Abstract
Tacrolimus is dependent on CYP3A5 enzyme for metabolism. Expression of the CYP3A5 enzyme is controlled by several alleles including CYP3A5*1, CYP3A5*3, CYP3A5*6 and CYP3A5*7. African Americans (AA) have on average higher tacrolimus dose requirements than Caucasians; however, some have requirements similar to Caucasians. Studies in AA have primarily evaluated the CYP3A5*3 variant; however, there are common nonfunctional variants in AA (CYP3A5*6 and CYP3A5*7) which do not occur in Caucasians. These variants are associated with lower dose requirements and may explain why some AA are metabolically similar to Caucasians. We created a tacrolimus clearance model in 354 AA using a development and validation cohort. Time posttransplant, steroid and antiviral use, age, CYP3A5*1, *3, *6 and *7 alleles were significant towards clearance. This study is the first to develop an AA specific genotype-guided tacrolimus dosing model to personalize therapy.
Keywords: tacrolimus, kidney transplant, pharmacokinetics, personalization, pharmacogenomics
Introduction
Kidney transplantation is a common and effective treatment for end stage renal disease. African Americans (AA) represent around 34% of the candidates on the kidney transplant waiting list. (1, 2) Long-term graft survival rates are lower and all-cause mortality rates are higher in AA than in Caucasians or Asians.(3–6) There are several reasons cited for poor outcomes including greater variation in HLA, immunological differences, higher medical non-adherence, socio-economic barriers and pharmacokinetic differences of the immunosuppressive agents including tacrolimus.(7, 8)
Tacrolimus has a narrow therapeutic index (9–13) with wide interindividual variability in pharmacokinetics resulting in unpredictable blood concentrations.(14–16) This necessitates therapeutic drug monitoring to avoid subtherapeutic and supratherapeutic concentrations, which places the recipient at risk of rejection and toxicity, respectively.(17, 18) There is a significant difference in tacrolimus pharmacokinetics by race where AAs have 20–50% lower bioavailability, higher clearance and lower blood concentrations as compared to Caucasians.(19–23) To achieve target tacrolimus trough concentrations some AA require ~1.5 to 2 times higher doses than Caucasians.(24–29) However, not all AA will require a higher dose and these individuals may have nonfunctional genetic variants that lead to reduced metabolic capacity similar to Caucasians.
Tacrolimus is metabolized by hepatic and intestinal CYP3A4 and CYP3A5 enzymes.(14, 30) CYP3A5 is a more efficient catalyst of tacrolimus metabolism as compared to CYP3A4.(31) Tacrolimus is also a substrate of P-glycoprotein which is an efflux transporter expressed on enterocytes.(32, 33) Genetic variants associated with CYP3A5, CYP3A4, P450 (cytochrome) oxidoreductase (POR) and P-glycoprotein have been studied for their influence on tacrolimus clearance, although only CYP3A5 variants have demonstrated major clinical relevance.(23, 30, 34–44)
CYP3A5*3 is an intronic variant which generates a cryptic splice site resulting in a nonfunctional enzyme.(45–47) The presence of the CYP3A5*3 allele is associated with lower oral tacrolimus clearance (Cl/F) whereas the CYP3A5*1 allele is associated with high Cl/F (CYP3A5*1/*1 individuals ~1 L/hr/kg, CYP3A5*1/*3 ~ 0.8 L/hr/kg vs CYP3A5*3/*3 ~ 0.5 L/hr/kg).(14, 48, 49) Therefore, the dose requirements for CYP3A5*1/*1 or *1/*3 carriers are about 1.5–1.7 fold higher than CYP3A5*3/*3 carriers. (23, 40, 42, 50, 51) These genotypes are also associated with delays in achieving therapeutic concentrations.(43, 52)
CYP3A5*6 is a missense mutation that codes for a splicing defect, deleting exon 7 resulting in absence of CYP3A5 enzyme and activity.(47) CYP3A5*7 is a frame shift mutation due to an insertion within codon 346 and termination of protein synthesis.(46, 47, 53) Few studies have evaluated the association between CYP3A5*6 and *7 alleles and tacrolimus pharmacokinetics. (54–59) Brazilian transplant recipients carrying two CYP3A5 variant alleles (*3, *6 or *7) had higher tacrolimus trough concentrations compared to those who did not (p<0.0001).(57) However no clearance models with dosing algorithms have been developed to account for these common AA variants. Algorithms that do not account for these alleles may incorrectly approximate clearance and dosing requirements. The objective of this study was to develop an AA dosing model which comprehensively includes the common AA specific CYP3A5 variants.
Methods
Subjects
The data for this analysis was obtained from our multicenter observational trial (DEKAF Genomics, clinicaltrials.gov NCT00270712). The study was approved by Institutional Review Board and an informed consent was obtained from each subject prior to the study. African American kidney transplant recipients (n=354) ≥18 years who received tacrolimus maintenance immunosuppression from 6 centers in the United States and Canada were studied. Tacrolimus was administered orally once or twice daily. The initial dose was based on weight and doses adjusted to achieve each institution’s target trough concentrations. Trough blood concentrations (n=6037) were measured at each center and, in general, concentrations of 8–12 ng/mL were targeted for the first 3 months and 6–10 ng/mL for 3–6 months posttransplant. A median (range) of 18 (1–24) concentrations were obtained from each subject in the first 6 months posttransplant, and if available, concentrations were obtained twice each week for the first 2 months, and then twice in each month up to 6 months. The concentrations were quantified in each center by their standard analysis technique. The majority (92.9%) of concentrations were measured by liquid chromatography with mass spectroscopy in CLIA certified labs.
Genotypes
Genotyping was performed on recipient DNA isolated from peripheral blood. Single nucleotide polymorphisms CYP3A5*3(rs776746, g.6986A>G), CYP3A5*6 (rs10264272, g.14690 G>A) and CYP3A5*7 (rs41303343, g.27131-27132insT) were found to be significant in our previous GWAS analysis and therefore were chosen for this analysis.(60) In addition POR*28 (rs1057868, g.1058C>T) and CYP3A4*22 (rs35599367, g.15389 C>T) were also evaluated based on data from our previous analyses in a mixed race populations suggesting their importance.(61) Genotypes were determined using a custom exome-plus Affymetrix TxArray SNP chip described elsewhere. (62) The allele frequency of CYP3A5*3 (G allele), CYP3A5*6 (T allele), CYP3A5*7 (A allele), POR*28 (T allele) and CYP3A4*22 (A allele) were 29.0%, 12.3%, 8.8%, 19.0%, 2.4%, respectively.
Population modeling of trough concentrations
The 354 subjects were randomly divided into a development (60%) and a validation cohort (40%). The data from the development cohort (212 subjects with 3704 troughs) was used to build the apparent oral tacrolimus clearance (Cl/F) model and subsequent dosing equation. The validation cohort (142 subjects with 2333 troughs) was used to evaluate the developed model. To assess differences in demographics, clinical and genotype distributions a two-sample t-test (for continuous factors) and sample proportion test (for categorical factors) were performed using R software package. Nonlinear mixed effect modeling was used to develop the Cl/F model with NONMEM (version 7.2, ICON development solutions, Maryland, USA) software on a Visual Fortran compiler (90/95). The NONMEM execution, model diagnostics, covariate testing and bootstrapping were conducted with Perl Speaks NONMEM (PsN) toolkit and the Xpose4 package through Pirana workbench (version 2.7.2). R studio 3.0.3 was used for predictive performance checks. A steady-state infusion model was used to develop the pharmacokinetic base model using $PRED library in NONMEM. In absence of intravenous data for the tacrolimus, it was not possible to calculate oral bioavailability. Therefore tacrolimus apparent oral clearance (Cl/F), which is the ratio of total clearance (Cl) to the bioavailability (F), was used to regress steady state tacrolimus concentrations (Css,av) to the administered dose. Cl/F was related to tacrolimus trough concentrations by the following equation:
| (1) |
Due to the longer half-life of tacrolimus, steady-state trough concentrations were assumed to be approximately equivalent to average steady-state concentrations (Css). Actual apparent oral clearance may vary from this approximated Cl/F; however, this difference is negligible for drugs with longer half-lives, such as tacrolimus.
An exponential error model was used to explain the inter-individual variability in Cl/F as shown in the following equation:
| (2) |
where, η(1) is the estimate of deviation of individual Cl/F from TVCl/F. η(1) is assumed to be normally distributed mean of zero and variance ω2.
An additive error model adequately explained the residual unexplained variability.
| (3) |
where Cij is the jth observed tacrolimus trough concentrations in the ith individual, Cpred,ij is the jth predicted tacrolimus trough concentrations in the ith individual and εij is the residual unexplained variability and where ε ~N(0,σ2). FOCE interaction was used as the NONMEM estimation method.
Covariate analysis
Clinical factors and genotypes were tested for their influence on tacrolimus TVCl/F. Covariates tested were recipient and donor age, gender, days posttransplant, steroid use (prednisone, methylprednisolone) at each trough measurement, calcium channel blocker use at each trough measurement, ACE-inhibitor use at each trough measurement, CMV sero-status at time of transplant (antibody positive or negative), anti CMV viral drug (as prophylaxis) use at each trough measurement, diabetes diagnosis at time of transplant, glomerular filtration rate calculated by the Modification of Diet in Renal Disease equation as a time varying covariate, body mass index (kg/m2), actual body weight (kg) at baseline (time of transplant), and actual body weight (kg) at time of trough measurement as a time varying covariate. Alleles tested were CYP3A5*3, CYP3A5*6, CYP3A5*7, POR*28, and CYP3A4*22. Recipients who did not carry any CYP3A5*3, *6 or *7 alleles were designated as CYP3A5*1/*1 genotype and those who carried one CYP3A5*3, *6 or *7 allele were designated CYP3A5*1/*3, *1/*6 or *1/*7 genotype, respectively. Recipients were classified into one of nine CYP3A5 genotypes (CYP3A5 *3/*3, *3/*6, *3/*7, *6/*7, *6/*6, *1*3, *1*6, and *1*7 and *1/*1). Recipients were also classified based on POR (POR*1/*1, *1*28 or *28/*28) and CYP3A4 (CYP3A4*1/*1 or *1/*22) genotype. No subjects had the CYP3A5*7/*7 or CYP3A4*22/*22 genotype. Recipient age, donor age and days posttransplant were tested both as continuous (using linear, exponential and power models) and categorical covariates. All other clinical factors were tested as categorical covariates. A strategy of forward inclusion and backward elimination was tested for inclusion of the covariates. In NONMEM, minimization of −2 log likelihood is used as a model statistic and is given by the objective function value (OFV); measure of goodness of fit similar to sum of squares. The significance of inclusion of each covariate was tested based on likelihood ratio test that follows a chi square distribution. A lower OFV is considered to be a better fit and a decrease in the OFV by 3.8 (p<0.05) or more was considered significant for forward inclusion and an increase in OFV by 6.6 (p < 0.01) was chosen for backward elimination.
Model evaluation
To evaluate the precision of the parameter estimates, a non-parametric bootstrap approach was performed using the development cohort. The method used random sampling with replacement to generate 1000 bootstrapped datasets using PsN toolkit. The final model developed with NONMEM was fit to each of the bootstrapped datasets and the parameters were obtained with their 5th and 95th prediction intervals. The model was also validated by using subjects in the validation cohort. The final model parameters were fixed in NONMEM (the estimation method was set to MAXEVAL=0 with the POSTHOC option) and were used to predict trough concentrations in validation cohort subjects. Population predicted trough concentrations (PRED) were obtained for each observed concentration (the dependent variable, DV) given their actual administered dose, the time after transplant, significant clinical covariates and genotypes (those identified from the development model). Median prediction error (MPE) and median percentage prediction error (MPPE) was then used to calculate the bias in model predictions and median absolute prediction error (MAPE) was used to calculate the imprecision. The following equations were used:
Results
Characteristics of the subjects in the development and validation cohorts are shown in Table 1. The median (range) daily dose and trough concentrations did not differ between the cohorts. The median tacrolimus concentrations were low during the first week post transplant and slowly increased over time until month 2 (2.8, 5.3, 6, 6.3, 6.9, 6.9, 7, 7.1, ng/mL in weeks 1–8 and 7.4, 7.2, 6.9 and 7 ng/mL in months 3–6, respectively). Tacrolimus TVCl/F was 54.6 L/hr and was significantly influenced by recipient age, steroid and antiviral coadministration, days posttransplant and CYP3A5*1/*3, *3/*3, *1/*6, *1/*7, *3/*6, *6/*6, *6/*7 and *3/*7 genotypes. All other tested covariates were not significant. The effect of genotypes and clinical covariates on tacrolimus TVCl/F and final parameter estimates in the model development cohort and in the bootstrap analysis are shown in Table 2. The inter-individual variability in TVCl/F after inclusion of covariates was 48.6%. Days posttransplant was the most important covariate where TVCl/F was 33% higher in the first 9 days posttransplant compared to after 9 days. Days post-transplant was first tested as continuous covariate however the model failed to converge and hence modeled as a categorical covariate. The plot of dose normalized trough concentrations over time showed a general increase in concentrations early posttransplant (up to day 9) and stabilized later. Several cut points were tested to understand the effect of time. There was also a break point in Cl/F at day 9 similar to that observed for concentrations. Addition of a third ordered category for days post transplant was not significant, hence only categorized as a bivariate. Tacrolimus TVCl/F increased by 23% with concomitant steroid use and reduced by 8% with concomitant antiviral use. Tacrolimus TVCl/F was 24% greater in subjects under the age of 34 years vs older subjects. Similar to days post-transplant, age as a continuous covariate, had problems with model convergence giving unrealistic parameter estimates. Hence age was categorized based on clinical definition of young (18–34 years), middle age (35–64 years) and older age (>64 years). In the current study, only 6% of AA patients were older than 64 years, and therefore we were unable to test the effect of the older age group and therefore was combined with age group 35–64 years.
Table 1.
Patient demographics
| All subjects | Development Cohort subjects | Validation Cohort subjects | P-valuea | |
|---|---|---|---|---|
|
| ||||
| No. of subjects | 354 | 212 | 142 | |
|
| ||||
| No. of male subjects (%) | 227(64) | 140(63) | 87(61) | 0.35 |
|
| ||||
| Daily dose (mg)b | 8(0.50–36) | 8(0.5–36) | 8(1–30) | 0.17 |
|
| ||||
| No. of troughs | 6037 | 3704 | 2333 | 0.09 |
|
| ||||
| Tacrolimus trough (ng/mL)b | 6.50(0.10–65.60) | 6.50 (0.10–65.60) | 6.40(0.70–50.00) | 0.34 |
|
| ||||
| Weight at baseline (kg)b | 85(42–140) | 85(42–140) | 83(47–137) | 0.34 |
|
| ||||
| GFR by MDRD mL/min/1.73m2 b,d | 55.89(6.18–168.28) | 55.88(6.18–168.28) | 55.24(14.25–122.71) | 0.08 |
|
| ||||
| No recipients in age category (%) | ||||
| 18–34 years | 66 (19) | 36 (17) | 30 (21) | 0.32 |
| 35–64 years | 268 (76) | 163(77) | 105 (74) | 0.52 |
| >64 years | 20 (6) | 13 (6) | 7 (5) | 0.63 |
|
| ||||
| Age at transplantb | 48(20–73) | 47 (20–73) | 49 (21–72) | 0.57 |
|
| ||||
| No. receiving dialysis at time of transplant (%) | 56(16) | 34(16) | 22(15) | 0.50 |
|
| ||||
| No. with diabetes at transplant (%) | 129(36) | 79(37) | 50(35) | 0.69 |
|
| ||||
| No. of troughs with calcium channel blocker (%) | 2944(49) | 1838(50) | 1106(53) | 0.01 |
|
| ||||
| No. of troughs with ACE inhibitor (%) | 905(15) | 522(14) | 383(16) | 0.01 |
|
| ||||
| No. of troughs with antiviral drug (%) | 3441(57) | 2128(57) | 1313(56) | 0.001 |
|
| ||||
| No. of troughs with steroid (%) | 3283(54) | 1941(52) | 1342(58) | 0.46 |
|
| ||||
| Simultaneous pancreas and kidney transplant (%) | 16(5) | 11(5) | 5(4) | 0.64 |
|
| ||||
| No. with living donor (%) | 172(31) | 108(30) | 64(31) | 0.27 |
|
| ||||
| No. with prior transplant (%) | 34(10) | 22(10) | 12(8) | 0.54 |
|
| ||||
| Primary cause of kidney disease (%) | ||||
| Diabetes | 94(27) | 58(27) | 36(25) | 0.67 |
| Glomerular nephritis | 50(14) | 28(13) | 22(15) | 0.54 |
| Hypertension | 148(42) | 93(44) | 55(39) | 0.34 |
| Polycystic kidney disease | 11(3) | 4(2) | 7(5) | 0.1 |
| Other | 44(12) | 26(12) | 18(13) | 0.91 |
| Unknown | 7(2) | 3(1) | 4(3) | 0.35 |
|
| ||||
| No. of individuals with genotype (%) | ||||
| CYP3A5*1/*3 | 96 (27) | 65 (31) | 31 (22) | 0.07 |
| CYP3A5*3/*3 | 34 (10) | 20 (9) | 14 (10) | 0.89 |
| CYP3A5*1/*7 | 36 (10) | 14 (7) | 22 (15) | 0.006 |
| CYP3A5*7/*7 | 0 | 0 | 0 | |
| CYP3A5*1/*6 | 47 (13) | 30 (14) | 17 (12) | 0.55 |
| CYP3A5*6/*6 | 4 (1) | 1 (0.5) | 3 (2) | 0.15 |
| CYP3A5*3/*6 | 21() | 15 (7) | 6 (4) | 0.26 |
| CYP3A5*3/*7 | 15 (4) | 8 (4) | 7 (5) | 0.59 |
| CYP3A5*6/*7 | 11 (3) | 5 (2) | 6 (4) | 0.32 |
| CYP3A5*1*1 | 80 (23) | 49 (23) | 31 (21) | 0.77 |
| CYP Not determinedc | 10 | 5 | 5 | |
|
| ||||
| POR*1/*1 | 151 (43) | 91 (43) | 60 (42) | 0.90 |
| POR*1/*28 | 86 (25) | 55 (26) | 31 (22) | 0.37 |
| POR*28/*28 | 25 (7) | 15 (7) | 10 (7) | 0.99 |
|
| ||||
| CYP3A4*1/*1 | 229 (65) | 140 (66) | 89 (63) | 0.52 |
| CYP3A4*1/*22 | 17 (4) | 12 (6) | 5 (4) | 0.35 |
| CYP3A4*22/*22 | 0 | 0 | 0 | |
p-value is the comparison of model development and validation cohorts
data are median (range)
These individuals did not have one or more of the CYP3A5 genotypes available and were excluded from the all analyses
GFR is glomerular filtration rate calculated by Modification of Diet in Renal Disease (MDRD) equation
Table 2.
The effect of genotypes and clinical covariates on tacrolimus clearance (Cl/F) and final parameters estimates
| Parameter/Covariate | Model development cohort. Estimate (%RSEa) of the effect on TVCl/F | Bootstrap analysis. Median (95% confidence interval) |
|---|---|---|
| Typical Value of Cl/F (TVCl/F) in L/hr | 54.60 (10.0%) | 54.48 (44.51–66.63) |
| Two loss of function alleles (CYP3A5*3/*3 or *3/*7 or CYP3A5*3/*6 or *6/*7) | 0.53 (10.9%) | 0.53 (0.43–0.66) |
| One loss of function alleles (CYP3A5*1/*3 or CYP3A5*1/*6 or CYP3A5*1/*7) | 0.85 (9.7%) | 0.85 (0.70–1.04) |
| Less than day 9 posttransplant | 1.33 (4.2%) | 1.33 (1.23–1.45) |
| Steroid drug use | 1.23 (6.9%) | 1.24 (1.07–1.42) |
| Antiviral drug use | 0.92 (2.9%) | 0.91 (0.87–0.97) |
| Recipient age (18–34 yrs) | 1.24 (7.8%) | 1.24 (1.07–1.47) |
| Between subject variabilityb | 0.21 (18.1%) [CV%=48.6%] |
0.21 (0.14–0.28) [CV%= 46.7% (38.76–56.84%] |
| Residual unexplained variability in trough (ng/mL) | 2.76 (7.5%) | 2.75 (2.55–2.96) ng/mL |
RSE is relative standard error
0.21 represents the estimate of the variance of individual η(1). CV% is the coefficient of variance and represents interindividual variability in the population. CV% = sqrt {[exp (variance)]−1}
In subjects with CYP3A5*1/*3, *1/*6 or *1/*7 genotypes the tacrolimus TVCl/F decreased by 16.2%, 8.2%, and 24.1%, respectively, compared to CYP3A5*1/*1 genotype. For CYP3A5*3/*3, *3/*6, *3/*7 or *6/*7 the TVCl/F declined by 51%, 36.5%, 54.5% and 44.2%, respectively, relative to CYP3A5*1/*1. Only one subject had *6/*6 genotype in the development cohort and therefore *6/*6 was not evaluable independently. To build a parsimonious model and to improve the power, we combined the genotypes with similar effect sizes and overlapping confidence intervals on tacrolimus TVCl/F and re-ran the model. The tacrolimus TVCl/F decreased by 47% in subjects carrying two loss of function alleles (CYP3A5*3/*3 or *3/*6 or *3/*7 or *6/*7, or *6/*6) and by 15% in subjects carrying one loss of function allele (CYP3A5*1/*3, *1/*6 or *1/*7) compared to the CYP3A5*1/*1. The POR*28 and CYP3A4*22 genotypes did not influence TVCl/F.
To examine the goodness of fit, diagnostic plots were assessed during model development. Histograms of η(1)s and Cl/F satisfied conditions of normal and log-normal distribution, respectively. Figures 1A and 1B shows the plots of observed concentration vs population predicted concentration, observed concentrations vs individual predicted concentrations. Figures 1C and 1D show the conditional weighted residuals (CWRES) vs independent variables, population predicted concentration and time. Although the model under-predicted slightly at higher concentrations, most of the data are evenly distributed across the line of unity. Also the CWRES do not show any specific trends of model misspecification. Thus the model adequately explains the observed data. The final tacrolimus TVCl/F model with clinical factors and genotypes is as follows:
Figure 1. Goodness of fit plots for the final tacrolimus model.
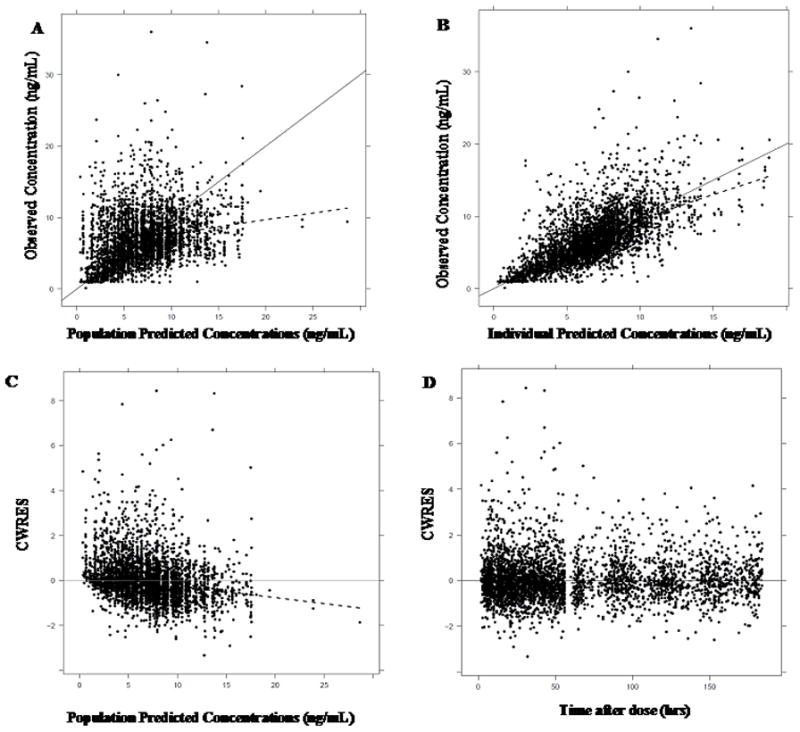
(A) observed concentrations (ng/mL) vs population predicted concentrations (ng/mL) and (B) observed conc. (ng/mL) vs individual predicted concentrations (ng/mL). The black dots represent the observed tacrolimus trough concentrations, the solid line represents the line of unity and the dashed line represents the loess smooth.
(C) conditional weighted residuals (CWRES) vs population predicted concentrations (ng/mL) and (D) CWRES vs time after dose (hrs). The dots represent the observed tacrolimus trough concentrations, the solid line is the line at y=0 and the dashed line represents the loess smooth.
Tacrolimus TVCl/F (L/hr)=54.6 L/hr x (1.33, if days less than 9 posttransplant) x [(0.53, if CYP3A5*3/*3 or CYP3A5*3/*7 or CYP3A5*3/*6 or CYP3A5*6/*7or CYP3A5*6/*6)] x (0.85, if CYP3A5*1/*3 or CYP3A5*1/*6 or CYP3A5*1/*7) x (1.23, if receiving a steroid) x (0.92, if receiving an anti CMV viral drug) x (1.24, if recipient age 18–34 years)
Using the TVCl/F calculated using the model above and a desired target tacrolimus trough concentration; the daily tacrolimus dose can be calculated by: Daily dose (mg/day) = [TVCl/F x target tacrolimus trough concentration (ng/ml) x 24hrs]/1000
Model Evaluation Using Bootstrap
Table 2 shows the median of the parameter estimates and their 95% prediction intervals obtained from 1000 bootstrap runs. Out of 1000 runs, 991 runs minimized successfully and the estimates from each bootstrap run were used to calculate the median and 95% interval. Parameter estimates for fixed and random effects obtained from the original dataset fell within the prediction interval of the estimates obtained from bootstrap therefore indicating that the model is robust and reproducible.
Model evaluation using the validation cohort
Table 3 shows the prediction performance of the tacrolimus TVCl/F model. The median prediction error with 95% CI was 0.48 (0.31–0.65) ng/mL and median percentage prediction error was 9.45% (6.44–12.45). Therefore, the model over-predicted the trough concentrations relative to the observed concentrations. Median absolute prediction error was 2.32 (2.21–2.44) ng/ml.
Table 3.
Predictive performance of the tacrolimus clearance model
| Predictive performance measure | Estimate |
|---|---|
| Median prediction error (MPE, 95% CI) | 0.48(0.31–0.65) |
| Median percentage prediction error (MPPE, 95% CI) | 9.45(6.44–12.45) |
| Median absolute prediction error (MAPE, 95% CI) | 2.32(2.21–2.44) |
Discussion
African Americans have poorer outcomes after transplantation and a possible contributory factor is high pharmacokinetic variability in immunosuppression leading to multiple dose changes and longer periods of time out of the therapeutic range.(3, 28) On average AA require higher tacrolimus doses than Caucasians to achieve the same target blood concentration and most centers administer higher initial doses to AAs. However, not all individuals require higher doses and therefore some may have elevated concentrations which lead to temporary cessation of therapy and/or dose reductions. Whereas others may require even higher doses of tacrolimus to avoid insufficient blood concentrations. Most tacrolimus pharmacogenomic studies in AAs and Caucasians have classified CYP3A5 metabolism based on the presence or absence of the nonfunctional CYP3A5*3 allele. The CYP3A5*3 allele frequency has a minor allele frequency of 18–35% in AA and 88–95% in Caucasians.(34, 47, 53, 65–67) However, AAs also carry CYP3A5*6 and/or *7 alleles which also encode for low activity or nonfunctional enzyme which have not been accounted for in most studies. CYP3A5*6 and *7 are common in AAs with a minor allele frequency of 16–18% and 10–12%, respectively, but absent in Caucasians.(47, 65, 66, 68, 69) We found that AAs who carry two nonfunctional alleles (*3, *6 or *7) have a tacrolimus clearance similar to Caucasians whereas those who carry no nonfunctional alleles have high clearance. Therefore, AAs have a broad range of CYP3A5 metabolism phenotypes. To develop personalized strategies to reduce pharmacokinetic variability, we evaluated the effect of these variants on tacrolimus clearance and developed the first genotype-guided dosing model for AAs.
We found that tacrolimus TVCl/F in AAs was significantly influenced by CYP3A5*1, *3, *6 and *7 alleles, days posttransplant, steroid and antiviral drug coadministration and age. The TVCl/F was 54.6 L/hr and higher than reported in non-AA studies (~22–40 L/hr) (14, 70–73) which is consistent with AAs being more likely to carry a *1 expresser allele than Caucasians. The CYP3A5*3, *6 and *7 alleles were each associated with a reduction in tacrolimus clearance. About 50% of our subjects carried one nonfunctional allele (CYP3A5*3/*1, *6/*1 or *7/*1) which decreased tacrolimus TVCl/F by 15%. Individually, the CYP3A5*1/*3, *1/*6 and *1/*7 genotypes, decreased TVCl/F by 16.2%, 8.2%, and 24.1%, respectively. In addition, about 24% of our subjects carried two nonfunctional alleles – primarily CYP3A5*3/*3, *3/*6 and *3/*7 and *6/*6. The effect of two variant alleles was large resulting in a decrease in tacrolimus TVCl/F by 47%. We did not observe any subject with more than two *3, *6 or *7 alleles. Based on our data and haplotype analyses by others the probability of this occurring is very low (<0.5%).(74, 75)
The CYP3A5*6 allele is thought to encode for nonfunctional enzyme; however, there is some uncertainty about its functionality and it may express low levels of enzyme. In our study tacrolimus TVCl/F was 24% lower in CYP3A5 *1/*7 carriers but only 8.2% lower in *1/*6 carriers relative to the *1/*1 carriers, supporting that *6 may express low levels of enzyme. Others found no difference in tacrolimus concentrations between CYP3A5*1/*1 and *1/*6 genotypes groups although the number of subjects was small.(56) In another study, CYP3A5*1/*1, *1/*3 or *1/*6 carriers had lower tacrolimus troughs than CYP3A5*3/*3 carriers but no difference in area under the curve although only one individual carried the CYP3A5*1/*6 genotype.(54) The influence of CYP3A5*6 and CYP3A5*7 alleles has been studied towards other CYP3A5 substrates and the effect may be substrate specific therefore our results may not be generalizable to other drugs. (75–81)
Day posttransplant was a significant covariate towards tacrolimus where TVCl/F is 33% higher in the first nine days posttransplant compared to after day 9 which is consistent with other studies.(14, 23, 70, 71, 82, 83) The higher TVCl/F may be due to early physiological changes such as fluid status, hepatic and kidney function and/or decreased bioavailability from dietary changes or concomitant medications. Concomitant steroid use was associated with a 23% higher tacrolimus TVCl/F most likely because steroids induce CYP3A enzymes.(84–87) We also found that younger subjects (18–34 years) had a 24% higher tacrolimus TVCl/F compared to older subjects. While some studies have not observed a significant association between tacrolimus Cl/F and age we previously showed in 1967 kidney recipients that age (18–34 vs 35–64 vs 65–84 years) had a highly significant effect on tacrolimus troughs.(14, 26, 70, 73, 88–90) We found that the co-administration of antivirals reduced tacrolimus TVCl/F but only by 8%. The mechanism of this effect is unknown. We did not find that calcium channel blockers were associated with TVCl/F. This is likely because amlodipine is the preferred agent at our centers and has a lower potential for an interaction than other calcium channel blockers.(91–93) Weight was not significant towards TVCl/F. Other studies have also not found weight to be significant.(94, 95)
The POR*28 and CYP3A4*22 variants have been previously associated with tacrolimus concentrations but we were unable to find an association in our AA population.(35–38, 42, 58, 96) One or two POR*28 alleles were present in ~30% of subjects whereas the CYP3A4*22 allele was infrequent (<5%). Our ability to detect an association with CYP3A4*22 was therefore limited.
A prospective trial, in a primarily Caucasian kidney transplant recipients, evaluated the effect of genotype guided tacrolimus dosing vs traditional weight based dosing.(97) The study tested an initial dose of 0.3 mg/kg/day PO in CYP3A5 expressors (CYP3A5*1) and 0.15 mg/kg/day PO for non-expressors (CYP3A5*3). The genotype guided group had a higher proportion of patients with tacrolimus troughs within the target, fewer dose modifications, and more rapid achievement of the target concentration. Although genotype guided dosing did not reduce major clinical outcomes it was an important study as it showed the value of genetic targeting in controlling systemic exposure. Data such as ours shows that race specific variants and clinical factors is necessary in future trials and may improve achievement of major clinical endpoints. The Clinical Pharmacogenetics Implementation Consortium recently published guidelines for initial tacrolimus dosing. The guidelines recommend increasing the starting dose by 1.5–2 times in extensive metabolizers (CYP3A5*1/*1) and intermediate metabolizers (CYP3A5*1/*3, *1/*6, *1/*7), and standard dose in poor metabolizers (CYP3A5*3/*3, *6/*6, *7/*7, *3/*6, *3/*7 and *6/*7).(98) Our data supports these recommendations where *6 and *7 allele carriers require lower doses.
One of the limitations of our study is that albumin, hematocrit and antifungal agents status was not available and not tested in our model.(14) Our study used clinical trough concentrations that were obtained as part of clinical care and draw times were not supervised by our study personnel but instead overseen by the clinicians. Compliance was also assessed by the clinical site and not through the study protocol.
To our knowledge this is the first study in which the effect of CYP3A5 alleles (*1, *3, *6, *7) common in AAs have been collectively studied towards tacrolimus clearance. We identified one or more nonfunctional CYP3A5 alleles (*3, *6 or *7) in 74.5 % of our AA study population whereas 90–95% of Caucasians will carry one or more CYP3A5*3 alleles.(53) This is considerably higher than what has been previously presumed in the AA population. If the *6 or *7 alleles had not been genotyped, 27% of our subjects would have been inappropriately categorized as carrying two CYP3A5 *1 alleles, and 10% categorized as carrying one CYP3A5*1 allele thereby overestimating tacrolimus Cl/F by nearly 50% in some individuals. Our data are consistent with a recent African study where only ~43% of individuals were considered CYP3A5 expressers since most carried one or more CYP3A5*3, *6 or *7 nonfunctional alleles.(74)
This is the first study to develop and validate an AA specific genotype guided dosing model using variants common and relevant in the AA population. This study demonstrates the importance of race specific genotypes to determine drug clearance. Using dosing models which account for the genotypes and clinical factors may lead to precision dosing of tacrolimus.
Acknowledgments
This project was supported by grants (U19-AI070119 and U01-AI058013) from the National Institute of Allergy and Infectious Disease. We acknowledge the dedication of our coordinators and generous patients.
DeKAF Genomics Investigators
J. Michael Cecka, M.D. UCLA Immunogenetics Center, Los Angeles, CA 90095, Email: mcecka@ucla.edu
Fernando G. Cosio, M.D. Division of Nephrology, Mayo Clinic, Rochester, MN 55905, Email: Cosio.Fernando@mayo.edu
Robert Gaston, M.D. University of Alabama, Division of Nephrology, Birmingham, AL 35294-0006, Email: rgaston@uab.edu
Sita Gourishankar M.D. Division of Nephrology and Immunology, University of Alberta, Edmonton, Alberta, Canada, Email: sitag@ualberta.ca
Lawrence Hunsicker, M.D. Nephrology Division, Iowa City, IA 52242-1082, Email: lawrence-hunsicker@uiowa.edu
Bertram Kasiske, M.D. Department of Medicine, Hennepin County Medical Center and the University of Minnesota, Minneapolis, MN 55415, Email: kasis001@umn.edu
David Rush, M.D. Health Sciences Center, Winnipeg MB, Canada, Email: drush@exchange.hsc.mb.ca
Footnotes
Conflict of Interest: None of the authors have any conflicts of interest
References
- 1.McCullough KP, Keith DS, Meyer KH, Stock PG, Brayman KL, Leichtman AB. Kidney and pancreas transplantation in the United States, 1998–2007: access for patients with diabetes and end-stage renal disease. Am J Transplant. 2009;9:894–906. doi: 10.1111/j.1600-6143.2009.02566.x. [DOI] [PubMed] [Google Scholar]
- 2.Matas AJ, Smith JM, Skeans MA, Thompson B, Gustafson SK, Schnitzler MA, et al. OPTN/SRTR 2012 Annual Data Report: kidney. Am J Transplant. 2014;14(Suppl 1):11–44. doi: 10.1111/ajt.12579. [DOI] [PubMed] [Google Scholar]
- 3.Fan PY, Ashby VB, Fuller DS, Boulware LE, Kao A, Norman SP, et al. Access and outcomes among minority transplant patients, 1999–2008, with a focus on determinants of kidney graft survival. Am J Transplant. 2010;10:1090–107. doi: 10.1111/j.1600-6143.2009.03009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gondos A, Dohler B, Brenner H, Opelz G. Kidney graft survival in Europe and the United States: strikingly different long-term outcomes. Transplantation. 2013;95:267–74. doi: 10.1097/TP.0b013e3182708ea8. [DOI] [PubMed] [Google Scholar]
- 5.Press R, Carrasquillo O, Nickolas T, Radhakrishnan J, Shea S, Barr RG. Race/ethnicity, poverty status, and renal transplant outcomes. Transplantation. 2005;80:917–24. doi: 10.1097/01.tp.0000173379.53347.31. [DOI] [PubMed] [Google Scholar]
- 6.Young CJ, Gaston RS. Renal transplantation in black Americans. N Engl J Med. 2000;343:1545–52. doi: 10.1056/NEJM200011233432107. [DOI] [PubMed] [Google Scholar]
- 7.Eckhoff DE, Young CJ, Gaston RS, Fineman SW, Deierhoi MH, Foushee MT, et al. Racial disparities in renal allograft survival: a public health issue? J Am Coll Surg. 2007;204:894–902. doi: 10.1016/j.jamcollsurg.2007.01.024. discussion -3. [DOI] [PubMed] [Google Scholar]
- 8.Martins D, Tareen N, Norris KC. The epidemiology of end-stage renal disease among African Americans. Am J Med Sci. 2002;323:65–71. doi: 10.1097/00000441-200202000-00002. [DOI] [PubMed] [Google Scholar]
- 9.de Jonge H, Naesens M, Kuypers DR. New insights into the pharmacokinetics and pharmacodynamics of the calcineurin inhibitors and mycophenolic acid: possible consequences for therapeutic drug monitoring in solid organ transplantation. Ther Drug Monit. 2009;31:416–35. doi: 10.1097/FTD.0b013e3181aa36cd. [DOI] [PubMed] [Google Scholar]
- 10.Kahan BD, Keown P, Levy GA, Johnston A. Therapeutic drug monitoring of immunosuppressant drugs in clinical practice. Clin Ther. 2002;24:330–50. doi: 10.1016/s0149-2918(02)85038-x. discussion 29. [DOI] [PubMed] [Google Scholar]
- 11.Kershner RP, Fitzsimmons WE. Relationship of FK506 whole blood concentrations and efficacy and toxicity after liver and kidney transplantation. Transplantation. 1996;62:920–6. doi: 10.1097/00007890-199610150-00009. [DOI] [PubMed] [Google Scholar]
- 12.McMaster P, Mirza DF, Ismail T, Vennarecci G, Patapis P, Mayer AD. Therapeutic drug monitoring of tacrolimus in clinical transplantation. Ther Drug Monit. 1995;17:602–5. doi: 10.1097/00007691-199512000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Monchaud C, Marquet P. Pharmacokinetic optimization of immunosuppressive therapy in thoracic transplantation: part I. Clin Pharmacokinet. 2009;48:419–62. doi: 10.2165/11317230-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staatz CE, Tett SE. Clinical pharmacokinetics and pharmacodynamics of tacrolimus in solid organ transplantation. Clin Pharmacokinet. 2004;43:623–53. doi: 10.2165/00003088-200443100-00001. [DOI] [PubMed] [Google Scholar]
- 15.Shaw LM, Holt DW, Keown P, Venkataramanan R, Yatscoff RW. Current opinions on therapeutic drug monitoring of immunosuppressive drugs. Clin Ther. 1999;21:1632–52. doi: 10.1016/S0149-2918(99)80044-7. discussion 1. [DOI] [PubMed] [Google Scholar]
- 16.Venkataramanan R, Swaminathan A, Prasad T, Jain A, Zuckerman S, Warty V, et al. Clinical pharmacokinetics of tacrolimus. Clin Pharmacokinet. 1995;29:404–30. doi: 10.2165/00003088-199529060-00003. [DOI] [PubMed] [Google Scholar]
- 17.Staatz C, Taylor P, Tett S. Low tacrolimus concentrations and increased risk of early acute rejection in adult renal transplantation. Nephrol Dial Transplant. 2001;16:1905–9. doi: 10.1093/ndt/16.9.1905. [DOI] [PubMed] [Google Scholar]
- 18.Undre NA, van Hooff J, Christiaans M, Vanrenterghem Y, Donck J, Heeman U, et al. Low systemic exposure to tacrolimus correlates with acute rejection. Transplant Proc. 1999;31:296–8. doi: 10.1016/s0041-1345(98)01633-9. [DOI] [PubMed] [Google Scholar]
- 19.Dirks NL, Huth B, Yates CR, Meibohm B. Pharmacokinetics of immunosuppressants: a perspective on ethnic differences. Int J Clin Pharmacol Ther. 2004;42:701–18. doi: 10.5414/cpp42701. [DOI] [PubMed] [Google Scholar]
- 20.Fitzsimmons WE, Bekersky I, Dressler D, Raye K, Hodosh E, Mekki Q. Demographic considerations in tacrolimus pharmacokinetics. Transplant Proc. 1998;30:1359–64. doi: 10.1016/s0041-1345(98)00275-9. [DOI] [PubMed] [Google Scholar]
- 21.Mancinelli LM, Frassetto L, Floren LC, Dressler D, Carrier S, Bekersky I, et al. The pharmacokinetics and metabolic disposition of tacrolimus: a comparison across ethnic groups. Clin Pharmacol Ther. 2001;69:24–31. doi: 10.1067/mcp.2001.113183. [DOI] [PubMed] [Google Scholar]
- 22.Hricik DE, Anton HA, Knauss TC, Rodriguez V, Seaman D, Siegel C, et al. Outcomes of African American kidney transplant recipients treated with sirolimus, tacrolimus, and corticosteroids. Transplantation. 2002;74:189–93. doi: 10.1097/00007890-200207270-00008. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson PA, Oetting WS, Brearley AM, Leduc R, Guan W, Schladt D, et al. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation. 2011;91:300–8. doi: 10.1097/TP.0b013e318200e991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews PA, Sen M, Chang RW. Racial variation in dosage requirements of tacrolimus. Lancet. 1996;348:1446. doi: 10.1016/S0140-6736(04)70087-2. [DOI] [PubMed] [Google Scholar]
- 25.Beermann KJ, Ellis MJ, Sudan DL, Harris MT. Tacrolimus dose requirements in African-American and Caucasian kidney transplant recipients on mycophenolate and prednisone. Clin Transplant. 2014;28:762–7. doi: 10.1111/ctr.12376. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson PA, Schladt D, Oetting WS, Leduc R, Guan W, Matas AJ, et al. Lower calcineurin inhibitor doses in older compared to younger kidney transplant recipients yield similar troughs. Am J Transplant. 2012;12:3326–36. doi: 10.1111/j.1600-6143.2012.04232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laftavi MR, Pankewycz O, Patel S, Nader N, Kohli R, Feng L, et al. African American renal transplant recipients (RTR) require higher tacrolimus doses to achieve target levels compared to white RTR: does clotrimazole help? Transplant Proc. 2013;45:3498–501. doi: 10.1016/j.transproceed.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Narayanan M, Pankewycz O, El-Ghoroury M, Shihab F, Wiland A, McCague K, et al. Outcomes in African American kidney transplant patients receiving tacrolimus and mycophenolic acid immunosuppression. Transplantation. 2013;95:566–72. doi: 10.1097/TP.0b013e318277438f. [DOI] [PubMed] [Google Scholar]
- 29.Vadivel N, Garg A, Holt DW, Chang RW, MacPhee IA. Tacrolimus dose in black renal transplant recipients. Transplantation. 2007;83:997–9. doi: 10.1097/01.tp.0000259248.60448.8a. [DOI] [PubMed] [Google Scholar]
- 30.Barbarino JM, Staatz CE, Venkataramanan R, Klein TE, Altman RB. PharmGKB summary: cyclosporine and tacrolimus pathways. Pharmacogenet Genomics. 2013;23:563–85. doi: 10.1097/FPC.0b013e328364db84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamdem LK, Streit F, Zanger UM, Brockmoller J, Oellerich M, Armstrong VW, et al. Contribution of CYP3A5 to the in vitro hepatic clearance of tacrolimus. Clin Chem. 2005;51:1374–81. doi: 10.1373/clinchem.2005.050047. [DOI] [PubMed] [Google Scholar]
- 32.Hebert MF. Contributions of hepatic and intestinal metabolism and P-glycoprotein to cyclosporine and tacrolimus oral drug delivery. Adv Drug Deliv Rev. 1997;27:201–14. doi: 10.1016/s0169-409x(97)00043-4. [DOI] [PubMed] [Google Scholar]
- 33.Jeong H, Chiou WL. Role of P-glycoprotein in the hepatic metabolism of tacrolimus. Xenobiotica. 2006;36:1–13. doi: 10.1080/00498250500485115. [DOI] [PubMed] [Google Scholar]
- 34.Birdwell KA, Grady B, Choi L, Xu H, Bian A, Denny JC, et al. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet Genomics. 2012;22:32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Jonge H, Metalidis C, Naesens M, Lambrechts D, Kuypers DR. The P450 oxidoreductase *28 SNP is associated with low initial tacrolimus exposure and increased dose requirements in CYP3A5-expressing renal recipients. Pharmacogenomics. 2011;12:1281–91. doi: 10.2217/pgs.11.77. [DOI] [PubMed] [Google Scholar]
- 36.Elens L, Hesselink DA, Bouamar R, Budde K, de Fijter JW, De Meyer M, et al. Impact of POR*28 on the pharmacokinetics of tacrolimus and cyclosporine A in renal transplant patients. Ther Drug Monit. 2014;36:71–9. doi: 10.1097/FTD.0b013e31829da6dd. [DOI] [PubMed] [Google Scholar]
- 37.Elens L, van Gelder T, Hesselink DA, Haufroid V, van Schaik RH. CYP3A4*22: promising newly identified CYP3A4 variant allele for personalizing pharmacotherapy. Pharmacogenomics. 2013;14:47–62. doi: 10.2217/pgs.12.187. [DOI] [PubMed] [Google Scholar]
- 38.Elens L, van Schaik RH, Panin N, de Meyer M, Wallemacq P, Lison D, et al. Effect of a new functional CYP3A4 polymorphism on calcineurin inhibitors’ dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenomics. 2011;12:1383–96. doi: 10.2217/pgs.11.90. [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Bravo MA, Salcedo M, Fondevila C, Suarez F, Castellote J, Rufian S, et al. Impact of donor and recipient CYP3A5 and ABCB1 genetic polymorphisms on tacrolimus dosage requirements and rejection in Caucasian Spanish liver transplant patients. J Clin Pharmacol. 2013;53:1146–54. doi: 10.1002/jcph.154. [DOI] [PubMed] [Google Scholar]
- 40.Hesselink DA, Bouamar R, Elens L, van Schaik RH, van Gelder T. The role of pharmacogenetics in the disposition of and response to tacrolimus in solid organ transplantation. Clin Pharmacokinet. 2014;53:123–39. doi: 10.1007/s40262-013-0120-3. [DOI] [PubMed] [Google Scholar]
- 41.Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmoller J, Johne A, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuypers DR, de Loor H, Naesens M, Coopmans T, de Jonge H. Combined effects of CYP3A5*1, POR*28, and CYP3A4*22 single nucleotide polymorphisms on early concentration-controlled tacrolimus exposure in de-novo renal recipients. Pharmacogenet Genomics. 2014;24:597–606. doi: 10.1097/FPC.0000000000000095. [DOI] [PubMed] [Google Scholar]
- 43.Macphee IA, Fredericks S, Mohamed M, Moreton M, Carter ND, Johnston A, et al. Tacrolimus pharmacogenetics: the CYP3A5*1 allele predicts low dose-normalized tacrolimus blood concentrations in whites and South Asians. Transplantation. 2005;79:499–502. doi: 10.1097/01.tp.0000151766.73249.12. [DOI] [PubMed] [Google Scholar]
- 44.Pallet N, Jannot AS, El Bahri M, Etienne I, Buchler M, de Ligny BH, et al. Kidney transplant recipients carrying the CYP3A4*22 allelic variant have reduced tacrolimus clearance and often reach supratherapeutic tacrolimus concentrations. Am J Transplant. 2015;15:800–5. doi: 10.1111/ajt.13059. [DOI] [PubMed] [Google Scholar]
- 45.Busi F, Cresteil T. CYP3A5 mRNA degradation by nonsense-mediated mRNA decay. Mol Pharmacol. 2005;68:808–15. doi: 10.1124/mol.105.014225. [DOI] [PubMed] [Google Scholar]
- 46.Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–9. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 47.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 48.de Jonge H, de Loor H, Verbeke K, Vanrenterghem Y, Kuypers DR. Impact of CYP3A5 genotype on tacrolimus versus midazolam clearance in renal transplant recipients: new insights in CYP3A5-mediated drug metabolism. Pharmacogenomics. 2013;14:1467–80. doi: 10.2217/pgs.13.133. [DOI] [PubMed] [Google Scholar]
- 49.Passey C, Birnbaum AK, Brundage RC, Oetting WS, Israni AK, Jacobson PA. Dosing equation for tacrolimus using genetic variants and clinical factors. Br J Clin Pharmacol. 2011;72:948–57. doi: 10.1111/j.1365-2125.2011.04039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rojas L, Neumann I, Herrero MJ, Boso V, Reig J, Poveda JL, et al. Effect of CYP3A5*3 on kidney transplant recipients treated with tacrolimus: a systematic review and meta-analysis of observational studies. Pharmacogenomics J. 2014 doi: 10.1038/tpj.2014.38. [DOI] [PubMed] [Google Scholar]
- 51.Vannaprasaht S, Reungjui S, Supanya D, Sirivongs D, Pongskul C, Avihingsanon Y, et al. Personalized tacrolimus doses determined by CYP3A5 genotype for induction and maintenance phases of kidney transplantation. Clin Ther. 2013;35:1762–9. doi: 10.1016/j.clinthera.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 52.Roy JN, Barama A, Poirier C, Vinet B, Roger M. Cyp3A4, Cyp3A5, and MDR-1 genetic influences on tacrolimus pharmacokinetics in renal transplant recipients. Pharmacogenet Genomics. 2006;16:659–65. doi: 10.1097/01.fpc.0000220571.20961.dd. [DOI] [PubMed] [Google Scholar]
- 53.Lee SJ, Usmani KA, Chanas B, Ghanayem B, Xi T, Hodgson E, et al. Genetic findings and functional studies of human CYP3A5 single nucleotide polymorphisms in different ethnic groups. Pharmacogenetics. 2003;13:461–72. doi: 10.1097/00008571-200308000-00004. [DOI] [PubMed] [Google Scholar]
- 54.Haufroid V, Wallemacq P, VanKerckhove V, Elens L, De Meyer M, Eddour DC, et al. CYP3A5 and ABCB1 polymorphisms and tacrolimus pharmacokinetics in renal transplant candidates: guidelines from an experimental study. Am J Transplant. 2006;6:2706–13. doi: 10.1111/j.1600-6143.2006.01518.x. [DOI] [PubMed] [Google Scholar]
- 55.Hesselink DA, van Schaik RH, van der Heiden IP, van der Werf M, Gregoor PJ, Lindemans J, et al. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003;74:245–54. doi: 10.1016/S0009-9236(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 56.Lopez-Montenegro Soria MA, Kanter Berga J, Beltran Catalan S, Milara Paya J, Pallardo Mateu LM, Jimenez Torres NV. Genetic polymorphisms and individualized tacrolimus dosing. Transplant Proc. 2010;42:3031–3. doi: 10.1016/j.transproceed.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Santoro A, Felipe CR, Tedesco-Silva H, Medina-Pestana JO, Struchiner CJ, Ojopi EB, et al. Pharmacogenetics of calcineurin inhibitors in Brazilian renal transplant patients. Pharmacogenomics. 2011;12:1293–303. doi: 10.2217/pgs.11.70. [DOI] [PubMed] [Google Scholar]
- 58.Santoro AB, Struchiner CJ, Felipe CR, Tedesco-Silva H, Medina-Pestana JO, Suarez-Kurtz G. CYP3A5 genotype, but not CYP3A4*1b, CYP3A4*22, or hematocrit, predicts tacrolimus dose requirements in Brazilian renal transplant patients. Clin Pharmacol Ther. 2013;94:201–2. doi: 10.1038/clpt.2013.68. [DOI] [PubMed] [Google Scholar]
- 59.Zheng S, Tasnif Y, Hebert MF, Davis CL, Shitara Y, Calamia JC, et al. Measurement and compartmental modeling of the effect of CYP3A5 gene variation on systemic and intrarenal tacrolimus disposition. Clin Pharmacol Ther. 2012;92:737–45. doi: 10.1038/clpt.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oetting W, Schaldt D, Guan W, Israni A, Remmel R, Dorr C, et al. Identification of Genetic Variants Associated With Variation of Tacrolimus Levels in African Americans Using GWAS. Am J Transplant; American Transplantation Congress Abstracts; 2015. p. 259. [Google Scholar]
- 61.Pulk R, Schladt D, Guan W, Oetting W, Israni A, Matas A, et al. Multi-Gene Pharmacogenomics of Tacrolimus Troughs in Kidney Transplant Recipients. doi 10.2217/PGS.15.42 [Epub ahead of print] Pharmacogenomics. 2014 [Google Scholar]
- 62.Li YR, van Setten J, Verma SS, Lu Y, Holmes MV, Gao H, et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med. 2015;7:90. doi: 10.1186/s13073-015-0211-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asselbergs F, Lord G, Synder M, Birdwell K, Schadt E, Oetting W, et al. The International Genomics & Translational Research in Transplantation Network (iGeneTrain). Transplantation; World Transplantation Congress Abstracts; 2014. pp. 2014pp. 871–905. [Google Scholar]
- 64.Keating B, Nair N, Holmes M, Oetting W, Jacobson P, Israni A, et al. Concept, Design and Implementation of a Sedicated Genome-Wide Transplant SNP Array With > 780,000 Markers. Transplantation; 2014 World Transplantation Congress Abstracts; 2014. p. 889. [Google Scholar]
- 65.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75:1059–69. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J, Zhang X, Liu L, Tong W. Value of CYP3A5 genotyping on determining initial dosages of tacrolimus for Chinese renal transplant recipients. Transplant Proc. 2010;42:3459–64. doi: 10.1016/j.transproceed.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 68.Roy JN, Lajoie J, Zijenah LS, Barama A, Poirier C, Ward BJ, et al. CYP3A5 genetic polymorphisms in different ethnic populations. Drug Metab Dispos. 2005;33:884–7. doi: 10.1124/dmd.105.003822. [DOI] [PubMed] [Google Scholar]
- 69.Lee SJ, Goldstein JA. Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests. Pharmacogenomics. 2005;6:357–71. doi: 10.1517/14622416.6.4.357. [DOI] [PubMed] [Google Scholar]
- 70.Bergmann TK, Hennig S, Barraclough KA, Isbel NM, Staatz CE. Population pharmacokinetics of tacrolimus in adult kidney transplant patients: impact of CYP3A5 genotype on starting dose. Ther Drug Monit. 2014;36:62–70. doi: 10.1097/FTD.0b013e31829f1ab8. [DOI] [PubMed] [Google Scholar]
- 71.Staatz CE, Willis C, Taylor PJ, Lynch SV, Tett SE. Toward better outcomes with tacrolimus therapy: population pharmacokinetics and individualized dosage prediction in adult liver transplantation. Liver Transpl. 2003;9:130–7. doi: 10.1053/jlts.2003.50023. [DOI] [PubMed] [Google Scholar]
- 72.Staatz CE, Willis C, Taylor PJ, Tett SE. Population pharmacokinetics of tacrolimus in adult kidney transplant recipients. Clin Pharmacol Ther. 2002;72:660–9. doi: 10.1067/mcp.2002.129304. [DOI] [PubMed] [Google Scholar]
- 73.Han N, Ha S, Yun HY, Kim MG, Min SI, Ha J, et al. Population pharmacokinetic-pharmacogenetic model of tacrolimus in the early period after kidney transplantation. Basic Clin Pharmacol Toxicol. 2014;114:400–6. doi: 10.1111/bcpt.12176. [DOI] [PubMed] [Google Scholar]
- 74.Bains RK, Kovacevic M, Plaster CA, Tarekegn A, Bekele E, Bradman NN, et al. Molecular diversity and population structure at the Cytochrome P450 3A5 gene in Africa. BMC Genet. 2013;14:34. doi: 10.1186/1471-2156-14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin Y, Wang YH, Miao J, Li L, Kovacs RJ, Marunde R, et al. Cytochrome P450 3A5 genotype is associated with verapamil response in healthy subjects. Clin Pharmacol Ther. 2007;82:579–85. doi: 10.1038/sj.clpt.6100208. [DOI] [PubMed] [Google Scholar]
- 76.Dennison JB, Mohutsky MA, Barbuch RJ, Wrighton SA, Hall SD. Apparent high CYP3A5 expression is required for significant metabolism of vincristine by human cryopreserved hepatocytes. J Pharmacol Exp Ther. 2008;327:248–57. doi: 10.1124/jpet.108.139998. [DOI] [PubMed] [Google Scholar]
- 77.Floyd MD, Gervasini G, Masica AL, Mayo G, George AL, Jr, Bhat K, et al. Genotype-phenotype associations for common CYP3A4 and CYP3A5 variants in the basal and induced metabolism of midazolam in European- and African-American men and women. Pharmacogenetics. 2003;13:595–606. doi: 10.1097/00008571-200310000-00003. [DOI] [PubMed] [Google Scholar]
- 78.Miao J, Jin Y, Marunde RL, Gorski CJ, Kim S, Quinney S, et al. Association of genotypes of the CYP3A cluster with midazolam disposition in vivo. Pharmacogenomics J. 2009;9:319–26. doi: 10.1038/tpj.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mirghani RA, Sayi J, Aklillu E, Allqvist A, Jande M, Wennerholm A, et al. CYP3A5 genotype has significant effect on quinine 3-hydroxylation in Tanzanians, who have lower total CYP3A activity than a Swedish population. Pharmacogenet Genomics. 2006;16:637–45. doi: 10.1097/01.fpc.0000230411.89973.1b. [DOI] [PubMed] [Google Scholar]
- 80.Mukonzo JK, Waako P, Ogwal-Okeng J, Gustafsson LL, Aklillu E. Genetic variations in ABCB1 and CYP3A5 as well as sex influence quinine disposition among Ugandans. Ther Drug Monit. 2010;32:346–52. doi: 10.1097/FTD.0b013e3181da79d6. [DOI] [PubMed] [Google Scholar]
- 81.Roberts PJ, Rollins KD, Kashuba AD, Paine MF, Nelsen AC, Williams EE, et al. The influence of CYP3A5 genotype on dexamethasone induction of CYP3A activity in African Americans. Drug Metab Dispos. 2008;36:1465–9. doi: 10.1124/dmd.107.020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Antignac M, Hulot JS, Boleslawski E, Hannoun L, Touitou Y, Farinotti R, et al. Population pharmacokinetics of tacrolimus in full liver transplant patients: modelling of the post-operative clearance. Eur J Clin Pharmacol. 2005;61:409–16. doi: 10.1007/s00228-005-0933-6. [DOI] [PubMed] [Google Scholar]
- 83.Fukatsu S, Yano I, Igarashi T, Hashida T, Takayanagi K, Saito H, et al. Population pharmacokinetics of tacrolimus in adult recipients receiving living-donor liver transplantation. Eur J Clin Pharmacol. 2001;57:479–84. doi: 10.1007/s002280100331. [DOI] [PubMed] [Google Scholar]
- 84.Eeckhoudt SL, Horsmans Y, Verbeeck RK. Differential induction of midazolam metabolism in the small intestine and liver by oral and intravenous dexamethasone pretreatment in rat. Xenobiotica. 2002;32:975–84. doi: 10.1080/0049825021000012655. [DOI] [PubMed] [Google Scholar]
- 85.Hukkanen J, Vaisanen T, Lassila A, Piipari R, Anttila S, Pelkonen O, et al. Regulation of CYP3A5 by glucocorticoids and cigarette smoke in human lung-derived cells. J Pharmacol Exp Ther. 2003;304:745–52. doi: 10.1124/jpet.102.038208. [DOI] [PubMed] [Google Scholar]
- 86.Ogg MS, Williams JM, Tarbit M, Goldfarb PS, Gray TJ, Gibson GG. A reporter gene assay to assess the molecular mechanisms of xenobiotic-dependent induction of the human CYP3A4 gene in vitro. Xenobiotica. 1999;29:269–79. doi: 10.1080/004982599238669. [DOI] [PubMed] [Google Scholar]
- 87.Schuetz EG, Wrighton SA, Barwick JL, Guzelian PS. Induction of cytochrome P-450 by glucocorticoids in rat liver. I. Evidence that glucocorticoids and pregnenolone 16 alpha-carbonitrile regulate de novo synthesis of a common form of cytochrome P-450 in cultures of adult rat hepatocytes and in the liver in vivo. J Biol Chem. 1984;259:1999–2006. [PubMed] [Google Scholar]
- 88.Miura M, Satoh S, Kagaya H, Saito M, Inoue T, Tsuchiya N, et al. No impact of age on dose-adjusted pharmacokinetics of tacrolimus, mycophenolic acid and prednisolone 1 month after renal transplantation. Eur J Clin Pharmacol. 2009;65:1047–53. doi: 10.1007/s00228-009-0721-9. [DOI] [PubMed] [Google Scholar]
- 89.Staatz CE, Tett SE. Pharmacokinetic considerations relating to tacrolimus dosing in the elderly. Drugs Aging. 2005;22:541–57. doi: 10.2165/00002512-200522070-00001. [DOI] [PubMed] [Google Scholar]
- 90.Stratta P, Quaglia M, Cena T, Antoniotti R, Fenoglio R, Menegotto A, et al. The interactions of age, sex, body mass index, genetics, and steroid weight-based doses on tacrolimus dosing requirement after adult kidney transplantation. Eur J Clin Pharmacol. 2012;68:671–80. doi: 10.1007/s00228-011-1150-0. [DOI] [PubMed] [Google Scholar]
- 91.Bhatnagar V, Garcia EP, O’Connor DT, Brophy VH, Alcaraz J, Richard E, et al. CYP3A4 and CYP3A5 polymorphisms and blood pressure response to amlodipine among African-American men and women with early hypertensive renal disease. Am J Nephrol. 2010;31:95–103. doi: 10.1159/000258688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu Y, Wang F, Li Q, Zhu M, Du A, Tang W, et al. Amlodipine metabolism in human liver microsomes and roles of CYP3A4/5 in the dihydropyridine dehydrogenation. Drug Metab Dispos. 2014;42:245–9. doi: 10.1124/dmd.113.055400. [DOI] [PubMed] [Google Scholar]
- 93.Zuo XC, Ng CM, Barrett JS, Luo AJ, Zhang BK, Deng CH, et al. Effects of CYP3A4 and CYP3A5 polymorphisms on tacrolimus pharmacokinetics in Chinese adult renal transplant recipients: a population pharmacokinetic analysis. Pharmacogenet Genomics. 2013;23:251–61. doi: 10.1097/FPC.0b013e32835fcbb6. [DOI] [PubMed] [Google Scholar]
- 94.Passey C, Birnbaum AK, Brundage RC, Schladt DP, Oetting WS, Leduc RE, et al. Validation of tacrolimus equation to predict troughs using genetic and clinical factors. Pharmacogenomics. 2012;13:1141–7. doi: 10.2217/pgs.12.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Press RR, Ploeger BA, den Hartigh J, van der Straaten T, van Pelt J, Danhof M, et al. Explaining variability in tacrolimus pharmacokinetics to optimize early exposure in adult kidney transplant recipients. Ther Drug Monit. 2009;31:187–97. doi: 10.1097/FTD.0b013e31819c3d6d. [DOI] [PubMed] [Google Scholar]
- 96.de Jonge H, Elens L, de Loor H, van Schaik RH, Kuypers DR. The CYP3A4*22 C>T single nucleotide polymorphism is associated with reduced midazolam and tacrolimus clearance in stable renal allograft recipients. Pharmacogenomics J. 2015;15:144–52. doi: 10.1038/tpj.2014.49. [DOI] [PubMed] [Google Scholar]
- 97.Thervet E, Loriot MA, Barbier S, Buchler M, Ficheux M, Choukroun G, et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin Pharmacol Ther. 2010;87:721–6. doi: 10.1038/clpt.2010.17. [DOI] [PubMed] [Google Scholar]
- 98.Birdwell KA, Decker B, Barbarino JM, Peterson JF, Stein CM, Sadee W, et al. Clinical pharmacogenetics implementation consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther. 2015 doi: 10.1002/cpt.113. [DOI] [PMC free article] [PubMed] [Google Scholar]


