Abstract
Candida haemulonii is now considered a complex of two species and one variety: C. haemulonii sensu stricto, Candida duobushaemulonii and the variety C. haemulonii var. vulnera. Identification (ID) of these species is relevant for epidemiological purposes and for therapeutic management, but the different phenotypic commercial systems are unable to provide correct species ID for these emergent pathogens. Hence, we evaluated the MALDI-TOF MS performance for the ID of C. haemulonii species, analyzing isolates/strains of C. haemulonii complex species, Candida pseudohaemulonii and Candida auris by two commercial platforms, their databases and softwares. To differentiate C. haemulonii sensu sctricto from the variety vulnera, we used the ClinProToolsTM models and a single-peak analysis with the software FlexAnalysisTM. The BiotyperTM database gave 100% correct species ID for C. haemulonii sensu stricto, C. pseudohaemulonii and C. auris, with 69% of correct species ID for C. duobushaemulonii. Vitek MSTM IVD database gave 100% correct species ID for C. haemulonii sensu stricto, misidentifying all C. duobushaemulonii and C. pseudohaemulonii as C. haemulonii, being unable to identify C. auris. The Vitek MSTM RUO database needed to be upgraded with in-house SuperSpectra to discriminate C. haemulonii sensu stricto, C. duobushaemulonii, C. pseudohaemulonii, and C. auris strains/isolates. The generic algorithm model from ClinProToolsTM software showed recognition capability of 100% and cross validation of 98.02% for the discrimination of C. haemulonii sensu stricto from the variety vulnera. Single-peak analysis showed that the peaks 5670, 6878, or 13750 m/z can distinguish C. haemulonii sensu stricto from the variety vulnera.
Keywords: MALDI-TOF mass spectrometry, Candida haemulonii complex species, Microflex LTTM, Vitek MSTM, ClinProToolsTM
Introduction
The taxonomy of the pathogenic Candida species such as Candida albicans, Candida parapsilosis, and Candida glabrata has suffered significant modifications due to the description of closely related species (Turner and Butler, 2014). Likewise, the taxonomy of Candida haemulonii has changed over the years. In the early 90s, Lehmann et al. (1993), studying strains of C. haemulonii from distinct geographic origins and clinical sources, divided this species into two genetically distinct groups, named group I and II. This concept that C. haemulonii was indeed a complex of different species, was later confirmed by Cendejas-Bueno et al. (2012) that proposed a new classification: C. haemulonii (former group I), Candida duobushaemulonii (former group II) and the new variety, C. haemulonii var. vulnera. The correct identification (ID) of the C. haemulonii complex species is clinically relevant, since resistance to azole derivatives are commonly reported among the isolates of this species complex and amphotericin B has poor in vitro activity against C. duobus haemulonii isolates (Cendejas-Bueno et al., 2012; Ramos et al., 2015).
This increasing number of Candida species causing human infections has created a challenge for clinical laboratories to provide reliable and fast ID, especially for closely related species (Merseguel et al., 2015). Recently, the widespread commercial system Vitek 2TM (bioMérieux, Marcy-L’Etoile, France) was linked to mis-IDs of Candida auris as C. haemulonii (Kathuria et al., 2015). Alternatively, reliable Candida species ID can be achieved by the sequence analysis of the internal transcribed spacer (ITS) region from the ribosomal DNA (rDNA; Schoch et al., 2012). However, the whole process of molecular analysis remains time consuming and costly. Thus, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) emerges as a fast and accurate method for yeast’s ID in clinical microbiology laboratories (Bader, 2013; Jamal et al., 2014). This method produces species-specific protein fingerprints that can be compared with databases that contain reference or main mass spectra (MSP) of a great diversity of microorganisms, including yeasts (Bader, 2013). Conversely, the performance of different MALDI-TOF MS instruments in the ID of yeasts was found to vary considerably when compared to the conventional techniques as these systems often lack MSPs of cryptic Candida species in their databases (Jamal et al., 2014).
Initial evaluation using MALDI-TOF MS for the ID of C. haemulonii complex species provided promising results (Cendejas-Bueno et al., 2012), however, the discrimination of the variety vulnera from C. haemulonii sensu stricto was troublesome and the performance of the Vitek MSTM (bioMérieux) remains unevaluated. Hence, aiming to provide a more detailed evaluation of the MALDI-TOF MS performance for the ID of C. haemulonii species ID, we analyzed a set of different isolates and reference strains belonging to the C. haemulonii complex species with the two available platforms, their databases and softwares.
Material And Methods
Isolates and Strains
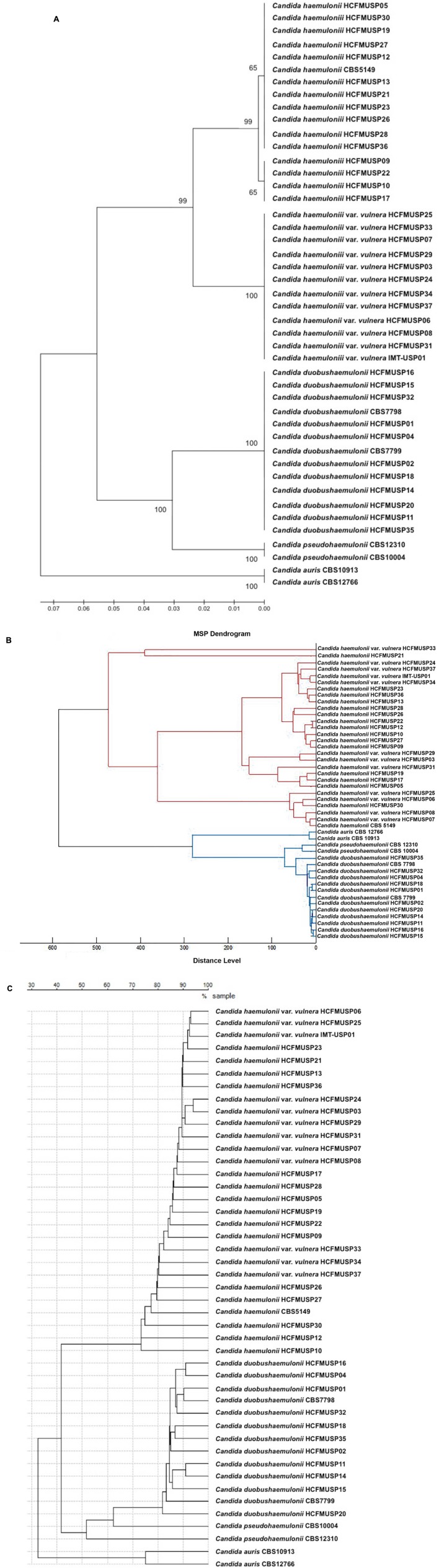
A total of 38 non-replicate clinical isolates belonging to the C. haemulonii complex species (15 C. haemulonii, sensu stricto, 12 C. haemulonii var vulnera, 11 C. duobushaemulonii) were analyzed in this study (Table 1). Species ID was carried out by ITS1 region sequence analysis with primers ITS1 and ITS4 and amplification parameters previously described (Luo and Mitchell, 2002). Both strands of purified ITS1 fragments were sequenced using the BigDye Terminator v.3.1 cycle sequencing kit on an ABI 3730 DNA analyzer (ABI, Foster City, EUA). Consensus sequence assembly and editing were performed using the software CondonCode Aligner, version 4.0 (CodonCode Corporation, Centerville, MA, USA), before being deposited in the GenBank database. In addition, a set of reference strains from the CBS-KNAW collection was also analyzed: C. haemulonii CBS5149T, C. duobushaemulonii CBS7798T, C. duobushaemulonii CBS7799. For specificity control, the reference strains belonging to the close related species Candida pseudohaemulonii (CBS10004 and CBS12370) and C. auris (CBS12766 and CBS10913) were also analyzed (Table 1). Phylogenetic analyses using UPGMA with 1,000 bootstrap simulations (Figure 1A) were conducted with Mega software, version 6.0 (Tamura et al., 2013).
Table 1.
Denominations and GenBank accession numbers of the isolates and strains analyzed in this study.
| Isolate/strain | GenBank acession no. | Species |
|---|---|---|
| HCFMUSP01 | KT236448.1 | Candida duobushaemulonii |
| HCFMUSP02 | KT968725.1 | C. duobushaemulonii |
| HCFMUSP03 | KT236449.1 | Candida haemulonii var. vulnera |
| HCFMUSP04 | KT968726.1 | C. duobushaemulonii |
| HCFMUSP05 | KT968713.1 | Candida haemulonii |
| HCFMUSP06 | KT968733.1 | C. haemulonii var. vulnera |
| HCFMUSP07 | KT968734.1 | C. haemulonii var. vulnera |
| HCFMUSP08 | KT968735.1 | C. haemulonii var. vulnera |
| HCFMUSP09 | KT968714.1 | C. haemulonii |
| HCFMUSP10 | KT257659.1 | C. haemulonii |
| HCFMUSP11 | KT968727.1 | C. duobushaemulonii |
| HCFMUSP12 | KT968715.1 | C. haemulonii |
| HCFMUSP13 | KT968716.1 | C. haemulonii |
| HCFMUSP14 | KT968728.1 | C. duobushaemulonii |
| HCFMUSP15 | KT968729.1 | C. duobushaemulonii |
| HCFMUSP16 | KT968730.1 | C. duobushaemulonii |
| HCFMUSP17 | KT968717.1 | C. haemulonii |
| HCFMUSP18 | KT968731.1 | C. duobushaemulonii |
| HCFMUSP19 | KT968718.1 | C. haemulonii |
| HCFMUSP20 | KT968732.1 | C. duobushaemulonii |
| HCFMUSP21 | KT968719.1 | C. haemulonii |
| HCFMUSP22 | KT968720.1 | C. haemulonii |
| HCFMUSP23 | KT968721.1 | C. haemulonii |
| HCFMUSP24 | KT968736.1 | C. haemulonii var. vulnera |
| HCFMUSP25 | KT968737.1 | C. haemulonii var. vulnera |
| HCFMUSP26 | KT968722.1 | C. haemulonii |
| HCFMUSP27 | KT257660.1 | C. haemulonii |
| HCFMUSP28 | KT968723.1 | C. haemulonii |
| HCFMUSP29 | KT968738.1 | C. haemulonii var. vulnera |
| HCFMUSP30 | KT968724.1 | C. haemulonii |
| HCFMUSP31 | KT968739.1 | C. haemulonii var. vulnera |
| HCFMUSP32 | KU365080.1 | C. duobushaemulonii |
| HCFMUSP33 | KU365081.1 | C. haemulonii var. vulnera |
| IMT-USP01 | KX118598.1 | C. haemulonii var. vulnera |
| HCFMUSP34 | KX118599.1 | C. haemulonii var. vulnera |
| HCFMUSP35 | KX118600.1 | C. duobushaemulonii |
| HCFMUSP36 | KX118601.1 | C. haemulonii |
| HCFMUSP37 | KX118602.1 | C. haemulonii var. vulnera |
| CBS5149T | AB118789.1 | C. haemulonii |
| CBS7798T | JX459765.1 | C. duobushaemulonii |
| CBS7799 | JX459766.1 | C. duobushaemulonii |
| CBS10004 | AB118791.1 | Candida pseudohaemulonii |
| CBS12310 | JX459678.1 | C. pseudohaemulonii |
| CBS10913 | KF689018.1 | Candida auris |
| CBS12766 | KC692039.1 | C. auris |
FIGURE 1.
Comparison of phylogenetic tree and MALDI-TOF dendrograms of the isolates and strains belonging to Candida haemulonii complex and close related taxa: (A) Phylogenetic tree obtained by UPGMA analyses and 1,000 bootstrap simulations based on ITS sequences; (B) Bruker BiotyperTM dendrogram clustering of the MSPs with distances displayed in relative units on the x axis; (C) SARAMISTM dendrogram clustering of representative spectra from each isolate/strain with distances displayed in % similarities on the x axis.
MALDI-TOF MS: Sample Preparation for Analysis
The isolates and strains were cultured on Sabouraud’s dextrose agar (SDA) plates and incubated for 48 h at 30°C before MALDI-TOF MS analysis. One loop of yeast biomass was transferred to a microtube containing 300 μl of purified water and final protein extraction protocol with absolute ethanol (Merck, Darmstadt, Germany) and formic acid 70% (Sigma-Aldrich, St. Louis, MO, USA) was carried out according to the Bruker’s recommendations.
One microliter of the crude protein extract of each isolate/strain was spotted onto a target plate. After air-drying, each spot was overlaid with 1 μl of HCCA matrix solution (Sigma-Aldrich).
Bruker Daltonics MALDI-TOF MS Analysis
Measurements were performed on a Microflex LTTM (Bruker Daltonics, Bremen, Germany) instrument using the software FlexControlTM version 3.4 (Bruker Daltonics). Bacterial test standard (BTS, Bruker Daltonics) was used for mass calibrationTM instrument parameter optimization. Spectra were acquired in linear positive mode within a mass range from 2000 to 20,000 m/z with the manufacturer’s suggested settings using automated collecting spectra mode. Then, the obtained spectra were analyzed by standard pattern-matching algorithm using the MALDI BiotyperTM 3.1 software (Bruker Daltonics), which compared the raw spectra with the reference spectra of the Bruker library (database version 3.3.1, 5627 reference spectra) by using the default settings. ID criteria were used as recommended by the manufacturer: a score ≥ 2.000 indicated species level ID, a score between 1.700 and 1.999 indicated ID to the genus level and a score < 1.700 was interpreted as no ID. For MainSpectra (MSP) and dendrogram construction, flat-liners and bad quality spectra were removed and additional measurements were carried out to obtain 20 spectra from each isolate/strain. Spectra were then loaded into BiotyperTM 3.1 software (Bruker Daltonics) for MSP creation and dendrogram clustering construction with the default settings (distance measure: correlation; linkage: average; score oriented).
bioMérieux MALDI-TOF MS Analysis
Measurements were performed on a Vitek MSTM instrument (bioMérieux) equipped with both IVD and RUO (SARAMISTM) databases (bioMérieux). For the IVD analysis, spectra were obtained using the Vitek MSTM automation control and Myla software (bioMérieux) with the manufacturer’s suggested settings. For each acquisition group, a standard (Escherichia coli ATCC 8739) was included to calibrate the instrument and validate the run. The spectra were analyzed by the Vitek MSTM v3.2 IVD database (bioMérieux) that contains spectral profiles covering 3555 species. The software compares the spectrum obtained to the expected spectrum of each organism or organism group (e.g., bacteria or fungi) and high confidence level ID was considered when single species showed probability of ID ≥ 60%. For the RUO analysis, spectra were generated using the Launchpad v2.8 software and compared to the SARAMISTM v.4.12 database (bioMérieux) that contains 720 SuperSpectra (each one corresponding to a spectral fingerprint with 15–20 species-specific biomarkers) of different fungal species. Peak matches that yield identification results with confidence values exceeding 75% were considered significant. For SuperSpectra and dendrogram construction, spectra of all isolates/strains were imported into the SARAMIS PremiumTM software package (bioMérieux). Then, SuperSpectra were calculated using the SARAMISTM SuperSpectrum tool (bioMérieux) according to the manufacturer’s instructions, and the specificity of the potential biomarker masses was determined by comparison against the whole SARAMISTM spectral archive (bioMérieux). Dendrogram was created based on whole spectra, with a single-link clustering algorithm and a binary mass list with an error of 800 ppm.
Differentiation of Candida haemulonii sensu sctricto from the Variety vulnera
ClinProToolsTM Models
The ClinProToolsTM (Bruker Daltonics) generates multiple mathematical algorithms to generate pattern recognition models for classification and prediction of different classes (e.g., C. haemulonii sensu stricto class 1, C. haemulonii var. vulnera class 2) from mass spectrometry based profiling data. Moreover, ClinProToolsTM provides a list of peaks sorted according to the statistical significance to differentiate between both classes (Ketterlinus et al., 2005). Thus, for recognition of mass spectra patterns and biomarkers between C. haemulonii sensu stricto and C. haemulonii var. vulnera, spectra peak analysis models with ClinProToolsTM software v.3.0 (Bruker Daltonics) were created from 320 mass spectra of the 16 C. haemulonii sensu stricto (10 high-quality mass spectra per isolate) and 12 C. haemulonii var. vulnera (≈14 high-quality mass spectra per isolate) isolates. Spectra were pretreated with a resolution of 800 ppm, a mass range of 2000–20000 Da, a top hat baseline subtraction with 10% minimal baseline width, enabling null spectra exclusion, and recalibration with 500 ppm maximal peak shift and 30% match calibrant peaks. ClinProToolTM smodels (Bruker Daltonics) were generated using three algorithms: Genetic Algorithm (GA), Supervised Neural Network (SNN), and Quick Classifier (QC). For each model, the recognition capability (RC) and cross validation (CV) percentage were generated to demonstrate the reliability and accuracy of the model. RC and CV percentages were indicators of the model’s performance and useful predictors of the model’s ability to classify test isolates. The model with the highest RC and CV values were used in the analysis.
Single-Peak Analysis
For each peak, the AUC for the discrimination of the groups was directly obtained from the ClinProToolsTM v.3.0 software (Bruker Daltonics). For the five peaks with the highest AUC, the detection performances were checked using FlexAnalysisTM v.3.4 (Bruker Daltonics). After smoothing and baseline subtraction, the mass lists for each isolate were obtained using the centroid algorithm with a signal-to-noise (SN) threshold of 0.5 and a maximum of 500 peaks and exported to Microsoft Excel. The SN ratios of the peaks with a tolerance of 1.000 ppm were exported to SPSS 18.0. ROC curves were constructed, and their optimal cutoff values were determined with the maximum Youden index.
Results
Bruker BiotyperTM
The Bruker BiotyperTM (Bruker Daltonics) gave correct species with scores ≥ 2.0 for all C. haemulonii sensu stricto isolates/strains (Table 2). For the species C. duobushaemulonii, correct species ID (score ≥ 2.0) was achieved for seven isolates and the two CBS strains (69%), while four isolates were assigned as C. duobushaemulonii with a score between 1.797 and 1.935. These isolates were analyzed at least two times and the results were confirmed. After the inclusion in the BiotyperTM (Bruker Daltonics) database of MSPs from two Brazilian isolates (HCFMUSP04 and HCFMUSP11), all mass spectra of that species had scores above 2.3. All reference strains of C. pseudohaemulonii and C. auris had correct species ID with scores ≥ 2.0. All C. haemulonii var. vulnera isolates were assigned as C. haemulonii sensu stricto with scores ≥ 2.0. The dendrogram generated by the BiotyperTM (Figure 1B) show the clustering of MSPs of the isolates/strains of C. haemulonii sensu stricto and C. haemulonii var. vulnera in the same node, exemplifying the similarity of these MSPs.
Table 2.
Performance of the different databases for the identification of C. haemulonii sensu stricto, C. haemulonii variety vulnera, C. duobushaemulonii, C. pseudohaemulonii, and C. auris isolates/strains.
| Species/variety | Bruker Microflex LTTM | VITEK MSTM | |||
| BiotyperTM v.3.1 (% with LS1 ≥ 2) | BiotyperTM v.3.1 with in-house MSPs2 (% with LS ≥ 2) | IVD (% with CI3 ≥ 60%) | RUO (% with CI ≥ 75%) | RUO with in-house SuperSpectra (% with CI ≥ 75%) | |
| Candida haemulonii sensu stricto (n = 16) | 16/16 (100) | 16/16 (100) | 16/16 (100) | 0/16 (0) | 16/16 (100) |
| C. haemulonii var. vulnera (n = 12) | 0/0 (0) | 0/12 (0)4 | 0/12 (0)4 | 0/12 (0) | 0/12 (0)4,5 |
| Candida duobushaemulonii (n = 13) | 9/13 (69) | 13/13 (100) | 0/13 (0)4 | 0/13 (0) | 13/13 (100) |
| Candida pseudohaemulonii (n = 2) | 2/2 (100) | 2/2 (100) | 0/2 (0)4 | 0/2 (0) | 2/2 (100) |
| Candida auris (n = 2) | 2/2 (100) | 2/2 (100) | 0/2 (0) | 0/2 (0) | 2/2 (100) |
1LS, logscore values; 2MSP, main spectrum profile; 3CI, confidence interval for a single species; 4misidentified as C. haemulonii sensu stricto; 5not able to find specific masses for the creation of SuperSpectra for the var. vulnera.
Vitek MSTM Species Identification
The bioMérieux Vitek MSTM IVD gave correct species with with 99.9% level of ID for all C. haemulonii sensu stricto isolates/strains (Table 2). However, all C. duobus haemulonii isolates/strains and the two reference strains of C. pseudohaemulonii were misidentified as C. haemulonii with 99.9% level of ID in at least two separate experiments, while the two strains of C. auris had no species ID. The Vitek MSTM RUO (SARAMISTM) analysis gave neither genus nor species ID for all isolates and strains (Table 2). After the upgrade of the SARAMISTM database with SuperSpectra of C. haemulonii, C. duobushaemulonii, C. pseudohaemulonii and C. auris, all isolates/strains belonging to these species had correct species assignment (Table 2). It was not possible to create a SuperSpectrum of C. haemulonii var vulnera with at least 15 masses that would have differentiated it from C. haemulonii sensu stricto. The dendrogram generated by the SARAMISTM (bioMérieux) software showed similar clustering results to the dendrogram generated by the BiotyperTM software, gathering C. duobushaemulonii, C. pseudohaemulonii, and C. auris into three species-specific nodes. However, C. haemulonii sensu stricto and the variety vulnera were found mixed inside the same node (Figures 1B,C).
Differentiation of Candida haemulonii sensu sctricto from the Variety vulnera
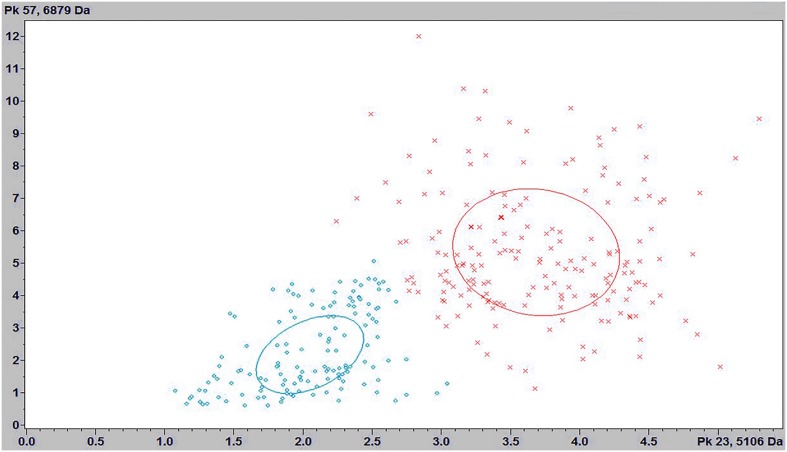
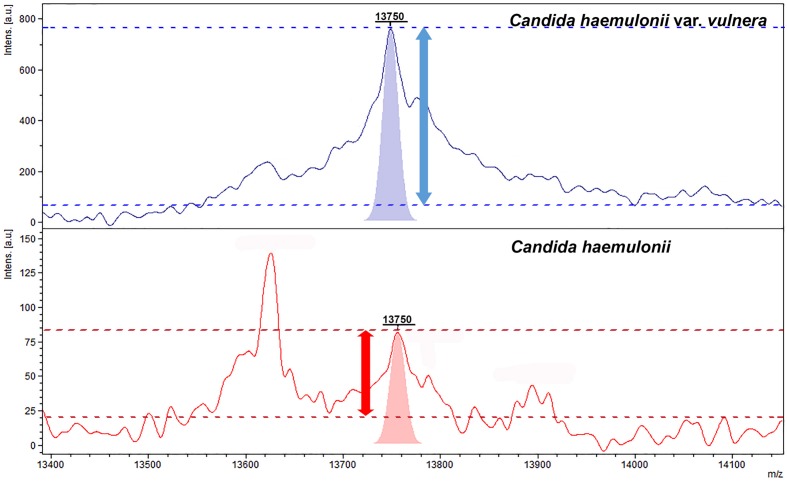
The three classification models from ClinProToolsTM showed values of RC above ≥90% for the discrimination of C. haemulonii sensu stricto and C. haemulonii var vulnera. The best results were provided by the GA model, with RC and CV of 100 and 98.02%, respectively. The strain distribution maps based on the GA clearly show that C. haemulonii sensu stricto and the variety vulnera can be divided into one of two categories based on their peptide mass fingerprints (Figure 2). The peaks that had the highest AUC (>0.9) for the discrimination of the C. haemulonii sensu strito and C. haemulonii var. vulnera by ClinProToolsTM were 5106, 5670, 6878, 13750, and 14046 m/z. However, the performances of these peaks for the discrimination of the two groups using the FlexAnalysisTM software showed that only the peaks 5670, 6878, and 13750 m/z had AUC > 0.9, with sensitivity and specificity of 94.7, 92.4, 94.1%, and 77.0, 94.5, 96.1%, respectively. The SN cut-off values of the peaks 5670, 6878, and 13750 m/z for the discrimination of the C. haemulonii (below cut-off) and C. haemulonii var. vulnera (above cut-off) were 3.1, 2.86, and 6.04, respectively. The ClinProToolsTM and single-peak analysis results for the differentiation of C. haemulonii sensu stricto from the variety vulnera are summarized in Table 3 and exemplified in Figure 3.
FIGURE 2.
Strain distribution map corresponding to C. haemulonii sensu stricto (green) and C. haemulonii var. vulnera (red).
Table 3.
Single-peak analysis for the discrimination of C. haemulonii sensu stricto and C. haemulonii variety vulnera.
| Peak (m/z) | ClinProToolsTM | FlexAnalysisTM | ||||||||
| AUC1 | Dave2 | PWKW3 | PAD4 | Ave15 | Ave2 | AUC6 | Cut-off6 | Sensitivity (%) | Specificity (%) | |
| 5106 | 0.95 | 1.82 | <0.0001 | <0.0001 | 10.46 | 17.88 | 0.81 | 2.77 | 96.5 | 74.86 |
| 5670 | 0.93 | 1.94 | <0.0001 | <0.0001 | 16.65 | 18.85 | 0.90 | 3.01 | 94.77 | 77.05 |
| 6878 | 0.92 | 3.51 | <0.0001 | <0.0001 | 12.89 | 15.53 | 0.95 | 2.89 | 92.44 | 94.54 |
| 13750 | 0.96 | 1.51 | <0.0001 | <0.0001 | 5.09 | 59.01 | 0.98 | 6.04 | 94.19 | 96.17 |
| 14046 | 0.95 | 0.27 | <0.0001 | 0.00335 | 23.37 | 26.06 | 0.77 | 2.01 | 70.35 | 77.60 |
Peaks with the best performances according to ClinProToolsTM and FlexAnalysisTM softwares.
1AUC, area under the curve; 2Dave, difference between the maximal and the minimal average peak area/ intensity of the groups; 3PWKW, p-value of Wilcoxon/Kruskal–Wallis test (range: 0–1; 0 = good); 4PAD: p-value of Anderson–Darling test, <0.05 indicates data not normally distributed; gives information about normal distribution (range: 0–1; 0 = not normal distributed); 5Ave, area/intensity average of a group from C. haemulonii sensu stricto (1) and C. haemulonii var. vulnera (2); 6AUCs and signal-to-noise cutoff values were obtained from ROC curve constructed by SPSS Version 18.0 with the using of FlexAnalysis.
FIGURE 3.
Representative mass spectra of C. haemulonii sensu stricto (red) and C. haemulonii var. vulnera (purple) between 13400 and 14100 Da showing the peak intensities and peak areas of the 13750 Da peak. Horizontal dashed lines depict the signal (above) and noise (below) intensity values used to calculate the signal-to-noise ratios.
Discussion
The taxonomy of the genus Candida has been suffered significant changes due to the description of new closely related species, with some species as C. haemulonii being considered as cryptic species complex (Brandt and Lockhart, 2012). The discrimination of these cryptic Candida species from their closest relatives is providing new information regarding their epidemiology, pathogenicity and clinical significance (Brandt and Lockhart, 2012; Cendejas-Bueno et al., 2012; Nobrega de Almeida et al., 2016).
In this context, MALDI-TOF MS has been successfully applied to discriminate cryptic species from the genus Candida, such as C. parapsilosis, Candida orthopsilosis, and Candida metapsilosis (Quiles-Melero et al., 2012; Nobrega de Almeida et al., 2014), C. glabrata, Candida nivariensis, and Candida bracariensis (Pinto et al., 2011), C. albicans and Candida dubliniensis (Pinto et al., 2011; Jamal et al., 2014), and more recently, it has been evaluated for the differentiation of the C. haemulonii species complex and other close related taxa (Cendejas-Bueno et al., 2012; Kathuria et al., 2015).
The Bruker’s BiotyperTM v.3.1 is currently the most adapted database for the ID of C. haemulonii species, presenting with 08, 04, and 07 MSPs of C. haemulonii, C. haemulonii var. vulnera and C. duobushaemulonii, respectively. However, four C. duobushaemulonii isolates, despite showing the best match results with the MSPs of the same species, had ID values below 2.0. After the expansion of the database with in-house MSPs, all isolates had ID values above 2.0. This illustrates the need for the Bruker database expansion with local well-identified isolates to reach optimal results, as previously reported by other authors (Cassagne et al., 2011; de Almeida Júnior et al., 2014).
The IVD database from the Vitek MSTM includes only the species C. haemulonii. Surprisingly, the different taxa C. duobushaemulonii and C. pseudohaemulonii were misidentified as C. haemulonii. Since these species have well-distinct spectral profiles, the misidentifications may be related to the inclusion of strains without updated species ID when this database was constructed. The update of the Vitek MSTM IVD database will certainly enhance its ability to correctly ascertain the different species from C. haemulonii complex and the close related taxa, since this system has proven to well-differentiate phylogenetically similar microorganisms, such as Streptococcus pneumoniae from species of the Streptococcus mitis group (Branda et al., 2013). The RUO database (SARAMISTM) has proven to be an auxiliary tool when the IVD database fails to provide correct yeast species ID (Chao et al., 2014). In the case of C. haemulonii complex species, the inclusion of SuperSpectra was necessary to optimize the performance of the Vitek MSTM. Nevertheless, we demonstrated the specificity of the SARAMIS database, since no mis-IDs were reported, which was not the case of the IVD database.
The ClinProToolsTM software is a biomarker analyzer that has been widely applied in bacteriology, providing rapid and cost-saving method for epidemiological clustering, strain typing (Xiao et al., 2014; Angeletti et al., 2015; Zhang et al., 2015), and for detection of staphylococcal Panton-Valentine leukocidin (Bittar et al., 2009). This software generates classification models from large numbers of spectra and detects small differences among different clusters, based on mass, signal-to-noise, intensity, peak heights and peak areas. Moreover, allying ClinProToolsTM and single-peak analysis with FlexAnalysisTM has proven to provide higher discriminatory power to detect bio-marker peaks (Angeletti et al., 2015; Zhang et al., 2015). However, ClinProtoolsTM was until now an unexplored tool for the differentiation of close taxa in the fungi kingdom.
In conclusion, we show that the BiotyperTM database 3.1 performs relatively well for discriminating C. haemulonii species and close related taxa, but requires addition of MPSs representing the local diversity to achieve optimal results, while Vitek MSTM databases perform less well and need major update. Moreover, we describe here the most discriminatory peaks along with the SN cut-off values that can differentiate C. haemulonii and the variety vulnera with a simple inspection of the mass spectra profile in routine basis software such as FlexAnalysisTM.
Author Contributions
JA: design the study, helped with acquisition and data analysis, drafted and revised the work, approved the final work and agrees with all the aspects of the work; RG, AS, GD: helped with acquisition and data analysis, revised the work, approved the final work, and agree with all the aspects of the work; VG, RM, DA, AR, AM: helped with acquisition of the data, revised the work, approved the final work and agree with all the aspects of the work; FR, LJ: helped with data analysis, revised the work, approved the final work and agree with all the aspects of the work; GB: helped with data analysis, drafted and revised the work, approved the final work and agrees with all the aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. JA was supported by grants from FAPESP (2014/10126-4).
References
- Angeletti S., Dicuonzo G., Lo Presti A., Cella E., Crea F., Avola A., et al. (2015). MALDI-TOF mass spectrometry and blakpc gene phylogenetic analysis of an outbreak of carbapenem-resistant K. pneumoniae strains. New Microbiol. 38 541–550. [PubMed] [Google Scholar]
- Bader O. (2013). MALDI-TOF-MS-based species identification and typing approaches in medical mycology. Proteomics 13 788–799. 10.1002/pmic.201200468 [DOI] [PubMed] [Google Scholar]
- Bittar F., Ouchenane Z., Smati F., Raoult D., Rolain J.-M. (2009). MALDI-TOF-MS for rapid detection of staphylococcal Panton-Valentine leukocidin. Int. J. Antimicrob. Agents 34 467–470. 10.1016/j.ijantimicag.2009.03.017 [DOI] [PubMed] [Google Scholar]
- Branda J. A., Markham R. P., Garner C. D., Rychert J. A., Ferraro M. J. (2013). Performance of the Vitek MSTM v2.0 system in distinguishing Streptococcus pneumoniae from nonpneumococcal species of the Streptococcus mitis group. J. Clin. Microbiol. 51 3079–3082. 10.1128/JCM.00824-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt M. E., Lockhart S. R. (2012). Recent taxonomic developments with Candida and other opportunistic yeasts. Curr. Fungal Infect. Rep. 6 170–177. 10.1007/s12281-012-0094-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassagne C., Ranque S., Normand A.-C., Fourquet P., Thiebault S., Planard C., et al. (2011). Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS ONE 6:e28425 10.1371/journal.pone.0028425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendejas-Bueno E., Kolecka A., Alastruey-Izquierdo A., Theelen B., Groenewald M., Kostrzewa M., et al. (2012). Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 50 3641–3651. 10.1128/JCM.02248-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q.-T., Lee T.-F., Teng S.-H., Peng L.-Y., Chen P.-H., Teng L.-J., et al. (2014). Comparison of the accuracy of two conventional phenotypic methods and two MALDI-TOF MS systems with that of DNA sequencing analysis for correctly identifying clinically encountered yeasts. PLoS ONE 9:e109376 10.1371/journal.pone.0109376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Júnior J. N., Figueiredo D. S. Y., Toubas D., Del Negro G. M. B., Motta A. L., Rossi F., et al. (2014). Usefulness of matrix-assisted laser desorption ionisation-time-of-flight mass spectrometry for identifying clinical Trichosporon isolates. Clin. Microbiol. Infect. 20 784–790. 10.1111/1469-0691.12502 [DOI] [PubMed] [Google Scholar]
- Jamal W. Y., Ahmad S., Khan Z. U., Rotimi V. O. (2014). Comparative evaluation of two matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems for the identification of clinically significant yeasts. Int. J. Infect. Dis. 26 167–170. 10.1016/j.ijid.2014.05.031 [DOI] [PubMed] [Google Scholar]
- Kathuria S., Singh P. K., Sharma C., Prakash A., Masih A., Kumar A., et al. (2015). Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by vitek 2, CLSI broth microdilution, and etest method. J. Clin. Microbiol. 53 1823–1830. 10.1128/JCM.00367-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterlinus R., Hsieh S.-Y., Teng S.-H., Lee H., Pusch W. (2005). Fishing for biomarkers: analyzing mass spectrometry data with the new ClinProToolsTM software. BioTechniques Suppl. 38 37–40. 10.2144/05386SU07 [DOI] [PubMed] [Google Scholar]
- Lehmann P. F., Wu L. C., Pruitt W. R., Meyer S. A., Ahearn D. G. (1993). Unrelatedness of groups of yeasts within the Candida haemulonii complex. J. Clin. Microbiol. 31 1683–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Mitchell T. G. (2002). Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 40 2860–2865. 10.1128/JCM.40.8.2860-2865.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merseguel K. B., Nishikaku A. S., Rodrigues A. M., Padovan A. C., e Ferreira R. C., de Azevedo Melo A. S., et al. (2015). Genetic diversity of medically important and emerging Candida species causing invasive infection. BMC Infect. Dis. 15:57 10.1186/s12879-015-0793-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega de Almeida J., Jr., Assy J. G. P. L., Levin A. S., Del Negro G. M. B., Giudice M. C., Tringoni M. P., et al. (2016). Candida haemulonii complex species, Brazil, January 2010-March 2015. Emerg. Infect. Dis 22 561–563. 10.3201/eid2203.151610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega de Almeida J. J., de Souza L. B., Motta A. L., Rossi F., Romano Di Gioia T. S., et al. (2014). Evaluation of the MALDI-TOF VITEK MSTMTM system for the identification of Candida parapsilosis, C. orthopsilosis and C. metapsilosis from bloodstream infections. J. Microbiol. Methods 105 105–108. 10.1016/j.mimet.2014.07.018 [DOI] [PubMed] [Google Scholar]
- Pinto A., Halliday C., Zahra M., van Hal S., Olma T., Maszewska K., et al. (2011). Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS ONE 6:e25712 10.1371/journal.pone.0025712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles-Melero I., García-Rodríguez J., Gómez-López A., Mingorance J. (2012). Evaluation of matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry for identification of Candida parapsilosis, C. orthopsilosis and C. metapsilosis. Eur. J. Clin. Microbiol. Infect. Dis. 31 67–71. 10.1007/s10096-011-1277-z [DOI] [PubMed] [Google Scholar]
- Ramos L. S., Figueiredo-Carvalho M. H. G., Barbedo L. S., Ziccardi M., Chaves A. L. S., Zancopé-Oliveira R. M., et al. (2015). Candida haemulonii complex: species identification and antifungal susceptibility profiles of clinical isolates from Brazil. J. Antimicrob. Chemother. 70 111–115. 10.1093/jac/dku321 [DOI] [PubMed] [Google Scholar]
- Schoch C. L., Seifert K. A., Huhndorf S., Robert V., Spouge J. L., Levesque C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U.S.A. 109 6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. A., Butler G. (2014). The Candida pathogenic species complex. Cold Spring Harb. Perspect. Med. 4:a019778 10.1101/cshperspect.a019778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Zhao F., Zhang H., Meng F., Zhang J. (2014). Novel strategy for typing Mycoplasma pneumoniae isolates by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry coupled with ClinProToolsTM. J. Clin. Microbiol. 52 3038–3043. 10.1128/JCM.01265-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Ding J., Rao X., Yu J., Chu M., Ren W., et al. (2015). Analysis of methicillin-resistant Staphylococcus aureus major clonal lineages by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS). J. Microbiol. Methods 117 122–127. 10.1016/j.mimet.2015.08.002 [DOI] [PubMed] [Google Scholar]