Abstract
MicroRNA (miRNA) deregulation in prostate cancer (PCa) contributes to PCa initiation and metastatic progression. To comprehensively define the cancer-associated changes in miRNA targeting and function in commonly studied models of PCa, we performed photoactivatable ribonucleoside-enhanced cross-linking immunoprecipitation of the Argonaute protein in a panel of PCa cell lines modeling different stages of PCa progression. Using this comprehensive catalogue of miRNA targets, we analyzed miRNA targeting on known drivers of PCa and examined tissue-specific and stage-specific pathway targeting by miRNAs. We found that androgen receptor is the most frequently targeted PCa oncogene and that miR-148a targets the largest number of known PCa drivers. Globally, tissue-specific and stage-specific changes in miRNA targeting are driven by homeostatic response to active oncogenic pathways. Our findings indicate that, even in advanced PCa, the miRNA pool adapts to regulate continuing alterations in the cancer genome to balance oncogenic molecular changes. These findings are important because they are the first to globally characterize miRNA changes in PCa and demonstrate how the miRNA target spectrum responds to staged tumorigenesis.
Introduction
Prostate cancer (PCa) is the most commonly diagnosed noncutaneous cancer and the second leading cause of cancer-related death in men in the United States [1]. The disease is primarily driven at all stages by activation of the androgen receptor (AR) [2], [3]. Various strategies designed to limit AR activity are the current standard of care for recurrent and metastatic PCa. Although androgen deprivation therapy often results in a substantial clinical response, the disease invariably recurs in a lethal, castrate-resistant manner in which AR is frequently reactivated in the absence of androgens [2], [3]. During intense antiandrogen therapy, a small percentage of men develop treatment-emergent AR-negative small cell/neuroendocrine PCa, a highly aggressive, androgen-independent tumor [4]. Various published studies have cataloged somatic point mutation, copy number aberration, and epigenetic and transcriptomic pathway alterations that occur during the clinical progression of PCa in tumors and model cell lines [5], [6], [7], [8], [9], [10], [11], [12]. Together, these analyses have defined the molecular alterations associated with PCa progression.
MicroRNAs (miRNAs) are ~ 22-nucleotide noncoding regulatory RNA molecules that exert posttranscriptional control over gene expression at the level of mRNA through translational inhibition and initiation of mRNA degradation [13]. In cancer, miRNAs have been shown to have broad oncogenic and tumor-suppressive roles across many tumor types [14], implicating them as key regulators of cancer biology. miRNA expression is broadly deregulated in PCa, and considerable evidence suggests that miRNAs play a role in PCa progression [15], [16], [17], [18], [19], [20], [21]. However, to date, the global changes in the miRNA target spectrum (targetome) present at various stages of PCa progression have yet to be comprehensively defined.
To define the global miRNA targetome in PCa, we performed photoactivatable ribonucleoside-enhanced cross-linking immunoprecipitation of the Argonaute protein (AGO-PAR-CLIP) [22], [23] to broadly map interactions between miRNAs and their cognate miRNA target sites across cell line models of PCa progression. We included the androgen-responsive, AR-positive models LNCaP and LAPC4 and the castrate-resistant PCa (CRPC) model 22Rv1 [3], [24], [25]. To model treatment-emergent small cell/neuroendocrine PCa, we also included the AR-negative lines DU145 and PC3.
We found that miRNAs persistently target primary drivers of PCa even in advanced tumor models. We noted an example of stage-specific driver targeting by miR-148a,which acts as an oncomiRNA in early PCa models by targeting CDKN1B but also acts as a metastatic suppressor by targeting CENPF. Globally, miRNAs reactively target the E2F and MYC pathways active in CRPC as well as the epithelial-to-mesenchymal transition (EMT) and glycolytic pathways active in AR-negative PCa. More broadly, we also found that miRNAs target components of the oxidative phosphorylation machinery that is known to be uniquely active in PCa [26]. Treatment of castrate-sensitive LNCaP cells with the AR antagonist MDV3100 (enzalutamide) led to global depletion of miRNA binding to the 3’UTR that corresponds with the well-known cytostatic properties of full AR blockade, suggesting plasticity in miRNA targeting of oncogenic pathways. Finally, we found that the target pathways we identified are associated with multiple clinical end points, including recurrence. In sum, miRNAs globally undergo a homeostatic response to driver pathways activated during stage-specific PCa progression. AGO-PAR-CLIP offers a novel approach to identify new stage-specific drivers of PCa.
Material and Methods
Cell Culture and Cell Line Acquisition
All cell lines in this study were obtained directly from the Baylor College of Medicine Tissue Culture Core. All cells had been regularly screened for infection and had undergone DNA fingerprint verification to determine authenticity. PC3, DU145, LAPC4, LNCaP, and 22Rv1 PCa cells were maintained in DMEM:F12, EMEM, IMDM, and RPMI 1640 (Invitrogen, Carlsbad, CA) cell growth medium, respectively. All growth media were supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). Cells were cultured in a humidified atmosphere at 37°C and in 5% CO2.
AGO-PAR-CLIP Dataset Production
AGO-PAR-CLIP was performed as a modified protocol similar to one previously described [27] using the Millipore 11A9 anti-AGO2 antibody, with one major modification: the Illumina TruSeq kit was used for indexed cDNA library synthesis. Samples were then multiplexed with up to eight samples per lane on an Illumina HiSeq 2000 machine (Supplementary Files 1–2, Supplementary Figure 1, A–C). To define cluster sites, we processed AGO-PAR-CLIP data through PARalyzer [28] and Piranha [29], and clusters were then mapped onto the GC19 transcriptome (Supplementary Figure 1D). We compared unique reads in our datasets with those reported in AGO-CLIP data and found similar percentages of unique reads (Supplementary Figure 1E). Additionally, we found enrichment in the T→C transitions (and complementary C→G transitions) expected in AGO-PAR-CLIP datasets (Supplementary Figure 1F).
Availability of Supporting Data
The raw reads of each data set supporting the results are available in the National Institutes of Health sequence read archive under the bioproject accession SRP075075, PRJNA321524. Processed AGO-CLIP data atlas files are uploaded in Synapse to the stable hyperlink Synapse ID syn5479902 (https://www.synapse.org/#!Synapse:syn5479902).
CLIP Dataset Mapping and Computational Analysis
CLIP datasets were processed through two of three pipelines. AGO-PAR-CLIP datasets were processed through Bowtie 1 [30] and Bowtie 2 [31], and clusters were called using the PARalyzer algorithm [28] for Bowtie 1 mapped reads and the Piranha algorithm [29] for Bowtie 2 mapped reads. AGO-HITS-CLIP datasets were processed through Bowtie 2 or Novoalign, and clusters were called using the Piranha algorithm for Bowtie 2 mapped reads and the CIMS [32] algorithm for Novoalign mapped reads. Unique reads were filtered using Samtools [33].
Clusters were then mapped onto the Gencode 19 transcriptome and processed through the TargetScan miRNA-seed motif-calling algorithm [34] and assembled into an atlas of miRNA target files in a manner similar to that previously described. Our CLIP atlas was updated to include new human and mouse CLIP data [14], [22], [23], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50]. For the CLIP atlas, PARalyzer clusters were used for AGO-PAR-CLIP data and Piranha clusters were used for AGO-HITS-CLIP data. CLIP binding data were visualized in Circos [51]. Read coverage in clusters was determined using Bedtools [52]. All read files used to determine cluster coverage were processed through the same mapping pipeline (Bowtie 2). Unique reads were used for cluster coverage, and complete read files were used for miRNA coverage.
Once assembled, coverage files were filtered for > 1 read in > 50% of analyzed datasets and then upper-quartile normalized in EdgeR [53] as counts per million in the genome segment of interest (e.g., miRNA, 3’UTR, all clusters). Gene set enrichment analysis (GSEA) [54] was used for read counts summed per gene to determine pathway enrichment. The Kyoto Encyclopedia of Genes and Genomes (KEGG) [55] and Panther [56] were used to perform analysis of binary peak calls. The AGO-CLIP atlas was mined for cluster data using custom scripts.
Clinical Correlation Analysis
Starting with the core-enriched gene signatures as defined by GSEA differential AGO-CLIP binding analysis, we evaluated the activity of each gene signature on a per-sample basis in the Taylor dataset [5]. To determine the activity score for each core-enriched gene signature, we first computed the z-score for the gene signature expression within the cohort, as previously described [5], and then computed the sum z-score for each patient. The z-scores of genes with low AGO-CLIP binding were subtracted from the z-scores of genes with high AGO-CLIP binding, resulting in a corresponding activity score for each specimen.
The association of the gene signatures with biochemical recurrence (BCR)–free survival was evaluated in a large PCa patient cohort for which gene expression profiles and clinical outcomes have been reported [5]. Gene signature activity for each patient was computed as described above. Within the patient cohort, specimens were ranked according to their gene signature activity score into quartiles (top 25% and bottom 75%, top 50% and bottom 50%, top 75% and bottom 25%), and association with BCR-free survival was evaluated using the log-rank test. The package “Survival” in the R statistical system was used to assess significance. Similar survival analysis was performed for CENPF using only CENPF gene expression data.
Following BCR-free survival analysis for all gene sets, we focused on the E2F and G2M gene sets for subsequent clinical associations, including Gleason score, ERG fusion status, tumor stage, patient race, extra-capsular extension, probability of lymph node metastasis, probability of seminal vesicle invasion, probability of organ-confined disease, prostate-specific antigen levels at diagnosis, and development of clinical metastasis. Of all analyzed associations, only the Gleason score was significantly correlated with the gene signatures. Gleason score groups were defined as follows: high > 7, intermediate = 7, low < 7. Significant associations were identified using an unpaired two-sided t test (P< .05 was considered statistically significant).
Proliferation Assays
Proliferation data were obtained in LNCaP cells transfected with miR-148a mimic and control mimic (Invitrogen), with Lipofectamine 2000 (Invitrogen) as the transfection reagent, using an Edu click-it flow cytometry assay kit and TOPRO-3 as a total DNA counterstain (Invitrogen). LNCaP proliferation experiments were performed in triplicate measurements of 50 nM and 100 nM. Proliferation was measured after 48 hours; concentrations yielded similar results and were pooled for final analysis (n= 6 per group). Samples were analyzed by flow cytometry (FACSCanto II, BD Biosciences, San Jose, CA), with a minimum of 20,000 events. Data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR). Statistical differences between treatment groups were evaluated using the Student t test.
Luciferase Assays
CDKN1B luciferase assays were performed in 293T cells, transfected once with 5 nM, once with 10 nM, and twice with 50 nM of miR-148a using Lipofectamine 2000 (6 replicates per group with pooled analysis, n= 24 per group). The CDKN1B plasmid was obtained from Addgene (Plasmid 21326: pGL3-Control-PTEN-3'UTR-Wt [29] and Plasmid 20881: pGL-p27UTR) [32]. Assays were performed twice using 50 nM of miR-148a mimic or control mimic (n= 6 and n= 4 per group). The CENPF 3’UTR luciferase construct was obtained from Switchgear Genomics (Carlsbad, CA) and performed in 293T cells using the Lightswitch assay kit according to manufacturer protocols. CENPF 3’UTR luciferase assays were performed twice at 20 nM and 50 nM (n= 4 and n= 7; one sample was excluded because of incomplete mixing) and twice at 50 nM (n= 4 and n= 4). Replicates were grouped for statistical analysis (n= 19 per group). Assays were performed twice in quadruplicate using 50 nM miR-148a mimic or control mimic. Comparisons were made using an unpaired two-sided Student’s t test. β-Galactosidase transfection controls were not used in the CENPF assay per the protocol defined by Addgene/Switchgear genomics for its vectors.
Site-directed mutagenesis was performed using a QuikChange II XL site-directed mutagenesis kit (Agilent Biosciences, Santa Clara, CA) with primer sequences:
- CENPF:
- Fwd: TGAAAAAGTTTGGAAGCATGAATCACCTGTTAGCATT
- Rev: AATGCTAACAGGTGATTCATGCTTCCAAACTTTTTCA
- CDKN1B:
- Fwd: GGACATCCTGTATAAGCTGAGAGAAAGCAAAAACAAT
- Rev: ATTGTTTTTGCTTTCTCTCAGCTTATACAGGATGTCC
Western Blotting
Primary antibodies included CENPF (DM1A; Santa Cruz Biotechnology, Santa Cruz, CA), AR (ab5; Abcam Inc., Boston, MA), CDKN1B/p27 Kip1 (D69C12) XP Rabbit mAb 3686 (Cell Signaling, Danvers, MS), and AGO2 (11A9, Millipore, Darmstadt, Germany). miR-148a Western blot analysis was performed twice in biological triplicate using transfection with 50nM miR-148a.
Quantitative polymerase chain reaction (PCR)
RNA was extracted from cells using the RNeasy Mini kit (Qiagen, Germantown, MD) or the miRvana total RNA extraction kit (Invitrogen). RNA was reverse-transcribed into cDNA using SuperScript III First-Strand Synthesis SuperMix (Invitrogen), and the cDNA was quantified using real-time PCR (StepOne PLUS Real-Time PCR System; Applied Biosystems, Foster City, CA) with the CENPF TaqMan Gene Expression Assays (Hs01118845_m1; Applied Biosystems) or has-miR-148a Taqman assay relative to U6 snoRNA Taqman assay (Invitrogen). Gene expression was normalized to GAPDH TaqMan Gene Expression Assays (Hs02758991_g1; Applied Biosystems) using the ΔΔCT method. Statistical differences between treatment groups were evaluated using the Student’s t test.
Migration Assay
Cell migration was determined by the wound-healing assay in PC3 cells. Transfected cells were incubated for 3 days to reach confluence, then streaked with a sterile pipette tip and allowed to recover for 24 additional hours. Five random fields per sample were visualized and photographed with a microscope at 4 × magnification immediately and 24 hours after wounding. Transfection of CENPF siRNA was performed twice in biological triplicate; replicates were combined for statistical analysis (n= 6 per group). Statistical differences between treatment groups were evaluated using a two-sided Student’s t test.
Invasion Assay
Invasion assays were performed using BD BioCoat Matrigel Invasion Chambers in PC3 cells (BD Biosciences). Three days after transfection, 1.5 × 105 cells were replated onto each Matrigel-coated insert. RPMI medium supplemented with 5% fetal bovine serum (Hyclone) was added to the lower chamber to act as a chemoattractant. Cells were allowed to invade through the Matrigel and 8-μm pore membrane for 24 hours. Noninvasive cells and matrix were removed using a cotton swab. Membranes were fixed in 4% paraformaldehyde, stained with 0.5% crystal violet, and air-dried, and six random fields per sample were then visualized and photographed with a microscope at 4 × and 10 × magnification. PC3 cells were transfected with miR-148a at 50 nM and 100 nM in triplicate; groups were combined for statistical analysis (n= 6 per group). Transfection of CENPF siRNA was performed twice in biological triplicate; replicates were combined for statistical analysis (n= 6 per group). Statistical differences between treatments were evaluated using an unpaired two-sided Student’s t test.
Results
AGO-PAR-CLIP miRNA Target Definition in PCa Cells
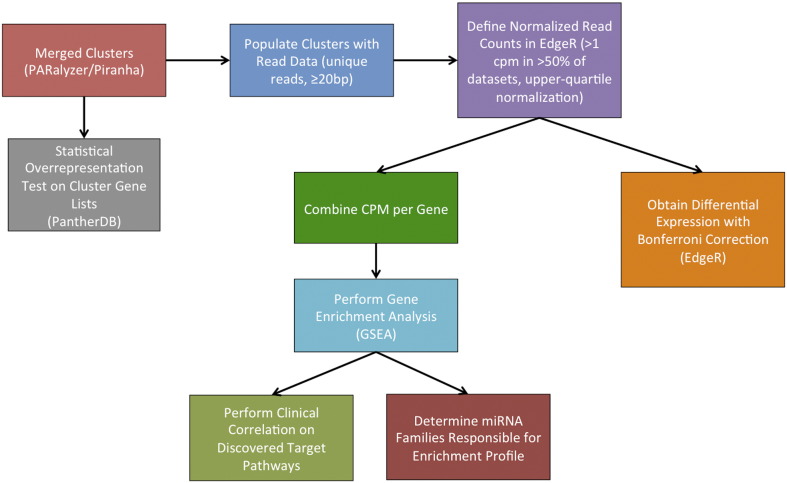
To experimentally characterize the miRNA targetome, we performed AGO-PAR-CLIP coupled with deep sequencing on the LNCaP, 22Rv1, LAPC4, DU145, and PC3 cell lines as described previously, with several modifications (see Methods, Supplementary Files 1–2). To identify differences in miRNA targetomes among the cell lines, we performed gene ontology analysis using transcripts that exhibit differential miRNA activity at individual clusters. We employed three independent approaches to characterize miRNA target activity: statistical overrepresentation of binary cluster gene lists (Panther and KEGG), statistically different read depths at miRNA target sites (EdgeR), and read depth enrichment in miRNA target sites of gene sets (GSEA; Figure 1).
Figure 1.
Workflow of the AGO-PAR- CLIPdata analysis. After transcriptome mapping, clusters were processed through three different algorithms (Panther, EdgeR, and GSEA) to determine altered miRNA target sites and pathways enriched in miRNA targeting between groups. The Panther database was used to analyze binary cluster calls in the form of gene lists. Target sites were analyzed in EdgeR to normalize read data at each target site and define target sites that underwent rigorous statistical change. Similarly, GSEA was used to analyze normalized read data at individual target sites (summed per gene) to identify statistical enrichment of pathways in which miRNA targeting was activated between cell types or treatments. Finally, once pathways enriched for miRNA targeting were discovered, the genes involved in those pathways were analyzed to identify associations with clinical end points in large publically available cancer datasets.
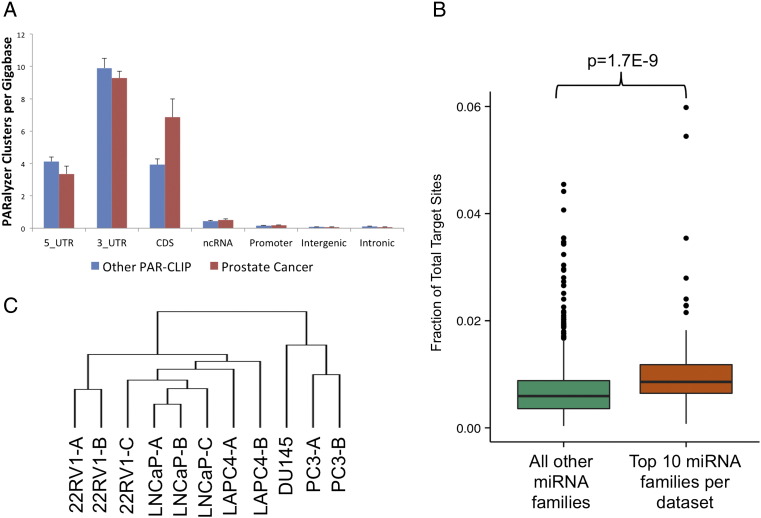
We found that our prostate AGO-PAR-CLIP datasets demonstrated similar levels of 3’UTR and coding sequence cluster enrichment, defined as clusters per gigabase, as those observed in an atlas containing published AGO-PAR-CLIP datasets (Figure 2A, Supplementary File 3) [14]. Our data also contained similar number of unique reads, T→C transitions, and clusters as previously published data (Supplementary File 3). Small RNA loading onto AGO can be determined from AGO-CLIP data, providing a description of which miRNAs are active in the cell. Highly expressed miRNA families, as defined by miRNA loading onto AGO (Supplementary File 4), had 50% (P= 1.7e-9) more identified target sites than all other families (Figure 2B), demonstrating that target discovery in CLIP data is proportionate to miRNA expression.
Figure 2.
AGO-PAR-CLIP-Seq defines the miRNA target spectrum (targetome) in PCa cell lines. (A) PCa CLIP datasets were enriched for PARalyzer clusters in the 3’UTR. CDS indicates coding sequence; ncRNA, noncoding RNA. Error bars are standard error of the mean. (B) Fractional enrichment of target sites in PCa CLIP datasets for miRNA families with the highest expression (top most expressed miRNA families) demonstrated the specificity of CLIP data based on the ability of the data to proportionately capture interactions of the most highly expressed miRNAs. (C) Hierarchical clustering of miRNA targets in PCa showed that the targetome in AR-positive cell lines was separate from that of AR-negative PCa cell lines.
AGO-CLIP sequencing (AGO-CLIP-Seq) data experimentally defined miRNA targets at all genomic locations, including the 3’UTR, 5’UTR, coding sequence, noncoding RNA, intron, promoter, and intergenic regions (Figure 2A). We found targeting in all of these regions, supporting the results of recent studies showing noncanonical AGO binding [46], [57], [58]. In the current analysis, we restricted our focus to canonical 3’UTR interactions to define miRNA target pairs that are likely to mediate mRNA regulation [59]. We focused on seed-compliments of conserved miRNA families defined by TargetScan [34], [60] because expression of conserved miRNAs is more detectable across tissues and less likely to represent a false positive [14]. Hierarchical clustering of read depth at 3’UTR target sites using this approach in PCa cell lines demonstrated that replicates from the same cell lines cluster together and those from AR-positive cell lines cluster separately from those from AR-negative cell lines (Figure 2C). This finding indicates that miRNA targeting of mRNA 3’UTRs is consistent across replicates and is tissue specific, with similar cell lines exhibiting similar miRNA targetomes.
MicroRNA Regulation of PCa Driver Genes
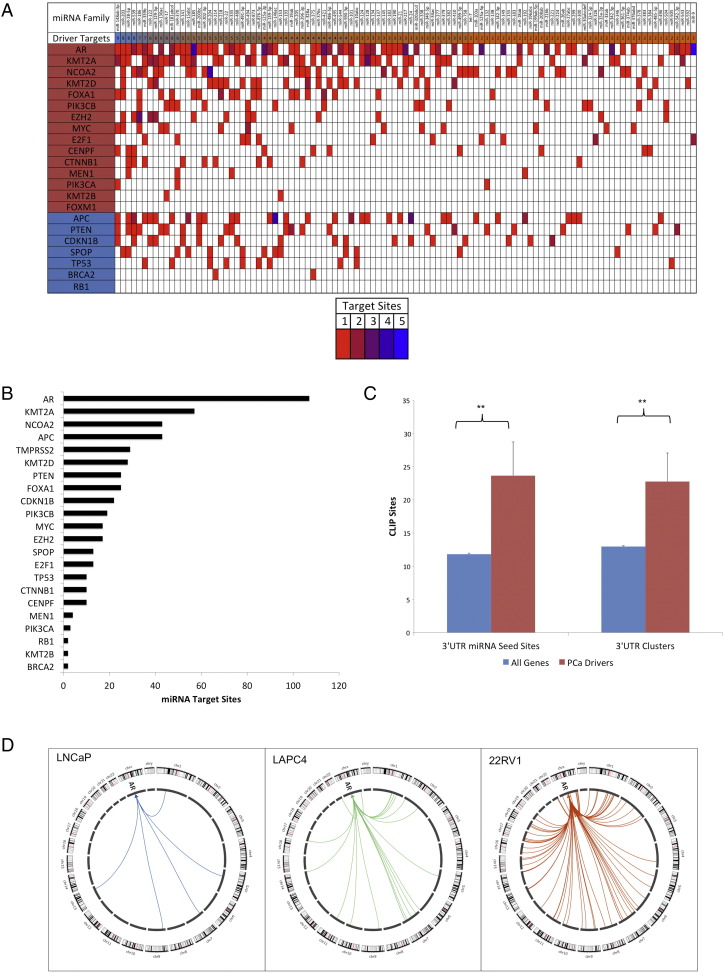
Because PCa undergoes discrete and stage-specific molecular alterations during progression, we first sought to define which miRNAs target known drivers of PCa. We curated a list of 22 previously described PCa drivers (Figure 3A) [3], [5], [6], [7], [10], [11], [12], [61], [62], [63], [64], [65], [66] and then analyzed the total amount and composition of miRNA binding on the 3’UTR of each driver mRNA (Figure 3A).
Figure 3.
MicroRNA-mediated targeting of drivers of PCa. (A) Intersections of PCa drivers and all miRNA families that target > 1 unique driver gene are shown. PCa tumor suppressors are represented in blue, and PCa oncogenes are represented in red. The total number of unique driver targets per miRNA is represented in blue (most unique targets) and orange (least unique targets) in the second line. The miR-148a family had the most unique driver targets in PCa. White cells indicate no targeting was present, and the remaining cells are color-coded to indicate the total number of miRNA target sites per target per miRNA family; red indicates one target site and blue indicates five (the highest number of recurrent target sites per miRNA identified in this analysis). Most miRNA targeting on PCa drivers involved only a single target site per 3’UTR. However, the miR-185 miRNA family targeted the AR at four different sites, and the miR-9 family targeted AR at five different sites along the 3’UTR. (B) miRNAs contributed to the regulation of almost all PCa drivers in our analysis. AR was the PCa driver most heavily bound by miRNAs. (C) PCa drivers were enriched for miRNA binding relative to all other genes. All drivers and genes with at least one CLIP target site are represented in the graph. **P< .0005 (t test). Error bars are standard error of the mean. (D) AR binding across AR-positive cell lines was variable; most binding occurred in the 22RV1 line. No AR binding was detected in AR-negative cell lines.
Most miRNA families targeted each PCa driver only once along its 3’UTR (Figure 3A). However, miR-185 and miR-9 each had greater than three target sites on the AR 3’UTR, suggesting that these miRNAs may strongly regulate AR (Figure 3A). miR-146 had four target sites on APC, and miR-204 had four target sites on NCOA2. The remaining miRNAs targeted each driver at less than four points along their 3’UTRs (Figure 3A). The miR-148a miRNA family targeted the most PCa drivers (nine unique targets), followed by the miR-203, miR-28, and miR-539 families, which each targeted eight unique PCa drivers (Figure 3A).
In total, 129 of 148 conserved miRNA families (87%) targeted at least one PCa driver, indicating convergent miRNA targeting even on a small number of genes. All PCa driver genes except FOXM1 had at least one miRNA target site identified in the PCa CLIP data (Figure 3B). Differential levels of miRNA targeting between PCa driver genes defined which driver pathways were most susceptible to miRNA regulation. Among tumor suppressors, APC, PTEN, and CDKN1B showed the greatest level of miRNA binding (Figure 3A). In the case of PTEN, our results are consistent with published data demonstrating that noncoding RNAs, including miRNAs, contribute to tumor progression by targeting PTEN [67], [68].
Overall, the PCa driver genes also had, on average, twice as many miRNA seed sites and twice as many CLIP clusters per 3’UTR as all other genes with at least one identified cluster (Figure 3C). This finding suggests preferential targeting of the miRNA pool to dominant oncogenic gene programs. AR had the most 3’UTR miRNA seed sites (147 total seed sites representing 71 unique miRNA families, Figure 3B), as well as the most 3’UTR clusters (73 total clusters, Supplementary Figure 2A), indicating that the AR transcript is under potent regulatory control by many different miRNAs. Consistent with these data, AR ranked in the top 5% of targeted genes across the AR-positive AGO-CLIP datasets, corroborating previous studies indicating that multiple miRNAs are capable of regulating AR expression, including the miR-135, miR-185, miR-34, miR-421, and miR-9 families [69]. We also observed the strongest AR targeting in 22RV1, suggesting differential targeting across the AR-positive cell lines (Figure 3D).
miR-148a Targeting of CDKN1B and CENPF
Because the miR-148ab family had the most PCa driver targets and is the highest expressed miRNA in PCa cell lines (Supplementary File 4), we selected miR-148a to validate our CLIP-based PCa driver analysis. The function of miR-148a in PCa remains controversial. Previous studies of PCa cell lines showed that miR-148a was weakly androgen responsive [17], [19] and exogenous miR-148a expression enhanced AR-positive LNCaP growth [19]. However, in the AR-negative PC3 cell line, miR-148a was shown to prevent proliferation, migration, invasion, changes in cellular polarity, and chemotherapy resistance [21]. Although there is no current consensus on the net tumorigenic or tumor-suppressive potential of miR-148a in PCa, the available data suggest that miR-148a activity in PCa may be context specific [15].
Clinical PCa data [5] indicate that miR-148a expression increases in primary tumors but is progressively silenced in metastatic disease (Figure 4A). In the PCa cell lines LNCaP and PC3, miR-148a mimicked its differential expression in prostate tumors: miR-148a expression relative to normal prostate cells was increased in LNCaP and decreased in PC3 cells (Supplementary Figure 2B). These expression differences in LNCaP and PC3 cells are consistent with the significant loss of miR-148a CLIP binding sites that we observed in PC3 and DU145 cells relative to LNCaP and LAPC4 cells (Figure 4B). Treatment of PC3 cells with miR-148a mimic was sufficient to rescue miR-148a CLIP targeting, demonstrating the specificity of these interactions (Figure 4B). In validation experiments, we confirmed that miR-148a is capable of increasing proliferation in LNCaP cells by enhancing the S-phase transition (Figure 4C, Supplementary Figure 2C). We also confirmed that miR-148a significantly reduced PC3 cellular migration, suggesting that miR-148a has potent antimetastatic properties (Figure 4D). These findings are consistent with a model wherein miR-148a contributes to early PCa tumorigenesis but hinders metastatic disease.
Figure 4.
miR-148a is a homeostatic regulator of PCa through CDKN1B and CENPF targeting. (A) In human tumors, miR-148a expression first increased in primary tumors and then was sequentially silenced in nodal and distant metastases. (B) Native miR-148a target sites occupied a larger percentage of total target sites in LNCaP and LAPC4 cell lines than in the more anaplastic PC3 and DU145 cell lines. The number of detectable miR-148a family binding sites was three times higher relative to the total binding sites in miR-148a-transfected PC3 cells. Error bars are standard error of the mean. (C) miR-148a increased proliferation in the LNCaP cell line by increasing the S-phase transition. (D) In the PC3 cell line, miR-148a greatly reduced cellular invasion through Matrigel. (E) 3’UTR-luciferase assays in 293T cells suggested that specific targeting of miR-148a occurs on the 3’UTR of CDKN1B, and transfection of miR-148a mimic into LNCaP cells was sufficient to reduce CDKN1B protein expression. (F) CENPF mRNA expression in human tumors was greatly increased in metastatic disease but only slightly increased in primary tumors. (G) Primary human tumors in the upper quartile of CENPF expression predicted poor BCR-free survival, indicating that CENPF may drive deadly high-grade disease. (H) Transfection of miR-148a into 293T cells specifically silenced a CENPF-3’UTR luciferase construct, demonstrating the ability of miR-148a to reduce CENPF expression through interaction with its 3’UTR. Transfection of PC3 cells with miR-148a reduced native CENPF protein expression, which may account for the effect of miR-148a on PC3 cellular migration and invasiveness. (I) siRNA-mediated specific knockdown of CENPF protein using siRNA cellular (J) migration and (K) invasion is shown. *P< .05, **P< .005, ***P< .0005 (t test). Error bars are standard error of the mean.
In our PCa driver target analysis, miR-148a targeted AR, KMT2A, FOXA1, MYC, CENPF, and PIK3CA oncogenes and APC, PTEN, and CDKN1B tumor suppressors. We hypothesized that targeting of both tumor-suppressive and oncogenic drivers of PCa may contribute to the contextual role of miR-148a. To determine which PCa drivers are most likely to be strongly regulated by miR-148a, we performed orthogonal computational analysis of miR-148a targeting using miR-148a overexpression arrays in LNCaP cells and miR-148b overexpression arrays in HeLa cells [17], [70], as well as correlative analysis of PCa tumors [5]. Specific examination of miR-148 targeting of PCa drivers discovered in PCa CLIP data revealed that six of the nine miR-148a PCa driver targets were decreased on average between the LNCaP and HeLa datasets and eight of the nine PCa driver targets were decreased specifically in the LNCaP dataset, demonstrating a preponderance of mRNA loss in miR-148a predicted targets (Table 1). In the Taylor PCa dataset [5], miR-148a negatively correlated with seven of nine driver targets (Table 1). Of the two driver targets miR-148a positively correlated with, one was AR, which has been shown to induce miR-148a expression [17], [19]. In this context, our data suggest that miR-148a and AR together may mediate a feedback loop on AR signaling. Taken together, the miR-148ab overexpression microarray and anticorrelation analysis results indicated that miR-148a has a predominantly inverse expression relationship with its CLIP-defined driver targets, indicative of miR-148a regulation over these genes.
Table 1.
Analysis of PCa Driver Genes Targeted by miR-148a in LNCaP and HeLa miR-148ab Overexpression Arrays
| Driver Target Gene | Type | Fold Change |
Pearson Correlate in Taylor [5] data | Fold Change + Correlate Average | ||
|---|---|---|---|---|---|---|
| HeLa-miR-148b [36] | LNCaP-miR-148a [17] | Array Average | ||||
| CENPF | Oncogene | − 0.988 | − 0.468 | − 0.728 | − 0.449 | − 0.588 |
| CDKN1B | Tumor suppressor | − 0.450 | − 0.242 | − 0.346 | − 0.114 | − 0.230 |
| APC | Tumor suppressor | − 0.187 | − 0.253 | − 0.220 | − 0.181 | − 0.200 |
| MLL | Oncogene | – | − 0.036 | − 0.036 | − 0.233 | − 0.135 |
| MYC | Oncogene | 0.068 | − 0.188 | − 0.060 | − 0.139 | − 0.100 |
| FOXA1 | Oncogene | 0.271 | − 0.028 | 0.121 | − 0.163 | − 0.021 |
| PTEN | Tumor suppressor | − 0.300 | 0.383 | 0.042 | − 0.070 | − 0.014 |
| PIK3CA | Oncogene | − 0.266 | − 0.506 | − 0.386 | 0.377 | − 0.005 |
| AR | Oncogene | 0.364 | − 0.265 | 0.050 | 0.334 | 0.192 |
On average, across both the overexpression array and the PCa anticorrelation analysis, miR-148a had the strongest negative association with the oncogene CENPF and the tumor-suppressor CDKN1B (Table 1). Interestingly, CDKN1B loss is an important initiator of PCa [61], and CDKN1B is commonly mutated or deleted early in PCa progression [5], [6]. In contrast, CENPF is a master regulator of metastasis in PCa [8], [9]. As such, we hypothesized that CDKN1B targeting by miR-148a drives PCa initiation and miR-148a targeting of the metastatic driver CENPF during PCa progression inhibits metastasis. This metastatic inhibition is lost when miR-148a is silenced during later stages of the disease. To test this hypothesis, we next focused on validating the putative interaction of miR-148a with CDKN1B and CENPF using luciferase constructs, with LNCaP as a model of early PCa and PC3 as a model of late PCa.
Transfection of miR-148a mimic into 293T cells was sufficient to specifically suppress CDKN1B 3’UTR-luciferase reporter activity by 55.7%, and miR-148a transfection into LNCaP cells was sufficient to suppress CDKN1B protein levels demonstrating the specificity of the miR-148a:CDKN1B-3’UTR interaction (Figure 4E). miR-148a mimic did not change luciferase expression relative to a control mimic in the same CDKN1B 3’UTR construct with the miR-148a seed ablated (Figure 4E). Silencing of CDKN1B by miR-148a is consistent with the increase in proliferation (Figure 4C) through increased S-phase transition (Supplementary Figure 2C) that we observed after miR-148a transfection into LNCaP cells.
In human tumors, CENPF mRNA expression was modestly (14%) increased in primary tumors but greatly increased in nodal and distant metastases (75% increase in nodal metastases and 127% increase in distant metastases; Figure 4F). CENPF mRNA expression was also associated with poor BCR-free survival in primary tumors, indicating that CENPF expression in the primary tumor predicts the development of terminal disease (Figure 4G).
Despite data indicating that CENPF apparently supports PCa metastasis [8], [9], the in vitro function of CENPF in PC3 has largely not been described. First, we investigated the miR-148a-CENPF interaction. Similar to CDKN1B, miR-148a transfection into 293T cells suppressed CENPF-3’UTR luciferase reporter activity by 28.7%, and miR-148a transfection into PC3 cells suppressed CENPF protein expression demonstrating specific 3’UTR targeting (Figure 4H). There was no difference between miR-148a and control transfection in the same CENPF 3’UTR construct with the miR-148a seed ablated (Figure 4H). Next, we determined whether CENPF silencing could account for the known ability of miR-148a to greatly reduce PC3 cellular migration and invasion. We found that siRNAs directed against CENPF robustly silenced its protein expression (Figure 4I), which corresponded with reduced cellular migration (Figure 4J) and invasion (Figure 4K). Taken together, these findings suggest that CENPF is most active in driving PCa cell line motility and EMT, and suppression of CENPF mirrored the effect of miR-148a in PC3 cells.
In sum, our findings suggest a model of tumor progression whereby miR-148a targeting may contribute to PCa initiation and growth by targeting CDKN1B but inhibit metastasis by targeting CENPF.
Homeostatic Response to Dominant Tumor Driver Pathways
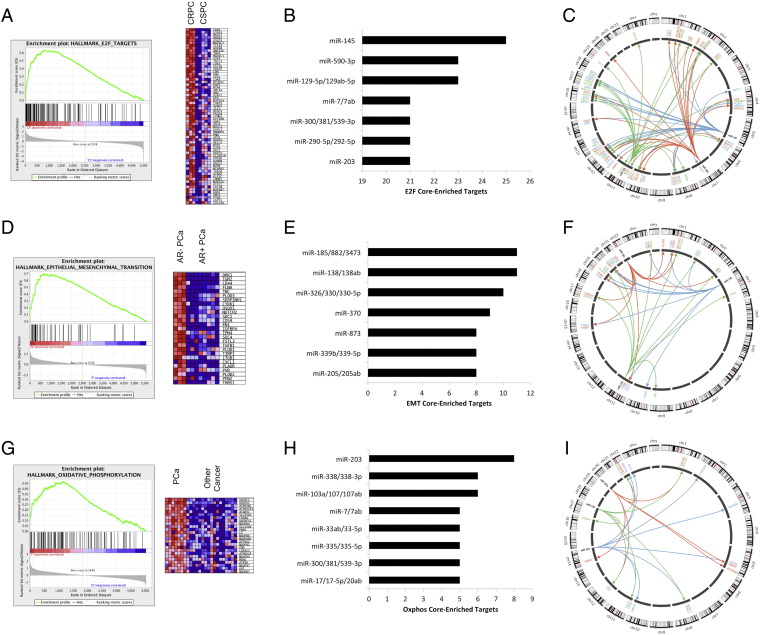
The significant enrichment of PCa driver targeting by miRNAs in our CLIP data led us to hypothesize that global tissue-specific miRNA targeting involves a homeostatic response to dominant driver pathways. To test this hypothesis, we performed pathway analysis on the panel of PCa cell lines to determine whether miRNA target sites were enriched for dominant signaling pathways at different stages of tumor progression. To examine stage-specific alterations, we performed GSEA analysis of read depth differences at individual target sites. Following gene ontology analysis, we mapped miRNAs responsible for binding core pathway genes and thus identified the miRNAs that, by targeting multiple core-enriched genes in a given pathway, may be most responsible for regulation of that pathway.
First, we modeled miRNA targeting in CRPC in the 22Rv1 cell line, which expresses the constitutively active ligand-independent AR splice isoform AR-V7 (Supplementary Figure 1D and Supplementary File 5 contain data describing significant changes in individual target sites). Expression of AR-V7 has previously been shown to allow persistent AR activity following castration [66] and drive a novel AR-mediated gene program that differs from native AR [67]. GSEA pathway analysis of miRNA 3’UTR targeting in 22Rv1 cells relative to LNCaP and LAPC4 cells showed that the E2F response was the strongest (most enriched), followed by the G2/M DNA damage checkpoint and MYC-V1/2 targets (Figure 5A, Table 2). Interestingly, the E2F and MYC pathways are associated with progression to CRPC [10], [11]. The fact that E2F signaling was the most heavily enriched miRNA targeting pathway in AR-V7–expressing 22Rv1 cells relative to LNCaP and LAPC4 cells suggests a model in which the miRNA pool reacts in a homeostatic fashion to equilibrate dominant signaling pathways active in CRPC.
Figure 5.
MicroRNA targets are enriched in AR-negative PCa cell line genes related to the EMT pathway, and PCa cells, compared with cells from other tumors, show enrichment of miRNAs targeting genes involved in oxidative phosphorylation. (A) Gene enrichment graph and heat map showing that CRPC had greatly enriched miRNA targeting of the E2F pathway compared with castrate-sensitive PCa. (B–C) E2F pathway targeting was driven by the miR-145, miR-590, and miR-129 miRNA families, suggesting that these miRNAs may regulate unique pathways involved in castration resistance. (D) Gene enrichment graph and heat map showing that genes relating to EMT had the most significantly different miRNA target pathways between AR-positive and AR-negative cell lines, reflecting increased metastatic potential in AR-negative cell lines as well as the tendency of miRNAs to reactively target uniquely activated gene pathways. (E–F) The miR-185, miR-138, and miR-326 families disproportionately targeted genes involved in EMT in PCa, suggesting that these miRNAs may contribute to metastasis in PCa. miR-205, a known metastatic regulator of PCa, also selectively targeted EMT-related genes. (G) Gene enrichment graph and heat map of genes involved in oxidative phosphorylation that were enriched in PCa cell lines compared with other cancer cell lines. Enrichment of genes involved in oxidative phosphorylation in PCa cell lines indicates resistive activation of miRNA in response to transcriptional upregulation of unique pathways. (H–I) The miR-203, miR-338, and miR-103/107 families were most responsible for targeting of core-enriched genes related to oxidative phosphorylation in PCa.
Table 2.
GSEA Analysis of miRNA Targeting in PCa
| Comparison | Gene Sets | NES | NOM P val | FDR Q val |
|---|---|---|---|---|
| Castrate-resistant vs castrate–sensitive cells | HALLMARK_E2F_TARGETS | 1.85 | .00 | .02 |
| HALLMARK_G2M_CHECKPOINT | 1.81 | .00 | .02 | |
| HALLMARK_MYC_TARGETS_V2 | 1.38 | .05 | .29 | |
| HALLMARK_MYC_TARGETS_V1 | 1.35 | .11 | .26 | |
| AR-negative vs AR-positive cells | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 2.18 | .00 | .01 |
| HALLMARK_GLYCOLYSIS | 1.94 | .00 | .01 | |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 1.91 | .00 | .01 | |
| HALLMARK_HYPOXIA | 1.87 | .00 | .01 | |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 1.83 | .03 | .01 | |
| HALLMARK_INFLAMMATORY_RESPONSE | 1.72 | .00 | .03 | |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 1.70 | .00 | .03 | |
| HALLMARK_IL2_STAT5_SIGNALING | 1.68 | .00 | .03 | |
| HALLMARK_APICAL_JUNCTION | 1.65 | .00 | .04 | |
| HALLMARK_ALLOGRAFT_REJECTION | 1.63 | .00 | .04 | |
| HALLMARK_UV_RESPONSE_DN | 1.60 | .03 | .04 | |
| HALLMARK_COMPLEMENT | 1.60 | .02 | .04 | |
| HALLMARK_ESTROGEN_RESPONSE_EARLY | 1.56 | .00 | .05 | |
| HALLMARK_REACTIVE_OXIGEN_SPECIES_PATHWAY | 1.54 | .01 | .06 | |
| HALLMARK_APOPTOSIS | 1.51 | .04 | .07 | |
| HALLMARK_COAGULATION | 1.51 | .08 | .07 | |
| HALLMARK_ESTROGEN_RESPONSE_LATE | 1.47 | .01 | .08 | |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.45 | .00 | .09 | |
| HALLMARK_MITOTIC_SPINDLE | 1.44 | .04 | .09 | |
| HALLMARK_WNT_BETA_CATENIN_SIGNALING | 1.37 | .08 | .13 | |
| HALLMARK_MYOGENESIS | 1.36 | .11 | .14 | |
| HALLMARK_PROTEIN_SECRETION | 1.32 | .07 | .18 | |
| HALLMARK_MTORC1_SIGNALING | 1.31 | .14 | .17 | |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | 1.29 | .08 | .19 | |
| HALLMARK_TGF_BETA_SIGNALING | 1.25 | .20 | .22 | |
| HALLMARK_MYC_TARGETS_V2 | 1.24 | .16 | .22 | |
| HALLMARK_XENOBIOTIC_METABOLISM | 1.22 | .16 | .24 | |
| PCa vs other cancer cells | HALLMARK_OXIDATIVE_PHOSPHORYLATION | 1.66 | .01 | .21 |
| LNCaP vs LNCaP-MDV3100 | HALLMARK_P53_PATHWAY | 1.81 | .01 | .02 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 1.49 | .06 | .10 | |
| HALLMARK_MTORC1_SIGNALING | 1.34 | .12 | .15 | |
| HALLMARK_MYC_TARGETS_V1 | 1.22 | .21 | .21 |
NES indicates normalized enrichment score; FDR, false discovery rate. The P value represents a normal distribution.
To determine which miRNAs are active in regulating the dominant E2F signaling pathway, we tallied how many core-enriched genes (genes defined as enriched in each pathway by GSEA) each miRNA is capable of targeting to best determine which individual miRNA families may have a strong influence over the entire signaling pathway. We found that miR-145, miR-590, and miR-129 had the most E2F core-enriched targets out of 126 miRNA families predicted to target at least one core-enriched gene in the CLIP data (Figure 5, B and C). Together, these three miRNAs are capable of targeting 69.7% of 66 E2F core-enriched genes with some overlap in their targetomes. miR-145 has been previously demonstrated to be RB1 responsive and regulate E2F signaling [73].
We next analyzed differences between the AR-positive LNCaP, LAPC4, and 22Rv1 cell lines and the AR-negative PC3 and DU145 cell lines. The AR-negative PCa cell lines had 27 significantly upregulated (false discovery rate < 0.25) GSEA gene sets relative to the AR-positive cell lines. EMT was the most significantly enriched concept, followed by glycolysis, IFN-γ response, and hypoxia (Figure 5D, Table 2).
Because EMT is a seminal process contributing to the ability of tumors to undergo metastasis, we sought to determine which miRNAs contributed to targeting EMT in the AR-negative PCa cell lines. We found that the miR-138, miR-185, and miR-326 miRNA families were most strongly associated with the EMT signature, targeting 73.1% of 26 EMT core-enriched targets (Figure 5, E and F describes all EMT core targeting by these miRNA families). miR-138 is a known suppressor of EMT [74]. Notably, miR-205, a well-described regulator of EMT and tumor suppressor of PCa, was also high on the list (Figure 5E) [16].
Finally, we analyzed potential differences in miRNA targeting between PCa cell lines and other tumorigenic cell lines in our CLIP atlas. Genes active in oxidative phosphorylation were the most significantly enriched (false discovery rate < 0.25) gene set in PCa cell lines relative to all other cell lines (Figure 5G, Table 2). miRNA targeting of genes related to oxidative phosphorylation in PCa cell lines is of particular interest because of the unique metabolic reprogramming of PCa. In contrast with many other cancers, PCa lipid synthesis and anaplerosis support an oxidative metabolic program [26], [75], [76].
We investigated which miRNA families were actively targeting oxidative phosphorylation by isolating miRNA families targeting the 29 significantly altered genes contributing to the core enrichment of the oxidative phosphorylation pathways. In total, 126 separate conserved miRNA families contained at least one predicted binding site in the core genes. Of these 126 miRNA families, 99 were predicted to target ≥ 2 of the core enriched genes, but only 8 miRNA families were predicted to target ≥ 5 core genes (Figure 5H). Among these 8 miRNA families, the miR-203 family targeted 8 of 29 enriched genes (27.6%), and the miR-103 and miR-338 families each targeted 6 of 29 enriched genes (20.7%). Together, these 3 miRNA families targeted 14 of 29 unique target genes (48.2%) with 6 overlapping targets (Figure 5I), suggesting that these miRNAs may exert substantial control over oxidative phosphorylation in PCa. All three of these miRNA families are known regulators of energy metabolism; knockdown of miR-203 blocks oxidative phosphorylation [77], miR-103 is a critical regulator of insulin signaling [78], and miR-338 directly suppresses oxidative phosphorylation in neurons [79].
Androgen Blockade Suppresses miRNA Targeting
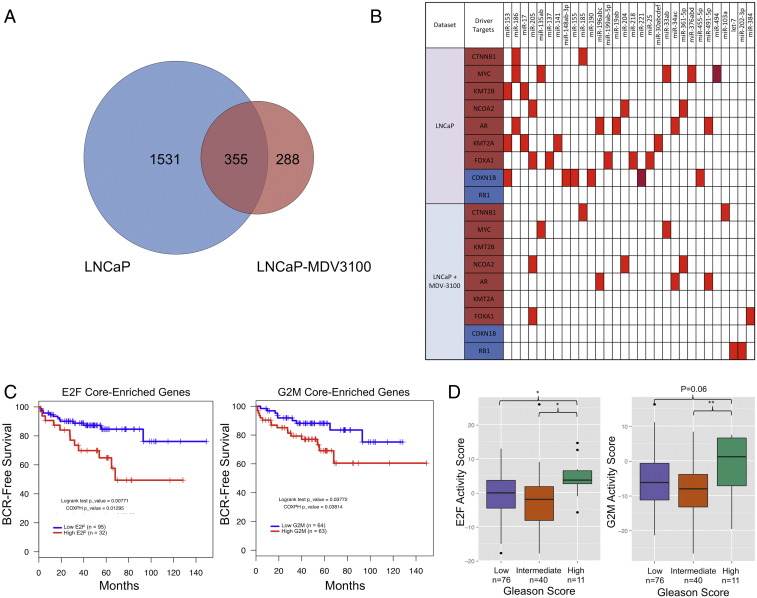
To further investigate the finding that miRNA targeting produces a homeostatic response to flux in cellular pathways, we performed AGO-CLIP on LNCaP cells treated with the AR antagonist enzalutamide. The Venn diagram in Figure 6A illustrates the overlap among unique mRNA targets in untreated LNCaP cells compared with LNCaP cells treated with enzalutamide. A total of 2174 3’UTRs with at least 1 cluster were discovered. Of these, 1531 3’UTR clusters were exclusive to untreated LNCaP cells, 288 3’UTR clusters were exclusive to cells treated with enzalutamide (LNCaP-MDV3100), and 355 3’UTR clusters appeared in both (Figure 6A, Supplementary File 5). These data demonstrate globally decreased targeting of individual mRNAs in the enzalutamide-treated samples, corroborated by the fact that treatment with enzalutamide globally reduced miRNA reads on the 3’UTR by ~ 25% (Supplementary Figure 1E).
Figure 6.
MicroRNA targeting in the 3’UTR is globally reduced after treatment with enzalutamide. (A) Inhibition of the AR by enzalutamide (LNCaP-MDV3100 cells) globally decreased the total number of discovered miRNA cluster sites while simultaneously activating a novel, specific set of miRNA targets. (B) 3’UTR read enrichment in the 3’UTR was reduced by ~ 25% upon treatment with enzalutamide, reflecting either global reduction in miRNA binding ability or global reduction in 3’UTR expression. (C) E2F and G2M pathway core-enriched genes were associated with poor BCR-free survival. (D) E2F and G2M pathway component expression was associated with a high-grade Gleason score upon prostate biopsy. *P< .05, **P< .005 (t test).
Global loss of targeting in the treated cells was reflected in miRNA driver target analysis as well. In total, cells treated with enzalutamide had 70% fewer total mRNA targets and 60% fewer total driver target interactions than untreated LNCaP cells (Figure 6B shows all drivers with at least one miRNA-target interaction discovered in the untreated LNCaP and treated LNCaP datasets).
Both KEGG and GSEA analyses showed that the TP53 pathway was significantly enriched in untreated LNCaP cells compared with treated LNCaP cells (Table 2, Supplementary File 6). This likely reflects known cooperative functionality between TP53 and AR signaling [75], [76] and the known activation of TP53 by AR-induced miRNAs [20]. Interestingly, three of five KEGG pathways gained after treatment with enzalutamide were related to PI3K signaling (Supplementary File 6). This finding could reflect the fact that in PTEN-deficient PCa cells such as LNCaP cells, loss of AR signaling leads to PI3K activation mediated by PI3KCD [77]. As such, it is apparent that the miRNA interactome is not simply lost upon deprivation of host transcripts but shifts to host transcripts that are activated after AR loss.
Activated Gene Pathways are Associated with Human PCa
To substantiate our findings in human PCa data, we analyzed correlations of the core-enriched pathway driver genes targeted by miRNAs in our analysis with clinical outcomes in human PCa. To do this, we assigned each sample in the Taylor PCa dataset [5] an activity score for the core-enriched genes based on the mRNA expression of each core component. We first performed quartile analysis of the top two significant gene sets for each GSEA comparison (EMT and glycolysis in AR-positive cells compared with AR-negative cells, E2F and G2M in castrate-sensitive PCa compared with CRPC, and oxidative phosphorylation in PCa compared with other cancers) identified in our analysis to determine clinical correlations with BCR-free survival.
We found that the upper quartile of core-enriched genes in the E2F pathway and the upper half of core-enriched genes in the G2M pathway were associated with poor BCR-free survival (Figure 6C). The EMT, glycolysis, and oxidative phosphorylation core enrichments were not associated with BCR-free survival, likely because the EMT and glycolysis pathways reflect AR-negative disease, which is not present in the Taylor data, and the oxidative phosphorylation pathway is constitutively active across PCa specimens.
Because the EMT and G2M pathways enriched in CRPC were associated with BCR-free survival, we further analyzed clinical end points associated with the enrichment of these pathways. We found that E2F and G2M core-enriched genes were also associated with poor combined Gleason score at biopsy (Figure 6D). The activity scores of the E2F and G2M pathway components were significantly correlated with each other (Supplementary Figure 1F), which likely reflect components of the E2F pathway driving cellular movement through the G2M checkpoint.
These results are consistent with the known role of the E2F pathway in driving castration resistance and BCR. These results suggest that primary tumors express core-enriched E2F pathway components and are more likely to undergo rapid BCR than tumors with reduced expression of core-enriched E2F pathway components. Finally, these results demonstrate that, in models of CRPC, the miRNA pool responds to mRNAs that are predictive of poor outcome and high-grade initial disease in human tumors.
Discussion
In the current study, we used AGO-CLIP-Seq technology to define miRNA target interactions in common models of human PCa progression. PCa is a leading cause of cancer-related death in men. Furthermore, miRNA deregulation in cancer is known to drive PCa progression. As such, in-depth understanding of miRNA targeting at different stages of PCa is of considerable value to the research community.
Initial analysis of miRNA targeting of known drivers of PCa demonstrated enrichment in driver targeting that is indicative of continued miRNA-mediated modulation of oncogenic pathways, even in models of advanced PCa. The AR 3’UTR underwent the most miRNA-mediated regulation of any driver gene. AR is also a well-described positive regulator of RNA transcription, and the apparent regulation of AR by miRNAs is indicative of a negative feedback loop initiated by many miRNA families. The heavy targeting of AR has broad implications for understanding the role of miRNA in PCa because of the important role AR plays in tumorigenesis and therapeutic resistance.
We found that miR-148a and miR-203 may play critical roles in the development and progression of PCa. We found that miR-148a targeted the most PCa drivers and was, on average, the most expressed miRNA in PCa cell lines. Previous research into the role of miR-148a in PCa has proven controversial, with divergent phenotype results depending on the cell line. We further defined the miR-148a targetome by examining its extensive targeting of drivers of PCa and analyzing its expression in human tumor progression. Our results elucidate the role of miR-148a; we found that it undergoes stage-specific regulation in PCa, targeting both known PCa oncogenes and known PCa tumor suppressors. Although we focused specifically on the interaction of miR-148a with CDKN1B and CENPF, miR-148a also contained > 300 targets in our PCa CLIP datasets, and its full function is likely driven by the integration of these interactions.
miR-203 was also among the miRNAs with the most driver targets, and it is well described as a tumor suppressor in PCa [15], [16], [18]. Most of the PCa drivers (seven of eight) that were targeted by miR-203 were oncogenes. Furthermore, miR-203 was also a primary regulator of the E2F pathways identified in our analysis of CRPC (Figure 5B), and the core-enriched genes targeted by miR-203 were part of a signature that was associated with poor BCR-free survival and high-grade initial disease (Figure 6C). Taken together, these findings indicate that miR-203 regulates PCa at multiple levels by targeting primary PCa oncogenes, disrupting pathways that contribute to castration resistance, and possibly interfering with oxidative phosphorylation, upon which PCa is uniquely dependent. These findings further clarify the mechanisms behind the known tumor-suppressive activity of miR-203 [16], [18].
After observing strong targeting of PCa drivers across tumor cells lines, we hypothesized that the miRNA pool may regulate dominant driver pathways. Primary PCa is primarily driven by increased fatty acid oxidation and electron transport chain activity [26]. However, in advanced and distant metastatic disease, glycolytic pathways are activated, potentially as a result of a hypoxic response [83]. Activation of these pathways is associated with aggressive tumors and poor prognosis [83]. Furthermore, the E2F and Myc pathways are specifically activated during castration resistance and directly contribute to advanced, therapy-resistant PCa. This stage-specific pathway alteration was directly mirrored in our PCa CLIP data in various models of tumor progression. The rediscovery of pathways known to be important drivers in PCa progression via an orthogonal approach using AGO-CLIP-Seq validates the use of a CLIP-based approach to identify dominant driver pathways and possibly nominate novel pathways in an unbiased manner, such as the G2M checkpoint barrier that we identified in the analysis of CRPC. The discovery of E2F, Myc, EMT, and glycolytic pathway involvement in PCa in our CLIP analysis demonstrates how the miRNA targetome can perfectly adapt to the vast differences in molecular physiology that occur during staged PCa progression.
As such, a primary finding of this research is that the miRNA pool performs remarkably specific homeostatic tracking of active mRNA pathways both globally and on the level of individual miRNAs. These findings demonstrate that the miRNA targetome adapts to accommodate and repress newly active molecular pathways during clinical progression. Such adaptation by the miRNA pool in response to gene changes is documented [84], [85], but our results define the consequence of such adaptation during tumorigenesis. This observation is important because it suggests that even in advanced tumors, in which expression of oncogenic miRNAs is greatly increased, the miRNA pool is still binding and suppressing tumor-promoting programs. These findings are especially enlightening because recently published studies have reported that components of the effector RISC complex themselves undergo regulation [86], [62], thereby suggesting that global miRNA binding activity is modifiable. In this circumstance, latent tumor-suppressive miRNA activity could be reactivated to increase suppression of key tumor programs, which could prove to be a viable future therapeutic mechanism.
Conclusions
In the current study, we implemented AGO-PAR-CLIP to analyze the miRNA interactome in multiple PCa cell lines. We used our novel, prostate-specific datasets as well as our larger CLIP atlas to characterize multiple aspects of miRNA binding in PCa. We then analyzed clinical associations between the identified pathways and human PCa. These findings offer a unique insight into how miRNAs function in a tissue-specific manner. Finally, we used our CLIP data to define potential tumor-driving interactions of miR-148a, and we found that CDKN1B targeting by miR-148a could promote PCa initiation but CENPF targeting by miR-148a could inhibit PCa invasion.
By identifying miRNA families that have enriched targeting of tumor-driving pathways as defined in the CLIP data, we were able to directly identify which miRNAs are responsible for the observed pathway enrichments. This type of progressive analysis of molecular pathways active at different stages of PCa is critical to the future of PCa research because new, highly aggressive tumors occur more often after long-term treatment with next-generation antiandrogens. In PCa, the primary unique miRNA targeting pathways active in advanced disease relative to low-grade disease and in PCa tissue relative to other tissues, such as the E2F pathway in CRPC, EMT and glycolysis pathways in AR-negative PCa, and oxidative phosphorylation in PCa cell lines (relative to other cancer cell lines), are precisely the type of tumor-driving programs that would be ideal to suppress therapeutically.
The following are the supplementary data related to this article.
Supplementary figures and tables.
AGO CLIP protocol.
AGO-CLIP atlas dataset inventory, read data, cluster data, and T-->C transition data.
CLIP defined values for miRNA loading onto the AGO protein.
EdgeR defined changes in individual miRNA target sites.
LNCaP versus LNCaP-MDV-3100 microRNA target analysis, Panther analysis, and KEGG analysis.
Competing Financial Interests
There are no competing financial interests to disclose.
Acknowledgements
This work was supported by the Caroline Weiss Law Foundation (S. E. M.), the Baylor Research Advocates for Student Scientists, and the Robert and Janice McNair Foundation (M. P. H.). S. E. McGuire is a Dan L. Duncan Scholar and member of the Dan L. Duncan Cancer Center supported by National Cancer Institute Cancer Center Support Grant (P30CA125123). We acknowledge the joint participation by the Diana Helis Henry Medical Research Foundation through its direct engagement in the continuous active conduct of medical research. The National Institutes of Health (K01DK096093 to S. M. H., 1F30CA196108-01 to D. A. B.), the American Heart Association Beginning Grant-in-Aid (15BGIA25850025 to S. M. H.), the Cancer Prevention Research Institute of Texas (CPRIT RP150232 to C. C. and K. I. R.), and the Alkek Center for Molecular Discovery also funded portions of the work.
Footnotes
This work was supported by the Caroline Weiss Law Foundation (S. E. M.), the Baylor Research Advocates for Student Scientists, and the Robert and Janice McNair Foundation (M. P. H.). S. E. McGuire is a Dan L. Duncan Scholar and member of the Dan L. Duncan Cancer Center supported by National Cancer Institute Cancer Center Support Grant (P30CA125123). We acknowledge the joint participation by the Diana Helis Henry Medical Research Foundation through its direct engagement in the continuous active conduct of medical research. The National Institutes of Health (K01DK096093 to S. M. H., 1F30CA196108-01 to D. A. B.), the American Heart Association Beginning Grant-in-Aid (15BGIA25850025 to S. M. H.), the Cancer Prevention Research Institute of Texas (CPRIT RP150232 to C. C. and K. I. R.), and the Alkek Center for Molecular Discovery also funded portions of the work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio A, Tzelepi V, Araujo JC, Guo CC, Liang S, Troncoso P, Logothetis CJ, Navone NM, Maity SN. Neuroendocrine prostate cancer xenografts with large-cell and small-cell features derived from a single patient's tumor: morphological, immunohistochemical, and gene expression profiles. Prostate. 2011;71:846–856. doi: 10.1002/pros.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitrofanova A, Aytes A, Zou M, Shen MM, Abate-Shen C, Califano A. Predicting drug response in human prostate cancer from preclinical analysis of in vivo mouse models. Cell Rep. 2015;12:2060–2071. doi: 10.1016/j.celrep.2015.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, MacArthur S, Stark R, Warren AY, Mills IG. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton MP, Rajapakshe K, Hartig SM, Reva B, McLellan MD, Kandoth C, Ding L, Zack TI, Gunaratne PH, Wheeler DA. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat Commun. 2013;4:2730. doi: 10.1038/ncomms3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene. 2014;33:135–147. doi: 10.1038/onc.2013.54. [DOI] [PubMed] [Google Scholar]
- 16.Boll K, Reiche K, Kasack K, Morbt N, Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn F. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene. 2012;32:277–285. doi: 10.1038/onc.2012.55. [DOI] [PubMed] [Google Scholar]
- 17.Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, Janne OA, Seppala J, Lahdesmaki H, Tammela TL, Visakorpi T. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31:4460–4471. doi: 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- 18.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y, Dahiya R. Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin Cancer Res. 2011;17:5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 19.Murata T, Takayama K, Katayama S, Urano T, Horie-Inoue K, Ikeda K, Takahashi S, Kawazu C, Hasegawa A, Ouchi Y. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010;13:356–361. doi: 10.1038/pcan.2010.32. [DOI] [PubMed] [Google Scholar]
- 20.Rokhlin OW, Scheinker VS, Taghiyev AF, Bumcrot D, Glover RA, Cohen MB. MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther. 2008;7:1288–1296. doi: 10.4161/cbt.7.8.6284. [DOI] [PubMed] [Google Scholar]
- 21.Fujita Y, Kojima K, Ohhashi R, Hamada N, Nozawa Y, Kitamoto A, Sato A, Kondo S, Kojima T, Deguchi T. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem. 2010;285:19076–19084. doi: 10.1074/jbc.M109.079525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–2117. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dakubo GD, Parr RL, Costello LC, Franklin RB, Thayer RE. Altered metabolism and mitochondrial genome in prostate cancer. J Clin Pathol. 2006;59:10–16. doi: 10.1136/jcp.2005.027664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafner M, Lianoglou S, Tuschl T, Betel D. Genome-wide identification of miRNA targets by PAR-CLIP. Methods. 2012;58:94–105. doi: 10.1016/j.ymeth.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LO, Smith AD. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28:3013–3020. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 2014;9:263–293. doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing, S The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipchina I, Elkabetz Y, Hafner M, Sheridan R, Mihailovic A, Tuschl T, Sander C, Studer L, Betel D. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev. 2011;25:2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 37.Haecker I, Gay LA, Yang Y, Hu J, Morse AM, McIntyre LM, Renne R. Ago HITS-CLIP expands understanding of Kaposi's sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farazi TA, Ten Hoeve JJ, Brown M, Mihailovic A, Horlings HM, van de Vijver MJ, Tuschl T, Wessels LF. Identification of distinct miRNA target regulation between breast cancer molecular subtypes using AGO2-PAR-CLIP and patient datasets. Genome Biol. 2014;15 doi: 10.1186/gb-2014-15-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 42.Karginov FV, Hannon GJ. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013;27:1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kameswaran V, Bramswig Nuria C, McKenna Lindsay B, Penn M, Schug J, Hand Nicholas J, Chen Y, Choi I, Vourekas A, Won K-J. Epigenetic regulation of the DLK1-MEG3 MicroRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19:135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boudreau RL, Jiang P, Gilmore BL, Spengler RM, Tirabassi R, Nelson JA, Ross CA, Xing Y, Davidson BL. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron. 2014;81:294–305. doi: 10.1016/j.neuron.2013.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luna JM, Scheel TK, Danino T, Shaw KS, Mele A, Fak JJ, Nishiuchi E, Takacs CN, Catanese MT, de Jong YP. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schug J, McKenna Lindsay B, Walton G, Hand N, Mukherjee S, Essuman K, Shi Z, Gao Y, Markley K, Nakagawa M. Dynamic recruitment of microRNAs to their mRNA targets in the regenerating liver. BMC Genomics. 2013;14 doi: 10.1186/1471-2164-14-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kenehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas PD. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allo M, Agirre E, Bessonov S, Bertucci P, Gomez Acuna L, Buggiano V, Bellora N, Singh B, Petrillo E, Blaustein M. Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc Natl Acad Sci U S A. 2014;111:15622–15629. doi: 10.1073/pnas.1416858111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156:920–934. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cristofano AD, Acetis MD, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 62.Horman SR, Janas MM, Litterst C, Wang B, MacRae IJ, Sever MJ, Morrissey DV, Graves P, Luo B, Umesalma S. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell. 2013;50:356–367. doi: 10.1016/j.molcel.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malik R, Khan AP, Asangani IA, Cieslik M, Prensner JR, Wang X, Iyer MK, Jiang X, Borkin D, Escara-Wilke J. Targeting the MLL complex in castration-resistant prostate cancer. Nat Med. 2015;21:344–352. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao JC, Yu J, Runkle C, Wu L, Hu M, Wu D, Liu JS, Wang Q, Qin ZS, Yu J. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome Res. 2012;22:322–331. doi: 10.1101/gr.131508.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, Xiao C, Hu WY, Han J, Sun P. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–8557. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ostling P, Leivonen SK, Aakula A, Kohonen P, Makela R, Hagman Z, Edsjo A, Kangaspeska S, Edgren H, Nicorici D. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 70.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18:R183–R196. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, Edwards J, Isaacs WB, Nelson PS, Bluemn E. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marzi MJ, Puggioni EM, Dall'Olio V, Bucci G, Bernard L, Bianchi F, Crescenzi M, Di Fiore PP, Nicassio F. Differentiation-associated microRNAs antagonize the Rb-E2F pathway to restrict proliferation. J Cell Biol. 2012;199:77–95. doi: 10.1083/jcb.201206033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu X, Wang C, Chen Z, Jin Y, Wang Y, Kolokythas A, Dai Y, Zhou X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Costello LC, Franklin RB. The intermediary metabolism of the prostate: a key to understanding the pathogenesis and progression of prostate malignancy. Oncology. 2000;59:269–282. doi: 10.1159/000012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30:2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim H, Cho H, Alexander R, Patterson HC, Gu M, Lo KA, Xu D, Goh VJ, Nguyen LN, Chai X. MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes. 2014;63:4045–4056. doi: 10.2337/db14-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 79.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alimirah F, Panchanathan R, Chen J, Zhang X, Ho S-M, Choubey D. Expression of androgen receptor is negatively regulated by p53. Neoplasia. 2007;9:1152–1159. doi: 10.1593/neo.07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cronauer MV, Schulz WA, Burchardt T, Ackermann R, Burchardt M. Inhibition of p53 function diminishes androgen receptor-mediated signaling in prostate cancer cell lines. Oncogene. 2004;23:3541–3549. doi: 10.1038/sj.onc.1207346. [DOI] [PubMed] [Google Scholar]
- 82.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pertega-Gomes N, Felisbino S, Massie CE, Vizcaino JR, Coelho R, Sandi C, Simoes-Sousa S, Jurmeister S, Ramos-Montoya A, Asim M. A glycolytic phenotype is associated with prostate cancer progression and aggressiveness: a role for monocarboxylate transporters as metabolic targets for therapy. J Pathol. 2015;236:517–530. doi: 10.1002/path.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bosson AD, Zamudio JR, Sharp PA. Endogenous miRNA and Target Concentrations Determine Susceptibility to Potential ceRNA Competition. Mol Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gosline SJ, Gurtan AM, JnBaptiste CK, Bosson A, Milani P, Dalin S, Matthews BJ, Yap YS, Sharp PA, Fraenkel E. Elucidating MicroRNA regulatory networks using transcriptional, post-transcriptional, and histone modification measurements. Cell Rep. 2016;14:310–319. doi: 10.1016/j.celrep.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.La Rocca G, Olejniczak SH, Gonzalez AJ, Briskin D, Vidigal JA, Spraggon L, DeMatteo RG, Radler MR, Lindsten T, Ventura A. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci U S A. 2015;112:767–772. doi: 10.1073/pnas.1424217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.
AGO CLIP protocol.
AGO-CLIP atlas dataset inventory, read data, cluster data, and T-->C transition data.
CLIP defined values for miRNA loading onto the AGO protein.
EdgeR defined changes in individual miRNA target sites.
LNCaP versus LNCaP-MDV-3100 microRNA target analysis, Panther analysis, and KEGG analysis.