rbmx | Eye? (Frog) | Microphthalmia (frog) | Knockdown in Xenopus | 65 |
| Rbpms (Hermes) | GCL (Mouse, Rat, Frog), Conjunctiva (Human) | Axon branching defects | Knockdown in Zebrafish, Knockdown in Xenopus | 82–86,111 |
| Safb | GCL, ONL (Mouse) | Eye examination not reported | Knockout mouse mutants available | 177 |
| Sfrs1 | Retina (Mouse) | Death of retinal neurons differentiated in mouse embryogenesis | Knockout mouse mutants available | 95 |
| Stau1 | Lens (Mouse) | Eye examination not reported | Knockout mouse mutants available | 15 |
| Stau2 | Optic Vesicle (Chicken), Eye (Mouse) | Microphthalmia (chicken KD), Increased eye size (chicken KI) | Chicken KD, Chicken KI | 73,112 |
| Tdrd7 | Lens (Chicken, Mouse), RPE (Mouse) | Cataract (Human, Mouse, Chicken); Glaucoma (Human, Mouse) | Knockout mouse mutants examined (2 independent mutant lines available), Knockdown in Chicken | 15,41 |
| Tia1 | Lens (Mouse) | Eye examination not reported | Knockout mouse mutants available | 15 |
| Tial1 (Tiar) | Lens (Mouse) | Eye examination not reported | Knockout mouse mutants available | 15 |
| Tra2b (Sfrs10) | Retina (Mouse, Rat, Chicken) | Up-regulated in age-related macular degeneration, AMD (Human), Early embryonic lethality in mouse mutants | Knockout mouse mutants available | 94 |
Abbreviations for the retinal cell layers are the same as given in Fig. 3.
RBPs associated with vertebrate lens development and defects
Application of the expression profiling based gene discovery approach iSyTE (integrated Systems Tool for Eye gene discovery) has led to the identification of several RBPs that exhibit high absolute expression and/or high enriched expression in developing lens tissue.40 The candidate RBPs that are functionally characterized in lens development and its associated defects are summarized below.
Tdrd7 deficiency causes cataracts
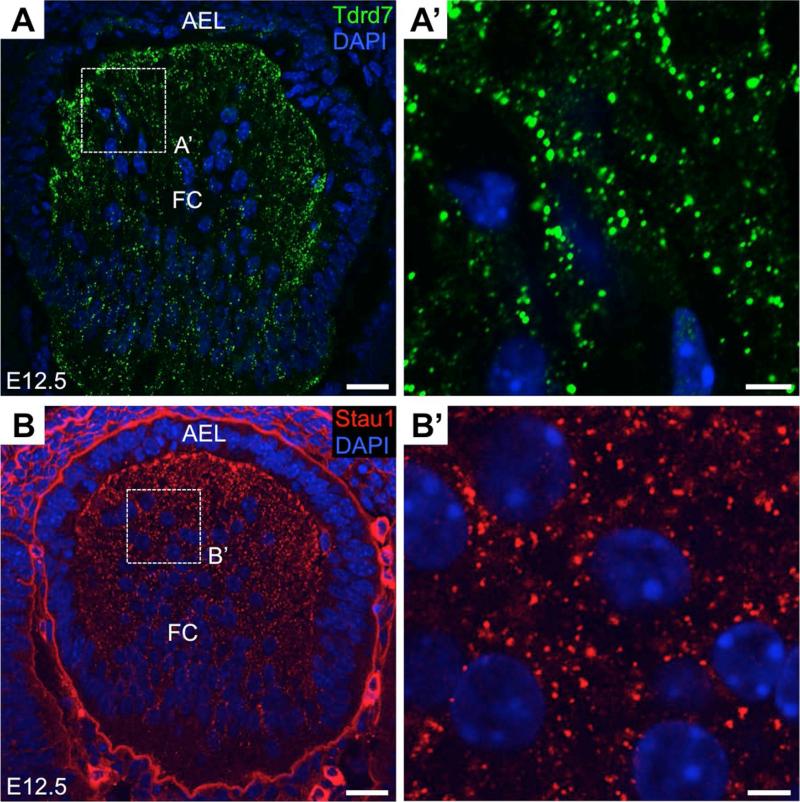
Initial evidence for the importance of post-transcriptional control in lens development comes from the findings that deficiency of a putative RBP and RNA granule component Tdrd7 (Tudor domain containing 7) causes juvenile cataracts in human, mouse and chicken.15,41 In Tdrd7 deficient mouse and human patients, elevated intraocular pressure, a feature of glaucoma was also observed.15 Cataract is the leading cause of blindness worldwide and can occur as an early onset pediatric defect or as a more common late-onset defect in aged individuals. An estimated 25% of congenital cataract cases are caused by genetic alterations.42 Congenital cataracts can cause permanent visual damage through sensory deprivation amblyopia43, and its treatment, cataract surgery, can present further eye complications in infants.44 The findings that TDRD7 mutations cause cataracts were unexpected because Tdrd family proteins were well-understood to play a highly conserved role in germ cell development across metazoa45, but their role in other tissues/organs was not well documented. Tdrd7 contains three Tudor domains, which are predicted to bind methylated arginine residues to mediate protein-protein interactions.46,47 It also contains three Lotus/OST-HTH domains that are predicted to bind RNA.48,49 Metazoan cells contain distinct cytoplasmic ribonucleoprotein (RNP) complexes – termed RNA granules – that function in multiple aspects of mRNA control, including its stabilization, degradation or localization within the cell.4,19,50 In differentiating sperm, Tdrd family proteins are known to associate with specialized RNA granules termed chromatoid bodies to mediate post-transcriptional regulation.45 In differentiating lens fiber cells, Tdrd7 forms RNA granules that partially co-localize with Stau1 proteins, which also exhibit a granular pattern (Fig. 5). To a lesser extent Tdrd7 granules also co-localize with P-bodies in these cells. Tdrd7 null mouse lenses exhibit reduced mRNA levels of RNA granule components such as Hsbp1 (Hsp27), as well as other important lens factors, some of which are found enriched in Tdrd7 pulldown assays.15 However, the detailed mechanism of Tdrd7-based regulation of these transcripts is yet to be determined.
Figure 5. Tdrd7 and Stau1 granules in mouse lens development.
(A) Tdrd7 protein is localized to cytoplasmic granules in lens fiber cells of mouse at embryonic day (E) 12.5. (A’) Broken line denoted area in “A” is shown at high magnification. Deficiency of Tdrd7, a Lotus/OST-HTH and tudor domain protein and RNA granule (RG) component, in human and mouse causes cataracts and glaucoma. (B) Stau1 cytoplasmic granules are observed in lens fiber cells at E12.5. (B’) Broken line denoted area in “B” is shown at high magnification. Stau1 (Staufen 1), a double stranded RBP and RG component, functions in the localization of mRNA in oocytes and neurons. Scale bar in A, B is 25 μm; in A’, B’ is 5 μm.
Caprin2 deficiency causes lens compaction defects and features of Peters anomaly
A second iSyTE-identified RBP and RNA granule component encoding gene Caprin2 exhibits highly enriched expression in chicken and mouse lens development, suggestive of a function in regulating this process.51,52 In support of this hypothesis, Caprin2 homozygous conditional knockout (Caprin2 cKO) mice exhibit two distinct lens defects.52 In majority of Caprin2 cKO mice, a defect in the central region of the lens (termed the “nucleus region” of the lens) is observed, which is characterized by a reduction in the zone of nuclear fiber cells. The aging process in humans can cause lens nucleus region reduction, which is termed as “nuclear compaction”.53 This process is linked to defects in the accommodation properties of the lens, as well as to cataracts.54,55 In addition to this defect, a subset of Caprin2 cKO mouse mutants exhibit a more severe phenotype characterized by the abnormal presence of a lenti-corneal stalk and cataracts. This phenotype results from defects in the separation of the developing lens and cornea tissues, which in turn likely result from defective cell-cell interactions involved in this process. In support of this hypothesis, Caprin2 protein is enriched in cells that are located at the rim of the lens vesicle, which may directly participate in this process. Further, Caprin2 protein is localized to granules in these cells, but the nature of these granules is not yet characterized (see section below on “Future challenges”). Interestingly, the Drosophila ortholog of Caprin2 is associated with RNA granules and is implicated in control of eye size in the fly (see section below on “RBPs and eye size control”).56–58 Caprin2 granules are known to be involved in inhibition of target mRNA translation in Drosophila. Therefore, it can be hypothesized that Caprin2 is a component of lens RNA granules that participate in localized post-transcriptional control of the proteome to co-ordinate the cellular re-arrangement morphogenesis events. Interestingly, the other Capr vertebrate ortholog Caprin1 is also expressed in the developing mouse lens and the retina but its function in these tissues has not been described.52 These findings suggest that Caprin2 has distinct functions in coordinating eye morphogenesis.
Expression of Rbm family proteins in vertebrate lens development
The Rbm (RNA binding motif) family gene Rbm24 (also known as Seb4 in Xenopus) encodes an RBP conserved among vertebrates. Rbm24 exhibits highly enriched expression in the lens and is also expressed in the developing heart and somite tissues in zebrafish, Xenopus and mouse. 59,60,40,61–63 Rbm24 has a single RNA recognition motif (RRM) near its N-terminus that has two sub-motifs RNP1 and RNP2. In zebrafish, there are two copies of Rbm24, called rbm24a and rbm24b.62 Morpholino induced rbm24b knockdown (KD) in zebrafish causes microphthalmia (abnormally small eye).62 Further, Rbm24-KD in frog leads to reduction in the transcript levels for the lens marker alpha-A-Crystallin in developing lens tissue. These findings suggest that the requirement of Rbm24 for proper eye development is conserved across multiple vertebrate species. Although a recent Rbm24 germline deletion mouse mutant model has been described, examination of eye tissue has not been reported.64 However, analysis of an independently derived Rbm24 targeted mouse knockout model reveals eye defects in these animals (Dash and Lachke, unpublished observations), suggesting that the requirement of this protein for development of the eye is conserved between fish and mammals.
Another Rbm family protein, Rbmx (also known as hnRNP G), has been identified in a cDNA library-based screen for genes expressed in neural plate development in Xenopus.65 Rbmx-KD led to microphthalmia in X. laevis. Examination of eye regulatory genes in these animals revealed abnormally high expression of Xrx1 transcripts in the region of the lens, where normally Xrx1 is absent suggesting that at least some genes in this tissue were mis-expressed. Further, Pax6 expression was restricted to a smaller region, correlating with the small size defect of the eye in these mutants. Together, these findings indicate that Rbmx may have an important function in Xenopus eye development, and provides the ground for investigating its requirement in this process in other vertebrate species. Indeed, analysis using the iSyTE tool indicates that Rbmx is expressed in mouse lens development (Lachke, unpublished observations).
Other RBP family proteins in vertebrate lens development
In addition to those discussed above, other RBPs that were previously described as “neuronal specific” have now been detected in the lens. Some of these RBPs exhibit the trend observed in differentiating neurons, where the ubiquitous isoform is expressed in undifferentiated progenitor cells, followed by expression of the neuron-specific isoform as these cells exit the cell cycle and commence neuronal differentiation. For example, while the ubiquitously expressed PTB (polypyrimidine tract binding protein, also known as Ptbp1) isoform is detected in nuclei of mouse lens epithelial cells, the neuronal isoform, nPTB (neuronal polypyrimidine tract binding protein, also known as Ptbp2) is localized to the cytoplasm of fiber cells.66 Both proteins themselves are involved in alternative splicing in neurons. Similarly, the Elav (Embryonic lethal abnormal vision) family RBP HuD (Elavl4), which is expressed in neurons and is associated with splicing, RNA metabolism, and translation, is also detected in lens fiber cells.66 HuD transgenic overexpression mouse mutants exhibit expression of the specific splice isoforms of HuD-target genes. Thus, it is proposed that lens fiber cells may support expression of neuron-specific splice variants of a subset of genes.67 Indeed, some evidence in support of this hypothesis has been gathered. For example, a neuron-expressed spice form of the RBP Fox-1 (Rbfox1) is also detected in mouse lens fiber cells.68
Another Elav family RBP, Celf1 (Cugbp1), which is involved in mRNA decay, splicing, and translational repression, is highly expressed in the developing lens across several vertebrates.69–72 Celf1 homozygous targeted knockout mice exhibit severe lens defects, suggesting that Celf1 has an important function in mediating post-transcriptional control of gene expression in vertebrate lens development (Siddam, Paillard, Lachke, unpublished observations). Another RBP conserved in vertebrates is Msi1 (Musashi family protein), which is detected in the epithelium, transition zone and fiber cells of the lens.73 Although Msi1 knockout mouse mutants have been generated, the lenses of these animals have not been examined.74 In addition to these RBPs, Pabp, Fxr1, HuR (Elavl1), Tia1, Tiar (Tial1) and Stau2 are also detected in mouse E12.5 embryonic lenses (Table 1).15,75,76 However, their function in lens tissues remains to be determined.
RBPs in the vertebrate retina
Expression of several RBPs has been well documented in the retina in several vertebrate species, and mutations in a few RBP genes have already been associated with distinct retina-related phenotypes such as retinitis pigmentosa, photoreceptor degeneration, delayed retinal cell differentiation, and age-related macular degeneration (AMD). Here, we examine expression of RBPs in specific cells within the retina and their implication in retinal disease.
Several RBPs containing RRM and other RNA binding domains are expressed in the ganglion cells within the vertebrate retina. These are Celf3-a, Cpeb1, Elavl2, Elavl3, Elavl4, hnRNPK, Igf2bp1, and Igf2bp3-a (Table 1).77–81 In particular, one RBP (Rbpms) has been recognized as an excellent marker for retinal ganglion cells (RGCs). Rbpms (RNA-binding protein with multiple splicing) was first identified in Xenopus as an RRM protein “Hermes” and is enriched in heart tissue and RGCs within the ganglion cell layer (GCL) in the retina. Subsequent studies have demonstrated that the gene Rbpms exhibits highly enriched expression in RGCs of several vertebrate species such as mouse, rat, rabbit.82–86 Moreover, Rbpms knockdown in zebrafish and Xenopus results in axon branching defects.86 Some of these RGC RBPs are also expressed in other retinal cell layers such as ONL (Celf3-a), INL (Elavl3, Elavl4, Igfbp1) as well as IPL and photoreceptors (Igf2bp1) (Table 1).77,80 Further, knockdown of some of these in fish or frog exhibit distinct phenotypes. For example, RGC-specific knockdown of hnRNPK in Xenopus causes inhibition of optic nerve regeneration.87
Retinitis pigmentosa is an inherited eye disorder that affects photoreceptor cells and causes a progressive loss of vision with age.88 Mutations in the gene encoding CERKL (Ceramide kinase like), which is considered to have RNA binding properties, cause retinitis pigmentosa and cone-rod dystrophy in humans.89,90 Interestingly, Cerkl is located to RNA granules in cell lines.91 Other genes linked to retinitis pigmentosa in humans are those encoding the pre-mRNA processing factor family proteins PRPF3, PRPF8 and PRPF31, which control splicing.92,93 Another splicing factor, Tra2b (Sfrs10, Serine-arginine rich splicing factor 10), is expressed in mouse, rat and chicken retina. Although TRA2B is not detected in normal human retina, its expression is elevated in AMD retina, suggesting its function in conditions of stress.94 Interestingly, a related Sfrs (SR) family protein, Sfrs1, controls alternative splicing in retina development and is necessary for survival of retinal neuron that were differentiated in embryogenesis.95 Another RBP, Fmr1 (Fragile X mental retardation protein 1) contains two KH domains required for binding to RNA. Fmr1 is expressed in the retina of mouse and chicken where its expression is modulated by light. Higher expression of Fmr1 is observed in retinas exposed to light compared to the dark adapted retinas, which implicates it in the maintenance of circadian rhythm.96 The Rbm family proteins Rbmx and Rbm38 are expressed in Xenopus retina (Table 1), and Rbmx-KD results in microphthalmia as described above.65 On the other hand, Rbm38 (XSeb4r)-KD in Xenopus retinal progenitor cells results in delayed differentiation of retinal tissue.97 Another RBP, Nrp1, is expressed in the optic vesicle in Xenopus and continues expression in the ciliary margin zone (CMZ), RPE and photoreceptors in the developing retina.77 Besides the lens, Msi1 is expressed in other eye tissues in mouse including the corneal epithelium, stroma and the retina, and in Xenopus it has been detected in the retina.73,98 However, in human it has so far been reported only in the iris.99,100 Other retina-expressed RBPs are Rbm4 (Xenopus), Nono (Xp54nrb) (Xenopus), and Puf60 (human and zebrafish), but their function is not yet defined.8,65,101,102
RBPs in other vertebrate eye tissues
In addition to the developing lens and retina, RBP expression has been described in eye components such as the cornea and the lacrimal gland. Among these RBPs are Elavl1 (HuR), Mbnl1, Pnn and Esrp2 (Table 1). Elavl1 is a member of the Elav protein family and contains three RRMs. In human, ELAVL1 expression changes are linked to a corneal disorder “keratoconus” in which thinning of the central epithelium of the corneal stroma results in a cone shaped cornea.103 Keratoconus corneal tissue obtained from human patients exhibited 3-fold downregulation in ELAVL1 expression compared to normal corneal tissue.104 This is accompanied by downregulation of ELAVL1's target, β-Actin, which results in inhibition of migration and proliferation of keratocytes in the corneal epithelium.105 Another corneal defect in which RBPs are implicated is Fuch's endothelial corneal dystrophy (FECD). FECD is an inherited disorder in humans, where swelling due to fluid accumulation (edema) is observed in corneal endothelium, which can result in the loss of vision. A strong association of FECD has been established with an intronic (CTG.CAG)n trinucleotide repeat expansion in the gene encoding TCF4.106,107 This results in poly(CUG)n (n > 150; normal n = 20) repeat containing RNA. These CUG repeats are unstable and aggregate in the nucleus to form abnormal RNA foci. In human FECD patients, MBNL1 (Muscleblind like splicing regulator 1) is sequestered to RNA foci. This sequestration results in differential splicing of transcripts implicated in epithelial-to-mesenchymal transition (EMT), that is considered to contribute to FECD.107 Animal models have also been studied to characterize RBPs in cornea. Conditional inactivation of the RBP gene Pnn (Pinin) in mouse corneal epithelium results in severe disruption of epithelium differentiation108, which is attributed to abnormal splicing in Pnn knockout cornea (Table 1).109 While its function has not been characterized, the Rbm protein Esrp2 (Rbm35B), containing three RRMs, is found to be expressed in the human lacrimal gland.110 Finally, in addition to RGC-enriched expression of RBPMS as described earlier, this gene is also found to be expressed in human conjunctiva tissue.111 All these findings suggest that several RBPs function in distinct eye tissues during development and present a case for their detailed functional characterization.
RBPs involved in eye size control
An RBP that is involved in regulating eye size is the vertebrate ortholog, Stau2, of Drosophila Staufen.112 Analysis of chicken eye development revealed that Stau2 transcripts and Stau1 protein are expressed in the retina at embryonic stage E4 and stage E6, respectively. Further, three distinct Stau2 isoforms were detected in embryonic chicken eyes. Interestingly, while over-expression of the long isoform of Stau2 led to an increase in eye size, down-regulation of all three Stau2 isoforms by miRNA-mediated silencing caused microphthalmia (small eye) in chicken embryos.112 The microphthalmia phenotype could be rescued by all three Stau2 isoforms and was not found to be associated with elevated apoptosis or premature neuronal differentiation, but rather with reduced cell proliferation. Moreover, transcripts of two important eye development transcription regulators, Sox2 and Hes1, were found to be reduced in Stau2 down-regulated chicken eyes. Interestingly, SOX2 mutations in humans are associated with microphthalmia (and anophthalmia), while Hes1 null mice exhibit defects in retinal cell proliferation.113–115 Thus, it will be important to pursue further the molecular mechanism of Stau2 mediated gene regulation in the eye. For example, is the Stau2 function described in chicken conserved in mammalian eye development? Because Stau2 is involved in distinct post-transcriptional control mechanisms such as mRNA localization, it will be important to investigate the nature of Stau2 control in regulating cell proliferation in eye tissue.
The Drosophila ortholog of mouse Caprin2 (and Caprin1) is Capr (see below). Recently, it was shown that Capr interacts with the UBA domain-containing protein Lig (Lingerer), and the RBPs Fmr1 and Rin (Rasputin; vertebrate ortholog is G3BP, a P-body component) to control cell number and eye size in Drosophila development.58 This study also demonstrated co-localization of Capr with Lig in subcellular punctate structures. Interestingly, Caprin2 expression has been shown to be cytoplasmic and granular in cells present near the rim of the lens pit in development (see section on “Future Challenges”).52 It will be interesting to investigate whether these granules share components with, or are part of, P-bodies in these developing tissues. Also interestingly, Fmr1 itself is regulated in the mouse retina by light.96 It is known that arginine methyl transferase enzymes can alter the binding capability of RBPs to their target mRNAs by methylating RGG motifs within these proteins.116–118 Interestingly, the RBPs Caprin2, Fmr1 and G3BP all have RGGs motifs. Thus, it is will be important to examine if the RGG motifs in these proteins are methylated in eye tissue. If so, further investigations can be pursued to test if these are targets of Tudor family proteins in the eye such as the eye defect-linked protein Tdrd7, which has Tudor domains that are predicted to bind methylated arginine residues within target proteins.
RNA binding proteins in Drosophila eye development
Over the past several decades, fly genetics has revealed many genes important in eye development.31,119 This process has often involved defining the genetic basis of the “rough eye” phenotype, which occurs due to perturbations in normal eye development. The rough eye is caused by changes in the ommatidial arrangement or fused ommatidia due to fusion of lens facets.120,121 Loss-of-function mutations (using RNAi or ENU mutagenesis) or gain-of-function mutations (using UAS-GAL4 ectopic expression) in several RBP genes have lead to a rough eye phenotype, indicating their function in this process (Table 2). We summarize these findings below.
Table 2.
RNA binding proteins in Drosophila eye development
| RNA binding protein gene | Expression in Eye Tissue | Linked to Eye Phenotype | Animal Mutant Model Tested/Available | References |
|---|---|---|---|---|
| Capr | Eye | Reduced eye size (capr mutant fly) Overgrown eyes (fly double mutants of capr and rin, and capr and fmr1) | Capr mutant fly, Fly Double mutants of Capr and Rin and Capr and FMR1 | 58 |
| Caz | Eye | Rough eye, cone defect | Fly KD | 122 |
| dNab2 | - | Rough-eye phenotype | Fly overexpression model | 128 |
| Elav | Eye imaginal disc | Abnormalities in photoreceptors | Fly mutant | 125 |
| Fmrp | Eye | Rough eye | Fly mutant | 127 |
| Hrp38 (Hrp98DE) | Eye | Rough-eye | Fly mutant and eye specific KD | 124 |
| Lark | - | Disorganized eye | Fly overexpression model | 127 |
| Mahe | Eye-antennal disc, optic lobes | Microphthalmia | Fly overexpression model | 132 |
| Mblc | - | Rough eye | Fly overexpression model | 129 |
| Musashi (Msi1 family protein) | Photoreceptors | Abnormal ommatidia with deformed rhabdomeres and/or irregular orientation | Fly mutant | 39,171 |
| Pabp2 | Eye | Small, disorganized, black eye | Fly mutant | 128 |
| Rbp6 (Vertebrate Msi1 ortholog) | Eye | No eye phenotype | Fly mutant | 185 |
| Rin | Eye imaginal disc | Mildly rough eye with defects in photoreceptor recruitment and ommatidial polarity in the eye (KO fly) Severe rough eye (Overexpression fly) |
Fly mutant Fly overexpression model |
121 |
| Rump | Eye | - | - | 186 |
Caz (Cabeza), encoding an RRM containing RBP, is the Drosophila ortholog of the human gene FUS (Fused in sarcoma), which is implicated in the human neurodegenerative disorder ALS (Amyotrophic lateral sclerosis). Caz-KD in Drosophila results in a rough eye phenotype, comprising of cone cell defects and abnormal ommatidia rotation.122 These defects are accompanied by apoptosis and abnormal cell differentiation and can be rescued by suppressing rhomboid-1.122 rhomboid-1 encodes a serine protease that cleaves a TGFα-like growth factor Spitz to activate EGFR signaling in normal flies.123 This suggests a genetic interaction between Caz and rhomboid-1.122
Hrp38 (Hrp98DE) encodes an RBP (ortholog of human hnRNP A1) that is known to regulate splicing. Hrp38 contains two RRMs, a poly(ADP-ribosyl)ation (pADPr) binding motif and a RGG domain. Hrp38 mutant flies exhibit a rough eye phenotype with disorganized ommatidia and downregulation of Drosophila E-cadherin (DE-cadherin).124 Parg (Poly (ADP-ribose) glycohydrolase) modulates the activity of RBPs. In the absence of Parg, Hrp38 undergoes post-translational modification by addition of pADPr at its pADPr-binding motif. Indeed, Parg-KD also results in rough eyes with disorganized lattice and lower numbers of photoreceptors due to the misregulation of DE-cadherin.124 This suggests that Parg and Hrp38 co-regulate DE-cadherin to maintain lattice structure in Drosophila eye.
Elav is a highly conserved RBP with three RRM domains.125 Loss of function mutations in Elav results in abnormalities in photoreceptors.126 Rin (Rasputin), which is an ortholog of vertebrate G3BP (found in P-bodies), contains two RRMs and a RGG domain. In Rin-KD flies,photoreceptor recruitment and orientation defects are observed.121 In Rin overexpression mutant fly, a rough eye phenotype was observed along with photoreceptor recruitment defects such as missing and extra photoreceptors. In these mutants, the ommatidia also display a non-chiral orientation similar to Ras mutants. Rin has genetic interactions with factors in the Ras and RhoA signaling pathways to regulate photoreceptor development in Drosophila. Loss of one copy of Rin in Sev (sevenless, which encodes a receptor tyrosine kinase) mutant flies suppresses the eye phenotype. Mutations in Rin exaggerate the photoreceptor differentiation defect in RhoA overexpressed mutant fly. As described in the section on “RBP control of eye size”, the RBP gene Capr is an ortholog of vertebrate Caprin1 and Caprin2, and its knockdown in flies causes reduced eye size. Reduced expression of Capr in single mutants of Fmr1 and Rin, or its reduced expression in Fmr1-Rin double mutants causes overgrown eyes, which suggests that these proteins likely function in the same pathway to regulate eye growth.
Fmr1 and Lark (a circadian rhythm clock output component protein) are two other RBPs that exhibit protein-protein as well as genetic interactions.127 Fmr1 and Lark (vertebrate Rbm4) are detected in a protein complex in adult fly head tissue. Overexpression of Lark in fly results in fused ommatidia and a rough eye phenotype that is enhanced by Fmr1-KD.127 Overexpression of dNab2, an ortholog of ZC3H14 whose mutation causes an intellectual disability disorder in humans, results in a rough eye phenotype in adult flies.128 dNab2 is responsible for maintaining the length of poly(A) tail in target mRNAs and has genetic interactions with two proteins of the polyadenylation machinery, namely the polymerase Hrg (hiiragi) and Pabp2 (Poly(A) binding protein 2).128 Loss of one allele of Hrg enhanced the dNab2 overexpression phenotype. Similarly, loss-of-function of Pabp2 in the fly results in a small and more disorganized blackened eye in dNab2 overexpression mutant flies. Interestingly, co-overexpression of both dNab2 and Pabp2 in fly eye suppresses the rough eye phenotype of dNab2 overexpression mutant fly, further confirming the antagonistic functions of these proteins in Drosophila eye.128 Overexpression of an isoform (MblC) of the RBP Mbl (Muscleblind) results in a rough eye phenotype.129 Although its function has not been studied in detail in the eye, Mbl regulates the sub-cellular localization of two other RBPs in sarcomeres, BSF (Bicoid stability factor) and TBPH (Tar binding protein 43 homolog).130 Interestingly overexpression of human TDP-43 in flies also results in abnormal spaces intervening ommatidia.131
Musashi-KD in flies results in deformed rhabdomeres with irregular orientation. 39 Interestingly photoreceptor recruitment in these mutant flies is not affected suggesting that Musashi functions in late stages of eye development. Along with Sina, Musashi regulates the expression of Tkk (Tramtrack) to control photoreceptor differentiation.39 Another Drosophila gene, Mahe (Maheshvara), encodes a putative DEAD box protein that belongs to the RNA helicase protein family. Overexpression of Mahe results in a reduced eye size in Drosophila similar to the Notch loss-of-function mutants. Interestingly, Mahe overexpression also rescues the Notch overexpression mediated hyper-proliferation of eye phenotype suggesting a genetic interaction between Mahe and Notch.132 Several RBPs studied in Drosophila have genetic interactions with other RBPs that either exaggerate or suppress the ocular phenotypes of the mutant flies, suggesting a co-ordination of RBPs to mediate mRNA processing. Several other RBPs are expressed in the Drosophila eye, but their function here is not yet defined (Table 2).
Future challenges in investigating RBPs in eye development
As appreciated from the previous sections, studies so far have led to the identification of several RBPs with important function in fly and vertebrate eye development. Interestingly, of the 42 RBPs that function or are expressed in vertebrate eyes, ~60% are recognized as components of RNA granules in other cell types (Table 3). Although some of these are associated with distinct types of RNA granules in the lens and the retina (Fig. 5, 6), for the majority of these RBPs, a clear connection with RNA granules in eye tissues remains to be investigated. Characterization of RBPs such as Tdrd7, Caprin2 and Stau2 has revealed their connections with clinically relevant ocular defects in human patients or animal models. However, this emerging field of post-transcriptional control in the eye presents numerous new challenges. In the sections below, we outline these by discussing, using the lens as an example, some of the key developmental events that may involve RBP function. There are challenges within other tissues and cell types in the eye, such as the retina, the neuronal cells of which may share RBP-based regulatory mechanisms similar to those identified in non-ocular neurons133, but we do not discuss them here due to space limitations. We conclude by highlighting some promising approaches for the continued discovery of RBPs in eye development, which involve application of new interdisciplinary methods, in addition to insights gained from fly ocular genetics.
Table 3.
Vertebrate eye RBPs that are known components of RNA granules
| RNA Binding Protein | Component of RNA Granule | References |
|---|---|---|
| Caprin1 | SG | 187 |
| Caprin2 | RG* | 57 |
| Celf1 (Cugbp1) | PB, SG | 188,189 |
| Cpeb | SG | 190 |
| Ddx6 | SG, PB | 15 |
| Elavl1 (HuR) | SG, PB, NG | 191 – 193 |
| Elavl4 (HuD) | SG, PB, NG | 194 |
| Fmr1 (Fmrp) | SG | 195 |
| Fxr1p | SG | 195 |
| hnRNPK | SG | 196 |
| Igf2bp1 | SG | 197,198 |
| Mbnl1 | SG | 199 |
| Msi1 (Musashi 1) | SG | 200 |
| Nono | NG | 201,202 |
| Nrp1 | SG, PB | 203 |
| Pabp | SG, PB, NG | 204,205 |
| Rbm4 | SG | 206 |
| Rbpms | NG | 201 |
| Rin | SG | 207 |
| Stau1 (Staufen 1) | SG, NG | 194 |
| Stau2 (Staufen 2) | NG | 164 |
| Tdrd7 | RG** | 15 |
| Tia1 | SG, PB | 194,208 |
| Tial1 | SG, PB | 194,208 |
RG refers to Caprin2 RNA granules that co-localization with FMRP and Poly(dT), but are not yet classified.
RG refers to Tdrd7 RNA granules that although exhibit partial co-localization with Stau1 and P-bodies in lens fiber cells, and with chromatoid bodies in differentiating sperm, are not yet classified.
Abbreviations are: SG, Stress granules; PB, Processing body; NG, Neuronal granule.
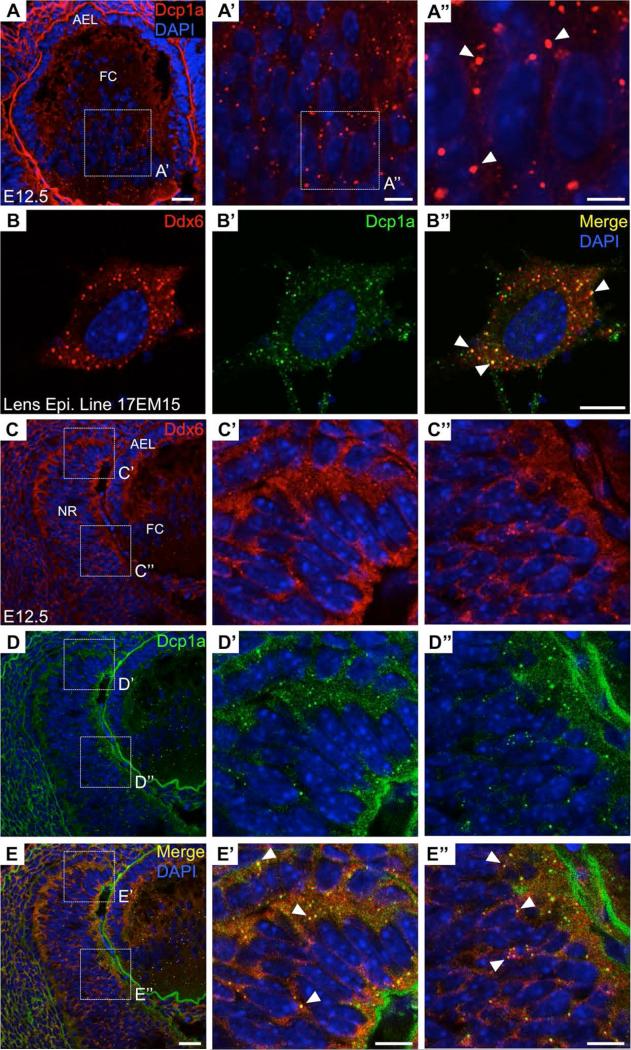
Figure 6. Processing bodies in mouse lens and retina development.
(A-A”) The Processing body (P-body) marker Dcp1a is observed to stain distinct granules in the mouse lens at embryonic day (E) 12.5. P-bodies are RNA granules that undertake mRNA storage, decay or silencing. (B-B”) A second P-body marker, Ddx6, stains distinct granules and co-localizes with Dcp1a in the mouse lens epithelial cell line 17EM15. (C-E’’) P-body markers Ddx6 and Dcp1a stain distinct granules and co-localize in the E12.5 mouse retina. Scale bar in A, E is 25 μm; in A’, B’’, E’, E’’ is 10 μm; in A’’ is 5 μm.
Cellular features that require post-transcriptional control in the eye
We first examine the evidence from specific lens cell biology and differentiation events that may represent potential sites of RBP-mediated post-transcriptional control (Fig. 7). As described earlier, the lens is made of two types of cells, the anteriorly located epithelial cells and the posteriorly located fiber cells (Fig. 3). Like stem cells in other tissues, anterior epithelial cells hold the capacity to proliferate and, within a specific region near the equator of the lens, to begin differentiation into elongated fiber cells. Epithelial cells exhibit a gene expression pattern distinct from fiber cells.134–139 While epithelial gene expression enables these cells to stay in the cell cycle, fiber gene expression is dedicated toward building the high levels of proteins such as crystallins, which are required for lens transparency. Thus, it is important to regulate the formation of distinct proteomes of these cells.140,141 The principle challenges recognized for post-transcriptional control in the lens are: (1) translational repression of some of the fiber transcripts that are also expressed in the epithelium, (2) decay of epithelial transcripts in early differentiating fiber cells, beyond the transition zone, (3) decay and/or translational repression of early differentiating fiber cell factors in late fiber cells, and (4) translation of usually high levels of proteins that are essential for rendering the lens its principle properties of transparency and high refractive index, and indeed whose deficiency or mutations cause congenital cataracts in humans.42 Below, we discuss the molecular evidence in support of these events.
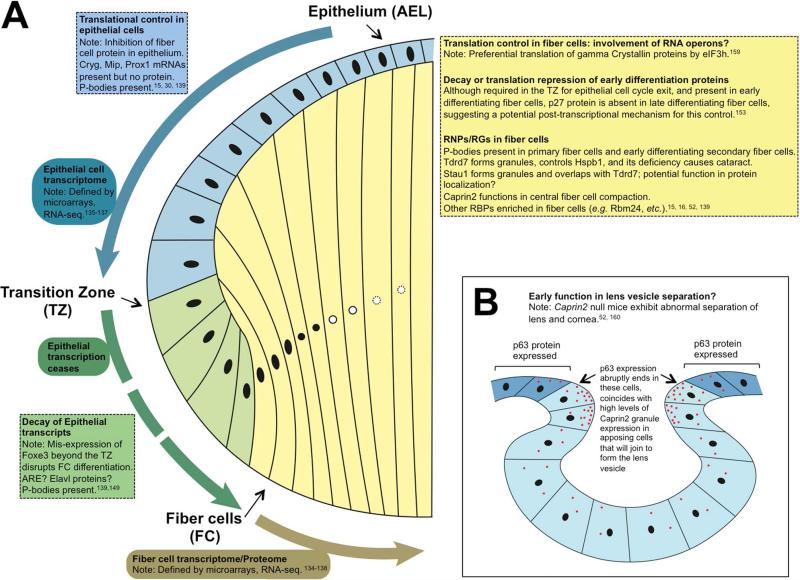
Figure 7. Mouse lens development events that may involve RBP function.
(A) Specific cellular and differentiation events in lens development where RBP-mediated post-transcriptional regulation may occur are outlined. (B) Potential function of Caprin2 in the separation of lens pit from the overlying surface ectoderm (future cornea) is outlined. p63 protein is expressed in cells that will contribute to the cornea. It is abruptly absent in adjacent to cells that separate out from the surface ectoderm and associate together to form the anterior epithelium of the lens vesicle. The absence of p63 in these cells coincides with enriched Caprin2 granular staining suggesting a potential relationship that may be the topic of future investigations.
Evidence for cell-specific translational control in the lens
Even as early as 1981, Beebe and Piatigorsky provided the initial evidence for translational control in the lens. They demonstrated that delta-crystallin mRNA was translated less efficiently in late chicken lens development compared to early stages.142 Further, experimentally increasing the levels of delta-crystallin mRNA in older lens cells did not result in an increase in delta-crystallin protein. Another example of potential translational control in the lens was provided by Cenedella who showed that HMGR (3-hydroxy-3-methylglutaryl coenzyme A reductase) protein levels could be increased in the lens without a similar increase in mRNA.143 More recently, Dr. Beebe's group showed that transcripts encoding gamma-crystallins, but not the proteins themselves, are present in mouse lens epithelial cells at birth.30 While they are translated in fiber cells at early stages 144,140, in the epithelium these mRNAs are translated only at later post-natal stages.30 Similarly, there is evidence that Prox1 and Sox1 mRNAs are transcribed in epithelial cells, but its protein is highly expressed in fiber cells and not in epithelial cells in late embryonic stages.145,146 Further, Dr. Beebe's findings have also shown that in addition to gamma-crystallin mRNAs, those encoding other fiber cell-enriched proteins such as Mip (Aquaporin 0) and certain transcription factors are expressed early in lens development prior to their translation.147,148 Indeed, Mip transcripts are present in lens epithelial cells from the placode stage through adulthood, but are only translated in fiber cells. Two general observations can be interpreted from the above data: (1) mRNAs that encode lens crystallins, lens membrane proteins and fiber cell “enriched” transcription factors accumulate early in lens development, before they are translated, and, in some cases, before fiber cells form, and (2) while their transcriptome is largely distinct from that of fiber cells, lens epithelial cells do express some fiber cell transcripts, but these are not translated into protein, at least until later stages in life.
Thus, these data suggest that post-transcriptional mechanisms are likely recruited for inhibiting translation in a spatio-temporal manner in the lens. Thus, it will be important to investigate whether this mechanism involves RBP function, miRNAs, or both. Interestingly, RNA granules such as P-bodies have been identified in embryonic as well as neonatal lens epithelial cells (Fig. 6).15,139 Because P-bodies are known to be involved in mRNA decay and/or translational silencing, it will be important to examine if they function in translational control in epithelial cells. Further, it will also be important to identify other mRNAs that are selectively translated in lens placode, vesicle, epithelial and fiber cells. These studies will involve comparisons between the transcriptome and the proteome of the developing lens with application of RNA-seq, 2-D gel electrophoresis with mass spectrometric analyses as well as polysome profiling, which may be challenging given the small nature of the tissue in developmental stages. However, once the differentially translated (silenced) mRNAs are defined, protein capture methods can be applied to identify the proteins that bind to them. Bioinformatics-based analysis can then be used to determine whether these differentially translated transcripts contain similar sequence motifs in their 5’- and 3’-UTRs to regulate their selective expression in lens cells.
Clues for mRNA Decay in lens development
mRNA decay has not been studied in detail in the lens. However, there are several molecular evidences that warrant its close examination, especially in the events wherein epithelial cells in the transition zone begin differentiation into fiber cells. There is evidence that down-regulation of epithelial gene transcription is critical during commitment to differentiation. For example, Peter Carlsson's group has demonstrated that mis-expression of an epithelial cell transcription factor Foxe3 just a few cells beyond its normal zone of down-regulation, results in defective fiber cell differentiation and abnormal epithelialization.149 Similarly, precise control of Pax6 levels in cells of the transition zone and beyond are necessary for proper fiber cell differentiation.150,151 A little further in the differentiation program, there is evidence for precise control of protein levels between early and late differentiating fiber cells. For example, the cell cycle kinase inhibitor p27 is necessary in the transition zone for epithelial cell cycle exit and commitment to fiber differentiation.152 However, just a few cell layers later p27 levels need to be reduced in late differentiating fiber cells for the proper degradation of fiber cell nuclei.153 Is the reduction in p27 levels manifested by just transcriptional control? It seems unlikely for the following reasons. Expression of high levels of crystallins and other fiber genes has been possible due to the evolution of specific transcription factor binding cis-regulatory elements (e.g. for Pax, Maf, Sox transcription factors) that allows combinatorial control by these regulatory proteins.154 As differentiation progresses and the fiber cell is further committed toward building the high levels of select transcripts, it is unlikely that this established combinatorial control will be dramatically changed for down-regulating a few select mRNAs. It is therefore plausible that differentiating fiber cells have recruited other regulatory mechanisms such as miRNA- or RBP-mediated control to decay transcripts (such as potentially Foxe3) or to inhibit their translation (such as potentially p27). Indeed, P-bodies are found in the transition zone and in early differentiating fiber cells and may contribute to this process, which needs to be investigated.139 As more RNA-seq data on epithelial and fiber cell transcriptomes becomes available it will be essential to analyze whether there are any spatio-temporal patterns in the expressed mRNAs that contain AU-rich elements (ARE)155 for their decay, or other RBP-motifs in their UTR regions that may suggest alternate regulatory mechanisms. Indeed different members of the Elav family proteins that are involved in ARE mediated decay have been identified in epithelial and fiber cells in lens development (Table 1). Future studies should examine the significance of these and other RBPs in specifying the proteomes of epithelial and fiber cells.
RNA granules and RNA regulons in lens fiber differentiation
Besides the spatio-temporal control of translation that is necessary in the lens, there are three other challenges posed by the fiber cell differentiation process: (1) fiber cells have to translate unusually high levels of specific proteins i.e. crystallins, whose concentrations reach ~450 mg/ml in the lens140,141, (2) while translating the abundant mRNA levels of various crystallins, differentiating fibers also have to translate sufficient levels of other key fiber proteins such as c-Maf, Mafg, Mafk, Sox1, Prox1 whose mRNAs are not as abundant as crystallins, and (3) because of their elongated nature and high protein levels, fiber cells may need to harbor mechanisms for transport of biomolecules to preferred locations. As a further requirement of forming a transparent tissue, lens fiber cells undergo a terminal differentiation program wherein their organelles and nuclei are degraded. However unusual they may seem, these lens fiber cell features are analogous to other specialized cells in the body. For example, lens fiber cells: 1) are long and therefore may share some of the cellular challenges faced by neurons, 2) become transcriptionally inactive analogous to differentiating sperm, and 3) analogous to migrating fibroblasts, exhibit polarity (e.g. Cdk5 is localized to fiber tips, and myosin IIB is localized to posterior fiber tips). Interestingly, neurons, differentiating sperm and migrating fibroblasts all involve an RNA granule-mediated post-transcriptional regulatory function in the generation of their specialized morphologies.
These findings present us with a novel hypothesis – do distinct RNA granules represent specialized subcellular domains for mediating post-transcriptional regulatory networks in fiber cell differentiation? Further, because lens fiber cells represent a cell type where the transcriptome and proteome composition is taken toward an extreme level of specialization (e.g. alpha-A-crystallin, alpha-B-crystallin and gamma-S-crystallin alone represent 12% of the total cDNA clones in an adult human lens library156), another hypothesis can be entertained: are RNA regulons3,17,18,157, involving RBP-mediated coordinate regulation of multiple functionally related mRNAs, a mechanism recruited by differentiating fiber cells? We discuss below some of the evidence gathered so far to address these hypotheses.
Cytoplasmic RNA granules are ribonucleoprotein (RNP) complexes that offer intracellular spatio-temporal control of mRNA and its translation into protein. There are several distinct classes of RNA granules, P-bodies, Stress granules, transport RNPs or neuronal granules and germ cell granules, which have been discussed in detail elsewhere.4,19,50 So far, four RNA granule components, namely Tdrd7, Caprin2, Celf1, and Rbm24, have been shown to have conserved expression in vertebrate lens development. Two of these, Tdrd7 and Caprin2, show a granular protein pattern in lens development, albeit in different stages.15,52 A careful protein-level study to address if Celf1 and Rbm24 form granules in the lens has yet to be reported. Tdrd7 and Caprin2 null mice exhibit distinct lens defects, that of cataract and Peters anomaly, respectively. Tdrd7 deletion results in down-regulation of key lens mRNAs, of which Hspb1 (Hsp27) and Crybb3 are enriched in Tdrd7 pulldowns. Because both Hspb1 and Crybb3 are involved in lens transparency, this may suggest regulation of functionally related mRNAs by Tdrd7, similar to RNA regulons. Other lens functionally related mRNAs (Epha2, Sparc) reduced in Tdrd7 null lenses are found to be among the mRNAs enriched in Stau1 pulldowns, albeit in non-lens cells.158 Because Tdrd7 partially co-localizes with Stau1 RNPs in the lens, this may suggest a combinatorial level of control by distinct RNA granule components and RBPs over functionally related RNAs. Genome-level interrogation of mRNAs that may directly bind to Tdrd7 and other lens RBPs will provide a more detail view of combinatorial control and RNA regulons in the lens. In addition to these regulatory mechanisms, recent exciting data has pointed to recruitment of the eIF3 non-core subunit eIF3h to directly facilitate translational up-regulation of a cohort of gamma-Crystallin 2d isoforms in the zebrafish lens.159 Future studies should address if analogous mechanisms exists in mammals and whether these are coordinated with RNA granule function in the lens.
In early stages of eye development, an enrichment of Caprin2 granules are observed in future lens epithelial cells that separate from overlying surface ectoderm, which will contribute to the future cornea. Interestingly, these future lens epithelial cells exhibit an abrupt down-regulation of specific proteins (e.g. p63) which are highly expressed in closely located future corneal cells.160 Because its Drosophila ortholog Capr is known to be important for translational repression, it will be interesting to investigate if Caprin2 granules are involved in p63 translational inhibition in the future lens epithelial cells (Fig. 7).
In addition to Tdrd7 and Caprin2, Stau1 and Stau2 are expressed in the lens. Stau1 forms RNPs in the lens and is also present at the apical region of fiber cells (Fig. 5).15 Considering their role in mRNA localization in oocytes and neurons, a provocative hypothesis can be presented: are Staufen proteins important for transport of mRNAs for localized translation in elongated fiber cells? Of course, a related hypothesis needs to be addressed first, namely: is there localized protein synthesis in fiber cells? Indeed, there is evidence for preferential localization of proteins to fiber cell tips. Both Abi2 and Cdk5 proteins are located to apical and basal fiber tips, while myosin IIB is located at the posterior tips.161–163 Also, interestingly, Stau2 was found to be present in a complex that contains other RNA granule proteins such as the translational repressor Pum2 in mouse embryonic radial glial precursor cells where its targets include the mRNAs for Prox1 and beta-actin, both of which are important in lens fiber cells.164 In radial glial precursors, Stau2 protein exhibits enriched localization at the apical region and negatively controls their differentiation into neurons by directly regulating the localization of Prox1 mRNA. As noted earlier, Stau2 down-regulation in chicken causes microphthalmia (discussed in section on Eye size control).112 Thus, further studies on Stau1 and Stau2 will be necessary to define their function in eye development. Besides the lens, other eye tissues exhibit localized mRNAs. For example, in embryonic corneal tissue, beta-actin mRNA is localized to specific sub-cellular regions, and in locations, even to distinct puncta.165 Thus, it will also be important to investigate the factors in other eye tissues that control the localization of these mRNAs. Toward this goal, the function Igf2bp1 (Zbp1) in mRNA localization and protein translation has been investigated in RGCs.80,81
It will be necessary to identify the full repertoire of RNA granules and their composition in eye tissues. In addition to Tdrd7, Stau1 and Caprin2, other distinct RNA granules such as the P-bodies, have been identified in developing lens and retina (Fig. 5, 6). It will be important to investigate if P-bodies function in translational silencing or mRNA decay in the lens. Moreover, additional RNA granule components in embryonic, neonatal, and adult mouse lenses should be identified as well as their target transcripts. Although challenging, experiments to characterize RNA granules or their components by proteomic analysis have yielded important information into their composition and function.166–170 These experiments are feasible on both mouse lens epithelial-derived cell lines such as 21EM15, 17EM15, and alpha-TN4 that have been recently molecularly characterized and shown to express P-bodies (Fig. 5), as well on mouse lens tissue.139 These efforts will yield insights into the nature of the RNA granules and their associated RNA regulons in the eye. In addition to molecular experiments, live cell imaging should be applied to visualize the dynamics of RBPs in elongated fiber cells to provide cell biological insights.
Application of Drosophila data to identify eye RBPs in vertebrates
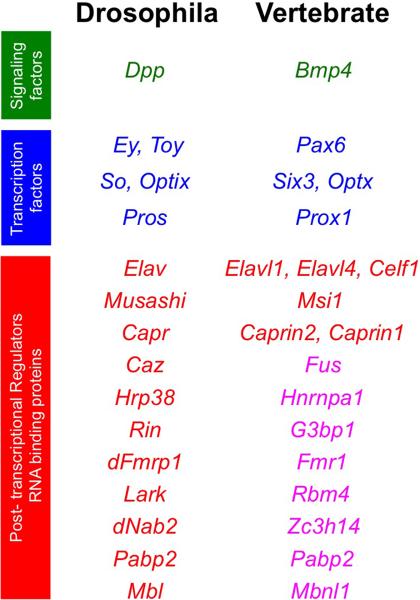
Several signaling and transcriptional regulators of eye development are conserved in Drosophila and vertebrates (Fig. 8). For example, orthologs or close family members of Bmp factors, Pax6, Six3, Eya1, Prox1 are commonly required for eye development in flies and vertebrates.31,119 A few RBPs have now been identified as candidates representing common regulators required for eye development in these diverse organisms (Fig. 8). For example, Drosophila Elav and vertebrate Elav family members, Elavl1 (HuR), Elavl4 (HuD) and Celf1 are implied in eye development.77,104,125 Musashi is expressed in fly photoreceptors, and its close family member Msi1 is expressed in the retina of mouse and frog. While vertebrate Msi1 has not been studied in detail in the eye, absence of Musashi in the fly results in abnormal rhabdomere development. 39,171 Drosophila Capr mutants exhibit reduced eye size while Caprin2 eye specific conditional null mouse mutants exhibit nuclear compaction defect and features of Peters anomaly.52,58 We outline several other RBPs that are implicated in Drosophila eye development, whose orthologs or close family members are excellent candidates for functional characterization in vertebrate eye development (Fig. 8). Thus, similar to their impact in uncovering signaling and transcription factors, studies in Drosophila have the potential to impact RBP discovery in vertebrate eyes.
Figure 8. Conservation of regulatory factors in metazoan eye development.
Comparison of signaling molecules, transcription factors, and RBPs in Drosophila and vertebrate eye development. RBPs shown in magenta color are orthologs or protein family members of fly proteins that are expressed in the vertebrate eye and need to be investigated in detail.
Eye RBP discovery by “iSyTE” and other bioinformatics approaches
Disease gene discovery in the eye has always been challenging. For example, majority of the 26 genes associated with non-syndromic human pediatric cataract were identified over a period of 25 years. However, the development and application of a bioinformatics approach called iSyTE has impacted lens gene discovery leading to the identification of several new cataract associated genes (Tdrd7, Pvrl3, Sep15, Mafg/k) and has contributed to the understanding of many other important regulatory pathways (e.g. Sip1, CBP, p300, Prox1 etc.).15,40,52,147,145,172–175 iSyTE is based on innovative processing and presentation of whole genome expression datasets for specific eye tissues. In recent years, several RBP genes have been identified as promising candidates such as Tdrd7, Caprin2, Celf1, Rbm24 in the lens using iSyTE. These discoveries have initiated the investigation of other classes of RBPs that function in conserved post-transcriptional regulatory pathways in vertebrate eye development. It is anticipated that investigation of these RBPs will advance the etiology of pediatric cataracts, and the integration of this new regulatory information into iSyTE will further expand the lens GRN, in turn increasing its efficacy in future ocular disease gene discovery. These efforts can be combined with analysis of gene expression databases/atlases such as GenePaint.176 Indeed, a study that developed genome-wide in situ map of RBPs in the mouse brain has information on the eye as the sections used included this tissue.177 These efforts combined with resources such as READDB,178 which can provide tissue-specific expression and RBP binding-motif information, will generate new testable hypotheses on the function of RBPs in the eye.
Conclusion
For the past 40 years, eye development studies in vertebrate and Drosophila models have mainly focused on signaling and transcriptional regulation. Recent evidence suggests that RBPs have important regulatory functions in ocular development, which may be conserved across diverse species with morphologically different eyes. Some RBPs are already linked to ocular disorders such as cataract, microphthalmia, Peters anomaly, retinitis pigmentosa, age-related macular degeneration, cone-rod dystrophy, Fuch's corneal dystrophy, among others. In particular, the vertebrate lens is now leading the way toward understanding the RBP and RNA granule-mediated gene expression control in the eye. Indeed deficiency of the RNA granule components Tdrd7, Caprin2, and Stau2 is linked to cataracts, Peters anomaly, and microphthalmia, respectively. Use of the lens as a model will allow investigation of RNA regulons in vertebrate development. A specific hypothesis can be tested: are mRNAs encoding functionally related proteins such as crystallins or other lens fiber proteins co-regulated by RBPs, perhaps in cytoplasmic RNA granules? RNPs and RNA granules may offer specialized domains for subcellular spatio-temporal post-transcriptional control, adding layers of complexity beyond that of transcriptional regulation. This is plausible in differentiating fiber cells that face challenges such as translation of unusually high levels of proteins, movement of molecules across long distances, and eventual loss of transcriptional ability. It will also be interesting to investigate RNA granules/RNPs in other eye tissues, especially since majority of the RBPs presently associated with eye development are also found to be in distinct RNP complexes in other cell types. For example, does photo-oxidative stress initiate stress granules or require RBP-based anti-stress response in eye tissues such as the cornea, lens and the retina? Indeed, RBPs have been linked to a protective function in UV-induced stress.179 Similarly, it will be important to address the roles of P-bodies in eye development as they are shown to be present in developing mouse lens and retina tissue. Besides the regulatory events described here, largely focusing on translational control, RBPs may also be functional in other post-transcriptional regulatory events such as controlling Pax6 alternative splicing in various eye tissues. Future studies aimed at identifying new RBPs and their RNA targets will help define the post-transcriptional regulatory circuit in eye development and reveal how its perturbation causes eye disorders.
Acknowledgements
Research from the Lachke laboratory that is described in this article was supported by the National Eye Institute of the National Institutes of Health under Award Number R01EY021505 to SAL. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. SAL is a Pew Scholar in Biomedical Sciences. This article is dedicated to the memory of Dr. David C. Beebe, Ph.D., FARVO, Janet and Bernard Becker Professor of Ophthalmology and Visual Sciences, Washington University-Saint Louis. Dr. Beebe's formidable knowledge of eye development, and his enthusiasm and encouragement of young investigators continues to be an inspiration for the eye research community.
Footnotes
Further Reading
Baker NE, Li K, Quiquand M, Ruggiero R, Wang L-H (2014) Eye Development. Methods 68:252-259.
Chalupa LM and Williams RW (eds) (2008) Eye, Retina, and Visual System of the Mouse. Cambridge, MA: MIT Press.
Kolb H, Ripps H and Wu S (eds) (2004) Concepts and Challenges in Retinal Biology. Amsterdam, The Netherlands: Elsevier Science Press.
Lachke SA, Zhang X, Maas RL (2010) Photoreceptor Cell Development Regulation. In: Encyclopedia of Life Sciences (ELS). John Wiley & Sons, Ltd: Chichester. DOI: 10.1002/9780470015901.a0000833.pub2
Tsonis PA (ed.) (2008) Animal Models in Eye Research. San Diego, CA: Academic Press.
References
- 1.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lachke SA, Maas RL. Building the developmental oculome: systems biology in vertebrate eye development and disease. Wiley Interdiscip Rev Syst Biol Med. 2010;2:305–323. doi: 10.1002/wsbm.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 4.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 5.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 6.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 7.Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 8.Neelamraju Y, Hashemikhabir S, Janga SC. The human RBPome: from genes and proteins to human disease. J Proteomics. 2015;127:61–70. doi: 10.1016/j.jprot.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 9.Singh G, Pratt G, Yeo GW, Moore MJ. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem. 2015;84:325–354. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukong KE, Chang K, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv. Exp. Med. Biol. 2014;825:1–55. doi: 10.1007/978-1-4939-1221-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lachke SA, et al. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011;331:1571–1576. doi: 10.1126/science.1195970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachke SA, Maas RL. RNA Granules and Cataract. Expert Rev Ophthalmol. 2011;6:497–500. doi: 10.1586/eop.11.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simone LE, Keene JD. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr. Opin. Genet. Dev. 2013;23:35–43. doi: 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin. Cell Dev. Biol. 2014 doi: 10.1016/j.semcdb.2014.05.014. doi:10.1016/j.semcdb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol. Cell. 2014;54:547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 20.Donner AL, Lachke SA, Maas RL. Lens induction in vertebrates: variations on a conserved theme of signaling events. Semin. Cell Dev. Biol. 2006;17:676–685. doi: 10.1016/j.semcdb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Graw J. Eye development. Curr. Top. Dev. Biol. 2010;90:343–386. doi: 10.1016/S0070-2153(10)90010-0. [DOI] [PubMed] [Google Scholar]
- 22.Cvekl A, Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graw J. The genetic and molecular basis of congenital eye defects. Nat. Rev. Genet. 2003;4:876–888. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- 24.Raghunath A, Perumal E. Micro-RNAs and their roles in eye disorders. Ophthalmic Res. 2015;53:169–186. doi: 10.1159/000371853. [DOI] [PubMed] [Google Scholar]
- 25.Sundermeier TR, Palczewski K. The impact of microRNA gene regulation on the survival and function of mature cell types in the eye. FASEB J. 2016;30:23–33. doi: 10.1096/fj.15-279745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Piatigorsky J. Targeted deletion of Dicer disrupts lens morphogenesis, corneal epithelium stratification, and whole eye development. Dev. Dyn. 2009;238:2388–2400. doi: 10.1002/dvdy.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf L, et al. Identification and characterization of FGF2-dependent mRNA: microRNA networks during lens fiber cell differentiation. G3 (Bethesda) 2013;3:2239–2255. doi: 10.1534/g3.113.008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaham O, et al. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9:e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh S, et al. Iris hypoplasia in mice that lack the alternatively spliced Pax6(5a) isoform. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6812–6815. doi: 10.1073/pnas.102691299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Garcia CM, Shui Y-B, Beebe DC. Expression and regulation of alpha-, beta-, and gamma-crystallins in mammalian lens epithelial cells. Invest. Ophthalmol. Vis. Sci. 2004;45:3608–3619. doi: 10.1167/iovs.04-0423. [DOI] [PubMed] [Google Scholar]
- 31.Charlton-Perkins M, Brown NL, Cook TA. The lens in focus: a comparison of lens development in Drosophila and vertebrates. Mol. Genet. Genomics. 2011;286:189–213. doi: 10.1007/s00438-011-0643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar JP. Signalling pathways in Drosophila and vertebrate retinal development. Nat. Rev. Genet. 2001;2:846–857. doi: 10.1038/35098564. [DOI] [PubMed] [Google Scholar]
- 33.Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 34.Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–1336. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- 35.Schlichting K, Dahmann C. Hedgehog and Dpp signaling induce cadherin Cad86C expression in the morphogenetic furrow during Drosophila eye development. Mech. Dev. 2008;125:712–728. doi: 10.1016/j.mod.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- 37.Legent K, Treisman JE. Wingless signaling in Drosophila eye development. Methods Mol. Biol. 2008;469:141–161. doi: 10.1007/978-1-60327-469-2_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domínguez M. Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development. 1999;126:2345–2353. doi: 10.1242/dev.126.11.2345. [DOI] [PubMed] [Google Scholar]
- 39.Hirota Y, et al. Musashi and seven in absentia downregulate Tramtrack through distinct mechanisms in Drosophila eye development. Mech. Dev. 1999;87:93–101. doi: 10.1016/s0925-4773(99)00143-4. [DOI] [PubMed] [Google Scholar]
- 40.Lachke SA, et al. iSyTE: integrated Systems Tool for Eye gene discovery. Invest. Ophthalmol. Vis. Sci. 2012;53:1617–1627. doi: 10.1167/iovs.11-8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka T, et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10579–10584. doi: 10.1073/pnas.1015447108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiels A, Hejtmancik JF. Genetics of human cataract. Clin. Genet. 2013;84:120–127. doi: 10.1111/cge.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mansouri B, Stacy RC, Kruger J, Cestari DM. Deprivation amblyopia and congenital hereditary cataract. Semin Ophthalmol. 2013;28:321–326. doi: 10.3109/08820538.2013.825289. [DOI] [PubMed] [Google Scholar]
- 44.Kuhli-Hattenbach C, Lüchtenberg M, Kohnen T, Hattenbach L-O. Risk factors for complications after congenital cataract surgery without intraocular lens implantation in the first 18 months of life. Am. J. Ophthalmol. 2008;146:1–7. doi: 10.1016/j.ajo.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 45.Gao M, Arkov AL. Next generation organelles: structure and role of germ granules in the germline. Mol. Reprod. Dev. 2013;80:610–623. doi: 10.1002/mrd.22115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirose T, et al. Identification of tudor repeat associator with PCTAIRE 2 (Trap). A novel protein that interacts with the N-terminal domain of PCTAIRE 2 in rat brain. Eur. J. Biochem. 2000;267:2113–2121. doi: 10.1046/j.1432-1327.2000.01218.x. [DOI] [PubMed] [Google Scholar]
- 47.Hosokawa M, et al. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev. Biol. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 48.Anantharaman V, Zhang D, Aravind L. OST-HTH: a novel predicted RNA-binding domain. Biol. Direct. 2010;5:13. doi: 10.1186/1745-6150-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Callebaut I, Mornon J-P. LOTUS, a new domain associated with small RNA pathways in the germline. Bioinformatics. 2010;26:1140–1144. doi: 10.1093/bioinformatics/btq122. [DOI] [PubMed] [Google Scholar]
- 50.Buchan JR. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorén CE, Schrader JW, Ahlgren U, Gunhaga L. FGF signals induce Caprin2 expression in the vertebrate lens. Differentiation. 2009;77:386–394. doi: 10.1016/j.diff.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 52.Dash S, Dang CA, Beebe DC, Lachke SA. Deficiency of the RNA binding protein caprin2 causes lens defects and features of peters anomaly. Dev. Dyn. 2015;244:1313–1327. doi: 10.1002/dvdy.24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Augusteyn RC. On the growth and internal structure of the human lens. Exp. Eye Res. 2010;90:643–654. doi: 10.1016/j.exer.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Ghoul KJ, et al. Structural evidence of human nuclear fiber compaction as a function of ageing and cataractogenesis. Exp. Eye Res. 2001;72:199–214. doi: 10.1006/exer.2000.0937. [DOI] [PubMed] [Google Scholar]
- 55.Dubbelman M, Van der Heijde GL, Weeber HA, Vrensen GFJM. Changes in the internal structure of the human crystalline lens with age and accommodation. Vision Res. 2003;43:2363–2375. doi: 10.1016/s0042-6989(03)00428-0. [DOI] [PubMed] [Google Scholar]
- 56.Papoulas O, et al. dFMRP and Caprin, translational regulators of synaptic plasticity, control the cell cycle at the Drosophila mid-blastula transition. Development. 2010;137:4201–4209. doi: 10.1242/dev.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shiina N, Tokunaga M. RNA granule protein 140 (RNG140), a paralog of RNG105 localized to distinct RNA granules in neuronal dendrites in the adult vertebrate brain. J. Biol. Chem. 2010;285:24260–24269. doi: 10.1074/jbc.M110.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baumgartner R, Stocker H, Hafen E. The RNA-binding proteins FMR1, rasputin and caprin act together with the UBA protein lingerer to restrict tissue growth in Drosophila melanogaster. PLoS Genet. 2013;9:e1003598. doi: 10.1371/journal.pgen.1003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oberleitner S. Seb4: An RNA-binding Protein as a Novel Regulator of Myogenesis During Early Development in Xenopus Laevis. 2008 [Google Scholar]
- 60.Li H-Y, Bourdelas A, Carron C, Shi D-L. The RNA-binding protein Seb4/RBM24 is a direct target of MyoD and is required for myogenesis during Xenopus early development. Mech. Dev. 2010;127:281–291. doi: 10.1016/j.mod.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 61.Poon KL, et al. RNA-binding protein RBM24 is required for sarcomere assembly and heart contractility. Cardiovasc. Res. 2012;94:418–427. doi: 10.1093/cvr/cvs095. [DOI] [PubMed] [Google Scholar]
- 62.Maragh S, et al. Rbm24a and Rbm24b are required for normal somitogenesis. PLoS ONE. 2014;9:e105460. doi: 10.1371/journal.pone.0105460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grifone R, et al. The RNA-binding protein Rbm24 is transiently expressed in myoblasts and is required for myogenic differentiation during vertebrate development. Mech. Dev. 2014;134:1–15. doi: 10.1016/j.mod.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 64.Yang J, et al. RBM24 is a major regulator of muscle-specific alternative splicing. Dev. Cell. 2014;31:87–99. doi: 10.1016/j.devcel.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 65.Dichmann DS, Fletcher RB, Harland RM. Expression cloning in Xenopus identifies RNA-binding proteins as regulators of embryogenesis and Rbmx as necessary for neural and muscle development. Dev. Dyn. 2008;237:1755–1766. doi: 10.1002/dvdy.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bitel CL, Perrone-Bizzozero NI, Frederikse PH. HuB/C/D, nPTB, REST4, and miR-124 regulators of neuronal cell identity are also utilized in the lens. Mol. Vis. 2010;16:2301–2316. [PMC free article] [PubMed] [Google Scholar]
- 67.Frederikse PH, Kasinathan C, Kleiman NJ. Parallels between neuron and lens fiber cell structure and molecular regulatory networks. Dev. Biol. 2012;368:255–260. doi: 10.1016/j.ydbio.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 68.Bitel CL, et al. Evidence that ‘brain-specific’ FOX-1, FOX-2, and nPTB alternatively spliced isoforms are produced in the lens. Curr. Eye Res. 2011;36:321–327. [PubMed] [Google Scholar]
- 69.Suzuki H, et al. Vegetal localization of the maternal mRNA encoding an EDEN-BP/Bruno-like protein in zebrafish. Mech. Dev. 2000;93:205–209. doi: 10.1016/s0925-4773(00)00270-7. [DOI] [PubMed] [Google Scholar]
- 70.Gautier-Courteille C, et al. EDEN-BP-dependent post-transcriptional regulation of gene expression in Xenopus somitic segmentation. Development. 2004;131:6107–6117. doi: 10.1242/dev.01528. [DOI] [PubMed] [Google Scholar]
- 71.Day RC, Beck CW. Transdifferentiation from cornea to lens in Xenopus laevis depends on BMP signalling and involves upregulation of Wnt signalling. BMC Dev. Biol. 2011;11:54. doi: 10.1186/1471-213X-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blech-Hermoni Y, Stillwagon S, Ladd A. Diversity and conservation of CELF1 and CELF2 RNA and protein expression patterns during embryonic development. Dev Dyn. 2013;242:767–777. doi: 10.1002/dvdy.23959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raji B, et al. The RNA-binding protein Musashi-1 is produced in the developing and adult mouse eye. Mol. Vis. 2007;13:1412–1427. [PubMed] [Google Scholar]
- 74.Susaki K, et al. Musashi-1, an RNA-binding protein, is indispensable for survival of photoreceptors. Exp. Eye Res. 2009;88:347–355. doi: 10.1016/j.exer.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 75.Huot M-E, et al. The RNA-binding protein fragile X-related 1 regulates somite formation in Xenopus laevis. Mol. Biol. Cell. 2005;16:4350–4361. doi: 10.1091/mbc.E05-04-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zelus BD, Giebelhaus DH, Eib DW, Kenner KA, Moon RT. Expression of the poly(A)-binding protein during development of Xenopus laevis. Mol. Cell. Biol. 1989;9:2756–2760. doi: 10.1128/mcb.9.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amato MA, et al. Comparison of the expression patterns of five neural RNA binding proteins in the Xenopus retina. J. Comp. Neurol. 2005;481:331–339. doi: 10.1002/cne.20387. [DOI] [PubMed] [Google Scholar]
- 78.Lin AC, et al. Cytoplasmic polyadenylation and cytoplasmic polyadenylation element-dependent mRNA regulation are involved in Xenopus retinal axon development. Neural Dev. 2009;4:8. doi: 10.1186/1749-8104-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Y, Gervasi C, Szaro BG. A crucial role for hnRNP K in axon development in Xenopus laevis. Development. 2008;135:3125–3135. doi: 10.1242/dev.022236. [DOI] [PubMed] [Google Scholar]
- 80.Gaynes JA, et al. The RNA Binding Protein Igf2bp1 Is Required for Zebrafish RGC Axon Outgrowth In Vivo. PLoS ONE. 2015;10:e0134751. doi: 10.1371/journal.pone.0134751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kalous A, Stake JI, Yisraeli JK, Holt CE. RNA-binding protein Vg1RBP regulates terminal arbor formation but not long-range axon navigation in the developing visual system. Dev Neurobiol. 2014;74:303–318. doi: 10.1002/dneu.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gerber WV, et al. The RNA-binding protein gene, hermes, is expressed at high levels in the developing heart. Mech. Dev. 1999;80:77–86. doi: 10.1016/s0925-4773(98)00195-6. [DOI] [PubMed] [Google Scholar]
- 83.Piri N, Kwong JMK, Song M, Caprioli J. Expression of hermes gene is restricted to the ganglion cells in the retina. Neurosci. Lett. 2006;405:40–45. doi: 10.1016/j.neulet.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 84.Kwong JMK, Caprioli J, Piri N. RNA binding protein with multiple splicing: a new marker for retinal ganglion cells. Invest. Ophthalmol. Vis. Sci. 2010;51:1052–1058. doi: 10.1167/iovs.09-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodriguez AR, de Sevilla Müller LP, Brecha NC. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J. Comp. Neurol. 2014;522:1411–1443. doi: 10.1002/cne.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hörnberg H, et al. RNA-binding protein Hermes/RBPMS inversely affects synapse density and axon arbor formation in retinal ganglion cells in vivo. J. Neurosci. 2013;33:10384–10395. doi: 10.1523/JNEUROSCI.5858-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Yu H, Deaton SK, Szaro BG. Heterogeneous nuclear ribonucleoprotein K, an RNA-binding protein, is required for optic axon regeneration in Xenopus laevis. J. Neurosci. 2012;32:3563–3574. doi: 10.1523/JNEUROSCI.5197-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 89.Tuson M, Marfany G, Gonzàlez-Duarte R. Mutation of CERKL, a novel human ceramide kinase gene, causes autosomal recessive retinitis pigmentosa (RP26). Am. J. Hum. Genet. 2004;74:128–138. doi: 10.1086/381055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aleman TS, et al. CERKL mutations cause an autosomal recessive cone-rod dystrophy with inner retinopathy. Invest. Ophthalmol. Vis. Sci. 2009;50:5944–5954. doi: 10.1167/iovs.09-3982. [DOI] [PubMed] [Google Scholar]
- 91.Fathinajafabadi A, Pérez-Jiménez E, Riera M, Knecht E, Gonzàlez-Duarte R. CERKL, a retinal disease gene, encodes an mRNA-binding protein that localizes in compact and untranslated mRNPs associated with microtubules. PLoS ONE. 2014;9:e87898. doi: 10.1371/journal.pone.0087898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tanackovic G, et al. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum. Mol. Genet. 2011;20:2116–2130. doi: 10.1093/hmg/ddr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Graziotto JJ, et al. Three gene-targeted mouse models of RNA splicing factor RP show late-onset RPE and retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:190–198. doi: 10.1167/iovs.10-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karunakaran DKP, Banday AR, Wu Q, Kanadia R. Expression analysis of an evolutionarily conserved alternative splicing factor, Sfrs10, in age-related macular degeneration. PLoS ONE. 2013;8:e75964. doi: 10.1371/journal.pone.0075964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kanadia RN, Clark VE, Punzo C, Trimarchi JM, Cepko CL. Temporal requirement of the alternative-splicing factor Sfrs1 for the survival of retinal neurons. Development. 2008;135:3923–3933. doi: 10.1242/dev.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guimarães-Souza EM, Perche O, Morgans CW, Duvoisin RM, Calaza KC. Fragile X Mental Retardation Protein expression in the retina is regulated by light. Exp. Eye Res. 2015;146:72–82. doi: 10.1016/j.exer.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boy S, et al. XSEB4R, a novel RNA-binding protein involved in retinal cell differentiation downstream of bHLH proneural genes. Development. 2004;131:851–862. doi: 10.1242/dev.00983. [DOI] [PubMed] [Google Scholar]
- 98.Nickerson PEB, Myers T, Clarke DB, Chow RL. Changes in Musashi-1 subcellular localization correlate with cell cycle exit during postnatal retinal development. Exp. Eye Res. 2011;92:344–352. doi: 10.1016/j.exer.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 99.Seko Y, et al. Derivation of human differential photoreceptor-like cells from the iris by defined combinations of CRX, RX and NEUROD. PLoS ONE. 2012;7:e35611. doi: 10.1371/journal.pone.0035611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dhamodaran K, Subramani M, Ponnalagu M, Shetty R, Das D. Ocular stem cells: a status update! Stem Cell Res Ther. 2014;5:56. doi: 10.1186/scrt445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Neant I, Deisig N, Scerbo P, Leclerc C, Moreau M. The RNA-binding protein Xp54nrb isolated from a Ca2+-dependent screen is expressed in neural structures during Xenopus laevis development. Int. J. Dev. Biol. 2011;55:923–931. doi: 10.1387/ijdb.103253in. [DOI] [PubMed] [Google Scholar]
- 102.Dauber A, et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am. J. Hum. Genet. 2013;93:798–811. doi: 10.1016/j.ajhg.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davidson AE, Hayes S, Hardcastle AJ, Tuft SJ. The pathogenesis of keratoconus. Eye (Lond) 2014;28:189–195. doi: 10.1038/eye.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Joseph R, Srivastava OP, Pfister RR. Downregulation of β-Actin Gene and Human Antigen R in Human Keratoconus. Invest Ophthalmol Vis Sci. 2012;53:4032–4041. doi: 10.1167/iovs.11-9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Joseph R, Srivastava OP, Pfister RR. Downregulation of β-actin and its regulatory gene HuR affect cell migration of human corneal fibroblasts. Mol. Vis. 2014;20:593–605. [PMC free article] [PubMed] [Google Scholar]
- 106.Baratz KH, et al. E2-2 protein and Fuchs's corneal dystrophy. N. Engl. J. Med. 2010;363:1016–1024. doi: 10.1056/NEJMoa1007064. [DOI] [PubMed] [Google Scholar]
- 107.Du J, et al. RNA toxicity and missplicing in the common eye disease fuchs endothelial corneal dystrophy. J. Biol. Chem. 2015;290:5979–5990. doi: 10.1074/jbc.M114.621607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Joo J-H, Kim YH, Dunn NW, Sugrue SP. Disruption of mouse corneal epithelial differentiation by conditional inactivation of pnn. Invest. Ophthalmol. Vis. Sci. 2010;51:1927–1934. doi: 10.1167/iovs.09-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Joo J-H, et al. Transcriptomic analysis of PNN- and ESRP1-regulated alternative pre-mRNA splicing in human corneal epithelial cells. Invest. Ophthalmol. Vis. Sci. 2013;54:697–707. doi: 10.1167/iovs.12-10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McKown RL, et al. Lacritin and other new proteins of the lacrimal functional unit. Exp. Eye Res. 2009;88:848–858. doi: 10.1016/j.exer.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Turner HC, Budak MT, Akinci MAM, Wolosin JM. Comparative analysis of human conjunctival and corneal epithelial gene expression with oligonucleotide microarrays. Invest. Ophthalmol. Vis. Sci. 2007;48:2050–2061. doi: 10.1167/iovs.06-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cockburn DM, et al. The double-stranded RNA-binding protein Staufen 2 regulates eye size. Mol. Cell. Neurosci. 2012;51:101–111. doi: 10.1016/j.mcn.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 113.Schneider A, Bardakjian T, Reis LM, Tyler RC, Semina EV. Novel SOX2 mutations and genotype-phenotype correlation in anophthalmia and microphthalmia. Am. J. Med. Genet. A. 2009;149A:2706–2715. doi: 10.1002/ajmg.a.33098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schilter KF, et al. Whole-genome copy number variation analysis in anophthalmia and microphthalmia. Clin. Genet. 2013;84:473–481. doi: 10.1111/cge.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee HY, et al. Multiple requirements for Hes 1 during early eye formation. Dev. Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 117.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 2010;19:1314–1323. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gehring WJ. The evolution of vision. Wiley Interdiscip Rev Dev Biol. 2014;3:1–40. doi: 10.1002/wdev.96. [DOI] [PubMed] [Google Scholar]
- 120.Pichaud F, Desplan C. A new visualization approach for identifying mutations that affect differentiation and organization of the Drosophila ommatidia. Development. 2001;128:815–826. doi: 10.1242/dev.128.6.815. [DOI] [PubMed] [Google Scholar]
- 121.Pazman C, Mayes CA, Fanto M, Haynes SR, Mlodzik M. Rasputin, the Drosophila homologue of the RasGAP SH3 binding protein, functions in ras- and Rho-mediated signaling. Development. 2000;127:1715–1725. doi: 10.1242/dev.127.8.1715. [DOI] [PubMed] [Google Scholar]
- 122.Shimamura M, et al. Genetic link between Cabeza, a Drosophila homologue of Fused in Sarcoma (FUS), and the EGFR signaling pathway. Exp. Cell Res. 2014;326:36–45. doi: 10.1016/j.yexcr.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 123.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 124.Ji Y, Jarnik M, Tulin AV. Poly(ADP-ribose) glycohydrolase and poly(ADP-ribose)-interacting protein Hrp38 regulate pattern formation during Drosophila eye development. Gene. 2013;526:187–194. doi: 10.1016/j.gene.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lisbin MJ, Qiu J, White K. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 2001;15:2546–2561. doi: 10.1101/gad.903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Campos AR, Grossman D, White K. Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. J. Neurogenet. 1985;2:197–218. doi: 10.3109/01677068509100150. [DOI] [PubMed] [Google Scholar]
- 127.Sofola O, et al. The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J. Neurosci. 2008;28:10200–10205. doi: 10.1523/JNEUROSCI.2786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pak C, et al. Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12390–12395. doi: 10.1073/pnas.1107103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vicente-Crespo M, et al. Drosophila muscleblind is involved in troponin T alternative splicing and apoptosis. PLoS ONE. 2008;3:e1613. doi: 10.1371/journal.pone.0001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Llamusi B, et al. Muscleblind, BSF and TBPH are mislocalized in the muscle sarcomere of a Drosophila myotonic dystrophy model. Dis Model Mech. 2013;6:184–196. doi: 10.1242/dmm.009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ihara R, et al. RNA binding mediates neurotoxicity in the transgenic Drosophila model of TDP-43 proteinopathy. Hum. Mol. Genet. 2013;22:4474–4484. doi: 10.1093/hmg/ddt296. [DOI] [PubMed] [Google Scholar]
- 132.Surabhi S, et al. Regulation of Notch Signaling by an Evolutionary Conserved DEAD Box RNA Helicase, Maheshvara in Drosophila melanogaster. Genetics. 2015;201:1071–1085. doi: 10.1534/genetics.115.181214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ivanov D, Dvoriantchikova G, Pestova A, Nathanson L, Shestopalov VI. Microarray analysis of fiber cell maturation in the lens. FEBS Lett. 2005;579:1213–1219. doi: 10.1016/j.febslet.2005.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nakahara M, et al. Degradation of nuclear DNA by DNase II-like acid DNase in cortical fiber cells of mouse eye lens. FEBS J. 2007;274:3055–3064. doi: 10.1111/j.1742-4658.2007.05836.x. [DOI] [PubMed] [Google Scholar]
- 136.Hoang TV, et al. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Mol. Vis. 2014;20:1491–1517. [PMC free article] [PubMed] [Google Scholar]
- 137.Sun J, et al. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 2015;43:6827–6846. doi: 10.1093/nar/gkv589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.De Maria A, Bassnett S. Birc7: A Late Fiber Gene of the Crystalline Lens. Invest. Ophthalmol. Vis. Sci. 2015;56:4823–4834. doi: 10.1167/iovs.15-16968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Terrell AM, et al. Molecular characterization of mouse lens epithelial cell lines and their suitability to study RNA granules and cataract associated genes. Exp. Eye Res. 2015;131:42–55. doi: 10.1016/j.exer.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bassnett S, Wilmarth PA, David LL. The membrane proteome of the mouse lens fiber cell. Mol. Vis. 2009;15:2448–2463. [PMC free article] [PubMed] [Google Scholar]
- 141.Shang F, et al. Newborn mouse lens proteome and its alteration by lysine 6 mutant ubiquitin. J. Proteome Res. 2014;13:1177–1189. doi: 10.1021/pr400801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Beebe DC, Piatigorsky J. Translational regulation of delta-crystallin synthesis during lens development in the chicken embryo. Dev. Biol. 1981;84:96–101. doi: 10.1016/0012-1606(81)90374-2. [DOI] [PubMed] [Google Scholar]
- 143.Cenedella RJ. Role of transcription, translation, and protein turnover in controlling the distribution of 3-hydroxy-3-methylglutaryl coenzyme A reductase in the lens. Invest. Ophthalmol. Vis. Sci. 1995;36:2133–2141. [PubMed] [Google Scholar]
- 144.McAvoy JW. Cell division, cell elongation and the co-ordination of crystallin gene expression during lens morphogenesis in the rat. J Embryol Exp Morphol. 1978;45:271–281. [PubMed] [Google Scholar]
- 145.Audette DS, et al. Prox1 and fibroblast growth factor receptors form a novel regulatory loop controlling lens fiber differentiation and gene expression. Development. 2016;143:318–328. doi: 10.1242/dev.127860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Donner AL, Ko F, Episkopou V, Maas RL. Pax6 is misexpressed in Sox1 null lens fiber cells. Gene Expr. Patterns. 2007;7:606–613. doi: 10.1016/j.modgep.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wolf L, et al. Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 2013;41:10199–10214. doi: 10.1093/nar/gkt824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xie Q, et al. Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS ONE. 2013;8:e54507. doi: 10.1371/journal.pone.0054507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Landgren H, Blixt A, Carlsson P. Persistent FoxE3 expression blocks cytoskeletal remodeling and organelle degradation during lens fiber differentiation. Invest. Ophthalmol. Vis. Sci. 2008;49:4269–4277. doi: 10.1167/iovs.08-2243. [DOI] [PubMed] [Google Scholar]
- 150.Duncan MK, et al. Ectopic Pax6 expression disturbs lens fiber cell differentiation. Invest. Ophthalmol. Vis. Sci. 2004;45:3589–3598. doi: 10.1167/iovs.04-0151. [DOI] [PubMed] [Google Scholar]
- 151.Shaham O, et al. Pax6 is essential for lens fiber cell differentiation. Development. 2009;136:2567–2578. doi: 10.1242/dev.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang P, Wong C, DePinho RA, Harper JW, Elledge SJ. Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 1998;12:3162–3167. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lyu L, et al. Unfolded-protein response-associated stabilization of p27(Cdkn1b) interferes with lens fiber cell denucleation leading to cataract. FASEB J. 2015 doi: 10.1096/fj.15-278036. doi:10.1096/fj.15-278036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cvekl A, Duncan MK. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Beisang D, Bohjanen PR. Perspectives on the ARE as it turns 25 years old. Wiley Interdiscip Rev RNA. 2012;3:719–731. doi: 10.1002/wrna.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wistow G, et al. Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants. Mol. Vis. 2002;8:171–184. [PubMed] [Google Scholar]
- 157.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 158.Furic L, Maher-Laporte M, DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–335. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Choudhuri A, Maitra U, Evans T. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:9818–9823. doi: 10.1073/pnas.1302934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kuracha MR, et al. Spry1 and Spry2 are necessary for lens vesicle separation and corneal differentiation. Invest. Ophthalmol. Vis. Sci. 2011;52:6887–6897. doi: 10.1167/iovs.11-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Negash S, Wang H-S, Gao C, Ledee D, Zelenka P. Cdk5 regulates cell-matrix and cell-cell adhesion in lens epithelial cells. J. Cell. Sci. 2002;115:2109–2117. doi: 10.1242/jcs.115.10.2109. [DOI] [PubMed] [Google Scholar]
- 162.Grove M, et al. ABI2-deficient mice exhibit defective cell migration, aberrant dendritic spine morphogenesis, and deficits in learning and memory. Mol. Cell. Biol. 2004;24:10905–10922. doi: 10.1128/MCB.24.24.10905-10922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maddala R, Skiba N, Vasantha Rao P. Lens fiber cell elongation and differentiation is associated with a robust increase in myosin light chain phosphorylation in the developing mouse. Differentiation. 2007;75:713–725. doi: 10.1111/j.1432-0436.2007.00173.x. [DOI] [PubMed] [Google Scholar]
- 164.Vessey JP, et al. An asymmetrically localized Staufen2-dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell. 2012;11:517–528. doi: 10.1016/j.stem.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 165.Yeh B, Svoboda KK. Intracellular distribution of beta-actin mRNA is polarized in embryonic corneal epithelia. J. Cell. Sci. 1994;107(Pt 1):105–115. doi: 10.1242/jcs.107.1.105. [DOI] [PubMed] [Google Scholar]
- 166.Villacé P, Marión RM, Ortín J. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 2004;32:2411–2420. doi: 10.1093/nar/gkh552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Elvira G, et al. Characterization of an RNA granule from developing brain. Mol. Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 168.Chen C, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fritzsche R, et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013;5:1749–1762. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 170.Meikar O, et al. An atlas of chromatoid body components. RNA. 2014;20:483–495. doi: 10.1261/rna.043729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Siddall NA, Hime GR, Pollock JA, Batterham P. Ttk69-dependent repression of lozenge prevents the ectopic development of R7 cells in the Drosophila larval eye disc. BMC Dev. Biol. 2009;9:64. doi: 10.1186/1471-213X-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lachke SA, et al. The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum. Genet. 2012;131:235–250. doi: 10.1007/s00439-011-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kasaikina MV, et al. Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J. Biol. Chem. 2011;286:33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Agrawal SA, et al. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum. Genet. 2015;134:717–735. doi: 10.1007/s00439-015-1554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Manthey AL, et al. Loss of Sip1 leads to migration defects and retention of ectodermal markers during lens development. Mech. Dev. 2014;131:86–110. doi: 10.1016/j.mod.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McKee AE, et al. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hashemikhabir S, Neelamraju Y, Janga SC. Database of RNA binding protein expression and disease dynamics (READ DB). Database (Oxford) 2015;2015:bav072. doi: 10.1093/database/bav072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang C, Carrier F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J. Biol. Chem. 2001;276:47277–47284. doi: 10.1074/jbc.M105396200. [DOI] [PubMed] [Google Scholar]
- 180.Mandal NA, et al. Expression and localization of CERKL in the mammalian retina, its response to light-stress, and relationship with NeuroD1 gene. Exp. Eye Res. 2013;106:24–33. doi: 10.1016/j.exer.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kaneko J, Chiba C. Immunohistochemical analysis of Musashi-1 expression during retinal regeneration of adult newt. Neurosci. Lett. 2009;450:252–257. doi: 10.1016/j.neulet.2008.11.031. [DOI] [PubMed] [Google Scholar]
- 182.Shibayama M, et al. Polypyrimidine tract-binding protein is essential for early mouse development and embryonic stem cell proliferation. FEBS J. 2009;276:6658–6668. doi: 10.1111/j.1742-4658.2009.07380.x. [DOI] [PubMed] [Google Scholar]
- 183.Licatalosi DD, et al. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Q, et al. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. Elife. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Siddall NA, et al. Drosophila Rbp6 is an orthologue of vertebrate Msi-1 and Msi-2, but does not function redundantly with dMsi to regulate germline stem cell behaviour. PLoS ONE. 2012;7:e49810. doi: 10.1371/journal.pone.0049810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.King MR, Matzat LH, Dale RK, Lim SJ, Lei EP. The RNA-binding protein Rumpelstiltskin antagonizes gypsy chromatin insulator function in a tissue-specific manner. J. Cell. Sci. 2014;127:2956–2966. doi: 10.1242/jcs.151126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Solomon S, et al. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 2007;27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fujimura K, Kano F, Murata M. Dual localization of the RNA binding protein CUGBP-1 to stress granule and perinucleolar compartment. Exp. Cell Res. 2008;314:543–553. doi: 10.1016/j.yexcr.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 189.Cui Y-H, et al. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol. Biol. Cell. 2012;23:151–162. doi: 10.1091/mbc.E11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell. Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 191.Guo X, Wu Y, Hartley RS. Cold-inducible RNA-binding protein contributes to human antigen R and cyclin E1 deregulation in breast cancer. Mol. Carcinog. 2010;49:130–140. doi: 10.1002/mc.20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tsai N-P, Tsui Y-C, Wei L-N. Dynein motor contributes to stress granule dynamics in primary neurons. Neuroscience. 2009;159:647–656. doi: 10.1016/j.neuroscience.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gallouzi IE, et al. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mazroui R, et al. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 196.Fukuda T, Naiki T, Saito M, Irie K. hnRNP K interacts with RNA binding motif protein 42 and functions in the maintenance of cellular ATP level during stress conditions. Genes Cells. 2009;14:113–128. doi: 10.1111/j.1365-2443.2008.01256.x. [DOI] [PubMed] [Google Scholar]
- 197.Wächter K, Köhn M, Stöhr N, Hüttelmaier S. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol. Chem. 2013;394:1077–1090. doi: 10.1515/hsz-2013-0111. [DOI] [PubMed] [Google Scholar]
- 198.Bley N, et al. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 2015;43:e26. doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Onishi H, et al. MBNL1 associates with YB-1 in cytoplasmic stress granules. J. Neurosci. Res. 2008;86:1994–2002. doi: 10.1002/jnr.21655. [DOI] [PubMed] [Google Scholar]
- 200.ErLin S, et al. Musashi-1 maintains blood-testis barrier structure during spermatogenesis and regulates stress granule formation upon heat stress. Mol. Biol. Cell. 2015;26:1947–1956. doi: 10.1091/mbc.E14-11-1497. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 201.Furukawa MT, Sakamoto H, Inoue K. Interaction and colocalization of HERMES/RBPMS with NonO, PSF, and G3BP1 in neuronal cytoplasmic RNP granules in mouse retinal line cells. Genes Cells. 2015;20:257–266. doi: 10.1111/gtc.12224. [DOI] [PubMed] [Google Scholar]
- 202.Sury MD, McShane E, Hernandez-Miranda LR, Birchmeier C, Selbach M. Quantitative proteomics reveals dynamic interaction of c-Jun N-terminal kinase (JNK) with RNA transport granule proteins splicing factor proline- and glutamine-rich (Sfpq) and non-POU domain-containing octamer-binding protein (Nono) during neuronal differentiation. Mol. Cell Proteomics. 2015;14:50–65. doi: 10.1074/mcp.M114.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kedersha N, et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hoyle NP, Castelli LM, Campbell SG, Holmes LEA, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lin J-C, Hsu M, Tarn W-Y. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2235–2240. doi: 10.1073/pnas.0611015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Van der Laan AMA, et al. mRNA cycles through hypoxia-induced stress granules in live Drosophila embryonic muscles. Int. J. Dev. Biol. 2012;56:701–709. doi: 10.1387/ijdb.103172al. [DOI] [PubMed] [Google Scholar]
- 208.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]