Highlights
-
•
Modelling and simulation can streamline decision making in drug safety testing.
-
•
Computational cardiac electrophysiology is a mature technology with a long heritage.
-
•
There are many challenges and opportunities in using in silico techniques in future.
-
•
We discuss how models can be used at different stages of drug discovery.
-
•
CiPA will combine screening platforms, human cell assays and in silico predictions.
Abstract
On the tenth anniversary of two key International Conference on Harmonisation (ICH) guidelines relating to cardiac proarrhythmic safety, an initiative aims to consider the implementation of a new paradigm that combines in vitro and in silico technologies to improve risk assessment. The Comprehensive In Vitro Proarrhythmia Assay (CiPA) initiative (co-sponsored by the Cardiac Safety Research Consortium, Health and Environmental Sciences Institute, Safety Pharmacology Society and FDA) is a bold and welcome step in using computational tools for regulatory decision making. This review compares and contrasts the state-of-the-art tools from empirical to mechanistic models of cardiac electrophysiology, and how they can and should be used in combination with experimental tests for compound decision making.
In this article we present how in silico cardiac modelling has matured into a decision making tool in drug discovery, contrast the different approaches being proposed and show the opportunities and challenges that lie ahead for its acceptance by regulators.
What is modelling and what are models?
Scientific models, although only reflecting simplified reality, help us to integrate our knowledge, to quantify a phenomenon and to predict outcomes; hence these models can facilitate evidence-based decision making. They can act as a repository of information for the modelled biological system allowing the viewer, researcher or modeller to understand the underlying assumptions of the model, for example which biological molecules are represented and what concentration and what association with other molecules are present. The next step is to take those static pictures and make them dynamic. For example, what happens to those biological molecules over time given a set of assumptions (model equations), initial conditions (model parameters) and concentrations of biological molecules (model variables)? This could be achieved by formulating and solving differential equations that simulate, for example, binding events, enzyme kinetics or the gating properties of cardiac ion channels.
Before we describe how models are used in drug discovery, it is worth reflecting upon the term ‘model’, which can have different meanings for different communities. At a high level, there are principally two main types of (in silico) models: statistical models and mechanistic models. Statistical (or empirical) models, sometimes referred to as ‘black-box models’, are built on historical data, and are ‘trained’ to imitate the trend of the data and to capture the relation between datasets. Mechanistic models are built based on our pre-knowledge of the system and the physical laws determining the system's output; they offer a descriptive advantage over the black-box approach by articulating more explicitly what is being represented by the model. Often a model that is mechanistic at one scale is black-box at another scale (e.g. we might model cell membrane potential with an electric circuit analogy but ion channel gating voltage-dependence is encoded by a statistical line of best fit equation fitted to measured data points).
Statistical models (including machine-learning approaches) often need to be trained on large datasets to increase their predictive power; a good statistical model should capture what we do not understand yet, see for example [1]. If our pre-knowledge is adequate, a mechanistic model does not need to be trained and should be predictive in new situations. However, this is infrequently the case, thus training and/or calibration datasets are often used to optimise all or a subset of the model parameters. In these cases, the lines between statistical and mechanistic models blur. Any mismatch between the reality and the output of a mechanistic model is useful, because it highlights limitations of the model that might indicate gaps in our knowledge [2]. An example of this is the prediction of the stoichiometry of the sodium/calcium exchanger when Denis Noble and Dario DiFrancesco formulated a model with dynamic concentration changes [3]. Mechanistic modelling, particularly that from a biological or physiological background, has historically included more complexity as more knowledge is learned, whereas the empiricist tries to minimise complexity. Rather than being seen as competing and isolated approaches, there is a need for a more dialectic approach with increased iteration and crosstalk between these modelling methods; we can (and should) see this as a race towards more-productive models.
Current modelling in drug discovery and development
Modelling and simulation (M&S) forms an integral part of the drug discovery and development process and its systematic application has been readily adopted by regulatory agencies as well as pharmaceutical research organisations 4, 5. M&S is expected to improve efficiency and productivity of drug discovery and development with its ability to test numerous scenarios in silico and to select those with the highest probability of success. A broad range of mathematical models are applied, with varying complexity and predictive power. The degree of model complexity is determined by the available information, the specific questions that need to be addressed and the stage of drug development [6]. We see in Fig. 1 that different modelling efforts support decision making along the drug discovery and development pipeline. Additionally, M&S is integral to decision making within the pharmaceutical industry. Translational pharmacokinetic/pharmacodynamic (PK/PD; see Glossary) modelling of efficacy and safety robustly supports a drug development programme when implemented in early-stage development 5, 7. It has the potential to project the pharmacological response in humans based on the exposure–response relationship in animal species by accounting for species differences [8]. In the early clinical development phase, it predicts the range of efficacious and tolerable target exposure and supports the selection of the most favourable dosage regimen and study design elements, such as selection of predictive PD biomarkers and PK sampling time points [5].
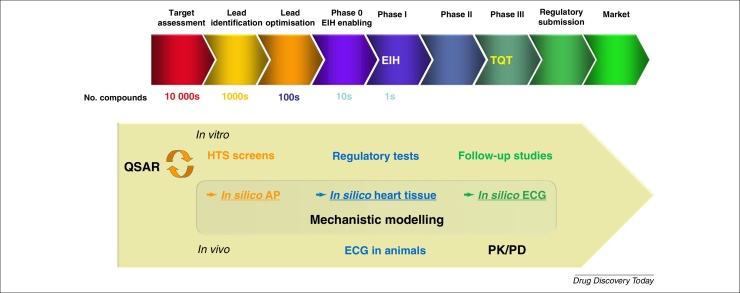
Figure 1.
Schematic of a typical drug discovery pipeline with how and when different in vitro, in vivo and in silico techniques could be applied for cardiac risk assessment. In the ideal state, each of the studies should provide sufficient information to support and aid decision making fully for the ascending milestone points along the drug discovery and development pipeline. Abbreviations: EIH, entry into human; TQT, thorough QT study; AP, Action Potential.
The impact of pharmacometrics, or modelling within clinical pharmacology, on approval and labelling decisions has greatly increased over the past decade [9]. These empirical, data-driven top-down approaches are applied to characterise the exposure–response relationship for efficacy and safety providing a quantitative assessment to guide dose selection and trial design decisions [10]. In recent years, approaches such as physiologically based pharmacokinetic (PBPK) models have been increasingly included in regulatory submissions, for example for the prediction of drug–drug interactions, drug-exposure predictions in paediatrics, in organ-impaired subjects and the effect of other patient factors [11]. Applications of PBPK specific to industry include lead optimisation and candidate selection, prediction of first-in-human PK and continue to support decision making in later phases [11]. These more mechanistic models provide a quantitative framework for prediction of systemic and tissue exposures with the distinct separation of physiology and drug-dependent information. PBPK models enable the extrapolation from in vitro to in vivo [12], from animal to human [13], from healthy volunteers to patient or special populations [11] and are applied at all stages of drug development [14]. Quantitative systems pharmacology (QSP) has emerged more recently [15] as a paradigm that combines elements of translational PK/PD and systems biology aiming to understand how the drug modulates cellular networks in space and time to predict how the pharmacological response affects the human pathophysiology [16]. This ‘middle-out’ approach (as opposed to ‘top-down’ or ‘bottom-up’), perhaps first described by Brenner et al. [17] and more recently by Vicini and van der Graaf [18], provides a repository of knowledge, which is powerful for mechanism-based extrapolation and presents the opportunity to predict unstudied scenarios throughout all stages of discovery and development [18]. It applies the concepts of systems engineering, systems biology and PK/PD to the study of complex biological systems through interaction between mathematical modelling and experimentation [19]. The rich heritage of mathematical cardiac electrophysiology modelling [20] makes this field one of the most mature examples of this middle-out (mechanistic) modelling approach, and as such offers a chance to define the pathway for wider uptake of QSP-type models.
Earlier in the drug discovery phase, the requirement is for an improved translation from early screening (or ideally even earlier computational chemistry) approaches to clinical cardiac outcomes, in the form of the thorough-QT (TQT) study or cardiac adverse events [21]. Key questions about compound progression are asked along this pipeline, and the value for M&S is to align and best support the decision making activities that take place. Figure 1 shows a typical drug discovery pipeline and how in silico cardiac modelling approaches (together with traditional experimental approaches) can support the decision points along the pathway. Ideally, more well established models such as QSAR models and simpler (e.g. classifier) models can be employed for many compounds (e.g. early chemistry-driven discovery). Later, when increasing amounts of experimental data are generated, for instance from automated patch-clamp systems, more mechanistic models can be utilised for the purpose of investigational and interpretation type work.
Introduction to cardiac models
The groundbreaking work of Hodgkin & Huxley on squid giant axon published in 1952 [22] laid the core foundation for the mechanistic modelling of electrophysiology. This work linked the kinetics of ion channel conformation change with ion fluxes across the membrane and the change of the transmembrane potential. In 1962, Denis Noble [23] successfully extended the Hodgkin–Huxley equations to model the electrophysiology of cardiac cells. Since then, with the emerging understanding of the underlying biology – for example discovery of new ion channels [24], exchangers 25, 26 and pumps [27] – cardiac cell models have been developed further to have more-detailed representations of cellular components. These models have been brought to a relatively mature state and have been used to guide further investigation of biology and underlying mechanisms [e.g. prediction of the sodium/calcium exchanger (NCX) characteristics] [26]; to quantify certain phenomena, for example adaptation to different pacing frequency [28]; or for understanding mechanisms related to atrial fibrillation 29, 30 and hypertrophic cardiomyopathy [31]; to refine experimental protocols [32]; and more recently, prediction of drug action (reviewed in more detail in later sections). Models have been developed for different cell types: for example pacemaker cells 33, 34, atrial cells 26, 35, 36, 37, Purkinje fibres 23, 38, 39, 40 and ventricular cells 41, 42, 43, 44, 45, 46, and for different species: for example rabbit 26, 35, 47, guinea pig 42, 43, 44, human 35, 41, 45, 46, 48.
The selection of ion channels represented in cellular models can be different. This can be as a result of biological differences between cell types (e.g. funny current is mainly expressed in pacemaker cells). It can also reflect advances in the understanding of cellular biology and physiology when new ion channels are identified and characterised: the earlier models might have fewer channels, exchangers and pumps included or they have ‘lumped’ currents (e.g. a generic delayed rectifier potassium current without the separate rapid and slow components, IKr and IKs) [42]. Models with ‘lumped’ currents can be inconvenient to use for drug-safety prediction, where the concept of the particular ion channel protein that is blocked by a compound needs to be linked to an individual ion current.
Ion currents can also be modelled differently. Not only can the parameter values differ (e.g. current conductance, inactivation or activation time constants) but the formulation can also differ. For example, the L-type calcium (ICaL) current can be modelled using Hodgkin–Huxley formulation (which assumes independence of different ion channel gating kinetics) [47] or a more generic Markov model [49]. One can also use Markov models with different numbers of transition states to model the same current, for example four states [50], five states [51] and six states [52] in Markov models for IKr. An exciting new development is the derivation of Markov models from the energy landscape produced by molecular dynamics simulations [53]. At present this approach has been taken for IKs, as more ion channel structures become known this could be an excellent way to derive Markov models.
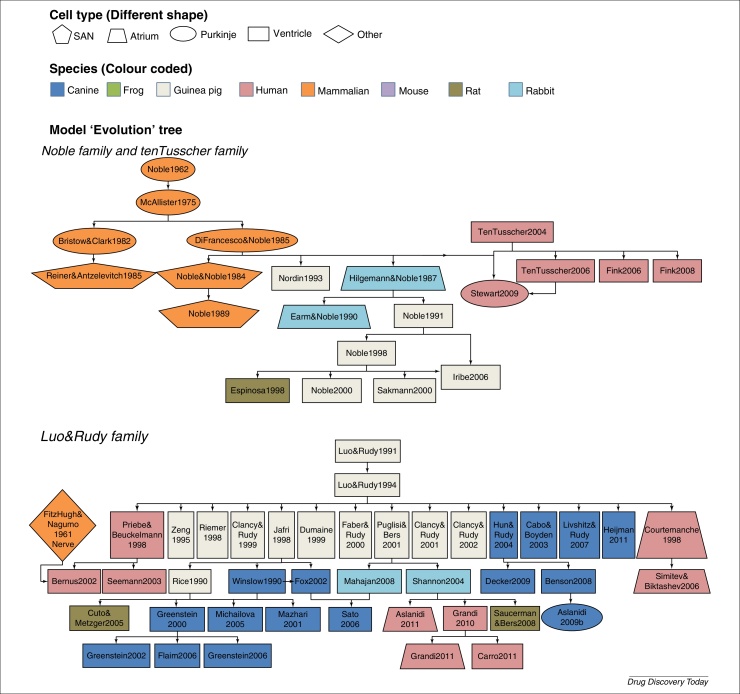
To address the complexity of intracellular Ca2+ handling in the cardiac cells, cardiac cellular models can have several intracellular compartments with different calcium handling processes (e.g. Ca2+ release or Ca2+ uptake) in each compartment and Ca2+ diffusion between the compartments. The number of intracellular compartments can differ from one model to another from the earlier single compartment models [42] to multicompartment models 49, 54. Although cardiac cell models differ from each other in many ways, they were often built as extensions to earlier models, with some channel formulations and parameter values inherited. A 2012 review paper [55] nicely summarises this inheritance between different models, which can be viewed as a family-tree diagram in Fig. 2.
Figure 2.
Complex heritage and interrelation between some frequently used cardiac electrophysiological cell models. Lines indicate how models inherit formulations from ‘parent’ models, whereas node colour indicates the reported species type, and shape is the reported cell type of the model. The inheritance shown in this figure was adapted, with permission, from [55].
In Fig. 2, we see a complex interrelation and heritage between cardiac cell models built for different cell types or different species. One could question to what degree a model is really cell-type- and species-specific. A meta-analysis was performed [56] on two frequently used human models: ten Tusscher 2004 [41] and Iyer 2004 [57]. The authors found that although these two models were fitted to different datasets, both models were based on data obtained from multiple species: ∼50% from human, ∼25% from guinea pig and ∼25% from other species (including frog). The experimental conditions were also diverse with measurement temperatures varying from 10°C to 37°C. One needs to consider these limitations when performing a species-specific simulation or prediction. Owing to the differences in the model parameterisation and structure, even models built to simulate the same phenomenon (e.g. same cell type and same species) can behave very differently under perturbation (e.g. drug block) [58]. This is an issue (perhaps comfortingly) not solely in the domain of cardiac models. Therefore consideration for how mathematical approaches have been applied elsewhere for determining model selection could be worthwhile [59]. Awareness of the assumptions and underlying experimentation and data used for the calibration of such models is essential. One must partner that awareness with careful evaluation and validation before using these models for prediction.
Cardiac physiology is modelled not only at the single-cell level but also in multiple tissue dimensions from 1D (representing a string of cells), 2D (representing a sheet or layer of cells) through to 3D models of the whole heart and torso. To couple the single cardiac cell models together to reflect the physiology, the electrical propagation from one cell to another is modelled as the diffusion of charge throughout space, with the cardiac action potential models providing sources of charge. The equations for representing this diffusion are termed either the mono- or bi-domain equations. In the bidomain model intracellular and extracellular charges can diffuse independently, whereas the monodomain equation assumes intracellular and extracellular diffusion operate proportionally but in the same directions. The monodomain equation is a special case of the bidomain equation, and can help to reduce the computational demand of spatial models at the expense of being unable to represent changes in just extracellular currents (e.g. during defibrillation). In both cases, the complex fibre direction in the heart is represented by the diffusion of charge operating more strongly in these equations in some directions than others [60]. Although the single-cell-level is sufficient to explore the impact of ion current changes on the action potential or calcium cycling, scaling up from the single-cell- to whole-organ- and whole-body-level will be a crucial step in supporting our understanding of how the effects at the ion-channel-level translate to changes in the electrocardiogram (ECG) and phenomena such as re-entry and arrhythmia. Models at these higher scales have been reviewed elsewhere [61], in the majority of this review we focus on the cellular-level models that are becoming routinely used in drug development.
How to handle variation in experimental data
Most cardiac models use a fixed set of parameters 33, 43, 49 – or a few distinct sets of parameters for different cell populations such as epicardial, midmyocardial and endocardial cells 62, 63 – without considering biological and experimental variability. Experimental variation has been recently demonstrated in the context of ion channel screening [64] and how to respond to this uncertainty is an important challenge for electrophysiology simulations. This variability can become important when making a prediction that accounts for intra- and inter-individual variation or expansion for an entire population where distinct subcategories of patients such as those with genetic channelopathies or underlying comorbidities (e.g. heart failure, atrial fibrillation) cause significant differences in the cellular action potentials. Population-based approaches, which can be used to fit a model to multiple datasets, have been applied to different types of modelling, for example PBPK modelling [65] and cardiac modelling 66, 67, 68. Statistical techniques, such as Bayesian inference, have also been used to parameterise models and to quantify the variability and uncertainty (with probabilities) 69, 70 which could be very beneficial for risk prediction.
Modelling and its application to cardiac risk assessment
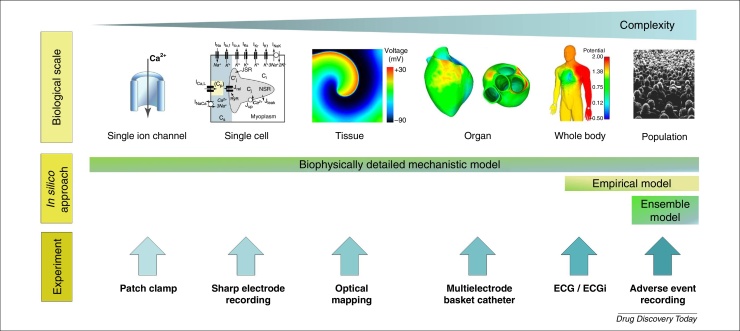
Figure 3 sets out the different scales at which measurements can be taken and in silico models can be used, a more detailed discussion on the opportunities for novel experimental techniques occurred recently at an interdisciplinary workshop [71]. The cardiac modelling field has advanced in the sense that in silico approaches and experimental measurements are now developed to the point where an assessment of the translation between different scales (e.g. between ion channel to whole cell or between in vitro to in vivo) is feasible.
Figure 3.
Schematic diagram showing that cardiac models (top) and corresponding experimental platforms (bottom) have been developed for use at different scales and levels of complexity. From left to right: single ion channel, single cell, 2D/3D tissue, whole organ and whole body (ECG). The biophysically detailed model has been used across scales from single ion channel to torso ECG (bottom-up modelling), whereas the empirical models tend to focus on modelling in vivo data such as ECG (top-down modelling). Ensemble approaches offer an opportunity to represent variants (virtual subjects) within a population. Images within the figure were adapted, with permission, from 49, 75, 81, and from StockSnap (https://stocksnap.io).
At the lower scales, investigators have described the pharmacology of single ion channels and single myocardial cells with focus on mechanistic approaches. Tissue, organ and whole-body biophysical models have been employed to understand the electrophysiology of cardiac activity. At the higher scales, modelling efforts are geared towards describing ECG characteristics in the population and explaining the link to adverse cardiac events [72]. As in any application, modelling should be pragmatic and its complexity fit for purpose, often ranging from cellular models with tens of equations and parameters to simple concentration–effect relationships. It is useful to recognise that observations (that can support, for example, model calibration and validation exercises) can be made along all the different scales (e.g. from isolated ion channel recordings) through single cell recordings, whole heart wave patterns via techniques such as ECGi [73] up to consequential observations via clinical trials and the emerging discipline of real-world data via adverse event recordings and electronic health records (EHRs) [74]. As a result, academic and commercial tools, such as the preDiCT project 75, 76, the UT-Heart initiative [77] and the Cardiac Safety Simulator™ [78], that include aspects of 3D structure considerations as well as PK, single and multicellular models are being developed. These tools offer a chance to bring these multiscale measurements, after careful calibration and validation, into a single framework for decision making. Evaluating predictive power for different modelling approaches is essential for understanding how the additional complexity that is introduced into the models affects overall predictive capacity. To minimise discordance between the different scales, there is a tendency to add increasing complexity to models. This can be reflected by observing that the number of parameters in models correlates well with the CPU transistor count over time (Davies et al., unpublished data). It is here that an iterative approach that can simplify and add the necessary complexity should be applied; bigger is not necessarily better 79, 80.
A challenge facing drug developers in the assessment of cardiac safety is the interpretation of preclinical findings and their translation to human. The pharmaceutical industry typically evaluates a palette of in silico, in vitro and in vivo assays for potential cardiovascular risk before testing in human. ECG and blood pressure recordings in animal models are often used to bolster confidence in a drug candidate's in vivo cardiovascular effects and support decision making on the compound's suitability for progression. Various publications describe the top-down analysis of such data and PK/PD models in support of the interpretation of in vivo findings 82, 83, 84. To that purpose, the models attempt to elucidate and reproduce the relationship between drug exposure and observed changes in various cardiovascular endpoints (e.g. QRS, QTc, T-wave morphology, heart rate, blood pressure and contractility) 85, 86. M&S techniques enable developers and researchers to gain key insights for the assessment of a compound's therapeutic index, for instance through the extrapolation of exposures associated with the onset of cardiovascular effects or specific effect magnitudes (e.g. 10 ms QTc prolongation). These exposure–response analysis techniques have recently formed a component of data presented to support a TQT study waiver and a key part of Phase I clinical studies [87]. Often, semi-mechanistic, empirical or statistical approaches are preferred over mechanistic models, owing to the lack of, or partial understanding of, the mechanism of pharmacological action. This in turn could hinder the translatability of findings across species, and indeed discordance is sometimes observed between predictions and clinical outcomes [80].
Naturally, opportunities exist to combine bottom-up and top-down approaches to use their strengths and potentially link pharmacological effects at a cellular level to cardiac observations and clinical outcomes. A mechanistic simulation of cellular processes should allow prediction of cardiac toxicity potential in a detail unsurpassed by empirical concentration–effect relationships, particularly for compounds affecting ion channels in novel ways. Cardiac myocyte modelling holds the promise of enabling truly translational in vitro investigations and in silico extrapolations from animal to human. By contrast, the complexity of most biophysical models does not readily permit their widespread use for high-throughput risk assessment and compound prioritisation in early development. It has been suggested that these models might not yet have matured enough to add predictive power over more pragmatic approaches [79]. Further efforts can be devoted to linking cardiac myocyte model readouts for the purpose of defining the mechanisms behind the cardiac disturbance (e.g. APD90, triangulation, EAD propensity, upstroke velocity) to biomarkers of preclinical and clinical significance (e.g. TQT, QTc, T-wave morphology, beat-to-beat variability) 58, 88, 89, 90, 91, 92. Such attempts to link cardiac safety endpoints across multiple scales have the potential to provide a more human-relevant assessment of proarrhythmic risk earlier in drug development. Here, the mechanism-based models together with physiological experiments should be used to integrate the findings and to reduce the incidence of discordance with some drugs by providing mechanistic insights for these observations.
Existing in silico evaluations for drug development decision making
The ideal scenario for drug developers and regulators is to identify that a minimal set of measurements needs to be made experimentally to predict accurately the propensity for causing arrhythmia. Preferably, these measurements would be made in early discovery and in a reproducible and high-throughput format. The outputs can then be integrated into an algorithm to provide a risk score for decision making. To date, various measurements, models and algorithms, and risk scores have been applied to this task within the pharmaceutical industry 58, 67, 79, 80, 82, 93, 94. Five of these studies use the IC50 score as the input values into the algorithm, whereas two of the studies 82, 94 use in vivo cardiovascular endpoints or gene expression signatures, respectively, as alternative strategies for the purpose of cardiac safety risk scoring.
The first study [82] demonstrated a PK/PD modelling approach for assessing cardiovascular safety when in vivo data are already known. In this study, the cardiovascular safety data, such as QRS complex, QTc interval, heart rate and blood pressure, could be correlated to the plasma concentrations to predict across species and to provide an estimate of therapeutic window. Whereas some ion channel data are measured in this study, the integration of this information was used only for qualitative purposes. A holistic approach was used to identify compounds with a likely cardiovascular risk [94]. This study suggests gene expression profiles can be used as a surrogate for hERG inhibition based on the premise that, although hERG inhibition is independent of structurally diverse chemicals, it is dependent on a conserved cell physiological response that can be independent of chemical diversity. They cluster gene expression fingerprints then identify those clusters where hERG channel inhibition has been previously described. Therefore de novo gene fingerprints that co-cluster with these hERG inhibitor enriched fingerprints are more likely to show hERG current block.
The remaining studies have all used ion channel screening data as an input with varying abstractions of statistical or mechanistically based models. Each of these studies attempts to emulate or improve on the inherent modelling performed within the mind of an experienced electrophysiologist or safety pharmacologist (i.e. in cerebro modelling). Variance in this strategy for predicting cardiac safety concern is also present, and referred to as a ‘matter of opinion’. A seminal study [95] was the first major attempt to classify potential risk of Torsade de Pointes (TdP) based on hERG IC50 data only, from 100 drugs. In their analysis the authors concluded a correlation of hERG activity with incidence of TdP, yet it is not absolute and in general a threshold of 30-fold difference between a free plasma concentration and hERG IC50 was proposed as sufficient to mitigate risk (except in the cases of multichannel drugs). Another study to use the Redfern categories as a surrogate of TdP risk [93] was also the first study to benchmark a mechanistic in silico model simulation against a drug's clinical risk. For the 31 drugs profiled, an improved prediction (i.e. Redfern categorisation) was observed when using multichannel (hERG, hNaV1.5 and hCaV1.2) data from combining literature reports with patch-clamp data versus using a ratio of hERG IC50 and effective free therapeutic plasma concentration (EFTPC) alone. A parallel study [67] also used a mechanistic model to integrate multichannel data to predict action potential changes from a canine cardiomyocyte assay. Here, they used an additional two channels (hKv4.3/hKChIP2.2 and hKv7.1/hminK, corresponding to the Ito1 and IKs currents) for a set of 53 compounds that had all been profiled using the IonWorks® automated patch-clamp system. This study also included an ensemble of parameter sets to represent 19 different dogs from which the model had been parameterised. A further study [96] evaluated the accuracy of an in silico model when presented with data from QSAR predictions or from different experimental platforms, either the PatchXpress® (77 compounds) or a combination of using data from IonWorks®/FLIPR assays (121 compounds) for predicting QT prolongation obtained from a rabbit ventricular wedge assay.
Reemphasising the importance of considering hERG, Nav1.5 and Cav1.2 channels was a study [80] that integrated the data obtained from PatchXpress® and QPatch platforms for 55 compounds divided between compounds that were either torsadogenic (32) or nontorsadogenic (23) (determined from either the Redfern study or the AZCERT database), and showed that a statistical (logistic regression) model based on multichannel data is superior to models based only on hERG data. In another study [58], ion channel screening datasets were combined (for up to five currents) from two pharmaceutical companies to evaluate the prediction of the outcome of the TQT study using simulations of APD90 at the EFTPC. The study importantly highlights discordance for some drugs between simulated action potential effects at EFTPC and clinical observations at EFTPC that cannot readily be explained.
The most recent study [79] shows that cardiac liability is essentially a balance between the predicted depolarising and repolarising effects. They simplified the risk assessment algorithm, similar to that described in [80], to just integrate three key cardiac ion currents: IKr, INa and ICaL, as the input values for an empirical, classifier model. Other emerging areas of prediction not reliant on mechanistic modelling include machine learning approaches that attempt to use the knowledge of well-studied compounds to parameterise an empirical model that accepts ion channel data as the differentiating data between compounds (see for example: https://cardiotox-predictor.com).
Considerations for improving the reproducibility and accuracy of cardiac risk prediction
The in silico approach has attracted a lot of attention and interest and has shown some promising results. However, it is vital to highlight that different models can produce very different predictions, see particularly [98] and also the Web Lab tool [97] (see https://chaste.cs.ox.ac.uk/FunctionalCuration). Understanding these differences and assessing the performance of any given model for individual context is important to avoid drawing incorrect conclusions. Modelling relies not only on diligence in building models but on a thorough and honest assessment of its limitations to avoid inappropriate predictions. In this section, we discuss some of the aspects of these models that can lead to divergence in predictions, what these divergences mean and what can be done to mitigate them.
Selection of training and validation data
Because each study has used different input parameters (measurements and different outputs) it is not currently possible to evaluate one versus the other easily. Therefore, a carefully balanced and highly characterised evaluation set that can be routinely applied to these different systems is essential. The CiPA initiative has defined a set of 28 compounds (Table 1) that have a range of proarrhythmic potential as defined by the CredibleMeds® score (http://www.crediblemeds.org). Therefore, each model system should be evaluated against a standard set of compounds to collect data across the scale continuum (Fig. 3) enabling a more thorough understanding of the translatability of these models.
Table 1.
Compounds selected for CiPA proarrhythmia testing, ranked by torsadogenic risk assessmenta
| High risk | Intermediate risk | Low risk |
|---|---|---|
| Azimilide | Astemizole | Diltiazem |
| Bepridil | Chlorpromazine | |
| dl-Sotalol | Cisapride | Loratadine |
| Dofetilide | Clarithromycin | Metoprolol |
| Ibutilide | Clozapine | Mexiletine |
| Methadone | Domperidone | Nifedipine |
| Quinidine | Droperidol | Nitrendipine |
| Vandetanib | Ondansetron | Ranolazine |
| Pimozide | Tamoxifen | |
| Risperidone | Verapamil | |
| Terfenadine |
Source: Table 1 from https://dx.doi.org/10.1038/nrd.2015.34.
Careful validation of a statistical or mechanical model's predictive performance involves testing the predictions against unseen validation data. The validation set defines the performance of a model, and should be chosen carefully to represent a context of use. In a cardiac safety situation there are various aspects to this. First, the training and validation datasets should be gathered in the same way as the data that are intended to be used for predictions when in production. Table 2 shows predictions are seen to depend strongly on: (Table 2a) the mathematical model that is used (O’Hara, ten Tusscher '06 or Grandi) when the data source (fully automated ‘Q’) is fixed; and (Table 2b) the basis of the validation dataset – using the same compounds but ion channel screening was performed using fully automated (Q) or automated and manual (M&Q) methods when the model is fixed (O’Hara model). In both cases, the concentration at which predictions are made, often unknown in early safety testing, is flexible to account for the apparent discordance between prediction and clinical observation. To provide an accurate estimate of future performance, the relevant production protocols should be used for validation data gathering. Second, the compounds that are evaluated should be representative of those that are to be assessed in future – predicting the effects of strong specific hERG blockers is necessary but not sufficient when we consider that most of the compounds that are likely to be encountered today are multichannel blockers.
Table 2.
Differences in predictive power for TQT study results when varying in silico models (a) and validation data (b)
| Concentration range | O’Hara | TenTusscher-06 | Grandi |
|---|---|---|---|
| (a) | |||
| At TQT conc. | 62% | 50% | 59% |
| 10-fold TQT conc. | 76% | 71% | 68% |
| 100-fold TQT conc. | 88% | 79% | 71% |
| Concentration range | Q-Patch | Manual and Q-Patch |
|---|---|---|
| (b) | ||
| At TQT conc. | 62% | 71% |
| 10-fold TQT conc. | 76% | 91% |
| 100-fold TQT conc. | 88% | 91% |
Data adapted, with permission, from [58].
In addition, it is important to know which data were used in the calibration of a statistical model (or, equally, in the selection of a mechanistic model), and these should be avoided in the validation dataset. Otherwise, a spuriously accurate validation assessment will be made for cases that are actually part of the training set; and the model will subsequently perform worse than expected on a future set of compounds. An example would be where a mechanistic model is trained, such that the model provides accurate predictions of the effect of 100 nm dofetilide on APD. Clearly, dofetilide should then be omitted from any validation dataset, because its effect will be correctly predicted by definition and hence skew the performance. Therefore users should demand a clearly defined training procedure from open- and closed-source models and software to avoid this situation.
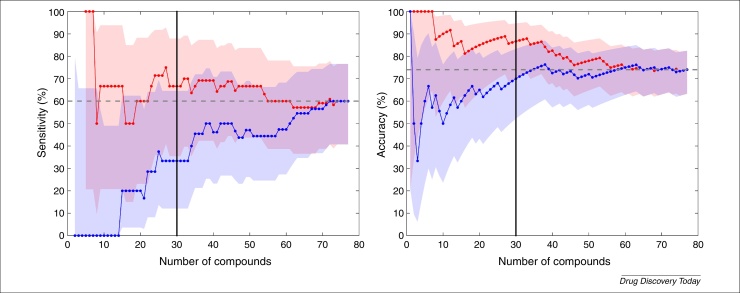
There should also be confidence estimates on our assessments of predictive power. In Fig. 4 we show how an assessment of sensitivity and accuracy of a binary classifier depends on the number of compounds in a validation set, as well as the order of the compounds in this set. The real predictive power of an assay (if it were to be evaluated on an extremely large dataset) can be very different to the reported value from a single validation study. By calculating a confidence interval, in this case based on Wilson's Score Interval [99] (as previously used in this contextin [96]), we see an estimate of how much confidence we should place in the performance statistics, and see how this changes with different numbers of validation compounds. Thirty compounds might appear a reasonable number to make a judgement as to the validity of a model (black vertical line from Fig. 4); but, in this example, precisely which 30 compounds have been randomly selected can markedly impact the predictive score of the model, we see 70% versus 86.7% accuracy in the example shown.
Figure 4.
An illustration of the dependence of performance statistics, and the uncertainty in these, on the number of compounds used in a validation study. Here, we compare whether compounds caused 10% prolongation of QT interval in a rabbit left-ventricular wedge with simulations based on multiple ion channel automated PatchXPress® screens. We plot the (left) sensitivity and (right) accuracy of the assay as a function of the number of compounds that are considered in the validation set (n = 77 available in total). The blue and red data are from the same compounds but considered in a different (randomly permuted) order, note that both measures have to start at either 0% or 100% (the first classification is right or wrong), and that the entries for n = 77 must be identical. 95% confidence intervals on these statistics are generated using Wilson's Score Interval, and are shown with the shaded regions. The data shown are taken, with permission, from [96] (available to download from: http://www.cs.ox.ac.uk/chaste/download.html – Jptm2013Beattie project).
Table 3 shows how the composition of compound sets across several in silico evaluation studies is variable and, therefore, the importance of a defined, well-balanced set of compounds (e.g. from the CiPA initiative) is of considerable value for making a genuine comparison possible. Although 28 compounds have appeared in more than one study, no compounds have been used in every study and only seven of those compounds have appeared in more than two studies, therefore meaning that evaluation across different studies is difficult.
Table 3.
Comparison of compound assessment from previous in silico studies
| Study | Compounds in common with at least one other studya | No. uniquein silicostudy compounds | Compounds common to CiPA listb |
|---|---|---|---|
| Mirams 2014 (39 cmpds) | 11 | 28 | 1 |
| Kramer 2013 (55 cmpds) | 28 | 27 | 20 |
| Davies 2012 (53 cmpds) | 6 | 47 | 5 |
| Mirams 2011 (31 cmpds) | 18 | 13 | 13 |
Seven compounds have appeared in more than two studies: amiodarone, cisapride, dofetilide, nifedipine, pimozide, quinidine and terfenadine. No compounds have been used in every study.
Eight compounds (azimilide, clarithromycin, domperidone, metoprolol, ondansetron, ranolazine, tamoxifen, vandetanib) from the CiPA list have not previously been the subject of an in silico cardiac study.
Finally, the example in Fig. 4 shows how a model scores for predicting 10% prolongation of QT interval. Multiple, equally valid markers (e.g. 5 ms change in QT-interval) could have been chosen that would have influenced the model's performance. This becomes more pronounced when the comparator is more subjective in nature, such as a Redfern category or AZCERT ranking. These values can and do change, based on medical observation, therefore a drug with ‘no TdP risk’ cannot always be viewed as safe, see for example recent modifications of drugs such as propofol, ciprofloxacin from ‘conditional’ to ‘known TdP’ (https://www.crediblemeds.org/blog/changes-made-crediblemeds-lists1/). An opportunity therefore exists to translate cardiac models not to ‘incidence of TdP’ but rather to cardiac adverse events. This is emphasised in a study [74] that shows that, although TdP is an exemplar adverse event, a low incidence of TdP from their observational study and a lack of ICD-10 coding options mean that some important drug-induced arrhythmia events could be missed or recorded as other ventricular fibrillation events.
Model version control
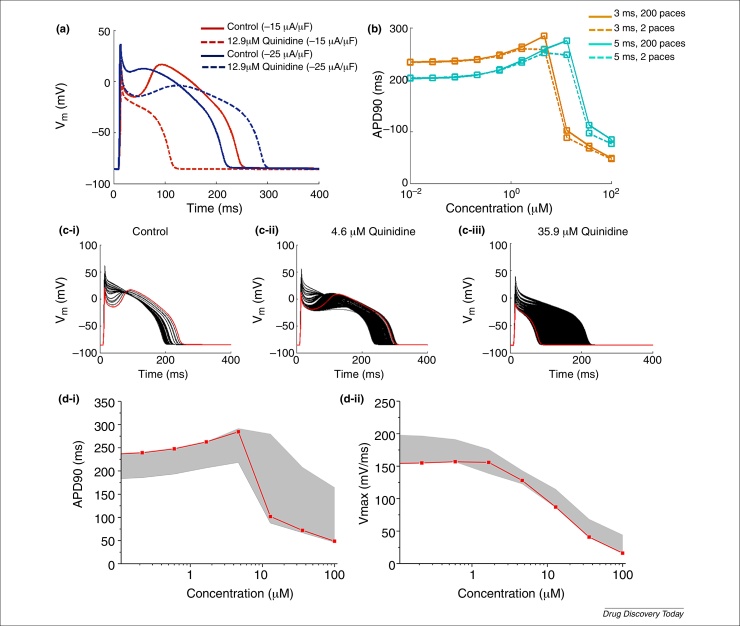
Models can be, and are, modified relative to the published version; often this can be to correct typographical errors [55] or to test alternative parameterisations. All of which emphasises the absolute requirement to standardise and version control such modifications, so that a simulation run in different labs, by different modellers, can produce mathematically identical results. An equally important aspect of ensuring reliable results is to ensure that simulation codes are well tested or verified 100, 101. To illustrate the issues that can arise, we use the Hund & Rudy 2004 model to demonstrate the importance of parameter control. Protocol-related parameters (number of paces, amplitude and duration of stimulation pulse) were varied to test the impact on the output. To see whether the protocol-related parameters can also influence the cardiac risk prediction, quinidine was taken as a case study, with IC50 values extracted from the literature – IKr: 1.5 μm [67], IKs: 50 μm [102], INa: 9.4 μm [67], ICaL: 15.5 μm [67] – and an AP was simulated with increasing concentration of quinidine (ten logarithmically, equally-spaced concentration steps between 0 and 100 μm), and with varied protocol parameters [number of paces: 1–200 paces; stimulation pulse duration: 3 (original value from the publication), 4 or 5 ms; stimulation pulse amplitude: −15 (original value from the publication), −18, −20, −22 or −25 μA/μF]. From Fig. 5a,b, one can observe that the stimulation pulse duration, amplitude and the number of pulses all influence the prediction. Although in this case the number of pulses has a smaller influence on the outcomes compared with the stimulation pulse amplitude and duration, some other models such as Mahajan model [49] are very sensitive to the number of pulses. In Fig. 5c (ci to ciii: each subfigure shows AP traces at one concentration point of quinidine), the AP traces simulated with 1–200 paces and with different stimulation pulse amplitude and duration are plotted (black); AP traces simulated using original stimulation pulse specification and after 200 paces are plotted in red. APD90 and maximal upstroke velocity were calculated from all AP traces simulated (with different stimulation pulse and after different number of paces) and are plotted against quinidine concentration (Fig. 5di,ii, respectively). Increasing variation is seen (Fig. 5c,d) as concentration of quinidine increases.
Figure 5.
The impact of the protocol-related parameters when simulating an increasing concentration of quinidine. (a) The effect of stimulation pulse amplitude on AP. The solid lines show control AP and the dashed lines show the AP with 12.9 μm quinidine. The AP traces simulated with −15 μA/μF stimulation pulse and −25 μA/μF stimulation pulse, coloured in red and blue, respectively. (b) The APD90 dose–response (with ascending concentration of quinidine) simulated using 3 ms (coloured in orange) and 5 ms (coloured in turquoise) stimulation pulse and after two (shown in dashed line) or 200 (shown in solid line) paces. (ci–iii) Simulated AP with different concentration of quinidine when applying various pacing protocols (stimulation pulse amplitude: −15, −18, −20, −22 or −25 μA/μF; stimulation pulse duration: 3, 4 or 5 ms; number of pulses: 1–200 pulses). The red line shows the AP simulated using stimulation protocol published with the original model and after 200 pulses and black line shows the AP simulated using other variants of stimulation protocols as mentioned above. (di,ii) shows dose–response of the APD90 and maximum upstroke velocity simulated using the variants of stimulation protocol used for subfigure (ci–iii). The red line shows the simulation result obtained using the stimulation protocol published with the original model, the grey shade marks out the variability induced by applying the variants (as above) of stimulation protocols.
Integration of PBPK modelling and cardiac modelling
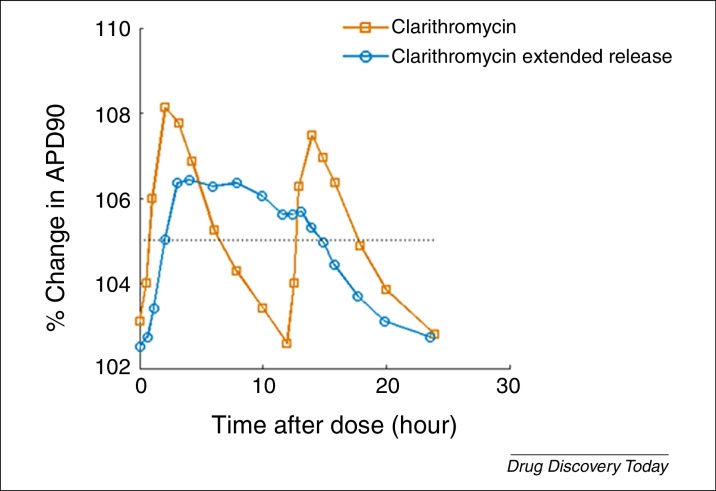
Prediction of cardiovascular effects for different formulations and dosing schedules requires a solid understanding of in vivo PK in the target patient population, which is where a PK model can prove a valuable tool. In particular, PBPK modelling techniques can also support the prediction of concentration time courses in the tissue of interest, namely the myocardium, and PK drug–drug interactions. A goal in coupling these modelling approaches is to support the link from bench to bedside in a more informative and accurate way. An opportunity exists to collect and collate experimental or observational information from across the continuum (Fig. 3) to instruct the models on how they should be parameterised and assess how they predict (at least for that limited sample). In doing so we can better understand the experimental variability that exists for, for example, patch-clamp data and therefore which value (or range of values) to use in simulations [60]. In Fig. 6 we show an example of linking PK with a cardiac model to simulate two related drug formulations with different clearance rates (one fast and one slow). It exemplifies the importance of applying a QSP approach, which looks to integrate our understanding of drug PK profile over time to these mechanism-based cardiac models. PK profiles of two different forms of clarithromycin were taken from the literature [103]; AP simulation was performed at data points along the PK profiles using the O’Hara–Rudy model [46]. The hERG IC50 (∼30 μm) of clarithromycin was taken from the literature [95]. The PK profiles and predicted APD90 and APD at 50% repolarisation (APD50) are shown in Fig. 6.
Figure 6.
Integrating an understanding of PK allows translation into a cardiac biomarker, in this case changes to action potential duration (APD50 and APD90) for two formulations of clarithromycin.
Contribution of in silico tools to reduce animal usage
In silico methodologies play an important part in the 3Rs: the refinement, reduction and replacement of tests in live animals. The in silico approach helps to identify safety liabilities and to remove toxic molecules from the pipeline long before animal experiments are conducted. This allows limiting drug testing to compounds with less toxic effects and higher chances for success. Computer models are, therefore, becoming an important tool for development of alternative testing methods that efficiently integrate complementary information derived from experimental data. The Toxicology Testing in the 21st Century document released by the US National Academy of Sciences contains recommendations on how to move the risk identification from descriptive to mechanistically based safety assessment including the use of computer-based technologies for the assessment of toxicities [104]. The FDA's Critical Path Initiative pipeline contains various computational science activities including a project on building an in silico tool for screening new drugs for QT prolongation potential, using human clinical trial data [105]. And there are several examples where in silico cardiac approaches have reduced animal tests. At AstraZeneca, although extensive screening of all key ventricular ion channels has significantly reduced overall cardiac ion channel liability, the implementation of an in silico cardiac model [67] for interpreting those remaining actives meant that an isolated myocyte study could be scaled down (from approximately 23 to one dog per year) via the simulation approach (C. Pollard, personal communication). Likewise, at GlaxoSmithKline in silico models are helping to profile and select an increased number of compounds while reducing the ex vivo rabbit wedge model by approximately 50% (J. Louttit, personal communication). Replacement and reduction of animal testing methods in different drug safety areas is not only being used in research but has also been taken up into EU law and the OECD regulations (https://eurl-ecvam.jrc.ec.europa.eu/).
A prerequisite for regulatory acceptance is validation of an alternative method showing that it can provide the same or better protection of human health when compared to traditional methods. Validation includes demonstration of relevance and reliability of a method for a specific purpose and it serves to facilitate and/or accelerate the international (regulatory) acceptance [106]. In general, the following aspects have to be considered when developing alternative methods using a combination of experimental data, computer models and integrative approaches:
-
•
Scientific relevance, characterisation, standardisation and affordability of a biological model.
-
•
The physicochemical diversity in the training and validation sets to cover a variety of mechanisms of action.
-
•
Strategic fit (e.g. prioritisation of approaches for further compound testing in lead identification and optimisation and support in decision making for selected drug candidates at later stages).
To facilitate the acceptance of mechanistically based models for regulatory purposes, new in silico tools should be associated with the following information according the OECD principles for QSAR validation 106, 107:
-
•
A defined endpoint.
-
•
An unambiguous algorithm.
-
•
A defined domain of applicability.
-
•
Appropriate measures of goodness of fit, robustness and predictivity.
-
•
A mechanistic interpretation, if possible.
No internationally accepted in silico alternative currently exists for a full replacement for all testing of a specific hazard. In silico tools alone are not yet sufficiently developed to replace the standard animal tests completely. Therefore, successful development and application of computational tools in drug safety is strongly dependent on their integration and interplay with the experimental approaches. Models are established using either in vitro or in vivo data or a combination of both to obtain predictions of system-level behaviour. Embracing more in silico tools at various levels in our current approaches is a logical consequence of trying to mimic an organ(ism). The higher level of complexity needs to be recapitulated through the use of multiple complementary tests and computational models, which translate the results of scientific research into valid tools that can significantly reduce animal testing while still ensuring the highest level of public health protection.
The practical use of mechanistic mathematical models for safety assessment by regulators is a rather new concept with no specific regulatory guidance in place. Computational safety data are usually submitted on a voluntary basis and are not required. New tools are emerging fast and changing continuously as new data become available – an intrinsic feature of the technology that makes it challenging for potential users to gain sufficient experience. The use of mechanistic models, therefore, must be seen in the context of their potential to model multiple mechanisms of compound toxicity. Customised computational platforms of understandable construction validated with appropriate compound sets provide advantages for practical implementation of in silico tools and for successfully integrating in vitro, in silico and in vivo information.
Concluding remarks
The current paradigm of cardiac safety screening for, primarily, hERG activity to eliminate TdP-causing drugs from reaching the market has reduced incidence of adverse drug-induced TdP. However, this is potentially at the cost of excluding many effective therapies where multichannel effectors might have mitigated hERG-induced cardiac risk. Therefore, the initiative to reconsider the current approach is something to be applauded. However, whereas the aim to reduce the number of incorrectly labelled TdP-liable drugs (false positives) is an important step for accessing more potentially highly efficacious new medicines, it cannot be done (blindly) at the expense of increased false negatives.
In silico modelling approaches have great potential to support the drug discovery process through better systematic decision making, animal usage reduction and for providing mechanistic insights as to the compound's intentional (and unintentional) actions. These approaches are already being utilised in the industry and now more mechanistic, systems-pharmacology models are also being investigated for regulatory purposes. However, before wide-scale adoption, careful evaluation and validation need to be conducted on these models, to ensure that we are indeed improving decision making rather than confusing it. It is vitally important to recognise the end user of such tools, so that a consistent interpretation is overlaid to the simulations. For this purpose, development of user-friendly tools and further training of software and model developers and the end user of such simulations to make predictions more readily interpretable is essential for the successful implementation of new initiatives 16, 71, 97, 108. In the consideration of the most appropriate model(s) we advocate a need to consider how the models can respond to future predictions of the unknown compounds rather than simply retrospectively fitting past observations. The reality is probably a need for complementary and fit-for-purpose solution rather than a one-size-fits-all approach, which also aims to support decision making in the understanding of prediction.
Reconciling the demand for ever more detail with the demand that models be sufficiently tractable, mathematically and computationally, to be useful is an essential consideration. These demands could appear to be in opposition to each other. There cannot be any doubt that more detail will be required because there are still many factors that can be involved in arrhythmogenesis that are not yet represented, but could be. These include more detail on metabolic changes underlying forms of arrhythmia and more detail on the feedbacks by which electrical and ionic changes might be involved longer-term in controlling gene expression (electrotranscription coupling). These alternative mechanisms have been attempted elsewhere, for instance inclusion of binding kinetics [52], gene expression [109], safety pharmacology [110], influence of beta-adrenergic activity [111] and cell signalling 112, 113. Because we are already faced with problems of under-determination in the more-complex models, how can these additional demands be compatible with the need for mathematical reduction to simpler models? One aspect is improving the information content of experiments by using models to assist in the experimental design. When we treat experiments as informing us about the parameters in models it becomes possible to optimise the experiments to tell us more about the underlying processes [114]. But there is a paradox: it could be by incorporating more detail that we will eventually find it possible to derive the most useful mathematical reductions. This might be a case of exploring more to focus on less.
For in silico cardiac models, many studies and providers have evaluated the value of a different model or software for predicting cardiac proarrhythmic risk. Yet with a different emphasis on each approach and a different set of evaluation drugs for which the approach is scored, an overall assessment of which platform is most appropriate for onward prediction is difficult. In this review we have discussed approaches that have been previously implemented, how each might respond differently dependent upon the circumstances and the consequent need for caution in implementing a solution. With a careful and thorough approach, there is considerable value to be gained from in silico approaches, where the primary motivation is not only about mapping out cardiac biology to high accuracy but in providing a method for supporting decision making within drug discovery and development.
Glossary
- APD90
action potential duration at 90% repolarisation (time to return to the resting membrane potential)
- Bidomain equations
differential equations representing the time and space changes in charge in two compartments (intra- and extra-cellular), owing to diffusion of charges separately and/or differently in each compartment, and transmembrane currents linking the two
- Cardiac arrhythmia
irregular heartbeat
- Cardiac action potential
the change in the electrical membrane potential of the cardiomyocyte
- CiPA
the comprehensive in vitro proarrhythmia assay, a new paradigm for nonclinical assessment of cardiac risk, by a combination of in vitro, in silico and integrated human-cell-based assay techniques
- Ensemble models
an approach that uses several sets of model parameters, rather than a single set, to give a collection of predictions. Generally used to provide an indication of variance in the prediction or to represent a population
- hERG
human Ether-a-go-go-related gene encodes the (α-subunit of the) protein underlying IKr, a major repolarisation current in the cardiomyocyte during an action potential
- Hodgkin–Huxley model
a mathematical method, extended to represent the cardiac ion channel, that uses independent variables to describe the voltage and time-dependent gating properties of an ion channel
- In silico models
describes models that use computers and algorithm-based approaches for representing a system. Particularly used to contrast to, for example, in vitro (cell-based) models or in vivo (animal-based) models
- Markov state model
an alternative method to Hodgkin–Huxley approaches that represents the open, closing and inactivation of ion channels as a sequence of dependent states and transitions, which can be dependent upon drug binding and ion concentrations, voltage or time
- Model calibration
the process by which all or a subset of parameters or components of the model are adjusted or modified to best fit with a set of previously measured outcomes
- Model validation
the process of testing the performance of an in silico model against a set of measurements that are (usually) not part of the model calibration process
- Monodomain equations
differential reaction–diffusion equation representing the time and space changes in charge throughout a single spatial ‘domain’ – the inside of cells – and the contributions that the transmembrane currents make to this
- Parameter set
this term is used to describe the set of values that form components of the algorithm behind the in silico model that are fit to characterise the system. These parameters might or might not have a direct physical meaning (e.g. the expression level of a protein or the gating kinetics of an ion channel)
- PK/PD
the study of how the body responds to a dose of drug [i.e. the drug concentration over time (PK) and the observed effects on the body by the dose (PD)]
- Quantitative systems pharmacology (QSP)
a discipline that takes a more mechanism-systems-based approach at modelling the pharmacology of a drug and its effects on the body
- Top-down, bottom-up, middle-out modelling
these terms are used to describe the starting point for the construction of an in silico model to explain the phenomena of interest. Bottom-up describes approaches that start with the individual molecules, whereas top-down begins at the physiological level and attempts to include only the physiological elements necessary for explaining the phenomena. Middle out is a more pragmatic attempt to address the short comings of the top-down and bottom-up approaches
- Torsade de Pointes
a polymorphic ventricular arrhythmia that exhibits distinct characteristics on the electrocardiogram (ECG) and can potentially lead to sudden cardiac death
Biographies

Mark Davies is a consultant at QT-Informatics, a consultancy for in silico and data science techniques primarily in cardiac safety assessment. Mark gained his PhD in biological sciences at the University of Warwick and has previously worked at the interface between mathematics and biology as a senior postdoctoral researcher at the University of Oxford and, prior to this, at AstraZeneca where he led a team that developed the in silico action potential (isAP) tool for supporting decision making in safety pharmacology.

Ken Wang holds a RPF postdoctoral fellowship at Roche Research & Early Development in Basel working in the In Vitro Cardiac Safety Lab and Quantitative Systems Pharmacology group. Ken obtained her PhD from the University of Oxford. During her PhD, she worked on live cardiac tissue slices and dual voltage calcium optical mapping as well as computational modelling of cardiac electrophysiology. Ken joined Roche in 2015 after her PhD and she is currently working on ion channel assays and developing and validating mechanistic in silico models for cardiac safety.

Gary Mirams is an applied mathematician at the University of Oxford working on problems in biology, particularly in the area of cardiac electrophysiology and drug safety. He is working with a number of pharmaceutical companies to embed simulation in their safety work, and to develop free open software to perform these simulations. He is currently funded by the Wellcome Trust & Royal Society on a Sir Henry Dale Research Fellowship. Gary has a PhD in mathematics from the University of Nottingham.

Liudmila Polonchuk is a Principal Scientist at Roche Research & Early Development in Basel. She has a PhD in natural sciences from the Swiss Federal Institute of Technology. A key area of her scientific expertise is cardiovascular biology. Liudmila joined Roche in 2001 as a Postdoctoral Fellow to develop a heart failure animal model for drug safety application and she has led a safety pharmacology lab since 2003. Her current research area involves implementation of in-silico-based integrative platforms for the cardiac safety assessment in drug development.
References
- 1.Sood S. A novel multi-tissue RNA diagnostic of healthy ageing relates to cognitive health status. Genome Biol. 2015;16:185. doi: 10.1186/s13059-015-0750-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendriks B. Negative modeling results: a dime a dozen or a stepping stone to scientific discovery? CPT Pharmacometrics Syst. Pharmacol. 2013;2:e48. doi: 10.1038/psp.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiFrancesco D., Noble D. A model of cardiac electrical activity incorporating ionic pumps and concentration changes. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 1985;307:353–398. doi: 10.1098/rstb.1985.0001. [DOI] [PubMed] [Google Scholar]
- 4.Gobburu J.V., Lesko L.J. Quantitative disease, drug, and trial models. Annu. Rev. Pharmacol. Toxicol. 2009;49:291–301. doi: 10.1146/annurev.pharmtox.011008.145613. [DOI] [PubMed] [Google Scholar]
- 5.Suryawanshi S. The current role of model-based drug development. Expert Opin. Drug Discov. 2010;5:311–321. doi: 10.1517/17460441003713470. [DOI] [PubMed] [Google Scholar]
- 6.Visser S.A. Implementation of quantitative and systems pharmacology in large pharma. CPT Pharmacometrics Syst. Pharmacol. 2014;3:e142. doi: 10.1038/psp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mould D.R. Developing exposure/response models for anticancer drug treatment: special considerations. CPT Pharmacometrics Syst. Pharmacol. 2015;4:e00016. doi: 10.1002/psp4.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mager D.E., Jusko W.J. Development of translational pharmacokinetic-pharmacodynamic models. Clin. Pharmacol. Ther. 2008;83:909–912. doi: 10.1038/clpt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J.Y. Impact of pharmacometric analyses on new drug approval and labelling decisions: a review of 198 submissions between 2000 and 2008. Clin. Pharmacokinet. 2011;50:627–635. doi: 10.2165/11593210-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Ette E.I., Williams P.J., editors. Pharmacometrics: The Science of Quantitative Pharmacology. Wiley-Blackwell; 2007. [Google Scholar]
- 11.Wagner C. Application of physiologically based pharmacokinetic (PBPK) modeling to support dose selection: report of an FDA public workshop on PBPK. CPT Pharmacometrics Syst. Pharmacol. 2015;4:226–230. doi: 10.1002/psp4.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caruso A. PK/PD assessment in CNS drug discovery: prediction of CSF concentration in rodents for P-glycoprotein substrates and application to in vivo potency estimation. Biochem. Pharmacol. 2013;85:1684–1699. doi: 10.1016/j.bcp.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 13.Parrott N. Application of full physiological models for pharmaceutical drug candidate selection and extrapolation of pharmacokinetics to man. Basic Clin. Pharmacol. Toxicol. 2005;96:193–199. doi: 10.1111/j.1742-7843.2005.pto960308.x. [DOI] [PubMed] [Google Scholar]
- 14.Jones H.M. Physiologically based pharmacokinetic modeling in drug discovery and development: a pharmaceutical industry perspective. Clin. Pharmacol. Ther. 2015;97:247–262. doi: 10.1002/cpt.37. [DOI] [PubMed] [Google Scholar]
- 15.Leil T.A., Ermakov S. Editorial: the emerging discipline of quantitative systems pharmacology. Front. Pharmacol. 2015;6:129. doi: 10.3389/fphar.2015.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorger P.K. National Institutes of Health; 2011. Quantitative and Systems Pharmacology in the Post-genomic Era: New Approaches to Discovering Drugs and Understanding Therapeutic Mechanisms. [Google Scholar]
- 17.Brenner S. Understanding complex systems: top-down, bottom-up or middle-out? In: Bock G., Goode J., editors. Vol. 239. John Wiley and Sons; 2001. pp. 150–159. (Novartis Foundation Symposium: Complexity in Biological Information Processing). [Google Scholar]
- 18.Vicini P., van der Graaf P.H. Systems pharmacology for drug discovery and development: paradigm shift or flash in the pan? Clin. Pharmacol. Ther. 2013;93:379–381. doi: 10.1038/clpt.2013.40. [DOI] [PubMed] [Google Scholar]
- 19.van der Graaf P.H., Benson N. Systems pharmacology: bridging systems biology and pharmacokinetics-pharmacodynamics (PKPD) in drug discovery and development. Pharm. Res. 2011;28:1460–1464. doi: 10.1007/s11095-011-0467-9. [DOI] [PubMed] [Google Scholar]
- 20.Noble D. Modeling the heart. Physiology. 2004;19:191–197. doi: 10.1152/physiol.00004.2004. [DOI] [PubMed] [Google Scholar]
- 21.Mirams G.R. Application of cardiac electrophysiology simulations to pro-arrhythmic safety testing. Br. J. Pharmacol. 2012;167:932–945. doi: 10.1111/j.1476-5381.2012.02020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgkin A.L., Huxley A.F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble D. A modification of the Hodgkin–Huxley equations applicable to Purkinje fibre action and pace-maker potentials. J. Physiol. 1962;160:317–352. doi: 10.1113/jphysiol.1962.sp006849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter H. The dependence of slow inward current in Purkinje fibres on the extracellular calcium-concentration. J. Physiol. 1967;192:479–492. doi: 10.1113/jphysiol.1967.sp008310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuter H., Seitz N. The dependence of calcium efflux from cardiac muscle on temperature and external ion composition. J. Physiol. 1968;195:451–470. doi: 10.1113/jphysiol.1968.sp008467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilgemann D.W., Noble D. Excitation–contraction coupling and extracellular calcium transients in rabbit atrium: reconstruction of basic cellular mechanisms. Proc. R. Soc. Lond. B: Biol. Sci. 1987;230:163–205. doi: 10.1098/rspb.1987.0015. [DOI] [PubMed] [Google Scholar]
- 27.Gadsby D.C. Activation of electrogenic Na+/K+ exchange by extracellular K+ in canine cardiac Purkinje fibers. Proc. Natl. Acad. Sci. U. S. A. 1980;77:4035–4039. doi: 10.1073/pnas.77.7.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pueyo E. Mechanisms of ventricular rate adaptation as a predictor of arrhythmic risk. Am. J. Physiol. Heart Circ. Physiol. 2010;298:H1577–H1587. doi: 10.1152/ajpheart.00936.2009. [DOI] [PubMed] [Google Scholar]
- 29.Colman M.A. Pro-arrhythmogenic effects of atrial fibrillation-induced electrical remodelling: insights from the three-dimensional virtual human atria. J. Physiol. 2013;591:4249–4272. doi: 10.1113/jphysiol.2013.254987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courtemanche M. Ionic mechanisms underlying human atrial action potential properties: insights from a mathematical model. Am. J. Physiol. 1998;275:H301–H321. doi: 10.1152/ajpheart.1998.275.1.H301. [DOI] [PubMed] [Google Scholar]
- 31.Passini E. Mechanisms of pro-arrhythmic abnormalities in ventricular repolarisation and anti-arrhythmic therapies in human hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohl P., Noble D. Systems biology and the virtual physiological human. Mol. Syst. Biol. 2009;5:292. doi: 10.1038/msb.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noble D., Noble S.J. A model of sino-atrial node electrical activity based on a modification of the DiFrancesco–Noble (1984) equations. Proc. R. Soc. Lond. B: Biol. Sci. 1984;222:295–304. doi: 10.1098/rspb.1984.0065. [DOI] [PubMed] [Google Scholar]
- 34.Reiner V.S., Antzelevitch C. Phase resetting and annihilation in a mathematical model of sinus node. Am. J. Physiol. 1985;249:H1143–H1153. doi: 10.1152/ajpheart.1985.249.6.H1143. [DOI] [PubMed] [Google Scholar]
- 35.Earm Y.E., Noble D. A model of the single atrial cell: relation between calcium current and calcium release. Proc. R. Soc. Lond. B: Biol. Sci. 1990;240:83–96. doi: 10.1098/rspb.1990.0028. [DOI] [PubMed] [Google Scholar]
- 36.Grandi E. Human atrial action potential and Ca2+ model: sinus rhythm and chronic atrial fibrillation. Circ. Res. 2011;109:1055–1066. doi: 10.1161/CIRCRESAHA.111.253955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simitev R.D., Biktashev V.N. Conditions for propagation and block of excitation in an asymptotic model of atrial tissue. Biophys. J. 2006;90:2258–2269. doi: 10.1529/biophysj.105.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAllister R.E. Reconstruction of the electrical activity of cardiac Purkinje fibres. J. Physiol. 1975;251:1–59. doi: 10.1113/jphysiol.1975.sp011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart P. Mathematical models of the electrical action potential of Purkinje fibre cells. Philos. Trans. A: Math. Phys. Eng. Sci. 2009;367:2225–2255. doi: 10.1098/rsta.2008.0283. [DOI] [PubMed] [Google Scholar]
- 40.Aslanidi O.V. Virtual tissue engineering of the human atrium: modelling pharmacological actions on atrial arrhythmogenesis. Eur. J. Pharm. Sci. 2012;46:209–221. doi: 10.1016/j.ejps.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 41.ten Tusscher K.H. A model for human ventricular tissue. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H1573–H1589. doi: 10.1152/ajpheart.00794.2003. [DOI] [PubMed] [Google Scholar]
- 42.Luo C.H., Rudy Y. A model of the ventricular cardiac action potential. Depolarization, repolarization, and their interaction. Circ. Res. 1991;68:1501–1526. doi: 10.1161/01.res.68.6.1501. [DOI] [PubMed] [Google Scholar]
- 43.Luo C.H., Rudy Y. A dynamic model of the cardiac ventricular action potential. II. Afterdepolarizations, triggered activity, and potentiation. Circ. Res. 1994;74:1097–1113. doi: 10.1161/01.res.74.6.1097. [DOI] [PubMed] [Google Scholar]
- 44.Luo C.H., Rudy Y. A dynamic model of the cardiac ventricular action potential. I. Simulations of ionic currents and concentration changes. Circ. Res. 1994;74:1071–1096. doi: 10.1161/01.res.74.6.1071. [DOI] [PubMed] [Google Scholar]
- 45.Grandi E. A novel computational model of the human ventricular action potential and Ca transient. J. Mol. Cell. Cardiol. 2010;48:112–121. doi: 10.1016/j.yjmcc.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Hara T. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput. Biol. 2011;7:e1002061. doi: 10.1371/journal.pcbi.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shannon T.R. A mathematical treatment of integrated Ca dynamics within the ventricular myocyte. Biophys. J. 2004;87:3351–3371. doi: 10.1529/biophysj.104.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ten Tusscher K.H., Panfilov A.V. Cell model for efficient simulation of wave propagation in human ventricular tissue under normal and pathological conditions. Phys. Med. Biol. 2006;51:6141–6156. doi: 10.1088/0031-9155/51/23/014. [DOI] [PubMed] [Google Scholar]
- 49.Mahajan A. A rabbit ventricular action potential model replicating cardiac dynamics at rapid heart rates. Biophys. J. 2008;94:392–410. doi: 10.1529/biophysj.106.98160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oehmen C.S. Mathematical model of the rapidly activating delayed rectifier potassium current I(Kr) in rabbit sinoatrial node. J. Cardiovasc. Electrophysiol. 2002;13:1131–1140. doi: 10.1046/j.1540-8167.2002.01131.x. [DOI] [PubMed] [Google Scholar]
- 51.Mazhari R. Molecular interactions between two long-QT syndrome gene products, HERG and KCNE2, rationalized by in vitro and in silico analysis. Circ. Res. 2001;89:33–38. doi: 10.1161/hh1301.093633. [DOI] [PubMed] [Google Scholar]
- 52.Di Veroli G.Y. High-throughput screening of drug-binding dynamics to HERG improves early drug safety assessment. Am. J. Physiol. Heart Circ. Physiol. 2013;304:H104–H117. doi: 10.1152/ajpheart.00511.2012. [DOI] [PubMed] [Google Scholar]
- 53.Silva J.R. A multiscale model linking ion-channel molecular dynamics and electrostatics to the cardiac action potential. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11102–11106. doi: 10.1073/pnas.0904505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pasek M. A model of the guinea-pig ventricular cardiac myocyte incorporating a transverse-axial tubular system. Prog. Biophys. Mol. Biol. 2008;96:258–280. doi: 10.1016/j.pbiomolbio.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 55.Noble D. How the Hodgkin–Huxley equations inspired the Cardiac Physiome Project. J. Physiol. 2012;590:2613–2628. doi: 10.1113/jphysiol.2011.224238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niederer S.A. A meta-analysis of cardiac electrophysiology computational models. Exp. Physiol. 2009;94:486–495. doi: 10.1113/expphysiol.2008.044610. [DOI] [PubMed] [Google Scholar]
- 57.Iyer V. A computational model of the human left-ventricular epicardial myocyte. Biophys. J. 2004;87:1507–1525. doi: 10.1529/biophysj.104.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mirams G.R. Prediction of thorough QT study results using action potential simulations based on ion channel screens. J. Pharmacol. Toxicol. Methods. 2014;70:246–254. doi: 10.1016/j.vascn.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silk D. Model selection in systems biology depends on experimental design. PLoS Comput. Biol. 2014;10:e1003650. doi: 10.1371/journal.pcbi.1003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vigmond E.J. Solvers for the cardiac bidomain equations. Prog. Biophys. Mol. Biol. 2008;96:3–18. doi: 10.1016/j.pbiomolbio.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yarov-Yarovoy V. Computational models for predictive cardiac ion channel pharmacology. Drug Discov. Today: Dis. Models. 2014;14:3–10. doi: 10.1016/j.ddmod.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ten Tusscher K.H., Panfilov A.V. Alternans and spiral breakup in a human ventricular tissue model. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H1088–H1100. doi: 10.1152/ajpheart.00109.2006. [DOI] [PubMed] [Google Scholar]
- 63.Benson A.P. The canine virtual ventricular wall: a platform for dissecting pharmacological effects on propagation and arrhythmogenesis. Prog. Biophys. Mol. Biol. 2008;96:187–208. doi: 10.1016/j.pbiomolbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Elkins R.C. Variability in high-throughput ion-channel screening data and consequences for cardiac safety assessment. J. Pharmacol. Toxicol. Methods. 2013;68:112–122. doi: 10.1016/j.vascn.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ball K. Comparing translational population-PBPK modelling of brain microdialysis with bottom-up prediction of brain-to-plasma distribution in rat and human. Biopharm. Drug Dispos. 2014;35:485–499. doi: 10.1002/bdd.1908. [DOI] [PubMed] [Google Scholar]
- 66.Britton O.J. Experimentally calibrated population of models predicts and explains intersubject variability in cardiac cellular electrophysiology. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E2098–E2105. doi: 10.1073/pnas.1304382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davies M.R. An in silico canine cardiac midmyocardial action potential duration model as a tool for early drug safety assessment. Am. J. Physiol. Heart Circ. Physiol. 2012;302:H1466–H1480. doi: 10.1152/ajpheart.00808.2011. [DOI] [PubMed] [Google Scholar]
- 68.Sanchez C. Inter-subject variability in human atrial action potential in sinus rhythm versus chronic atrial fibrillation. PLOS ONE. 2014;9:e105897. doi: 10.1371/journal.pone.0105897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robert C.P., Casella G., editors. Monte Carlo Statistical Methods. Springer; New York: 2004. [Google Scholar]
- 70.Johnstone R.H. Uncertainty and variability in models of the cardiac action potential: can we build trustworthy models? J. Mol. Cell. Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez B. Human-based approaches to pharmacology and cardiology: an interdisciplinary and intersectorial workshop. Europace. 2015 doi: 10.1093/europace/euv320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sadrieh A. Multiscale cardiac modelling reveals the origins of notched T waves in long QT syndrome type 2. Nat. Commun. 2014;5:5069. doi: 10.1038/ncomms6069. [DOI] [PubMed] [Google Scholar]
- 73.Ghanem R.N. Noninvasive electrocardiographic imaging (ECGI): comparison to intraoperative mapping in patients. Heart Rhythm. 2005;2:339–354. doi: 10.1016/j.hrthm.2004.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sarganas G. Epidemiology of symptomatic drug-induced long QT syndrome and Torsade de Pointes in Germany. Europace. 2014;16:101–108. doi: 10.1093/europace/eut214. [DOI] [PubMed] [Google Scholar]
- 75.Zemzemi N. Computational assessment of drug-induced effects on the electrocardiogram: from ion channel to body surface potentials. Br. J. Pharmacol. 2013;168:718–733. doi: 10.1111/j.1476-5381.2012.02200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Obiol-Pardo C. A multiscale simulation system for the prediction of drug-induced cardiotoxicity. J. Chem. Inf. Model. 2011;51:483–492. doi: 10.1021/ci100423z. [DOI] [PubMed] [Google Scholar]
- 77.Okada J.-I. Screening system for drug-induced arrhythmogenic risk combining a patch clamp and heart simulator. Sci. Adv. 2015;1:e1400142. doi: 10.1126/sciadv.1400142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glinka A., Polak S. QTc modification after risperidone administration – insight into the mechanism of action with use of the modeling and simulation at the population level approach. Toxicol. Mech. Methods. 2015;25:279–286. doi: 10.3109/15376516.2015.1025346. [DOI] [PubMed] [Google Scholar]
- 79.Mistry H.B. A new classifier-based strategy for in-silico ion-channel cardiac drug safety assessment. Front. Pharmacol. 2015;6:59. doi: 10.3389/fphar.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kramer J. MICE models: superior to the HERG model in predicting Torsade de Pointes. Sci. Rep. 2013;3:2100. doi: 10.1038/srep02100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mirams G.R. Chaste: an open source C++ library for computational physiology and biology. PLoS Comput. Biol. 2013;9:e1002970. doi: 10.1371/journal.pcbi.1002970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caruso A. Translational PK/PD modeling for cardiovascular safety assessment of drug candidates: methods and examples in drug development. J. Pharmacol. Toxicol. Methods. 2014;70:73–85. doi: 10.1016/j.vascn.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 83.Gotta V. Sensitivity of pharmacokinetic-pharmacodynamic analysis for detecting small magnitudes of QTc prolongation in preclinical safety testing. J. Pharmacol. Toxicol. Methods. 2015;72:1–10. doi: 10.1016/j.vascn.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 84.Gotta V. Inter-study variability of preclinical in vivo safety studies and translational exposure-QTc relationships – a PKPD meta-analysis. Br. J. Pharmacol. 2015;172:4364–4379. doi: 10.1111/bph.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johannesen L. Differentiating drug-induced multichannel block on the electrocardiogram: randomized study of dofetilide, quinidine, ranolazine, and verapamil. Clin. Pharmacol. Ther. 2014;96:549–558. doi: 10.1038/clpt.2014.155. [DOI] [PubMed] [Google Scholar]
- 86.Vicente J. Comprehensive T wave morphology assessment in a randomized clinical study of dofetilide, quinidine, ranolazine, and verapamil. J. Am. Heart Assoc. 2015;4:e001615. doi: 10.1161/JAHA.114.001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pastino G. Annual Meeting of the American College of Clinical Pharmacology. 2015. Lemborexant does not affect the QT interval: high-precision QT analysis from early clinical studies. [Google Scholar]
- 88.Abrahamsson C. Assessment of ventricular repolarization variability with the DeltaT50 method improves identification of patients with congenital long QT syndromes. Ann. Noninvasive Electrocardiol. 2013;18:240–250. doi: 10.1111/anec.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abrahamsson C. DeltaT50 – a new method to assess temporal ventricular repolarization variability. J. Electrocardiol. 2011;44:e471–e479. doi: 10.1016/j.jelectrocard.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 90.Jacobson I. Beat-by-beat QT interval variability, but not QT prolongation per se, predicts drug-induced Torsades de Pointes in the anaesthetised methoxamine-sensitized rabbit. J. Pharmacol. Toxicol. Methods. 2011;63:40–46. doi: 10.1016/j.vascn.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 91.Heijman J. Determinants of beat-to-beat variability of repolarization duration in the canine ventricular myocyte: a computational analysis. PLoS Comput. Biol. 2013;9:e1003202. doi: 10.1371/journal.pcbi.1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson D.M. I(Ks) restricts excessive beat-to-beat variability of repolarization during beta-adrenergic receptor stimulation. J. Mol. Cell. Cardiol. 2010;48:122–130. doi: 10.1016/j.yjmcc.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 93.Mirams G.R. Simulation of multiple ion channel block provides improved early prediction of compounds’ clinical torsadogenic risk. Cardiovasc. Res. 2011;91:53–61. doi: 10.1093/cvr/cvr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Babcock J.J. Integrated analysis of drug-induced gene expression profiles predicts novel hERG inhibitors. PLOS ONE. 2013;8:e69513. doi: 10.1371/journal.pone.0069513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Redfern W.S. Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and Torsade de Pointes for a broad range of drugs: evidence for a provisional safety margin in drug development. Cardiovasc. Res. 2003;58:32–45. doi: 10.1016/s0008-6363(02)00846-5. [DOI] [PubMed] [Google Scholar]
- 96.Beattie K.A. Evaluation of an in silico cardiac safety assay: using ion channel screening data to predict QT interval changes in the rabbit ventricular wedge. J. Pharmacol. Toxicol. Methods. 2013;68:88–96. doi: 10.1016/j.vascn.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Williams G., Mirams G.R. A web portal for in-silico action potential predictions. J. Pharmacol. Toxicol. Methods. 2015 doi: 10.1016/j.vascn.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cooper J. The cardiac electrophysiology web lab. Biophys J. 2016;110:292–300. doi: 10.1016/j.bpj.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson E.B. Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 1927;22:209–212. [Google Scholar]
- 100.Pathmanathan P., Gray R.A. Ensuring reliability of safety-critical clinical applications of computational cardiac models. Front. Physiol. 2013;4:358. doi: 10.3389/fphys.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pathmanathan P., Gray R.A. Verification of computational models of cardiac electro-physiology. Int. J. Numer. Methods Biomed. Eng. 2014;30:525–544. doi: 10.1002/cnm.2615. [DOI] [PubMed] [Google Scholar]
- 102.Tamargo J. Pharmacology of cardiac potassium channels. Cardiovasc. Res. 2004;62:9–33. doi: 10.1016/j.cardiores.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 103.Gao P. Recent advances in materials for extended-release antibiotic delivery system. J. Antibiot. 2011;64:625–634. doi: 10.1038/ja.2011.58. [DOI] [PubMed] [Google Scholar]
- 104.Council N.R., editor. Toxicity Testing in the 21st Century: A Vision and A Strategy, National Academy Press; 2007. [Google Scholar]
- 105.Valerio L.G., Jr. In silico toxicology models and databases as FDA Critical Path Initiative toolkits. Hum. Genomics. 2011;5:200–207. doi: 10.1186/1479-7364-5-3-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.OECD . 2004. Principles for the Validation, for Regulatory Purposes, of (Quantitative) Structure-Activity Relationship Models. Available at: http://www.oecd.org/chemicalsafety/risk-assessment/37849783.pdf. [Google Scholar]
- 107.OECD . 2007. Guidance document on the validation of (quantitative) structure-activity relationships [(Q)SAR] models. Available at: http://www.oecd.org/env/guidance-document-on-the-validation-of-quantitative-structure-activity-relationship-q-sar-models-9789264085442-en.htm. [Google Scholar]
- 108.ABPI . 2015. Bridging the Skills Gap in the Biopharmaceutical Industry. Available at: http://www.abpi.org.uk/industry-info/education/Documents/Skills_Gap_Industry_Executive_Summary.pdf. [Google Scholar]
- 109.Sivagangabalan G. Regional ion channel gene expression heterogeneity and ventricular fibrillation dynamics in human hearts. PLOS ONE. 2014;9:e82179. doi: 10.1371/journal.pone.0082179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bowes J. Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat. Rev. Drug Discov. 2012;11:909–922. doi: 10.1038/nrd3845. [DOI] [PubMed] [Google Scholar]
- 111.Amanfu R.K., Saucerman J.J. Modeling the effects of beta1-adrenergic receptor blockers and polymorphisms on cardiac myocyte Ca2+ handling. Mol. Pharmacol. 2014;86:222–230. doi: 10.1124/mol.113.090951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heijman J. Local control of beta-adrenergic stimulation: effects on ventricular myocyte electrophysiology and Ca2+-transient. J. Mol. Cell. Cardiol. 2011;50:863–871. doi: 10.1016/j.yjmcc.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Heijman J. Function and regulation of serine/threonine phosphatases in the healthy and diseased heart. J. Mol. Cell. Cardiol. 2013;64:90–98. doi: 10.1016/j.yjmcc.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 114.Beattie K. Voltage protocol design for determining hERG channel kinetics. J. Pharmacol. Toxicol. Methods. 2015;75:164. [Google Scholar]