ABSTRACT
Hepatocyte-like cells (HLCs) are derived from human pluripotent stem cells (hPSCs) in vitro, but differentiation protocols commonly give rise to a heterogeneous mixture of cells. This variability confounds the evaluation of in vitro functional assays performed using HLCs. Increased differentiation efficiency and more accurate approximation of the in vivo hepatocyte gene expression profile would improve the utility of hPSCs. Towards this goal, we demonstrate the purification of a subpopulation of functional HLCs using the hepatocyte surface marker asialoglycoprotein receptor 1 (ASGR1). We analyzed the expression profile of ASGR1-positive cells by microarray, and tested their ability to perform mature hepatocyte functions (albumin and urea secretion, cytochrome activity). By these measures, ASGR1-positive HLCs are enriched for the gene expression profile and functional characteristics of primary hepatocytes compared with unsorted HLCs. We have demonstrated that ASGR1-positive sorting isolates a functional subpopulation of HLCs from among the heterogeneous cellular population produced by directed differentiation.
KEY WORDS: Hepatocytes, Human pluripotent stem cells, FACS, MACS, Transcriptomics
Highlighted article: Expression of cell surface protein ASGR1 can be used to isolate a subpopulation of more mature, homogenous hepatocyte-like cells derived during differentiation from pluripotent stem cells.
INTRODUCTION
The hepatocyte mediates many liver functions by carrying out a multitude of activities at the cellular level. Despite the regenerative capacity of the liver in vivo, primary human hepatocytes (PHHs) are not viable in vitro, and they are in limited supply. Although micropatterned co-culture can extend the viability of PHHs ex vivo (Khetani and Bhatia, 2008), there is a substantial need for a renewable source of human hepatocytes for in vitro studies and the development of cell-based therapies. Human pluripotent stem cells (hPSCs) are a promising source of these cells (Schwartz et al., 2014).
There are well-established methods for the directed differentiation of hepatocytes from hPSCs using defined media and feeder-free culture conditions (Mallanna and Duncan, 2013). These protocols can be used to produce hepatocytes from hPSCs, generating a cellular population at least 70% positive for the hepatocyte-specific marker albumin. These cells also express other hepatocyte-specific genes and perform many of the hallmark cellular functions of hepatocytes, such as cytochrome activity and apolipoprotein secretion. However, hPSC-derived hepatocytes are not equivalent to primary adult human hepatocytes and are more accurately considered ‘hepatocyte-like cells’ (HLCs). Unlike adult hepatocytes, HLCs typically retain expression of the fetal hepatocyte marker alpha fetoprotein (AFP) and fall considerably short of mature hepatocytes in terms of quantifiable functional capabilities, such as albumin secretion and drug detoxification.
Substantial obstacles must be overcome before advanced disease modeling studies can be attempted with HLCs. One notable hurdle is the variability and inefficiency of differentiation (Bock et al., 2011; Osafune et al., 2008; Takayama et al., 2014). Evidence suggests that this characteristic variability stems from inherent differences in hPSC lines (Kajiwara et al., 2012). This problem poses a challenge for in vitro modeling of subtle phenotypes, as well as phenotypes that could be confounded by incomplete or inaccurate differentiation. Here we describe the validation of a strategy for the prospective isolation of HLCs differentiated from a variety of hPSC lines based on the expression of a liver-specific cell surface protein, ASGR1. ASGR1 has long been recognized as a hepatic surface marker (Ashwell and Morell, 1974; Schwartz et al., 1981) and has been used to identify circulating hepatocellular carcinoma cells (Li et al., 2014), purify hPSC-derived HLCs (Basma et al., 2009) and to demonstrate the efficiency of HLC differentiation from hPSCs (Takayama et al., 2014). Whereas the utility of ASGR1 as a marker of hepatocyte identity is well established, the subpopulation of cells expressing ASGR1 in hPSC-derived HLCs has not been rigorously studied on the transcriptional level. To improve our understanding of the ASGR1-positive subpopulation of hPSC-derived HLCs and in the interest of developing a strategy for the purification of functional HLCs, we extensively characterized ASGR1-positive cells. ASGR1 marks a subset of albumin-positive HLCs, which are more similar than unpurified cells to mature hepatocytes. Furthermore, we show that ASGR1-enriched HLCs can be replated for further functional analysis, while retaining hepatocyte marker expression and cellular functions for up to 72 hours after sorting. These purification strategies increase the utility of hPSC-derived HLCs by enabling the isolation of a homogeneous population of hepatocytes for functional studies.
RESULTS AND DISCUSSION
Directed differentiation of HLCs
Depending on the hPSC line used and other experimental variables, differentiation generally results in a mixture of HLCs (the desired cell type) and a variable number of other cell types (Fig. 1A). The specific composition of mixed HLC differentiation cultures has not been investigated. Our laboratory has developed an optimized HLC-directed differentiation protocol based on established methods (Pagliuca et al., 2014; Si-Tayeb et al., 2010) with modest modifications.
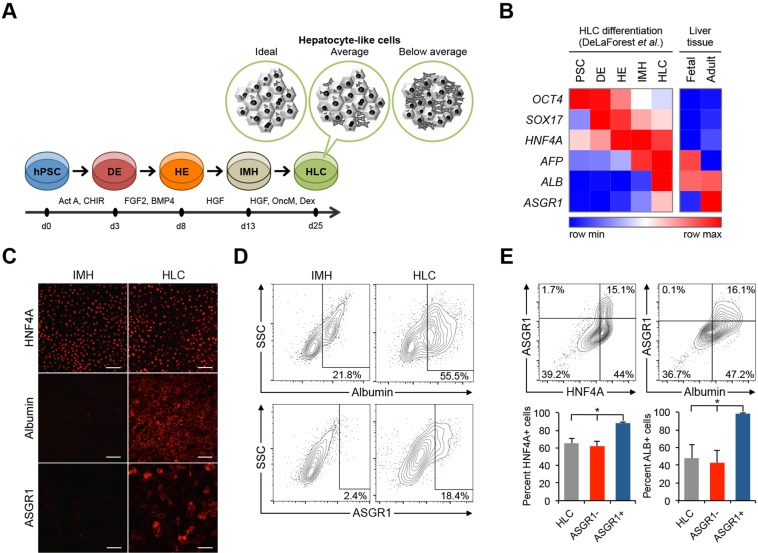
Fig. 1.
Directed differentiation of hPSCs to hepatocyte-like cells (HLCs). (A) Overview of optimized protocol for directed differentiation from hPSCs to HLCs. Non-hepatic cell types contaminate the cell culture in suboptimal differentiation conditions. (B) Heatmap showing gene expression level of representative markers during each stage of HLC differentiation in vitro and in normal liver tissue in vivo from published microarray expression data (DeLaForest et al., 2011; Su et al., 2004). Expression values are row normalized; red denotes higher than average expression and blue denotes lower than average expression for each gene. (C) Confocal microscopy images of immunocytochemical staining after the third (IMH) and fourth (HLC) differentiation stages. Scale bars: 100 μm. (D) Representative flow cytometry analyses of ALB and ASGR1 expression at the IMH and MH differentiation stages. Similar patterns were observed in multiple independent differentiations (Fig. S1D). (E) Representative flow cytometry analyses showing co-expression of ASGR1 with HNF4α or ALB. Results of two independent differentiations per analysis are shown in the accompanying bar charts. Error bars represent s.e.m. *P<0.05, Student's t-test.
We analyzed published gene expression data from human tissues as well as from HLC differentiation of hPSCs and found that ASGR1 is expressed in adult liver tissue, is not expressed or is present at an extremely low level in fetal liver, and is expressed most highly during HLC differentiation after the final differentiation stage – the ‘HLC’ stage (Fig. 1B). We confirmed this expression pattern during HLC differentiation by immunocytochemistry (Fig. 1C) and flow cytometry (Fig. 1D,E), which indicate that albumin (ALB) and ASGR1 are expressed at a very low level by a minority of cells at the end of the immature hepatocyte stage (IMH) and that they are both far more prevalent at the HLC stage of differentiation.
ASGR1 marks a subset of HLCs
Using our HLC differentiation protocol we found that ASGR1 is present in a small number of cells after the third differentiation stage (the IMH stage), and is more prevalent at the final stage of differentiation (Fig. 1D). This is in contrast to the expression pattern of the secreted protein ALB (a marker of functional hepatocytes), which is expressed at the IMH stage as well as the HLC stage (Fig. 1D).
The percentage of ASGR1-positive cells is almost always lower than that of ALB-positive cells in published reports as well as in our differentiations (Takayama et al., 2014). To confirm that ASGR1-positive cells present at the end of HLC differentiation are in fact a subset of HLCs, we performed intracellular flow cytometry by co-staining for ASGR1 and other markers. We found that ASGR1-positive cells occur within a subpopulation of differentiated cells that express the hepatocyte lineage marker hepatocyte nuclear factor 4 alpha (HNF4α), as well as ALB (Fig. 1E).
Enrichment of differentiated hepatocytes based on ASGR1 surface expression
We next investigated the utility of ASGR1 for prospective hepatocyte isolation. We differentiated multiple human embryonic stem cell (hESC) and human induced pluripotent stem cell (hiPSC) lines representing a range of HLC differentiation propensities and characterized the expression of hepatocyte markers among cells positive for surface ASGR1. Fluorescence-activated cell sorting (FACS) analysis following HLC differentiation of four hPSC lines showed that a large proportion of surface ASGR1-positive cells were also ALB positive, even when the overall differentiation efficiency was extremely low (2.97% ASGR1-positive cells, Fig. 2A, summarized in Fig. 2B). Similar results were obtained when the expression of alpha-1 antitrypsin (AAT; SERPINA1 – Human Gene Nomenclature Committee), an additional marker of functional hepatocytes, was assessed (Fig. S2B).
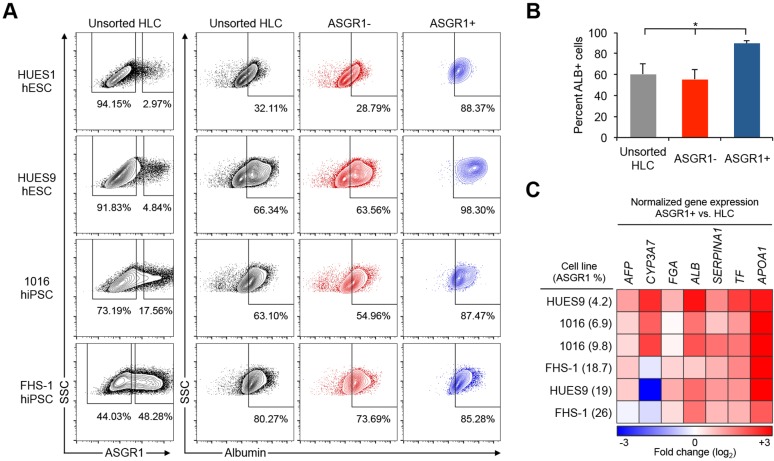
Fig. 2.
Enrichment of hepatocytes from HLC differentiation cultures by surface ASGR1 FACS. (A) Four different hPSC lines were differentiated to HLCs. The percentage of cells expressing the hepatocyte marker ALB among unsorted HLCs, surface ASGR1-negative cells, and surface ASGR1-positive cells was quantified by intracellular flow cytometry. (B) Summary of results in A showing the mean percentage of ALB-positive cells by flow cytometry among unsorted HLCs, surface ASGR1-negative cells and surface ASGR1-positive cells (n=4 differentiations). Error bars represent s.e.m. *P<0.05, Student's t-test. (C) Heatmap summarizing qRT-PCR results, showing relative expression levels in ASGR1-positive cells compared with matched unsorted HLCs.
Next, we analyzed the expression of several hepatocyte-specific genes in unsorted HLCs and FACS-isolated ASGR1-positive cells from multiple differentiations of three representative hPSC lines. The average differentiation efficiency in these experiments ranged from 4.2% to 26% as measured by the percentage of ASGR1-positive cells (Fig. 2C, Fig. S2C). Particular hPSC lines generally differentiated well (e.g. FHS-1 hiPSC), whereas others differentiated with a lower efficiency (e.g. 1016 hiPSC), as expected based on prior studies of HLC differentiation propensity (Takayama et al., 2014). Hepatocyte marker genes, representing a range of in vivo expression patterns, from fetal expression (AFP) to adult liver expression [apolipoprotein A-I (APOA1)], were analyzed by qPCR (Su et al., 2004). We found that expression of hepatocyte marker genes was significantly higher in ASGR1-positive cells than in matched unsorted HLCs. This result was particularly pronounced for inefficient differentiations.
Global transcriptional profiling of ASGR1-positive cells
We next sought to confirm the identity of ASGR1-positive cells by global gene expression profiling. We analyzed the transcriptional profiles of matched samples of ASGR1-positive cells and unsorted HLCs differentiated from an hESC line (HUES9) and an hiPSC line (1016), as well as PHHs and HepG2 hepatoma cells (Knowles et al., 1980). Based on unbiased hierarchical clustering of all expressed genes, the global expression profiles of ASGR1-positive cells were distinct from those of matched unsorted HLCs (Fig. 3A). There was greater correlation between the expression profiles of ASGR1-positive cells from different hPSC lines (r2=0.959) than there was between unsorted HLCs and ASGR1-positive cells from the same cell line (r2=0.944) (P<0.05, Fig. S3A,B).
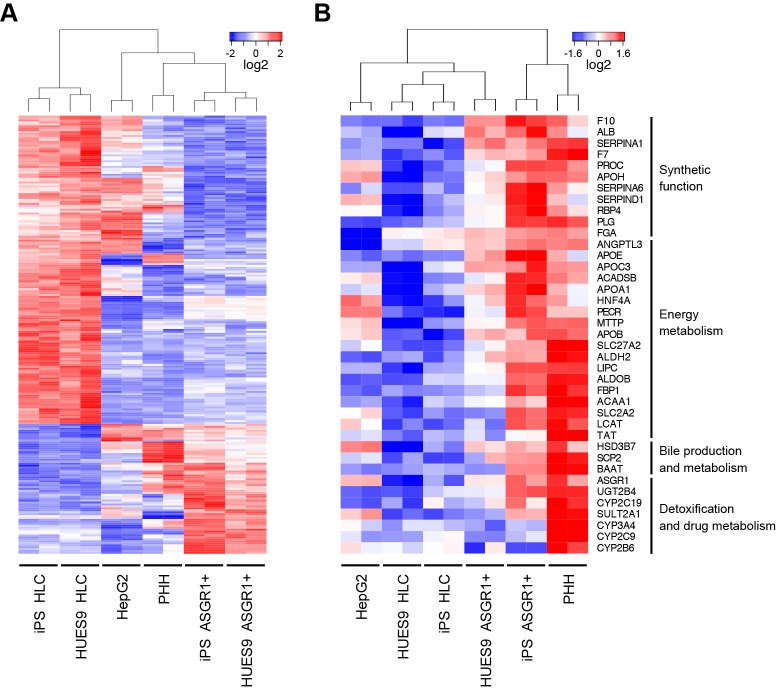
Fig. 3.
Global transcriptional analysis shows that ASGR1-positive cells share a distinct transcriptional profile and are more similar than unsorted HLCs to adult hepatocytes. (A) ASGR1-positive cells and HLCs differentiated from two different hPSC lines were compared with HepG2 hepatoma cells and adult primary human hepatocytes (PHHs) using microarrays. Shown is a heatmap of hierarchical clustering performed on genes differentially expressed more than 2-fold between ASGR1-positive cells and HLCs at a 5% FDR. 226 probe sets were differentially expressed greater than 2-fold between ASGR1-positive and unsorted HLCs at 5% FDR; 67 upregulated and 159 downregulated in ASGR1-positive versus unsorted HLCs. (B) Heatmap and hierarchical clustering of a panel of genes related to characteristic hepatic functions across the same samples as in A. Blue, below-average expression, red, above-average expression.
To further examine the effects of ASGR1 sorting, we performed differential expression analysis of the microarray data comparing ASGR1-positive cells with unsorted HLCs. We performed hierarchical clustering and heatmap visualization of all genes (probe sets) differentially expressed at a false discovery rate (FDR) of 5% (Fig. S3C) as well as of the most highly differentially expressed genes (>2-fold difference in expression between ASGR1-positive cells and HLCs, Fig. S2C). From this analysis we observed that ASGR1-positive cells cluster more closely to PHHs than to unsorted HLCs, whereas unsorted HLCs cluster more closely to HepG2 cells than PHHs.
A paired-sample design was used for differential expression analysis to characterize the gene expression profile of ASGR1-positive cells. This analysis identified genes differentially expressed in ASGR1-positive cells relative to matched HLCs differentiated from two different hPSC lines. 766 genes were differentially expressed more than 2-fold between ASGR1-positive cells and HLCs. Of these, 330 genes were significantly more highly expressed in ASGR1-positive cells versus unsorted HLCs (Fig. S3C). Functional enrichment analysis of the genes more highly expressed in ASGR1-positive cells was performed using the PANTHER classification system (Mi et al., 2013). Statistical overrepresentation analysis revealed overrepresentation of a number of hepatocyte-related gene ontology (GO) biological processes in ASGR1-positive cells versus unsorted HLCs (Fig. S3D). Many of the overrepresented processes related to key metabolic functions performed by the liver.
Next, we assembled a panel of hepatocyte genes representing important categories of hepatic function: synthetic function (including the production of coagulation factors), energy metabolism (including lipid and carbohydrate metabolism and lipoprotein processing), bile production and metabolism, and detoxification and drug metabolism (including metabolism of xenobiotics). The majority of these genes were expressed more highly in ASGR1-positive cells than in unsorted HLCs (Fig. 3B). Hierarchical clustering based on the expression of these hepatic functional genes suggested that ASGR1-positive cells are more similar than HLCs to PHHs (Fig. 3B). It should be noted that this trend is particularly pronounced for genes related to energy metabolism and systemic function in comparison with genes related to detoxification. Finally, we further verified that ASGR1-positive cells are more similar than unsorted HLCs to hepatocytes using gene set enrichment analysis (GSEA) (Subramanian et al., 2005). We found that 346 of 437 liver-enriched genes (79.2%) were more highly expressed in ASGR1-positive cells (Fig. S3E). We have detailed the subsets of liver-enriched genes that are both upregulated and downregulated in ASGR1-positive sorted HLCs versus unsorted HLCs in Table S1.
Replating ASGR1-enriched HLCs
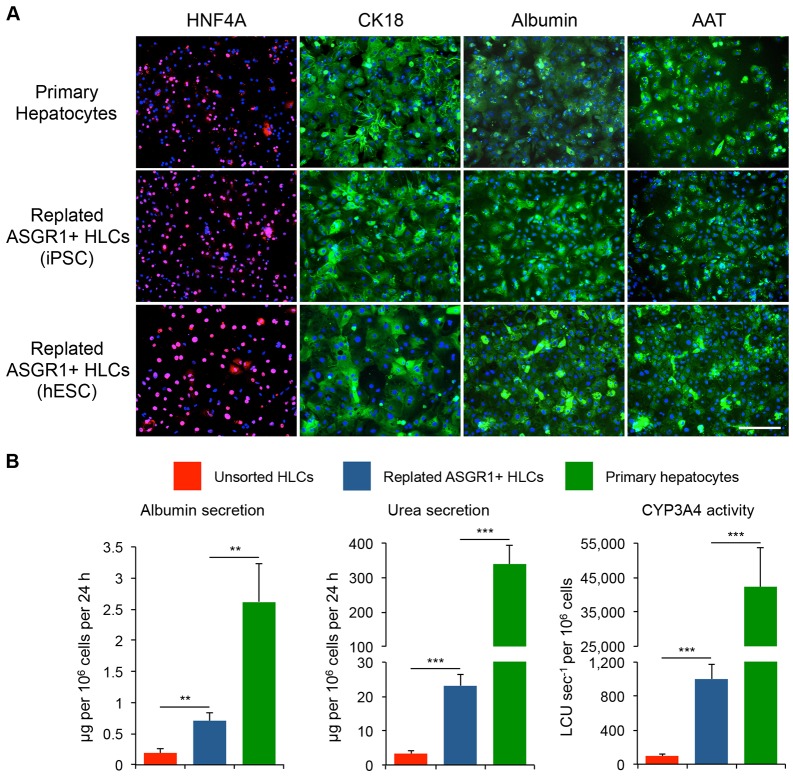
To facilitate functional studies of ASGR1-positive HLCs, we optimized replating HLCs following enrichment of ASGR1-positive cells by magnetic-activated cell sorting (MACS). ASGR1 MACS-enriched HLCs adhered to standard collagen-coated 24-well plates. We examined the expression of lineage and functional hepatocyte markers in replated HLCs differentiated from hESCs and hiPSCs and found that replated cells were positive for these markers as determined by immunofluorescence (Fig. 4A) and qPCR (Fig. S4A). This result was replicated with several hPSC lines. Finally, we assessed representative hepatocyte cellular functions in replated HLCs in comparison to unsorted HLCs and PHHs. ALB secretion, urea secretion and CYP3A4 activity (representing secretory and detoxification functions of the liver) were significantly increased in replated HLCs versus standard unsorted HLCs (Fig. 4B). As anticipated based on prior characterization of hPSC-derived HLCs, these activities were substantially higher in PHHs. As with the dedifferentiation of primary hepatocytes in cell culture (Miyazaki et al., 1981; Rowe et al., 2010), replated ASGR1-positive HLCs dedifferentiate after 96 h and at this point no longer express the marker genes AFP, FGA, AAT, ALB, TF or APOA1 (Fig. S4A) and secrete diminished quantities of ALB (Fig. S4B).
Fig. 4.
HLCs can be replated following ASGR1 MACS enrichment and retain hepatocyte characteristics. (A) Immunocytochemical staining for hepatocyte markers in PHHs and replated ASGR1 MACS-enriched HLCs. CK18, cytokeratin 18 (keratin 18, type I). Staining was performed 72 h after replating. Scale bar: 100 μm. (B) Representative assays of hepatocyte cellular functions. Assays were performed at the following time points: unsorted HLCs, day 28 of differentiation; replated HLCs, 72 h after replating on day 25 of differentiation; PHHs, 48 h after plating. n=4 per cell line for HLCs and n=6 per cell line for replated HLCs. Cryopreserved PHHs were thawed and plated (n=4). Error bars represent s.e.m. **P<0.01, ***P<0.001, Student's t-test.
Prospective isolation of hepatocytes from heterogeneous HLC cultures based on ASGR1 surface expression is a viable solution to the problem of variable and incomplete HLC differentiation. We have shown for the first time that ASGR1, an established liver-specific protein, is expressed by a subset of ALB-positive hepatocytes upon HLC differentiation. ASGR1 FACS can be used to isolate a population of cells that express hepatocyte functional markers (ALB and AAT) even when the overall differentiation efficiency is low. ASGR1-positive sorting addresses both the impurity of HLC differentiation cultures as well as inter-cell line variability in differentiation efficiency, both of which present limitations to performing in vitro genetic studies using HLCs. ASGR1 localization at the plasma membrane allows sorting without fixation, making this protocol ideal for the isolation of nucleic acid and protein from purified cells. MACS and replating of ASGR-positive HLCs eliminates the variability commonly encountered during functional studies of differentiated HLCs. This strategy ensures uniformity of cellular activities in comparison to unpurified cells. In conclusion, purification of ASGR1-positive cells could be generally applied to isolate HLCs differentiated from hPSCs.
MATERIALS AND METHODS
Cell culture
Two hESC lines (HUES1 and HUES9; Cowan et al., 2004) and two hiPSC lines (1016 and FHS-1) were used. HUES1 and HUES9 cells are part of the NIH hESC registry; 1016 and FHS-1 were derived by the Harvard University Induced Pluripotent Stem Cell Core Facility using retroviral and Sendai viral reprogramming, respectively. Cryopreserved PHHs (HMCPMS lot no. Hu8138) were cultured according to the manufacturer's instructions (Life Technologies). All cell lines were routinely tested for mycoplasma contamination. All ESC lines were maintained in accordance with ESCRO guidelines and iPSC lines were reprogrammed with full consent of donors.
Differentiation of hESCs and iPSCs into HLCs
Media
Basal differentiation medium (BDM): RPMI-1640 (Corning) plus B27 supplement plus penicillin/streptomycin (100 U/ml penicillin, 100 μg/ml streptomycin, final concentration) (Thermo Scientific). Definitive endoderm (DE) medium: BDM with 100 ng/ml activin A and 3 μM CHIR99021. Hepatic endoderm (HE) medium: BDM with 5 ng/ml basic fibroblast growth factor (bFGF, also known as FGF2), 20 ng/ml bone morphogenetic protein 4 (BMP4) and 0.5% DMSO. IMH medium: BDM with 20 ng/ml hepatocyte growth factor (HGF) and 0.5% DMSO. Mature hepatocyte (MH) medium: hepatocyte basal medium (HBM) (Lonza) with SingleQuots added (Lonza), as well as 20 ng/ml HGF, 20 ng/ml oncostatin M, 100 nM dexamethasone and 0.5% DMSO.
Plating and differentiation
Day 1: cultures of ESCs/iPSCs were split and plated at a density of 3×104 cells/cm2 in mTESR (Stem Cell Technologies) with 4 μM ROCK inhibitor Y27632. Plating density may have to be optimized for each cell line. Days 2-4: cells were treated with DE medium. Days 5-9: cells were treated with HE medium for 5 days. Days 10-14: cells were treated with IMH for 5 days. Days 15-27: cells were treated with MH medium for 10-12 days. The medium was changed daily throughout differentiation. All cell sorting and staining experiments for the HLC stage were performed at day 25 of differentiation, whereas experiments studying the IMH stage were performed at day 14 of differentiation.
Immunocytochemistry
The following primary and secondary antibodies were used for immunocytochemical staining: AAT (MA1-90438, Thermo Scientific), ALB (A80-129A, Bethyl), ASGR1 (clone 8D7, BD Biosciences), CK18 (ab82254, Abcam) and HNF4α (ab92378, Abcam) primary antibodies at 1:250, and donkey anti-goat IgG Alexa Fluor 555 (A-21432, Thermo Scientific), donkey anti-mouse IgG Alexa Fluor 488 (A-21202, Thermo Scientific) and donkey anti-rabbit IgG Alexa Fluor 488 (A-21206, Thermo Scientific) secondary antibodies (all Life Technologies) at 1:1000. Hoechst (1:5000; Life Technologies) was used for nuclear staining. Staining in unsorted HLCs was visualized using an LSM 700 confocal microscope (Carl Zeiss) and an inverted Eclipse Ti microscope (Nikon).
Intracellular flow cytometry
For flow cytometry analysis, differentiated cells were fixed and stained using the Cytofix/Cytoperm Kit (BD) following the manufacturer's instructions. The primary and secondary antibodies described above were used at experimentally optimized dilutions. Additional antibodies used in flow cytometry experiments were mouse IgG1 isotype control (554121, BD; 1:20), donkey anti-mouse IgG Alexa Fluor 594 (A-21203, Thermo Scientific; 1:500), donkey anti-goat IgG Alexa Fluor 488 (ab150129, Abcam; 1:500) and donkey anti-goat IgG Alexa Fluor 647 (A-21447, Thermo Scientific; 1:500). Cells were analyzed using an LSR II cytometer (BD) and FlowJo software.
FACS of HLCs
After day 12 of feeding with MH medium, HLCs were washed with Dulbecco's phosphate-buffered saline (DPBS) and treated with 0.25% trypsin-EDTA. Cells were treated with a Stempro EZ Passage passaging tool (Life Technologies) and incubated at 37°C for 15 min. After dissociation, remaining cells were gently scraped from the dish and filtered through a 100 µm mesh. Cells were stained with a PE-conjugated ASGR1 antibody (8D7, BD, 563655) or a PE-conjugated mouse IgG1κ isotype control antibody (BD, 551436) according to manufacturer's dilution instructions. After incubation, cells were washed twice with PBS. Alternatively, in some experiments cells were stained with unconjugated ASGR1 primary antibody or mouse IgG1 isotype control, washed, and stained with donkey anti-mouse IgG Alexa Fluor 488 secondary antibody (as above). ASGR1-positive cells were purified by FACS using a FACSAria II (BD).
RNA isolation and qRT-PCR
RNA was isolated using Trizol (Life Technologies). qPCR was performed using TaqMan Gene Expression Master Mix and TaqMan gene expression assays (Life Technologies) for the following: RPLP0 (reference control gene), AFP, CYP3A7, FGA, ALB, AAT, TF and APOA1.
Microarray gene expression profiling
U219 gene expression arrays (Affymetrix) were used according to the manufacturer's instructions. These data have been uploaded to Gene Expression Omnibus (GEO) under accession number GSE77086.
Preprocessing and hierarchical clustering
Microarrays were normalized and background corrected using the robust multi-array (RMA) method in the Bioconductor affy package in R v3.1.2 (Gautier et al., 2004). Normalized array values were reported on a log2 scale and probe sets with low expression values across all samples (log2 intensity <2.5) were filtered out. Unsupervised hierarchical clustering was performed using the hclust function in R. The statistical significance of clustering results was estimated by multiscale bootstrap resampling using the pvclust function with 10,000 iterations.
Standard differential expression analysis and heatmap generation
Analysis was performed using Transcriptome Analysis Console software (Affymetrix). FDR-adjusted P-values were calculated based on the Benjamini-Hochberg method and 5% FDR cutoff was used. Heatmaps were created using the heatmap.2 function in R, with accompanying dendrograms drawn based on Euclidean distance.
Paired-sample differential expression analysis, functional enrichment testing of differentially expressed genes and GSEA
The R/Bioconductor limma package was used to fit linear models for each gene, utilizing a paired-sample design. Moderated t-statistics, log fold change and P-values were calculated. To correct for multiple hypothesis testing, FDR-adjusted P-values were calculated by the Benjamini-Hochberg method. Probe sets were filtered by FDR<5% and fold change >2 (1079 probe sets). Functional enrichment analysis was performed using the PANTHER resource. GSEA (Subramanian et al., 2005) was performed using GSEA software (Broad Institute, Cambridge, MA, USA) and a list of liver-enriched genes (Yang et al., 2011).
Analysis of published gene expression data
Published microarray gene expression data were obtained from BioGPS.org and the GEO database: accession numbers GSE25417 (DeLaForest et al., 2011) and GSE1133 (Su et al., 2004). Analysis and heatmap generation were conducted using GENE-E software (Broad Institute).
ASGR1 MACS and replating of HLCs
Cells were released as described for flow cytometry analysis. After blocking, anti-ASGR1 antibody (8D7, BD; 1:150) was added. After incubation on ice for 30 min, cells were pelleted (5 min, 233 g) and resuspended in cold MACS buffer with anti-mouse IgG microbeads (1:20; Miltenyi Biotec). After incubation on ice for 30 min, cells were washed and then passed through a 100 µM filter. Cells were then sorted using an autoMACS Pro separator (Miltenyi Biotec). After sorting, cells were pelleted and resuspended in HLC replating medium: hepatocyte basal medium with SingleQuots supplements, HGF (20 ng/ml), dexamethasone (100 nM), ROCK inhibitor (4 µM), penicillin/streptomycin and gentamicin (50 µg/ml). Cells were plated on collagen-coated 24-well culture dishes (A1142802, Thermo Scientific) by centrifugation of the plate at 100 RCF for 15 min at 37°C. Cells were replated at 15,000 cells per cm2 and thereafter maintained in HLC replating medium without ROCK inhibitor. Cells were used in functional assays no later than 72 h after replating, owing to cell dedifferentiation during prolonged culture.
Cellular assays of hepatocyte functions
Cellular assay results were normalized to the number of live cells as determined by hemocytometer cell counting with Trypan Blue staining. ALB secretion was quantified using an enzyme-linked immunosorbent assay (Bethyl Laboratories, E80-129) according to the manufacturer's instructions. Urea production was quantified using the QuantiChrom Urea Assay Kit (BioAssay Systems) following the manufacturer's instructions. Cytochrome activity was quantified using the P450-Glo CYP3A4 assay (Luc-PFBE) (Promega) following the manufacturer's instructions.
Statistical analysis
Statistical significance was assessed using standard Student's t-test (two-tail); P<0.05 was considered statistically significant. Experiments were performed in triplicate unless otherwise noted.
Acknowledgements
We acknowledge Jennifer Shay for management of the C.A.C. laboratory. We thank the staff of the HSCRB-HSCI flow cytometry core for assistance with cell sorting and analysis.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
D.T.P. and C.R.W. designed and performed experiments and wrote the manuscript. C.A.H. and M.F. designed and performed experiments. F.X. performed experiments. C.E.B. reprogrammed iPSC lines. K.M. supported D.T.P. C.A.C. designed experiments and guided the study.
Funding
This project was supported by National Institutes of Health grants from the National Heart, Lung, and Blood Institute (NHLBI) [U01HL100408, U01HL107440 and R21HL120781] and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) [R01DK097768] to C.A.C. C.A.C. and K.M. were supported by the Harvard Stem Cell Institute and Harvard University. Deposited in PMC for release after 12 months.
Data availability
Microarray data are available at Gene Expression Omnibus under accession number GSE77086.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.132209/-/DC1
References
- Ashwell G. and Morell A. G. (1974). The role of surface carbohydrates in the hepatic recognition and transport of circulating glycoproteins. Adv. Enzymol. Relat. Areas Mol. Biol. 41, 99-128. 10.1002/9780470122860.ch3 [DOI] [PubMed] [Google Scholar]
- Basma H., Soto-Gutiérrez A., Yannam G. R., Liu L., Ito R., Yamamoto T., Ellis E., Carson S. D., Sato S., Chen Y. et al. (2009). Differentiation and transplantation of human embryonic stem cell–derived hepatocytes. Gastroenterology 136, 990-999.e4. 10.1053/j.gastro.2008.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z. D., Ziller M., Croft G. F., Amoroso M. W., Oakley D. H. et al. (2011). Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439-452. 10.1016/j.cell.2010.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C. A., Klimanskaya I., McMahon J., Atienza J., Witmyer J., Zucker J. P., Wang S., Morton C. C., McMahon A. P., Powers D. et al. (2004). Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 350, 1353-1356. [DOI] [PubMed] [Google Scholar]
- DeLaForest A., Nagaoka M., Si-Tayeb K., Noto F. K., Konopka G., Battle M. A. and Duncan S. A. (2011). HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development 138, 4143-4153. 10.1242/dev.062547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B. M. and Irizarry R. A. (2004). affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307-315. 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- Kajiwara M., Aoi T., Okita K., Takahashi R., Inoue H., Takayama N., Endo H., Eto K., Toguchida J., Uemoto S. et al. (2012). Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 109, 12538-12543. 10.1073/pnas.1209979109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani S. R. and Bhatia S. N. (2008). Microscale culture of human liver cells for drug development. Nat. Biotechnol. 26, 120-126. 10.1038/nbt1361 [DOI] [PubMed] [Google Scholar]
- Knowles B. B., Howe C. C. and Aden D. P. (1980). Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 209, 497-499. 10.1126/science.6248960 [DOI] [PubMed] [Google Scholar]
- Li J., Chen L., Zhang X., Zhang Y., Liu H., Sun B., Zhao L., Ge N., Qian H., Yang Y. et al. (2014). Detection of circulating tumor cells in hepatocellular carcinoma using antibodies against asialoglycoprotein receptor, carbamoyl phosphate synthetase 1 and pan-cytokeratin. PLoS ONE 9, e96185 10.1371/journal.pone.0096185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna S. K. and Duncan S. A. (2013). Differentiation of hepatocytes from pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 26, 1G.4.1-1G.4.13. 10.1002/9780470151808.sc01g04s26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A. and Thomas P. D. (2013). PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377-D386. 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., Takaki R., Nakayama F., Yamauchi S., Koga A. and Todo S. (1981). Isolation and primary culture of adult human hepatocytes: ultrastructural and functional studies. Cell Tissue Res. 218, 13-21. 10.1007/BF00210087 [DOI] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R. J., Fitz-Gerald C. S., Sato Y., Cowan C. A., Chien K. R. and Melton D. A. (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26, 313-315. 10.1038/nbt1383 [DOI] [PubMed] [Google Scholar]
- Pagliuca F. W., Millman J. R., Gürtler M., Segel M., Van Dervort A., Ryu J. H., Peterson Q. P., Greiner D. and Melton D. A. (2014). Generation of functional human pancreatic beta cells in vitro. Cell 159, 428-439. 10.1016/j.cell.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe C., Goldring C. E. P., Kitteringham N. R., Jenkins R. E., Lane B. S., Sanderson C., Elliott V., Platt V., Metcalfe P. and Park B. K. (2010). Network analysis of primary hepatocyte dedifferentiation using a shotgun proteomics approach. J. Proteome Res. 9, 2658-2668. 10.1021/pr1001687 [DOI] [PubMed] [Google Scholar]
- Schwartz A. L., Marshak-Rothstein A., Rup D. and Lodish H. F. (1981). Identification and quantification of the rat hepatocyte asialoglycoprotein receptor. Proc. Natl. Acad. Sci. USA 78, 3348-3352. 10.1073/pnas.78.6.3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. E., Fleming H. E., Khetani S. R. and Bhatia S. N. (2014). Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol. Adv. 32, 504-513. 10.1016/j.biotechadv.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto F. K., Nagaoka M., Li J., Battle M. A., Duris C., North P. E., Dalton S. and Duncan S. A. (2010). Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51, 297-305. 10.1002/hep.23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G. et al. (2004). A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101, 6062-6067. 10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Morisaki Y., Kuno S., Nagamoto Y., Harada K., Furukawa N., Ohtaka M., Nishimura K., Imagawa K., Sakurai F. et al. (2014). Prediction of interindividual differences in hepatic functions and drug sensitivity by using human iPS-derived hepatocytes. Proc. Natl. Acad. Sci. USA 111, 16772-16777. 10.1073/pnas.1413481111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Ye Y., Wang G., Huang H., Yu D. and Liang S. (2011). VeryGene: linking tissue-specific genes to diseases, drugs, and beyond for knowledge discovery. Physiol. Genomics 43, 457-460. 10.1152/physiolgenomics.00178.2010 [DOI] [PubMed] [Google Scholar]