Abstract
Key message
The combination of three non-functional alleles of the flowering repressor VRN2 results in a spring growth habit in wheat.
Abstract
In temperate cereals with a winter growth habit, a prolonged exposure to low temperatures (vernalization) accelerates flowering. Before vernalization, the VRN2 locus plays a central role in maintaining flowering repression. Non-functional VRN2 alleles result in spring growth habit and are frequent in diploid wheat and barley. However, in hexaploid wheat, the effect of these non-functional VRN2 alleles is masked by gene redundancy. In this study, we developed a triple VRN2 mutant (synthetic vrn2-null) in hexaploid wheat by combining the non-functional VRN-A2 allele present in most polyploid wheats with a VRN-B2 deletion from tetraploid wheat, and a non-functional VRN-D2 allele from Aegilops tauschii (Ae. tauschii) (the donor of hexaploid wheat D genome). Non-vernalized vrn2-null plants flowered 118 days (P < 2.8E−07) earlier than the winter control, and showed a limited vernalization response. The functional VRN-B2 allele is expressed at higher levels than the functional VRN-D2 allele and showed a stronger repressive effect under partial vernalization (4 °C for 4 weeks), and also in non-vernalized plants carrying only a functional VRN-B2 or VRN-D2 in heterozygous state. These results suggest that different combinations of VRN-B2 and VRN-D2 alleles can be a used to modulate the vernalization response in regions with mild winters. Spring vrn2-null mutants have been selected repeatedly in diploid wheat and barley, suggesting that they may have an adaptative value and that may be useful in hexaploid wheat. Spring wheat breeders can use these new alleles to improve wheat adaptation to different or changing environments.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-016-2713-3) contains supplementary material, which is available to authorized users.
Introduction
Wheat is an important source of calories and protein worldwide, and within the cereals is the most widely cultivated. Wheat’s adaptability to different environments is favored by a large allelic diversity at the loci that regulate reproductive development in response to seasonal signals, including photoperiod and temperature. Wheat flowering is accelerated under long day photoperiods by the PHOTOPERIOD1 (PPD1) gene (Beales et al. 2007; Wilhelm et al. 2009; Diaz et al. 2012). However, even under inductive photoperiods, wheat varieties with a winter growth habit (winter wheats) require a prolonged exposure to cold temperatures (vernalization) to become competent to flower. Winter wheats are planted in the fall, and the vernalization requirement protects the fragile flowering meristems from being exposed to freezing temperatures during the winter. By contrast, spring wheats are usually planted in the spring, and they have either no vernalization requirement or a very limited response to vernalization.
The vernalization requirement in the temperate cereals is controlled by four loci (VRN1, VRN2, VRN3 and VRN-D4) and natural variation at these loci has been associated with different levels of vernalization requirement both in wheat and barley (Yan et al. 2003, 2004a, b, 2006 ; Fu et al. 2005; Kippes et al. 2014, 2015). Both VRN1 and its paralog VRN-D4 encode a protein with high similarity to Arabidopsis meristem identity protein APETALA1 (AP1) (Yan et al. 2003; Kippes et al. 2015). Polymorphisms in regulatory regions of the promoter or first intron of this MADS-box gene are associated with loss or reduction of the vernalization requirement (Yan et al. 2003, 2004b; Fu et al. 2005; Hemming et al. 2009; Kippes et al. 2015).
In both wheat and Arabidopsis, the VRN1/AP1 gene is directly up-regulated by the FLOWERING LOCUS T gene, which is designated as TaFT1 or VRN3 in hexaploid wheat (Yan et al. 2006). The protein encoded by this gene has been shown to travel through the phloem carrying the photoperiodic signal from the leaves to the shoot apical meristem (Corbesier et al. 2007; Tamaki et al. 2007). Once in the meristem, FT forms a protein complex with FD and 14-3-3 proteins that binds to the promoter of AP1 homologs OsMADS15 in rice and VRN1 in wheat, resulting in their transcriptional activation and the initiation of flowering (Taoka et al. 2011; Li et al. 2015). Natural TaFT1 alleles with high levels of TaFT1 expression overcome the vernalization requirement and are associated with spring grow habit (Yan et al. 2006). During the fall, TaFT1 transcript levels are repressed by the flowering repressor VRN2 (Yan et al. 2004a). However, the upregulation of VRN1 during the winter prevents the upregulation of VRN2 in the spring (Chen and Dubcovsky 2012). In the absence of VRN2, the increase in the length of the days during the spring results in the induction of TaFT1. TaFT1, VRN1 and VRN2 are part of a positive regulatory feedback loop that once induced promotes an irreversibly transition from the vegetative to the reproductive stage.
The VRN2 locus includes two tandemly repeated genes named ZCCT1 and ZCCT2 that encode proteins carrying a putative zinc finger and a CCT domain (Yan et al. 2004a). The CCT domain is a 43-amino acid region, first described in the Arabidopsis proteins CONSTANS (CO), CONSTANS-like (COL) and TIMING OF CAB1 (TOC1) (Putterill et al. 1995; Strayer et al. 2000; Robson et al. 2001), and is present in multiple regulatory proteins associated with light signaling, circadian rhythms and photoperiodic flowering (Wenkel et al. 2006). Deletions or mutations involving positively charged amino acids at the CCT domain are associated with recessive ZCCT1 and ZCCT2 alleles for spring growth habit in both diploid and tetraploid wheat (Yan et al. 2004a; Dubcovsky et al. 2005; Distelfeld et al. 2009).
The ZCCT genes act as long day repressors of flowering and their expression is down-regulated by vernalization and short days (Yan et al. 2004a; Trevaskis et al. 2006; Dubcovsky et al. 2006; Chen and Dubcovsky 2012). The GRAIN NUMBER, PLANT HEIGHT AND HEADING DATE 7 (GHD7) gene in rice and sorghum (SbGHD7, Ma6) is the closest homolog to VRN2 and also works as long day flowering repressor (Xue et al. 2008; Murphy et al. 2014).
A VRN2 homolog is present in Brachypodium, but it is not down-regulated by cold; indeed, a survey of several grasses demonstrated that the down-regulation of VRN2 by cold is unique to core Pooideae such as wheat and barley (Woods et al. 2016).
The VRN2 protein interacts in vitro with CO2 (CONSTANTS2) and with members of the NUCLEAR FACTOR-Y (NF-Y) transcription factor family, but the role of these interactions in the regulation of wheat flowering time are still not clear (Li et al. 2011).
Natural variation in VRN2 has been detected frequently in diploid wheat (T. monococcum L. Yan et al. 2004a) and barley (von Zitzewitz et al. 2005) but not in hexaploid wheat, where gene redundancy likely masks the effect of recessive vrn2 mutations. A tetraploid wheat line with no functional copies of VRN2 was previously developed (Distelfeld et al. 2009) by introgressing both a non-functional vrn-A m 2 allele from T. monococcum accession DV92 (Yan et al. 2004a) and a vrn-B2 allele with a large deletion including ZCCT-B1 and the two ZCCT-B2 copies present in this locus (Distelfeld et al. 2009). Using this genetic stock it was demonstrated that most tetraploid wheat accessions carry a non-functional VRN-A2 allele with mutations in critical positively charged amino acids of the CCT domain (Distelfeld et al. 2009). The only exception was T. turgidum ssp. dicoccoides accession 10-85 collected at Amiad (Israel), which showed no mutations in the CCT domain from ZCCT-A1.
Since hexaploid wheat is grown in a wider range of environments than tetraploid wheat and represents more than 95 % of the wheat currently grown in the world (Taylor and Koo 2015), a vrn2-null genetic stock in hexaploid wheat would be a useful tool to test the adaptative value of VRN2 loss-of-function mutations in the more diverse environments where hexaploid wheat is grown. In this study, we introgressed a non-functional vrn-D2 of Aegilops tauschii in hexaploid wheat, and combined it with vrn-A2 and vrn-B2 non-functional loci to generate a vrn2-null genetic stock in hexaploid wheat. Using this genetic stock we demonstrate that simultaneous mutations in all three VRN2 homologs result in a spring growth habit in hexaploid wheat. Using different combinations of functional and non-functional homoeologs we also show that functional VRN-B2 confers a stronger vernalization requirement than functional VRN-D2.
Methods
Plant materials
The non-functional vrn-D2 allele was obtained from the early flowering diploid Aegilops tauschii E1 (Dudnikov 2003). To confirm the presence of the recessive vrn-D2 allele, E1 was crossed with Ae. tauschii AS75, a winter accession previously described by Luo et al. (2009). The F1 was self-pollinated and an F2 segregating population including 71 individuals was generated. Heading data from this population was used to validate the recessive vrn-D2 allele from E1 and to characterize the dominance of the VRN-D2 allele from AS75. The degree of dominance was calculated using the formula: D = (2X 2 − X 1 − X 3)/(X 1 − X 3) (Falconer 1964), where X 1, X 2 and X 3 are the heading time values of plants homozygous for the non-functional vrn-D2 allele, the heterozygotes, and plants homozygous for the wild type late flowering VRN-D2 allele, respectively.
The diploid E1 line was then crossed with tetraploid wheat line #3089 (BC3F2-521 x Kronos, Distelfeld et al. 2009) which carries non-functional alleles for vrn-A2 and vrn-B2. Chromosome doubling of the triploid hybrid resulted in a synthetic hexaploid with no functional copies of VRN2, designated hereafter as “synthetic vrn2-null”, which was deposited in the National Small Grain Collection (PI 676269).
To confirm the effect of the non-functional vrn2 alleles the synthetic vrn2-null hexaploid was crossed with Goodstreak (PI 632434, Baenziger et al. 2004). Goodstreak is a hexaploid winter wheat developed by the University of Nebraska that carries alleles for winter growth habit at the three VRN1 homoeologs and functional copies of VRN-B2 and VRN-D2. The winter wheat line Triple Dirk C (TDC, Pugsley 1971) and a winter sister line of the synthetic vrn2-null carrying functional VRN-B2 and VRND2 alleles were used to compare the relative transcript levels of these genes by qRT-PCR.
Growth conditions
Plants were grown in plastic cones (656 ml, 25 cm (9.8 in.) deep × 6.4 cm (2.5 in.) diameter (Steuwe and Sons, USA). Evaluations of plant heading times without vernalization were conducted at UC Davis in a greenhouse with a temperature of 20–25 °C and a photoperiod of 16 h of light (long day). The natural daylight in the greenhouses was supplemented with incandescent lamps at night to extend the photoperiod to 16 h. Vernalization treatments were performed in a growth chamber at constant 4 °C under long day conditions.
For the partial vernalization experiment, plants were grown for one week in the greenhouse, and were then transferred to a 4 °C chamber (16 h of light) for 2, 4 or 6 weeks of vernalization treatment. Plants included in the different treatments were planted at intervals of 2 weeks, so all the plants completed their respective vernalization treatments at the same time. All plants were then transferred together to the same greenhouse (20–25 °C, 16 h of light). Heading time was recorded at the time when half of the spike emerged from the flag leaf.
Molecular markers
Since the recessive vrn-B2 allele is the result of deletion of all the ZCCT genes in this locus, we designed a cleavage amplification polymorphism (CAP) marker for the tightly linked SNF-B2 gene. Primers SNF-B2-3p-F1 and SNFB2-3p-R2 (Table S1) were used to amplify a 1000-bp product. Digestion of the amplified product with restriction enzyme HpyCH4IV yielded two fragments (816 and 184 bp) for the synthetic vrn2-null and three fragments (626-bp, 188 bp and 184 bp) for hexaploid winter wheat Goodstreak.
For screening of the non-functional vrn-D2 allele, we developed a CAP marker using primers ZCCT-D1-E1-CAPS-F1 and ZCCT-D1-E1-CAPS-R1 (Table S1). These primers amplified an 800 bp product including the first exon of ZCCT-D1. Digestion of the amplified product with restriction enzyme MboII yielded two fragments of 650 bp and 150 bp in hexaploid wheat Goodstreak and a single 800 bp product in Ae. tauschii E1. Digestion products were separated on 6 % polyacrylamide gels stained with 2 % ethidium bromide. PCR amplifications were performed at 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, annealing at 62 °C for 30 s, and extension at 72 °C for 1 min/kb. A final extension step was carried at 72 °C for 7 min.
For screening of the VRN-A1 alleles we used primers developed by Fu et al. (2005) that detect the presence or absence of a large deletion in the first intron of VRN-A1. These primers were used to differentiate the VRN-A1 allele for spring growth habit present in Kronos (intron deletion) from the vrn-A1 allele for winter growth habit (no intron deletion) present in Goodstreak.
Identification of mutations in ZCCT1 and ZCCT2 of Aegilopstauschii E1
Genomic sequences of ZCCT-1A m, ZCCT-2A m from Triticum monococcum, and ZCCT-A1, ZCCT-B1, ZCCT-D1, ZCCT-A2 from Triticum aestivum were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/). Additional sequences of ZCCT-B2 and ZCCT-D2 were retrieved from the genomic contigs database from the International Wheat Genome Sequencing Consortium website (IWGSC, http://wheat-urgi.versailles.inra.fr). Complete sequences of the ZCCT1 and ZCCT2 homologs including their 5′ and 3′ UTRs were aligned using T-COFFEE (www.tcoffee.org).
All ZCCT1 and ZCCT2 homologs have two exons separated by a large intron. The two exons from ZCCT1 and ZCCT2 were PCR-amplified separately from Ae. tauschii accession E1 using primers and conditions listed in Table S1. Genomic DNA sequences were obtained by Sanger sequencing using an Applied Biosystems 3730XL DNA Analyzer, and were assembled using GAP4 v1.5 (Bonfield et al. 1995).
To confirm different splicing variants detected for ZCCT-D1 in Ae. tauschii E1, RNA was extracted using the Spectrum™ Plant Total RNA kit (SIGMA) from leaves of two-week old plants grown without vernalization under long day photoperiod. cDNAs were produced using the High Capacity Reverse Transcription Kit (Applied Biosystems) according to manufacturer’s instructions. Specific primers ZCCT-D1-F, ZCCT-D1-R, ZCCT-D2-F, and ZCCT-D2-R and PCR annealing temperatures are listed in Table S1. Cloning was performed using NEB® PCR Cloning Kit (New England Biolabs, USA). Positive colonies were grown and used for colony PCR and the product confirmed by Sanger sequencing.
ZCCT2 transcript levels in hexaploid wheat
Plants were grown for 3 weeks in a PGR15 growth chambers (Conviron) under long day conditions (16 h light/8 h dark) at a temperature of 20 °C during light period and 18 °C during dark. Samples were collected 4 h after the lights were turned on (Zeitgeber Time 4). RNA was extracted from leaves using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). Quantitative PCR was performed using SYBR Green and a 7500 Fast Real-Time PCR system (Applied Biosystems). Primers for ACTIN are described in Distelfeld et al. (2009) and primers for ZCCT-B2 and ZCCT-D2 are described in Table S1. Transcript levels are expressed as linearized fold-ACTIN levels calculated by the formula 2(ACTIN CT – TARGET CT) ± SE of the mean. The resulting number indicates the ratio between the initial number of molecules of the target gene and the number of molecules of ACTIN.
Results
Validation of the non-functional vrn-D2 allele in Ae. tauschii E1
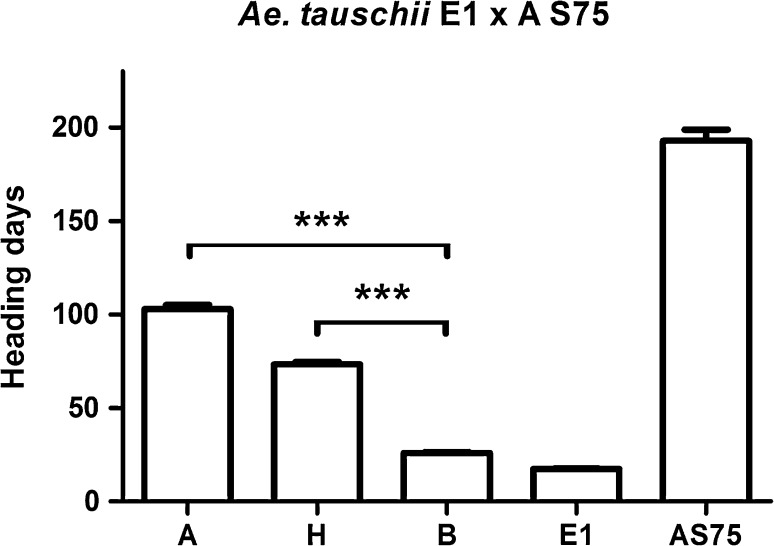
We first confirmed the presence of the non-functional vrn2 allele in the seeds received for Ae. tauschii accession E1. The F2 population from a cross between Ae. tauschii AS75 and E1 accessions showed the expected 1:2:1 segregation (16:33:22, χ2 P = 0.51) for VRN-D2. All the F2 plants homozygous for the E1 VRN-D2 allele headed very early (average 26.1 ± 0.5 d), and those homozygous for the AS75 allele headed very late (average 102.8 ± 2.4 d), confirming that the E1 accession has a recessive vrn-D2 allele (Fig. 1).
Fig. 1.
Effect of VRN2 mutations on flowering time in diploid Ae. tauschii. Heading time of 71 F2 plants from the cross between Ae. tauschii accessions E1 (early flowering) and AS75 (late flowering). A = AS75 allele, B = Ae. tauschii E1 allele and H = heterozygous. E1 = Ae. tauschii E1 and AS75 = Ae. tauschii AS75 indicate parental controls. Plants were grown under long days in the absence of vernalization. ***P < 0.001
The heterozygous plants showed an intermediate flowering time (average 73.3 ± 1.3 days), which was slightly closer to the average of the plants homozygous for the AS75 allele than to those homozygous for the E1 allele (Fig. 1). The degree of dominance of the functional VRN-D2 allele for heading time was 23 % in this population. A very similar degree of dominance (26 %) was reported before in a tetraploid population segregating only for the functional VRN-B2 allele (Distelfeld et al. 2009).
Molecular characterization of ZCCT-D1 and ZCCT-D2 sequences
To better understand the nature of the recessive vrn-D2 allele in E1, we sequenced the ZCCT-D1 and ZCCT-D2 genes from this accession and compared it to the sequence of winter lines AS75 and AL8/78. The primers used in this study are described in supplemental Table S1.
The predicted protein sequence of ZCCT-D1 from E1 (KM489155) showed two amino acid changes (F36C and Q45H) that differentiated E1 from the two winter lines. A two amino acid deletion at positions 101 and 102 was observed in both the early flowering E1 and the late flowering AL8/78, and therefore, is not likely to affect the VRN2 function (Fig. 2a, Fig. S1). According to the SIFT score (Ng and Henikoff 2001), the amino acid substitution in F36C has a high probability to alter protein structure or function (Fig. 2a, Fig. S1). The deletion at positions 101 and 102 (also found in ZCCT-D1, GenBank accession ACI00354) is absent in the ZCCT-A1 and ZCCT-B1 sequences (Fig. S1).
Fig. 2.
Mutations in the VRN 2 locus of Ae. tauschii E1. Multiple alignments of predicted coding sequences of ZCCT1 a and ZCCT2 b in Ae. tauchii E1 (KM489155 and KM489156), and winter controls AS75 (KM503042 and KM503043) and AL8/78 (ACI00354 and ACI00358). Numbers in parenthesis are GenBank accession numbers for ZCCT1 and ZCCT2, respectively. For the ZCCT1 protein from E1, a 35 amino acid deletion observed in 87.5 % of the cloned and sequenced cDNAs is underlined (the other 12.5 % of the clones include an intron region producing a premature stop codon). The CCT domain is indicated with asterisks in both genes. SNPs are highlighted in gray. Critical Arg residues in the CCT domain are highlighted in black
The genomic ZCCT-D1 sequence of E1 has a 24 bp deletion located 6 bp before the splicing acceptor site at the 3′ end of the single intron that is not observed in the two winter Ae. tauschii accessions sequenced in this study (Fig. S2). To test the effect of this intron deletion on splicing, we cloned and sequenced multiple ZCCT-D1 transcripts from Ae. tauschii E1 and the winter control AS75. In E1, 21 of the 24 sequenced clones (87.5 %) showed an alternative splicing variant that generated a deletion of 35 amino acids before the CCT domain (region underlined in Fig. 2a). The other three clones (12.5 %) included a 58-bp region of the intron producing a premature stop codon and disrupting the CCT domain. None of the clones obtained from E1 showed a normal and complete VRN2 transcript, indicating that different acceptor sites were used for all the ZCCT-D1 transcripts. By contrast, the winter control AS75 showed a single splicing product encoding the complete ZCCT-D1 gene.
The ZCCT-D2 genomic sequence showed one amino acid change and a 1 bp deletion that were present only in E1. The 1 bp deletion generates a change in the reading frame that disrupts the CCT domain (Fig. 2b). These changes were confirmed by sequencing 37 cDNA clones of ZCCT-D2 from E1. Taken together, the molecular and the genetic data indicate that E1 carries a non-functional VRN2 allele.
Development and characterization of a triple VRN2 mutant in hexaploid wheat
The E1 accession carrying the non-functional VRN2 allele was crossed with tetraploid line #3089 carrying non-functional VRN-A2 and VRN-B2 alleles (Distelfeld et al. 2009, Fig. 3). The triploid F1 hybrids were treated with colchicine (0.1 % for 7–7:30 h), but they were very susceptible to colchicine and most of them died. Fortunately, the triploid F1 hybrids naturally produced some fertile seeds. Cytogenetic characterization of these seeds confirmed the chromosome doubling (2n = 42), which likely occurred by meiotic restitution.
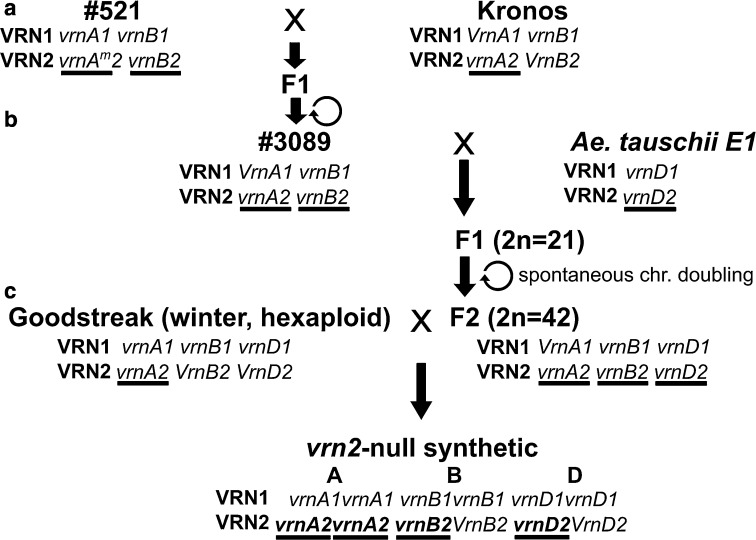
Fig. 3.
Development of synthetic vrn2-null hexaploid wheat. a Introgression of vrn-A2 and vrn-B2 in the tetraploid cultivar Kronos. b Production of synthetic aneuploid to introgress vrn-D2, self-pollination to recover chromosome number. c Backcross with winter hexaploid wheat to produce a segregating line for vrn-A2, vrn-B2 and vrn-D2 in a winter background. X represents a crossing step, arrow represents a progeny. Non-functional VRN2 alleles are underlined
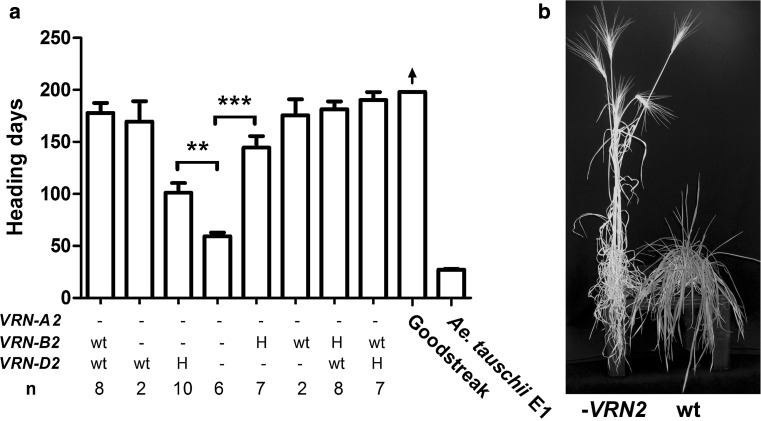
The resulting synthetic hexaploid carried non-functional copies of all three VRN2 homoeologs (synthetic vrn2-null), but it also carried a dominant Vrn-A1 allele for spring growth habit that complicated the analysis. To eliminate the VRN-A1 allele, the vrn2-null synthetic was crossed with the winter wheat variety Goodstreak that carries recessive vrn-A1, vrn-B1, and vrn-D1 alleles (Fig. 3). Both parental lines carry the non-functional VRN-A2 allele. The F1 hybrid was self-pollinated and the segregating population (93 F2 plants) was genotyped with markers for VRN-B2, VRN-D2 and VRN-A1. Only two plants were triple homozygous vrn-A1, vrn-B2 and vrn-D2 and both were early flowering. To increase the number of plants segregating for only for VRN-B2 and VRN-D2, we selected F2 plants homozygous for the vrn-A1 allele for winter growth habit and heterozygous for the two VRN2 genes. In this segregating F3 population (69 plants), we identified 6 plants homozygous for the non-functional vrn-A2, vrn-B2 and vrn-D2 plants. These plants headed more than 100 days earlier than those homozygous for at least one of the functional VRN2 alleles (P < 2.8E–07) in the absence of vernalization (Fig. 4).
Fig. 4.
Effect of mutations in VRN-B2 and VRN-D2 on heading time under non vernalizing conditions. a Heading times for the triple mutant. Bars represent mean and error bars represent the SEM. Dashes represent mutant alleles, wt: wild type alleles and H: heterozygous. Lower doses of functional VRN2 were enough to significantly delay flowering. **P < 0.005 ***P < 0.001. Arrow indicates that the experiment was stopped before heading. b Maturity differences between the triple VRN2 mutant (left) and the wild type (right)
Relative effect of VRN-B2 and VRN-D2 on flowering time and vernalization response
In the absence of vernalization, plants carrying only the functional VRN-B2 or the functional VRN-D2 allele in homozygous state exhibited a winter growth habit, and showed no significant differences in heading time with the control plants carrying both functional VRN2 genes (Fig. 4). Plants carrying only the functional VRN-D2 allele flowered six days earlier (169 days) than the plants carrying only the functional VRN-B2 allele (175 days), but the differences were not significant. Interestingly, plants carrying only one functional VRN2 allele in heterozygous stage flowered significantly earlier than plants homozygous for any of the functional VRN2 alleles (Fig. 4). Plants carrying only the functional VRN-B2 allele in heterozygous state headed 43 d later than plants carrying only the functional VRN-D2 allele in heterozygous state (144.5 d vs 101.2 d, P = 0.009, Fig. 4). These results suggest that the functional VRN-B2 allele has a stronger effect in repression of flowering under long days than the VRN-D2 allele (Fig. 4).
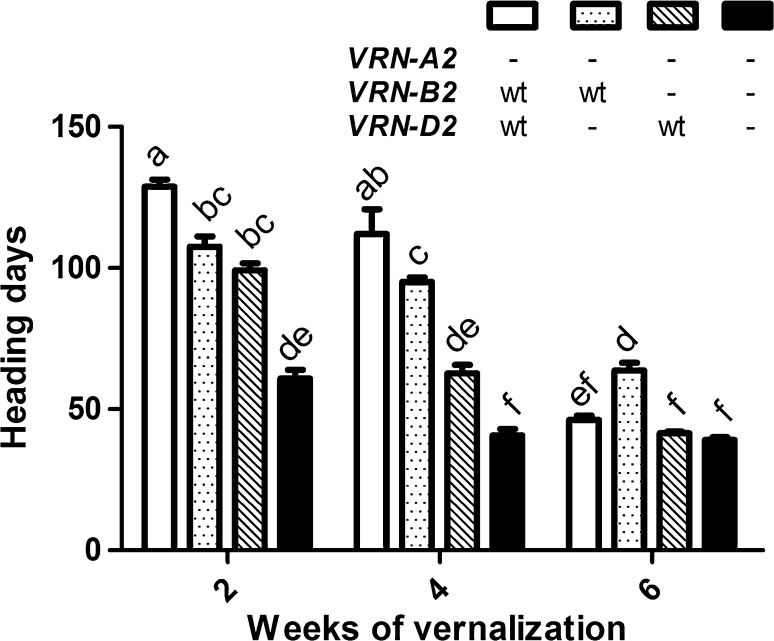
The stronger vernalization requirement of the functional VRN-B2 allele relative to VRN-D2 was also supported by a partial vernalization experiment. After 2 weeks of vernalization, the heading time differences between the functional VRN-B2 and VRN-D2 alleles were still significant, but plants with a single functional VRN2 allele flowered significantly earlier than the control with both functional alleles (Fig. 5). After 4 weeks of vernalization, plants carrying only the functional VRN-B2 allele headed 32 days later than those carrying the functional VRN-D2 allele (P = 1.34E−5).
Fig. 5.
Effects of mutations in VRN-B2 and VRN-D2 on heading time under different vernalization treatments. Error bars represent the SE of the mean of 5 biological replications. Dashes represent a non-functional allele and wt a wild type allele. For each treatment different letters indicate significant differences (Tukey’s test, P < 0.05)
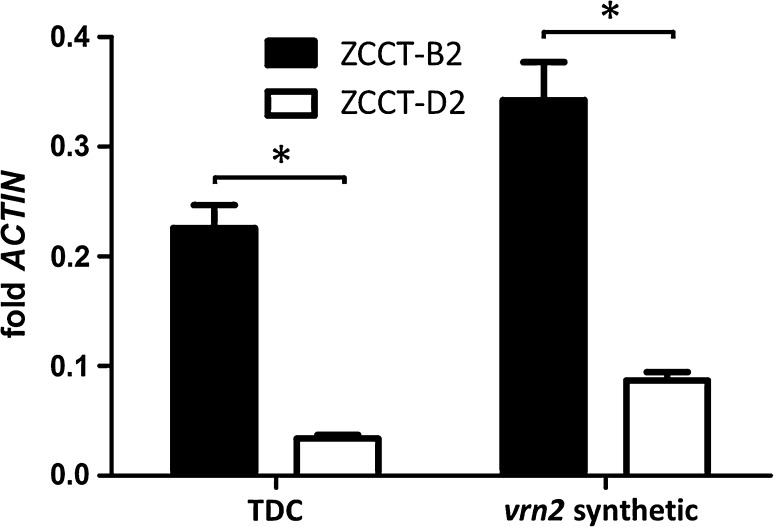
To test if the stronger repressive effect of VRN-B2 relative to VRN-D2 was associated with differences in expression, we compared their transcript levels in two winter wheat lines using qRT-PCR. In a sister line of the synthetic vrn2-null carrying functional VRN-B2 and VRN-D2 alleles, the transcript levels of ZCCT-B2 were fourfold higher than those of ZCCT-D2 (P < 0.001, Fig. 6). A similar result was observed in the winter line Triple Dirk C, where the transcript levels of ZCCT-B2 were sixfold higher than those of ZCCT-D2 (P < 0.001, Fig. 6).
Fig. 6.
Transcript levels of ZCCT-B2 and ZCCT-D2 in hexaploid wheat. ZCCT-B2 levels are 6 and 4 times higher than ZCCT-D2 in TDC and vrn2-synthetic, respectively. Triple Dirk C (TDC) is a winter hexaploid line and vrn2-synthetic corresponds to the wild type sister line of the synthetic vrn2-null developed in this study. Bars represent means of 5 biological and 2 technical replications. Plants were grown under long day conditions (16 h light) at a temperature of 20 °C day/18 °C night for 3 weeks. Samples were collected at ZT 4 (Zeitgeber Time). *P < 0.001
Finally, after 6 weeks of vernalization all genotypes flowered very early although the plants carrying the functional VRN-B2 allele flowered later than the other genotypes (Fig. 5). We do not know the cause of this difference.
As in the previous experiment with non-vernalized plants (Fig. 4), the synthetic vrn2-null showed the earliest heading time among the four genotypes for the plants exposed to 2 and 4 weeks of vernalization (Fig. 5). Interestingly, the synthetic vrn2-null showed a residual response to vernalization. When these plants were vernalized for 4 weeks, they flowered 20 days earlier (P = 0.001) than when they were vernalized for 2 weeks (Fig. 5). No significant differences in heading time were detected between synthetic vrn2-null plants vernalized for 4 or 6 weeks (P = 0.529, Fig. 5), or between those that were vernalized for 2 weeks (60.8 days) relative to non-vernalized plants (58.0 days, P = 0.433).
Discussion
VRN2 natural variation in polyploid wheat
In hexaploid wheat, most of the natural variation in growth habit is associated with dominant mutations in regulatory regions of the three VRN1 homoeologs (Yan et al. 2004b; Fu et al. 2005; Dubcovsky et al. 2006; Zhang et al. 2008; Chu et al.2011; Zhang et al. 2012; Muterko et al. 2015, 2016), and less frequently with dominant mutations in VRN3 (=TaFT1, Yan et al. 2006; Zhang et al. 2008) or VRN-D4 (a VRN1 paralogue; Kippes et al. 2015). However, no natural variation in growth habit has been associated so far with variation at any of the three VRN2 homoeologs in hexaploid wheat.
The limited effect of single VRN2 mutations on heading time in non-vernalized or fully vernalized hexaploid plants may explain why no vrn2-null spring varieties have been selected so far in polyploid wheat. By contrast, vrn2-null spring accessions are frequent in both T. monococcum (Yan et al. 2004b) and barley (von Zitzewitz et al. 2005), supporting the hypothesis that functional redundancy among VRN2 homoeologs in the polyploid wheat species may have masked the effect of recessive mutations on heading time from natural or human selection.
Although no vrn2-null spring accessions of hexaploid wheat have been reported so far, molecular and genetic studies have revealed allelic variation in ZCCT1 and ZCCT2 at each of the three VRN2 homoeologs. Since non-functional mutations in both tightly linked ZCCT1 and ZCCT2 genes are required to generate a recessive VRN2 locus, it is important to describe the allelic variation in both genes within each locus. The initial studies in T. monococcum identified ZCCT1 as the functional gene in the VRN-A m 2 locus (Yan et al. 2004a), but a subsequent study in tetraploid wheat showed that ZCCT2 was the functional gene in the VRN-B2 locus (Distelfeld et al. 2009).
Most of the VRN-A2 loci show non-functional mutations in the CCT domains of both ZCCT-A1 and ZCCT-A2 in tetraploid wheat. Only one accession of T. turgidum subsp. dicoccoides showed a wild type CCT domain for ZCCT-A1 in a collection of 78 tetraploid wheats including both wild and domesticated accessions (Distelfeld et al. 2009). For ZCCT-A2, all 78 accessions have a C16R mutation in the CCT domain. This mutation was also detected in 15 accessions of T. urartu and in two accessions of T. monococcum, suggesting that this mutation was already fixed (or frequent) before the divergence of these two closely related diploid species (Distelfeld et al. 2009). In tetraploid wheat combination of CCT mutations C39R in ZCCT-A1 and C16R in ZCCT-A2 result in a recessive vrn-A2 allele (Distelfeld et al. 2009). The same mutations are also present in the early flowering synthetic vrn2-null hexaploid generated in this study, confirming that this vrn-A2 allele is also non-functional in hexaploid wheat. Zhu et al. (2011) identified six accessions of hexaploid wheat where they were unable to amplify the promoter of ZCCT-A1, suggesting that these varieties may carry non-functional vrn-A2 alleles. However, ZCCT-A2 was not characterized in the previous study.
The VRN-B2 locus sequenced in tetraploid wheat carries a non-functional ZCCT-B1 protein (C39R mutation in 37 sequenced T. turgidum subsp. durum), and two recently duplicated copies of ZCCT-B2 (wild type CCT domain). The presence of these two ZCCT-B2 copies is sufficient to confer a strong vernalization requirement (Distelfeld et al. 2009). A complete deletion of all three ZCCT genes present in VRN-B2 was identified in one accession of T. turgidum subsp. dicoccon collected in Turkey (PI470739), and was associated with early flowering in a tetraploid wheat segregating population (Distelfeld et al. 2009). In this study, we confirmed that the VRN-B2 deletion is associated with early flowering and spring growth habit in the synthetic vrn2-null hexaploid. A similar deletion in VRN-B2 was recently reported in the hexaploid wheat variety Jagger, but no significant differences in heading time were detected for this locus in a segregating population tested in the field in Oklahoma (USA) under natural vernalization conditions (Tan and Yan 2015).
More limited allelic variation has been described for the VRN-D2 locus. Ae. tauschii accessions AS75 has a winter growth habit and the predicted ZCCT-D1 and ZCCT-D2 proteins show no mutations in the conserved amino acids of the CCT domains (Distelfeld et al. 2009; Tan and Yan 2015). Zhu et al. (2011) identified one accession from Macedonia (CItr 17383) with a deletion in the ZCCT-D1 promoter, but this accession was later reclassified in GRIN as T. turgidum (2n = 28), explaining the absence of the D genome locus. The limited variability in the D genome of hexaploid wheat observed in this locus prompted us to look for variation in Ae. tauschii to identify a non-functional VRN-D2 allele.
Ae. tauschii E1 carries a recessive vrn-D2 allele
The early flowering in Aegilops tauschii accession E1 was previously reported to be linked to isozyme ACO2 locus on chromosome arm 4DL (recombination fraction 0.32), suggesting that it may correspond to the VRN-D2 locus (Dudnikov 2003). The segregating population generated in this study from the cross between Ae. tauschii spring accession E1 and winter accession AS75 confirmed that the differences in heading time were completely linked to polymorphisms in VRN-D2, suggesting that E1 carries a recessive vrn-D2 allele.
The previous hypothesis was also supported by the presence of disruptive mutations in both ZCCT-D1 and ZCCT-D2 in E1 but not in the two Ae. tauschii winter accessions used for comparison.
For ZCCT-D1, two amino acid changes close to the predicted zinc finger were observed only in E1. One of these changes resulted in an additional cysteine in E1 (F36C), which has the potential to disrupt the zinc finger. Although the previous amino acid changes may affect the function of ZCCT1, the most drastic change in this gene was the presence of altered splice forms probably associated with a 24 bp deletion in the intron. Although this deletion did not include the AG splicing site, it is located only six bp from the splicing acceptor. The deleted region includes a polypyrimidine tract that is known to be important for correct splicing and it also alters the distance between the acceptor site and the putative branch point, which is also important for the selection of the correct splicing site (Black 2003). The analysis of the ZCCT-D1 transcripts confirmed that none of them use the canonical acceptor site. The alternative splice sites were associated either with a 35 amino acid deletion before the CCT domain or with the translation of part of the intron encoding a premature stop codon. For ZCCT-D2, we detected a frame shift mutation that was present only in E1, and is predicted to result in a protein lacking almost the complete CCT domain (Fig. 2b).
In summary, both ZCCT-D1 and ZCCT-D2 showed disruptive mutations in E1 that are expected to greatly reduce or eliminate VRN-D2 function. This hypothesis was confirmed by the early flowering of the diploid plants carrying the E1 vrn-D2 allele in the diploid segregating population, and also by the early flowering of the synthetic vrn2-null synthetic hexaploid including this allele. These results also confirmed that the protein region adjacent to the CCT domain is necessary for proper protein function, and that the intron region adjacent to the acceptor splicing site is critical for correct splicing of the ZCCT1 transcripts.
Recessive alleles at the three VRN2 homologs results in a spring growth habit
With the identification of the recessive vrn-D2-null allele it was possible to develop a triple vrn2-null hexaploid wheat. This vrn2-null hexaploid line was crossed with the winter hexaploid wheat Goodstreak to study the effect of the different vrn-2 alleles in the absence of spring alleles in the VRN1 homoeologs. This is important because dominant VRN1 alleles for early flowering are epistatic to VRN2 and can mask their effect (Tranquilli and Dubcovsky 2000; Chen and Dubcovsky 2012).
In the absence of dominant VRN1 alleles for spring growth habit, non-vernalized plants carrying non-functional vrn2-null alleles took roughly 60 days from sowing to heading, whereas plant carrying either the functional VRN-B2 or VRN-D2 alleles (or both) headed more than 100 days later then the vrn2-null plants (Fig. 5). These large differences in heading time disappeared when plants were vernalized for 6 weeks (Fig. 5), confirming that the late flowering was associated to a strong vernalization requirement.
Interestingly, synthetic vrn2-null hexaploid plants showed a residual response to the vernalization treatment. When these plants were vernalized for 4 or 6 weeks they flowered on average 20 days earlier than when they were vernalized for 2 weeks or were not vernalized at all. Although we cannot completely rule out the possibility that this residual vernalization response is caused by some functionality of the mutant VRN-A2 or VRN-D2 proteins (hypomorphic mutations), the presence of similar effects in other species supports an alternative explanation.
Vernalization of barley plants in which all the ZCCT genes have been deleted (vrn2-null) still accelerated heading time by 2 weeks (Hemming et al. 2008). In the same study, VRN1 expression was induced by vernalization in the presence and absence of VRN2, suggesting that this gene may contribute to the residual vernalization response of the vrn2-null plants. Although we do not disregard a possible contribution of VRN1, it was recently demonstrated that tetraploid wheat plants, carrying non-functional copies of both VRN1 and VRN2 genes (henceforth, vrn1-vrn2-null), still flower on average 23 days earlier when they are vernalized than when they are not (Chen and Dubcovsky 2012). Since the vrn1-vrn2-null plants lack any functional VRN1 and VRN2 genes, these results indicate that wheat has additional genes that are able to accelerate flowering in response to vernalization.
ZCCT homoeologs exhibit differential responses under partial vernalization and when present in different dosages
The stronger effect of the functional VRN-B2 allele relative to the functional VRN-D2 allele was evident in the non-vernalized plants carrying only one functional VRN2 allele in heterozygous state (Fig. 4). Plants carrying only one functional copy of VRN-B2 flowered in average 43 days later than those with only one functional copy of VRN-D2 (Fig. 4, P < 0.009). The stronger effect of the functional VRN-B2 allele relative to the VRN-D2 allele was also evident in the partial vernalization response experiments (Fig. 5).
The stronger effect of the VRN-B2 locus on heading time was correlated with four- to sixfold higher transcript levels of ZCCT-B2 relative to those of ZCCT-D2 (Fig. 6). Although these differences in expression are likely sufficient to explain the different effects of these alleles on heading time, we cannot rule out the possibility of additional differences in the strength of the encoded ZCCT-B2 and ZCCT-D2 proteins or the presence of residual activity of the protein encoded by the vrn-D2 recessive allele.
A previous study in tetraploid wheat showed that the ZCCT2 genes are expressed at 3- to 10-fold higher levels than the ZCCT1 genes (Distelfeld et al. 2009). This may contribute to the stronger effect of the VRN-B2 allele, which includes two recently duplicated copies of the wild type ZCCT-B2 gene (Distelfeld et al. 2009).
Although plants carrying only a single copy of the functional VRN-B2 allele in heterozygous state flowered more than 40 days later than those carrying only a single copy of the functional VRN-D2 allele in heterozygous state, the differences were no longer significant when these functional alleles were in the homozygous state. No further delays in heading time were observed when both the VRN-B2 and VRN-D2 functional alleles were combined. Taken together, these results suggest that the repressive effect of VRN2 on heading time may be saturated beyond two functional copies of any of the active alleles. The small phenotypic effect of the non-functional mutations in one of the VRN2 homoeologues limits the effects of natural or human selection on these alleles. This may explain why no natural vrn2-null mutants have been identified so far.
Conclusions and potential practical applications
Both the single and double VRN2 mutants generated in this study may have practical applications. The reduced vernalization requirement associated with the presence of single VRN-B2 and VRN-D2 mutants under partial vernalization suggests that these mutants may be useful to modulate the vernalization response in regions with mild winters (e.g., Mediterranean regions) or as global warming reduces the severity of winters in other locations.
It would be also important to study the effect of the absence of all functional VRN2 alleles (vrn2-null) on the duration of the different reproductive phases, and the effect of these changes on hexaploid wheat adaptation to different environments. We speculate that this new class of spring wheat varieties may have practical applications in some environments based on the relatively high frequency of vrn2-null spring accession in diploid wheat and barley species. The non-functional vrn2-null allele was detected in 80 % of the accessions in a collection of 65 cultivated T. monococcum ssp. monococcum accessions (Yan et al. 2004a) and is also frequent in barley. Barley varieties carrying the vrn2-null allele have been designated as “facultative winters” and have been recommended for some of the barley growing regions (Karsai et al. 2005; Szucs et al. 2007).
Seeds of the synthetic vrn2-null hexaploid wheat with no functional homoeologues of VRN2 have been deposited in the National Small Grain Collection (PI 676269). Seeds from this genetic stock are publicly available and can be used as a source of non-functional VRN-B2 and VRN-D2 alleles in common wheat breeding programs. These new VRN2 alleles provide common wheat breeders new tools to improve wheat adaptation to new or changing environments.
Author contribution statement
CA sequenced VRN2 in E1 and AS75 and performed E1xAS75 progeny test, LA and ZX produced the synthetic hexaploid wheat, KN sequenced coding regions of ZCCT1 and ZCCT2 in E1, cloned and sequenced ZCCT1 and ZCCT2 amplicons from E1 and controls, carried out the ZCCT2 qRT-PCR, performed the segregation analysis tests with and without vernalization, made the multiple sequence alignments analysis and produced the figures. KN and DJ analyzed data and wrote the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This project was supported by the National Research Initiative (grants 2011-68002-30029 and 2016-67013-24617) from the USDA National Institute of Food and Agriculture and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation Grant GBMF3031. We are grateful to Anna Dominique-Rodriguez for technical support and to Alexander Dudnikov from the Institute of Cytology and Genetics (Novosibirsk, Russia) for the Ae. tauschii E1 seeds.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-016-2713-3) contains supplementary material, which is available to authorized users.
A correction to this article is available online at https://doi.org/10.1007/s00122-018-3141-3.
Change history
7/12/2018
In the original version of this article, PCR fragments and digestion product sizes for the VRN-B2 and VRN-D2 markers were not accurate. The corrected sizes are detailed below.
Contributor Information
Nestor Kippes, Email: nfkippes@ucdavis.edu.
Andrew Chen, Email: a.chen2@uq.edu.au.
Xiaoqin Zhang, Email: xqzzhang@ucdavis.edu.
Adam J. Lukaszewski, Email: adam.lukaszewski@ucr.edu
Jorge Dubcovsky, Phone: 530 752 5159, Email: jdubcovsky@ucdavis.edu.
References
- Baenziger PS, Beecher B, Graybosch RA, Baltensperger DD, Nelson LA, Krall JM, Mcvey DV, Watkins JE, Hatchett JH, Chen MS. Registration of ‘Goodstreak’ wheat. Crop Sci. 2004;44:1473–1474. doi: 10.2135/cropsci2004.1473. [DOI] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theor Appl Genet. 2007;115:721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012;8:e1003134. doi: 10.1371/journal.pgen.1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CG, Tan CT, Yu GT, Zhong S, Xu SS, Yan L. A novel retrotransposon inserted in the dominant Vrn-B1 allele confers spring growth habit in tetraploid wheat (Triticum turgidum L.) G3-Genes Genom Genet. 2011;1:637–645. doi: 10.1534/g3.111.001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Diaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum) PLoS ONE. 2012;7:e33234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J. Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 2009;149:245–257. doi: 10.1104/pp.108.129353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Chen C, Yan L. Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breeding. 2005;15:395–407. doi: 10.1007/s11032-005-0084-6. [DOI] [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol. 2006;60:469–480. doi: 10.1007/s11103-005-4814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudnikov A. Allozymes and growth habit of Aegilops tauschii: genetic control and linkage patterns. Euphytica. 2003;129:89–97. doi: 10.1023/A:1021558628874. [DOI] [Google Scholar]
- Falconer DS. Introduction to quantitative genetics. Edinburgh: Oliver & Boyd; 1964. [Google Scholar]
- Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. Low-temperature and day length cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 2008;147:355–366. doi: 10.1104/pp.108.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B. Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Genet Genomics. 2009;282:107–117. doi: 10.1007/s00438-009-0449-3. [DOI] [PubMed] [Google Scholar]
- Karsai I, Szűcs P, Meszaros K, Filichkina T, Hayes PM, Skinner JS, Lang L, Bedo Z. The Vrn-H2 locus is a major determinant of flowering time in a facultative x winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet. 2005;110:1458–1466. doi: 10.1007/s00122-005-1979-7. [DOI] [PubMed] [Google Scholar]
- Kippes N, Zhu J, Chen A, Vanzetti L, Lukaszewski A, Nishida H, Kato K, Dvorak J, Dubcovsky J. Fine mapping and epistatic interactions of the vernalization gene VRN-D4 in hexaploid wheat. Mol Genet Genom. 2014;289:47–62. doi: 10.1007/s00438-013-0788-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippes N, Debernardi J, Vasquez-Gross HA, Akpinar BA, Budak H, Kato K, Chao S, Akhunov E, Dubcovsky J. Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc Natl Acad Sci USA. 2015;112:E5401–E5410. doi: 10.1073/pnas.1514883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Distelfeld A, Comis A, Dubcovsky J. Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 2011;67:763–773. doi: 10.1111/j.1365-313X.2011.04630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lin H, Dubcovsky J. Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 2015;84:70–82. doi: 10.1111/tpj.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Deal KR, Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, Crossman CC, Dubcovsky J, Gill BS, Gu YQ, Hadam J, Heo HY, Huo N, Lazo G, Ma Y, Matthews DE, McGuire PE, Morrell PL, Qualset CO, Renfro J, Tabanao D, Talbert LE, Tian C, Toleno DM, Warburton ML, You FM, Zhang W, Dvorak J. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA. 2009;106:15780–15785. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RL, Morishige DT, Brady JA, Rooney WL, Yang S, Klein PE, Mullet JE. Ghd7 (Ma6) represses sorghum flowering in long days: Ghd7 alleles enhance biomass accumulation and grain production. Plant Genome. 2014;7(2):1–10. doi: 10.3835/plantgenome2013.11.0040. [DOI] [Google Scholar]
- Muterko A, Balashova I, Cockram J, Kalendar R, Sivolap Y. The new wheat vernalization response allele Vrn-D1 s is caused by DNA transposon insertion in the first intron. Plant Mol Biol Rep. 2015;33:294–303. doi: 10.1007/s11105-014-0750-0. [DOI] [Google Scholar]
- Muterko A, Kalendar R, Salina E. Novel alleles of the VERNALIZATION1 genes in wheat are associated with modulation of DNA curvature and flexibility in the promoter region. BMC Plant Biol. 2016;16:9. doi: 10.1186/s12870-015-0691-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AT. A genetic analysis of the spring-winter habit of growth in wheat. Aust J Agric Res. 1971;22:21–31. doi: 10.1071/AR9710021. [DOI] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Robson F, Costa MM, Hepworth SR, Vizir I, Piñeiro M, Reeves PH, Putterill J, Coupland G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28:619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz T, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Szucs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen THH, Hayes PM. Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol Genet Genomics. 2007;277:249–261. doi: 10.1007/s00438-006-0195-8. [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tan C, Yan L. Duplicated, deleted and translocated VRN2 genes in hexaploid wheat. Euphytica. 2015;208(2):227–284. [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Taylor RD, Koo WW (2015) 2015 outlook of the U.S. and world wheat industries, 2015-2024, Agribusiness & Applied Economics, North Dakota State University, Dept. of Agribusiness and Applied Economics, Center for Agricultural Policy and Trade Studies, Fargo, North Dakota, pp 58108–6050. http://ageconsearch.umn.edu/bitstream/201310/2/AAE738.pdf
- Tranquilli GE, Dubcovsky J. Epistatic interactions between vernalization genes Vrn-Am 1 and Vrn-Am2 in diploid wheat. J Hered. 2000;91:304–306. doi: 10.1093/jhered/91.4.304. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to day length, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 2006;140:1397–1405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zitzewitz J, Szűcs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS. Molecular and structural characterization of barley vernalization genes. Plant Mol Biol. 2005;59:449–467. doi: 10.1007/s11103-005-0351-2. [DOI] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm EP, Turner AS, Laurie DA. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.) Theor Appl Genet. 2009;118:285–294. doi: 10.1007/s00122-008-0898-9. [DOI] [PubMed] [Google Scholar]
- Woods D, Mckeown M, Dong Y, Preston JC, Amasino RM. Evolution of VRN2/GhD7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol . 2016;170(4):2124–2135. doi: 10.1104/pp.15.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet. 2004;109:1677–1686. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XK, Xia XC, Xiao YG, Dubcovsky J, He ZH. Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1 and Vrn-B3 in Chinese common wheat cultivars and their association with growth habit. Crop Sci. 2008;48:458–470. doi: 10.2135/cropsci2007.06.0355. [DOI] [Google Scholar]
- Zhang J, Wang YY, Wu SW, Yang JP, Liu HW, Zhou Y. A single nucleotide polymorphism at the Vrn-D1 promoter region in common wheat is associated with vernalization response. Theor Appl Genet. 2012;125:1697–1704. doi: 10.1007/s00122-012-1946-z. [DOI] [PubMed] [Google Scholar]
- Zhu X, Tan C, Cao S, Yan L. Molecular differentiation of null alleles at ZCCT-1 genes on the A, B, and D genomes of hexaploid wheat. Mol Breeding. 2011;27:501–510. doi: 10.1007/s11032-010-9447-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.