Abstract
Prostate cancer is the second leading cancer in men world-wide. Due to its heterogeneous nature, a considerable amount of research effort has been dedicated in identifying effective clinical biomarkers with a focus on proteins, messenger RNA and microRNAs [1]. However, there is limited data on the role and expression of long noncoding RNAs (lncRNAs) in prostate cancer exosomes [2]. This array dataset which is linked to our publication describes the profiling of human lncRNAs in prostate cancer and their exosomes from five different cell lines [3]. From this dataset, we identified a list of statistically significant prostate cancer lncRNAs which are differentially expressed in the exosomes compared to their parent cell lines. This dataset has been deposited into Gene Expression Omnibus (GSE81034).
Keywords: Exosome, lncRNA, Prostate cancer
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/prostate cancer cell lines: LNCaP, PC3, DU145, VCaP/healthy prostate cell lines: PNT2 |
| Sex | Male |
| Sequencer or array type | Agilent Oligo nucleotide array |
| Data format | Raw text files |
| Experimental factors | Tumor vs normal cell line |
| Experimental features | Extraction of total RNA from exosomes of prostate cancer and healthy cell lines, followed by lncRNA profiling using the Human lncRNA array v2.0 (8 × 60 K, Arraystar) |
| Consent | NA |
| Sample source location | Sydney, Australia |
1. Direct link to deposited data
[http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81034].
2. Experimental design, materials and methods
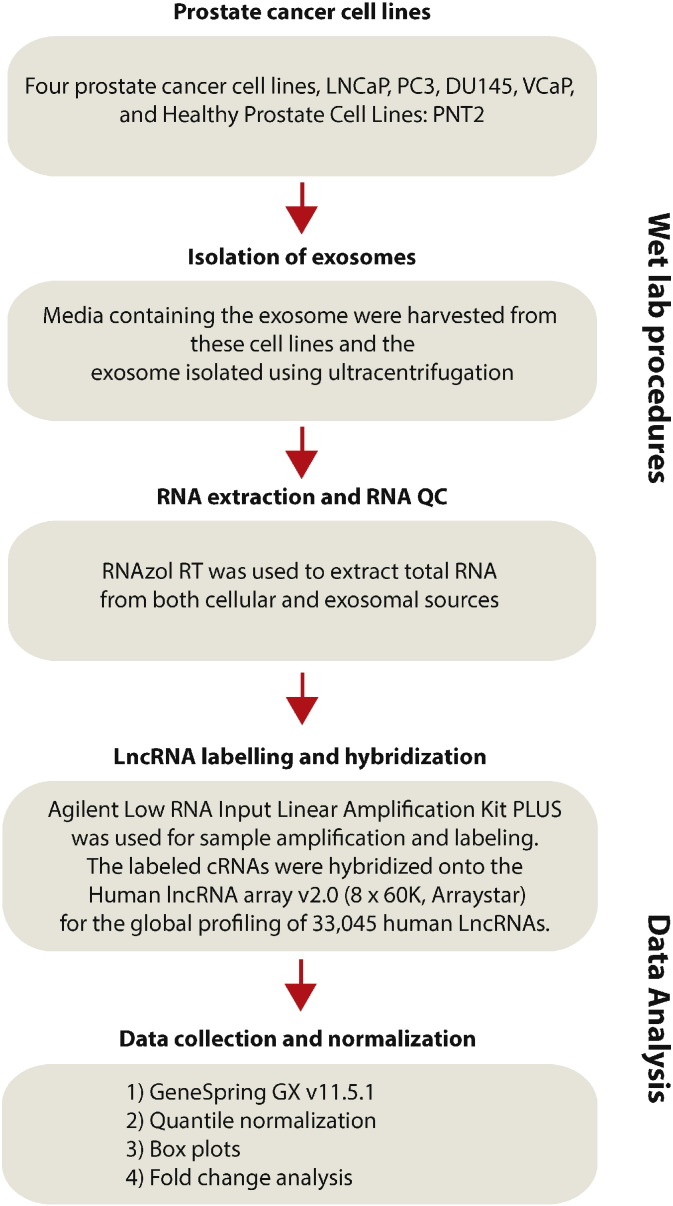
The experiment design and data analysis pipeline is shown in Fig. 1. In the following sections, we review the experiment design, quality assessment of the prepared data and the analysis pipeline for this dataset.
Fig. 1.
Experimental overview of the project and the data processing pipeline. Four difference prostate cancer cell lines were grown and the media harvested from these cell lines were used to isolate exosomes. Total RNA was then extracted from both cellular and exosomal sources. Integrity and concentration of RNA were assessed after RNA extraction and prior to sample labeling. Agilent low RNA input linear amplification kit PLUS was used for sample amplification and labeling. After washing, slides were scanned with the Agilent DNA Microarray Scanner. Data was extracted using Agilent feature extraction software. Normalization and further data analysis was performed using GeneSpring v11.5.1 software.
3. Tissue culture of prostate cancer cell lines
Prostate cancer cell lines, LNCaP, PC3, and DU145 were cultured in RPMI medium, VCaP cells were grown in F12:DMEM and PNT2 cell lines in defined KSFM (Invitrogen, USA). All cells were incubated at 37 °C in 5% CO2, and supplemented with 1% v/v penicillin, streptomycin, glutamine and 10% v/v fetal calf serum. As Fetal Calf Serum contains exosomes of bovine origin all media were depleted of these bovine exosomes. To remove bovine exosomes, we utilized the ultracentrifugation protocol devised by Théry, C. et al. [2].
4. Isolation of exosomes using ultracentrifugation
Harvested culture was centrifuged twice, firstly at 500g for 30 min, and then at 2000g for 10 min to remove dead cells and any debris. The resulting supernatant was then centrifuged at 10,000g for 30 min to separate micro-particles and other proteins from the exosomes. The supernatant was collected from this step and then centrifuged at 100,000g for 120 min to obtain an exosomal pellet. The supernatant was discarded, and the pellet retained. The exosomal pellet was then re-suspended in PBS and centrifuged at 100,000g for 90 min. This resulting pellet contains the exosomes which was subjected to RNA isolation.
5. RNA extraction procedures
RNAzol RT (Molecular Research Center, USA) was used to extract total RNA from both cellular and exosomal sources [4]. 1 mL of RNAzol was added to the exosomal pellet and homogenised using a blunt needle syringe. This homogenate is added to 0.4 mL of water and left to incubate on ice for up to 15 min. The sample was then centrifuged at 4 °C, at 12,000g for 15 min. 1 mL of the resulting supernatant was removed and purification performed via the addition of 5 μL of 4-bromoanisole. The sample was incubated for 3–4 min, and centrifuged at 12,000g, 4 °C, for 10 min. The supernatant was removed from the sample, and 1 volume of isopropanol was added with 5 μL of glycogen (25 mg/mL) to precipitate the RNA. This mixture was then incubate on dry ice for 15 min followed by centrifugation at 12,000g, 4 °C, for 10 min. The resulting RNA pellet was then washed twice in 75% ethanol and centrifuge at 8000g for 5 min to collect the pellet. After discarding the supernatant, the exosomal RNA pellet was solubilized with the addition of 50 μL of RNA/DNA free water.
6. Preparation of lncRNAs for hybridization to Agilent arrays
RNA Integrity and concentration was assessed post RNA extraction and prior to sample labeling. The sample RNA integrity number (RIN) was determined using a Bioanalyzer 2100 to assess RNA quality. RNA concentration, protein contamination and organic compound contamination was measured for all samples using the NanoDrop ND-1000. Agilent Low RNA Input Linear Amplification Kit PLUS was used for sample amplification and labeling. Hybridization was performed using the Agilent SureHyb Hybridization Chambers. The labeled cRNAs were hybridized onto the Human lncRNA array v2.0 (8 × 60 K, Arraystar) for the global profiling of 33,045 human lncRNAs. The lncRNAs in this array were collected from databases such as RefSeq, UCSC Known genes, and Ensembl. After washing the array slides, they were scanned using the Agilent Scanner G2505B. Agilent Feature Extraction software (version 10.7.3.1) was then used to analyze the acquired array images for 1) outlier analysis, 2) net signal statistics analysis, 3) local background statistical analysis, and 4) reproducibility analysis of the signals.
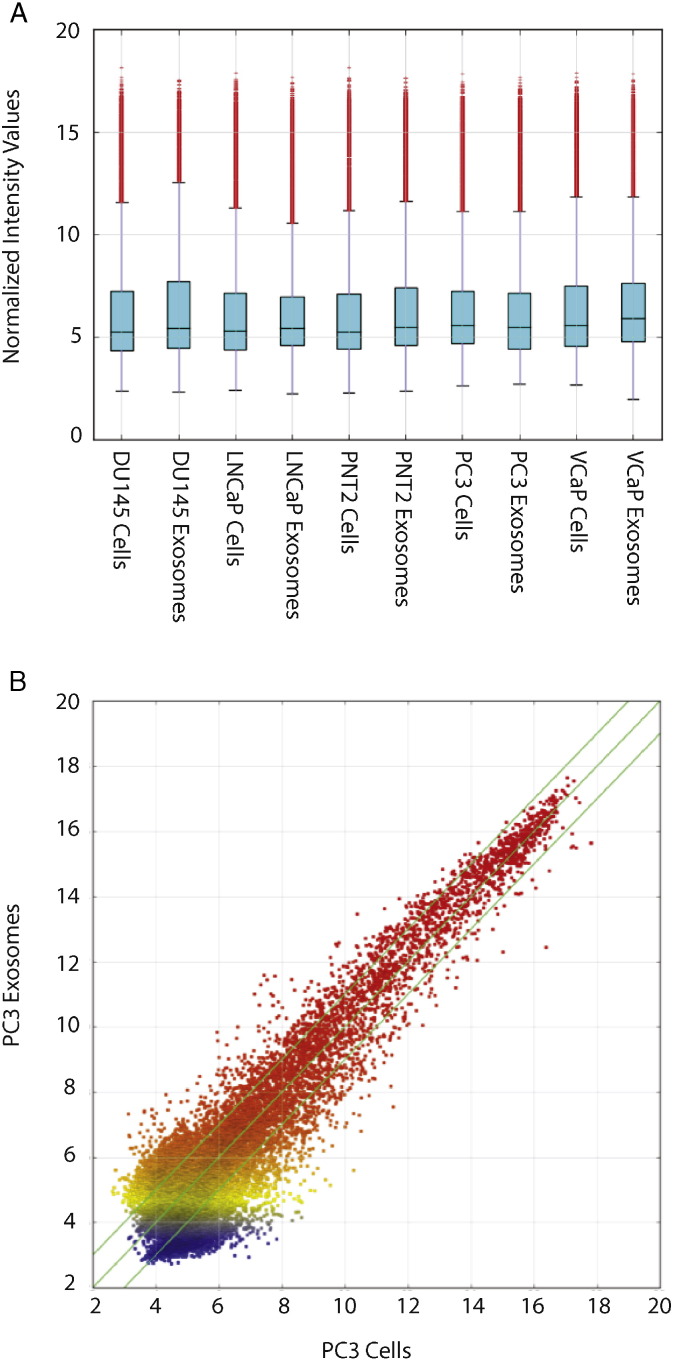
7. Data normalization and analysis
Quantile normalization was performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). After quantile normalization of the raw data, lncRNAs of which 6 out of 10 samples had flags in Present or Marginal (“All Targets Value”) were adjusted by Combat program to remove batch effects and were chosen for differentially expressed LncRNA screening. In the next step, quality assessment of lncRNA data for the different cell lines was performed by 1) investigating the box plot of the normalized intensity values to compare the distributions of intensities among different samples (Fig. 2A) and 2) inspecting the scatter plot of the log2 scaled values of the averaged normalized signal values of the exosomes versus the cells of each cell line to assess the lncRNA expression variations (Fig. 2B). To identify differentially expressed lncRNA, a fold change filtering with a threshold of fold change ≥ 2.0 was performed. Generation of the heatmaps and the hierarchical cluster analysis was performed using Agilent GeneSpring GX v11.5.1.
Fig. 2.
A) Box plot representing intensities distributions. B) A representative scatter plot of PC3 exosomes vs PC3 cell. The green lines indicate fold change lines whereby the default fold change value is 2.0.
8. Discussion
Our dataset represents a novel categorization of lncRNA levels in prostate cancer exosome and their parental lineages. We have profiled four common prostate cancer cell lines and compared these to the normal PNT2 cells. Our array data suggested that lncRNAs are present in abundance in both healthy and prostate cancer exosomes. We normalized the data using quantile normalization and then filter the data by using a two-fold threshold. From this the dataset, we also identified 26 down-regulated and 19 upregulated lncRNAs which were common to these exosomes. Furthermore, these exosomal lncRNAs appeared to be enriched for miRNA seeds with a preference for miR-17, miR-18a, miR-20a, miR-93, miR-106b and the let-7 family members [3]. With these observation, we put forward the notion that lncRNAs can act as miRNA sponges and their specific enrichment in exosomes is an important step in prostate cancer carcinogenesis. The heat maps clearly showed the similarities and differences in the lncRNA expression between cancer exosomes/cells versus the normal exosomes/cells (Fig. 3). Although there have been many studies using small ncRNAs for cancer diagnosis [1], with this dataset, it may be possible to discover potential exosomal lncRNAs which can be utilized for prostate cancer diagnosis.
Fig. 3.
A heatmap representing the differential lncRNA expression in the parental prostate cancer cell lineages and their exosomes. “red” indicates high relative expression and “blue” indicates low relative expression.
References
- 1.Khoury S., Tran N. Circulating microRNAs: potential biomarkers for common malignancies. Biomark. Med. 2015;9(2):131–151. doi: 10.2217/bmm.14.102. [DOI] [PubMed] [Google Scholar]
- 2.Walsh A.L., Tuzova A.V., Bolton E.M., Lynch T.H., Perry A.S. Long noncoding RNAs and prostate carcinogenesis: the missing 'linc'? Trends Mol. Med. 2014;20(8):428–436. doi: 10.1016/j.molmed.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Ahadi A., Brennan S., Kennedy P.J., Hutvagner G., Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci. Rep. 2016;6:24922. doi: 10.1038/srep24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoury S., Ajuyah P., Tran N. Isolation of small noncoding RNAs from human serum. J. Vis. Exp. 2014;88 doi: 10.3791/51443. [DOI] [PMC free article] [PubMed] [Google Scholar]