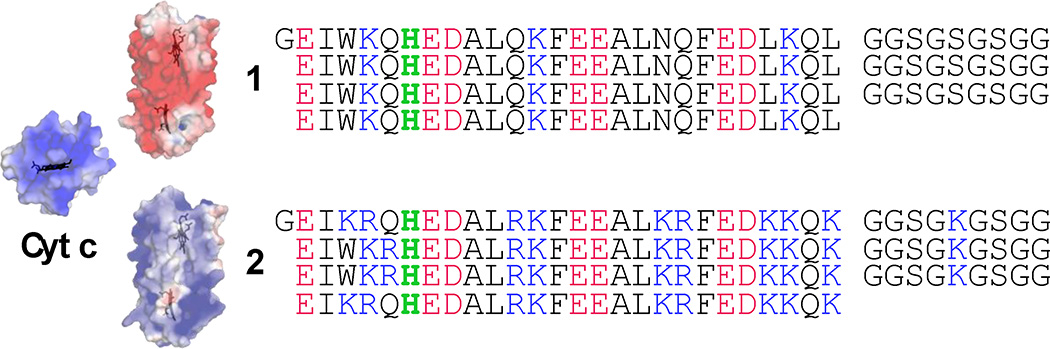Abstract
Maquettes are man-made cofactor-binding oxidoreductases designed from first principles with minimal reference to natural protein sequences. Here we focus on water-soluble maquettes designed and engineered to perform diffusive electron transport of the kind typically carried out by cytochromes, ferredoxins and flavodoxins and other small proteins in photosynthetic and respiratory energy conversion and oxido-reductive metabolism. Our designs were tested by analysis of electron transfer between heme maquettes and the well-known natural electron transporter, cytochrome c. Electron-transfer kinetics were measured from seconds to milliseconds by stopped-flow, while sub-millisecond resolution was achieved through laser photolysis of the carbon monoxide maquette heme complex. These measurements demonstrate electron transfer from the maquette to cytochrome c, reproducing the timescales and charge complementarity modulation observed in natural systems. The ionic strength dependence of inter-protein electron transfer from 9.7 × 106 M−1s−1 to 1.2 × 109 M−1s−1 follows a simple Debye-Hückel model for attraction between +8 net charged oxidized cytochrome c and −19 net charged heme maquette, with no indication of significant protein dipole moment steering. Successfully recreating essential components of energy conversion and downstream metabolism in man-made proteins holds promise for in vivo clinical intervention and for the production of fuel or other industrial products.
Keywords: Man-made redox proteins, Maquettes, Protein design, Cytochrome c, Inter-protein electron transfer, Electron-transfer kinetics, Charge complementarity, Electrostatics, Diffusion
1. Introduction
Small, diffusive water-soluble proteins, accommodating one or a few oxidation-reduction cofactors, connect larger and less mobile protein complexes in the electronic circuits of natural respiratory and photosynthetic energy conversion metabolism. Decades of research describes the design elements that confer recognition and binding specificity between the transporter and the electron-donor and acceptor-protein partners [1–4]. Short and long-range interactions between reacting protein pairs promote the formation of dynamic encounter complexes, bringing the electron-exchanging cofactors close enough, and for sufficient time, to support electron tunneling between the proteins [5]. Cytochrome c (cyt c), was the first such electron-transporter protein to be described [6] in a family that has since grown to encompass other cytochromes, flavodoxins, ferredoxins and metal-binding proteins. Despite the differences in cofactors, operating redox potentials, cellular compartments and protein partners, it seems clear that these diffusive transporters share the same broad operational characteristics and engineering tolerances that combine to support the millisecond rates typical of energy conversion and catalysis [7].
Table 1 summarizes key functional characteristics for a range of inter-protein electron transfers between diffusing c cytochromes and natural partners cyt c oxidase, cyt c peroxidase [8], cyt bc1 and photosynthetic P870 [9,10]. Driving forces for the interfacial electron-tunneling step are often modestly favorable, but near zero or energetically uphill electron transfer is possible if followed by a sufficiently downhill intra-protein step [11]. Reported cofactor separations in the electron-transfer complexes range from 4 to 14 Å, comparable to the range of intra-protein electron-tunneling distances between cofactors within the same protein [5]. Inter-protein electron-transfer turnover rates from ~100 to 1,000 s−1 are typically orders of magnitude slower than the intrinsic electron tunneling of the complex, suggesting that diffusion between redox partners or complex formation can be a limiting step.
Table 1.
Key engineering parameters for several well-characterized inter-protein c-type cytochrome electron-transfer complexes [10,12–15]. Minimum distance between cofactor edges are estimated from published structures [13,15]. Log of electron-tunneling rates estimated from empirical expression [5], assuming protein reorganization energy of 1 eV [34].
| Redox Pair | D (eV) |
Min. Cofactor Distance (Å) |
Kd (µM) | log kET (s−1) |
|---|---|---|---|---|
| cyt c → CcO | −0.07 (spheroides) |
11 (thermophilus) |
0.35 | 5.9 |
| cyt c → CcP | ~ −1.0 | 14 | 20 | 6.8 |
| cyt bc1 → cyt c | ~ 0 | 9 | 9.5 | 6.7 |
| cyt c2 → P870 | −0.16 | 8 | 1 | 8.2 |
| Isocyt c2 → P870 | −0.22 | 8 | 40 | 8.5 |
Maquettes are man-made proteins designed from first principles, sidestepping evolutionary complexity in sequence and structure but highly adaptable in supporting a range of oxidoreductase functions. These elementary α-helical proteins bind a range of redox-active cofactors, including b-type hemes [16] and many other natural and unnatural tetrapyrroles [17,18], FeS clusters [19,20], flavins [21], quinones [22] and a range of light activatable cofactors [17]. With modest sequence alterations, maquettes replicate a diverse range of natural oxidoreductase functions including the promotion of ultrafast intra-protein electron transfer in photo-activated charge separation [21], and the suppression of electron transfer into the minute time scale to promote stable heme dioxygen binding [23].
Inter-protein electron transfer offers a new functional frontier for designed proteins. So far, preliminary experiments have shown electron transfer from a heme maquette to natural bovine cyt c on a millisecond time scale (Figure 2) [17] and from ferredoxin-NADPH reductase to a heme a maquette [24]. In recent work, Ghirlanda and coworkers describe a related α-helix protein ligating 4Fe4S clusters that reduces cyt c [25]. Complimentary pairs of designed redox proteins also demonstrate inter-protein electron transfer [26]. Here, we deepen the enquiry to define the engineering principles governing critical electrostatic interactions between redox proteins, bringing cofactors within electron-transfer distance. We have improved the temporal resolution from the millisecond regime afforded by stopped-flow method [17] to the microsecond time domain using classic ferrous heme-CO flash photolysis [27]. This allows us to resolve rapid electron transfers from the maquette at rates approaching the diffusion limit. A series of time-resolved measurements of electron transfer from ferrous heme maquettes with contrasting surface charge patterns, to ferric bovine cyt c at various ionic strengths, reveal a process broadly similar to electron-transfer interactions between physiological redox partners.
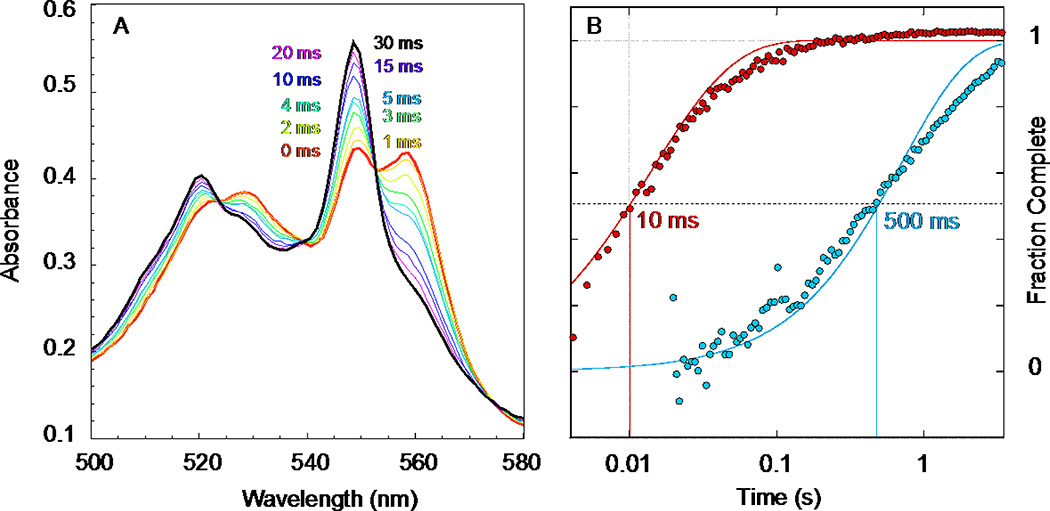
Figure 2.
A. Stopped-flow rapid mixing of 4.75 µM reduced maquette 1 with 8.5 µM oxidized cyt c showing the reduced heme alpha band moving from the maquette b heme at 560 to cyt c at 550 nm in 120 mM KCl, 50 mM HEPES pH 8, as described in [17] figure 4. B. A log time view of the cyt c reduction kinetics at 549 nm shows the negatively charged maquette 1 (red) is about 50 times faster than the positively charged maquette 2 (blue). Solid lines are theoretical time courses, second order in protein concentration, with rate constants 1.9 and 0.036 × 107 M−1s−1. These fits were computed as described in section 2.4.
Successful recreation of basic component parts of respiratory and photosynthetic energy conversion in maquettes confirms a practical understanding of the underlying engineering and construction of their natural counterparts. It also holds promise for a range of practical applications, ultimately in vivo. Examples include the interception and diversion of high or low potential electrons in living cells directly towards production of useful chemicals and fuels (analogous to synthetic biology’s goal of redirecting metabolic pathways), and amelioration of genetic or age-related failures in respiratory energy conversion in humans [28].
2. Materials and Methods
2.1 Maquette preparation
Reagent grade chemicals from Sigma-Aldrich were used, unless otherwise noted. Protein maquettes were expressed and purified as described in [17]. Heme cofactors were incorporated from DMSO stock solutions with concentrations quantified using the hemochrome method [29]. Maquette proteins were dissolved in CHES buffer (20 mM CHES, 100 mM NaCl, pH 9.0) and protein concentrations were measured via tryptophan absorbance at 280 nm (ε = 5600 M−1cm−1). 2.5 equivalents of heme stock solution were added and allowed to incubate at room temperature for 5 minutes. A PD-10 gravity column (GE Healthcare) was equilibrated with pH 7.9 phosphate buffer for subsequent electron-transfer experiments. The heme-bound protein was passed through the column to exchange buffer and remove unbound heme. Finally, the protein was diluted to the desired experimental concentration by following the 412nm oxidized heme Soret band of the bound cofactor (ε = 117000 M−1cm−1).
The redox difference spectra of heme bound to both maquette designs considered here can be compared with the redox difference spectrum of an equivalent amount of free heme (Supplementary figure 1). In both maquettes, absorption around 560 nm is dominated by a bis-His coordinated, cytochrome b-like alpha band, with a minor contribution (5 to 10%) of a single His coordinated, myoglobin-like absorption band. In the presence of either maquette, the broad free-heme spectrum contribution is less than 10% in this wavelength region, indicating that almost all the heme present is bound at the designed histidine sites.
2.2 Oxidation of cytochrome c
Bovine heart cytochrome c was dissolved in phosphate buffer (50 mM Na2HPO4, NaCl 110 mM, pH 7.9). 3 stoichiometric equivalents of K3Fe(CN)6 were added to guarantee that all cyt c was in the oxidized state prior to electron-transfer experiments. The oxidized protein solution was then passed through a PD-10 gravity column (GE Healthcare) to remove residual K3Fe(CN)6. Cyt c solutions were diluted to the desired concentration experimental concentration by following the 409 nm Soret band (ε = 107000 M−1cm−1).
2.3 Degassing and CO binding
Prior to electron-transfer experiments, several mL of heme-bound maquette solution and cyt c solution were placed in septum-sealed quartz cuvettes with stir bars. The maquette was placed under either an argon or carbon monoxide (CO) gas flow, while cyt c was placed under argon. After 30 minutes, an oxygen scrubbing mixture of 80 nM glucose oxidase, 12 nM catalase, and 1 mM glucose was added to the maquette solution. The maquette protein was reduced by careful titration of sodium dithionite as monitored at 318nm to minimize excess unreacted dithionite. For CO bound samples, dithionite was added until full CO binding was observed spectroscopically, with a dithionite excess of only a few µM. The maquette was then placed under argon flow for 5 min with stirring to remove excess CO. Cyt c and CO bound or CO free maquette were transferred under positive argon pressure to 10 mL gastight syringes (SGE Analytical) equipped with stainless steel Luer valves (Cadence Science) used as load syringes in the stopped-flow apparatus.
2.4 Electron-transfer measurements without CO photolysis
Electron-transfer measurements were run on an RSM-1000 UV-Vis stopped-flow spectrophotometer (OLIS, Inc.). For experiments without CO photolysis, the spectrophotometer was equipped with a rapid-scan monochromator sampling the Q-band spectral region at 1000 spectra per second. Roughly 3900 scans (3.9 seconds) were acquired after mixing the maquette and cytochrome c reactants. All experiments were performed at 20 C.
Cyt c reduction was monitored at 549 nm, corresponding to the reduced α band of cyt c. Note that a maquette binds two hemes, each potentially available to transfer an electron to cyt c. Static measurements of the reduced and oxidized forms of the maquette and cyt c provide an extinction coefficient change of 19040 M−1cm−1 per electron transferred. Although any anaerobic compromise during handling in the stop-flow may lessen the concentration of reduced maquette (but not oxidized cyt c) prior to mixing, the amount of reduced maquette oxidized by excess cyt c can be monitored using this redox extinction coefficient for cyt c at 549 nm. Reaction profiles were fit to second-order reaction trajectories; for comparison in Figure 2B, the trajectories were normalized to fraction completed to 0 at early time and 1 at late time.
2.5 Electron-transfer measurements with CO photolysis
All experiments were performed at 20 C. A 10 Hz Q-switched frequency-doubled Nd:YAG laser (532 nm, DCR-11 Spectra Physics) was focused with a cylindrical lens onto the 20 mm × 2 mm stopped-flow cell of an Olis RSM-1000 spectrophotometer. A mechanical shutter (Thor Labs) triggered by the stopped-flow TTL output selected a laser single pulse. A 600 µM fixed slit sampled an individual wavelength at 1 µs time resolution for 1 s, with the wavelength serially adjusted over the whole Soret region. Scattered laser flash artifact was minimized with a 450 nm short-pass filter on the sample photomultiplier tube (PMT). The reference PMT voltage was acquired using the standard OLIS data acquisition, while the reference PMT voltage was captured with a Tektronix MSO 2014 oscilloscope. Finally, the voltage response from 10 laser flashes was averaged, and the voltage response of the sample PMT was corrected by subtracting this flash artifact profile. A Python post-processing script computed absorbance changes from the raw data. Accuracy of the kinetic analysis method was confirmed by reproducing published results of azobenzene isomerization kinetics standards [30].
2.6 Kinetics computations for photolysis at high salt: 110 mM NaCl / 50 mM Na2HPO4
CO photolysis yield was significantly less than 50% in our pump-probe geometry. The effective concentration of CO free reduced maquette heme competent for reduction of cytochrome c was quantified by the laser induced absorbance changes at 420 nm. As seen in Figure 4, this wavelength is isosbestic for electron transfer but shows a large change for photolysis (εphotolysis = −44000 M−1cm−1 per heme). The initial rate for the electron-transfer process was monitored by the essentially linear post-flash absorbance change at 424 nm from 1 to 5 ms. This wavelength is isosbestic for CO recombination and has an extinction change of εET = −90000 M−1cm−1 (per electron) for electron transfer; the isosbestic point and extinction coefficient were determined from static spectra of the maqette and of cyt c. To avoid possible spectral contamination by sub-ms histidine recombination, the first 3 post-photolysis ms of this kinetics was not included in the initial rate estimate for electron transfer.
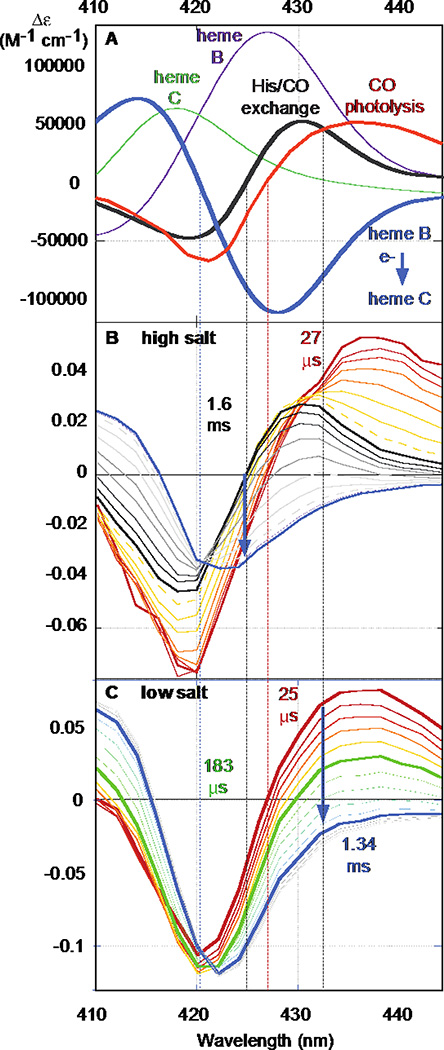
Figure 4.
A) Reaction difference spectra: Immediate CO photolysis (red); His-CO ligand exchange (black); heme b to c electron transfer (blue) computed from heme b redox spectrum (purple) and heme c redox spectrum (green). Isosbestic wavelengths: electron transfer (420 nm); His-CO ligand exchange (424 nm); ns CO photolysis (427 nm); His rebinding (432 nm). Transient absorption changes of photolysis and electron transfer at 110 mM NaCl, 50 mM Na2HPO4 pH 7.9 (B) and 11 mM NaCl, 5 mM Na2HPO4 pH 7.9 (C).
2.7 Kinetics computations for photolysis at lower salt concentrations
At lower salt concentrations, post-photolysis ligand exchange was comparable to electron-transfer rates and both were much faster than CO recombination. Thus the ligand exchange isosbestic at 432 nm was chosen to measure the rate of electron-transfer kinetics. At this wavelength, differences in static spectra for known concentrations of oxidized, reduced, and CO-bound maquette and cyt c indicate extinction changes of εphotolysis = 45000 M−1cm−1 per heme and εET = −102000 M−1cm−1 per electron. The initial electron transfer rate was computed by measuring the slope of the essentially linear region following the laser flash; this region ranged from 30 µS – 70 µS to 30 µS – 300 µS for the fastest and slowest reactions. The photolysis yield was computed by linearly extrapolating the absorbance to the time of the laser pulse.
2.8 Protein Surface Charge Estimates, Visualization, and theoretical ET rate calculations
Visualizations use Pymol 1.7.5. [31] Maquette structure figures are estimates generated from a 200 ps molecular dynamics trajectory in GROMACS [32] starting from four parallel helices connected by loops optimized in Modeller [33]. Surface charge figures use the APBS plug-in for Pymol [34]. Theoretical interprotein ET rate surfaces use a tunneling rate expression [35] embedded in a Pymol-integrated Python script.
3. Results
3.1 Charge complementarity dependent inter-protein electron transfer
Single chain heme-binding maquettes are highly tolerant of extensive changes in surface residues while maintaining interior heme site properties [17]. Figure 1 shows the sequences of two examples. The first sequence (1 in Figure 1) is dominated by surface aspartates and glutamates. Assuming all exposed ionizable residues to be charged, maquette 1 will have a net charge of −16 near neutral pH. The second sequence (2 in Figure 1) introduces 27 more lysines and arginines, bringing the predicted net charge to +11. Heme binding affinity remains high in both sequences (Kd oxidized heme <2 nM for 1 and 20 and 800 nM for 2 [17]) with no indication in the redox difference spectra of a significant population of unbound heme in either the oxidized or reduced spectra (Supplementary figure 1). After binding two reduced hemes with deprotonated surface exposed propionates, the net charges become −20 and +7. Because of the density of ionizable acidic residues in 1, some of these groups may be neutralized and the net charge may be somewhat smaller. Not surprisingly, the dramatic change in surface electrostatics influences the heme reduction potential, with the Em8 rising from −290 mV in 1 to −150 mV in 2 [17]. However, both maquettes are much lower in potential than bovine cyt c, Em8 ~ +260 mV, and thus maintain large favorable driving forces for cytochrome reduction.
Figure 1.
Sequences for molecules 1 and 2. Residues with positive and negative charges at pH 7.9 are shown in blue and red, and heme-ligating histidines are green. Electrostatic surface estimates are shown for each maquette and for bovine cyt c, generated using APBS [34].
Oxidized cyt c has a net charge of +8 at pH 7.9 [1]. Its natural redox protein partners exhibit complimentary surface electrostatics, either overall net negative molecular charge or negatively charged surface patches (Table 1). If the heme maquettes obey the same physical chemical principles as natural proteins, we expect cyt c to be more rapidly reduced by negatively charged maquette 1 than by positively charged maquette 2. This is verified via stopped-flow spectroscopy mixing reduced maquettes with oxidized cyt c (Figure 2). Upon stopped-flow mixing, the α–band maximum at 560 nm of the reduced heme b maquette decreases as the reduced α-band maximum of cyt c at 550 nm increases. Changing the net charge of the heme maquette from negative (1) to positive (2) raises the electron-transfer halftime by a factor of 50. Increasing the concentration of the reactants (not shown) raises the initial rate of the electron transfer on mixing, as expected for a simple second-order reaction.
3.2 Flash-induced interprotein electron transfer
Although the effect of charge complementarity is clear from the above experiment, the stopped-flow method does not resolve sub-ms details of the electron transfer between maquette 1 and cyt c. Furthermore, dithionite titration to reduce the maquette is prone to either over reduction leaving excess dithionite, or under reduction and subsequent oxidation of the maquette during handling in the stopped-flow. Each can introduce distortions to the electron-transfer time course. To circumvent these technical challenges we employed classic heme CO photolysis to initiate inter-protein electron transfer, much as Chance used CO to trap a reduced heme prior to photo-activated electron transfer in cytochrome oxidase [27]. CO binding also helps stabilize the reduced maquette by preventing small amounts of O2 from prematurely oxidizing the heme during handling.
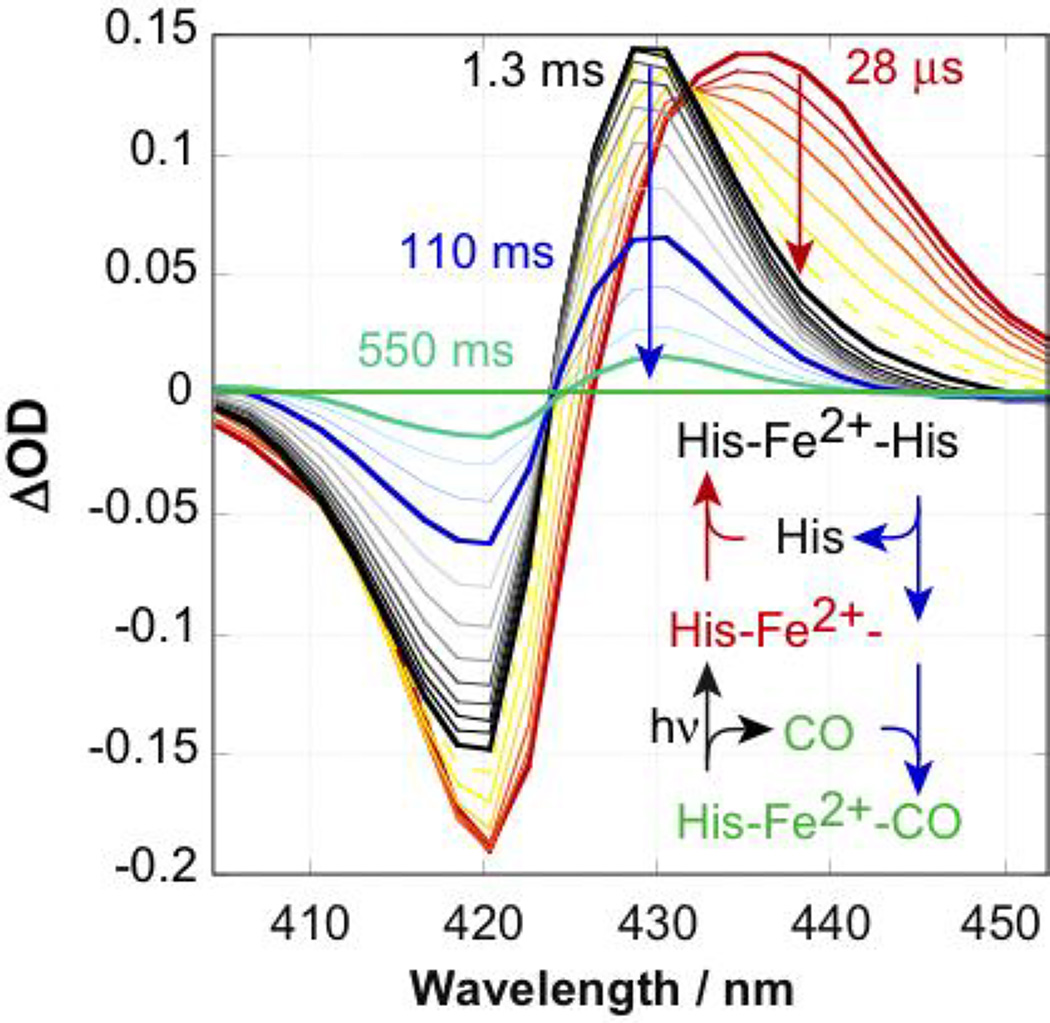
In the absence of a gaseous ligand, each reduced heme iron in 1 is axially ligated by the Nε atoms of two histidine sidechains. In the presence of either bound O2 or CO, the diatomic ligand displaces one of these histidine sidechains [17,23]. Thus, these heme maquettes function much like the natural hexacoordinate oxygen transport protein neuroglobin. Using a 532 nm laser pulse, a bound CO ligand rapidly photo-dissociates yielding a ferrous heme ligated by only a single histidine. This is seen as a red shift in the Soret difference spectrum (red spectrum Figure 3A) with a 435 nm peak and an isosbestic at 427 nm. At low CO concentrations, the second His rebinds in about a millisecond, characterized by a less extreme Soret bandshift (black spectrum in Figure 3B) with a peak around 429 nm. The isosbestic for this His rebinding event is 432 nm. Eventually, CO returns to displace this His. The His-CO ligand exchange isosbestic is 424 nm. The unimolecular rate constants kHis-on and kHis-off and the bimolecular CO rebinding rate kCO-on for maquette 1 are 210 s−1, 50 s−1, and 4.3 µM−1s−1 respectively [36].
Figure 3.
Soret region spectral changes following CO flash photolysis relative to the CO bound to reduced maquette 1 starting state. Immediately after photolysis, neither CO nor the second His are bound (red spectrum). His rebinds in a ms (black). At minimal CO concentrations of an argonpurged sample, CO slowly rebinds to restore the pre-flash state (green). Post-flash times are given for bold traces. 110 mM NaCl, 50 mM Na2HPO4 pH 7.9.
Flash photolysis provides a fast method to initiate interprotein electron transfer; CO release returns the maquette to a ~290 mV Em8, immediately available to reduce an acceptor such as cyt c. At the minimal CO concentrations in these experiments, CO recombination is significantly slower than the electron transfer. In the experiments that follow, the CO-bound ferrous maquette donor 1 mixes with oxidized cyt c in the stopped-flow apparatus before activation with a laser pulse to initiate CO photolysis and subsequent inter-protein electron transfer.
Figure 4 illustrates the photolysis-induced electron-transfer spectroscopy. Panel A shows the difference spectra expected for CO photolysis (red), His-CO ligand exchange (black), and maquette heme B to cyt c electron transfer (blue) computed from the redox difference spectra of maquette 1 (purple) and cyt c (green). Panel B shows observed photolysis, ligand exchange and electron-transfer kinetics under the high salt environment of 110 mM NaCl, 50 mM Na2HPO4 pH 7.9. This salt regime is comparable to in vivo conditions. Here, electron transfer is slower than His rebinding. We track photolysis at the electron-transfer isosbestic of 420 nm. The steady state spectral extinction coefficients of CO-free and CO-bound reduced maquette 1 at this wavelength determines the yield of flash photolysis, typically around 10%. Subsequent electron transfer is followed at the His-CO ligand exchange isosbestic of 424 nm (blue arrow). Panel C shows electron transfer at the low salt condition of 11 mM NaCl, 5 mM Na2HPO4 pH 7.9. Here electron transfer is at least as fast as the characteristic ms time for His rebinding, so the electron transfer is best monitored at the His binding isosbestic of 432 nm.
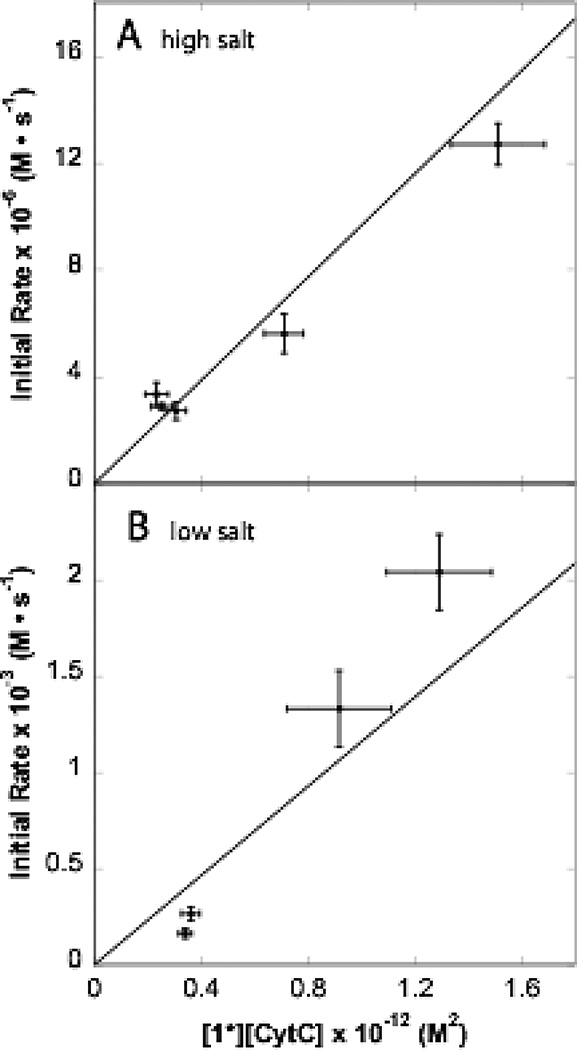
Figure 5A shows the initial electron-transfer rates as a function of the product of the concentrations of cyt c and flash photolyzed 1. The essentially single exponential kinetics show initial rates under high salt conditions that as a group fit to a second-order rate constant of 9.7 × 106 M−1s−1. Under the low salt conditions of Figure 5B, kinetics are much faster, with essentially single exponential time courses at higher cyt c concentrations. Considering just initial rates provides a second-order rate constant estimate of 1.2 × 109 M−1s−1. However, at lower cyt c concentrations a slower phase begins to appear (Supplementary figure 2), suggesting that some cyt c may be bound to unactivated maquettes, needing to dissociate before reacting with activated maquettes. This would tend to lower the observed initial rate at low concentrations, as seen in Figure 5B.
Figure 5.
Initial rate of electron transfer at high and low salt conditions are roughly second-order in concentration of reactants. [1*] is denotes the concentration of photolyzed maquette 1. High salt rates measured at 424 nm and low salt rates at 432 nm.
3.3 Ionic strength dependence of inter-protein electron transfer
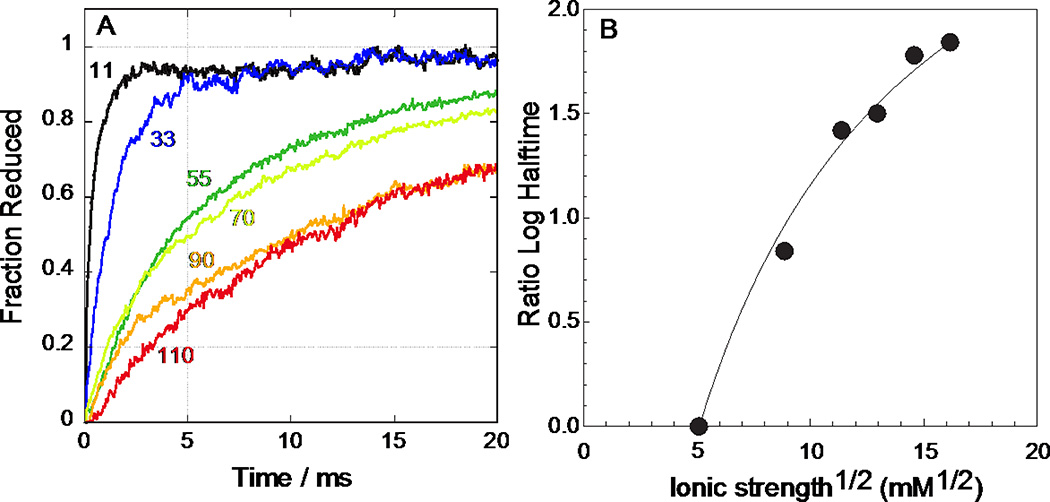
Figure 6 shows the cyt c reduction time courses after photolysis of maquette 1 for a range of ionic strengths. Kinetics are normalized to nearly complete reduction observed at 100 ms. At the lowest ionic strength (11 mM NaCl, 5 mM Na2HPO4), the halftime for the electron transfer is about 160 µs and the reaction is essentially complete in 2 ms. The ionic strength in the photolysis experiment at 55 mM NaCl and 25 mM K2HPO4 (Figure 6A, dark green trace) is similar to that in the mixing experiments at 120 mM NaCl and 50 mM HEPES (Figure 2). The second-order rate of 1.9 × 107 M−1s−1, shown in Figure 2B, is consistent with the photolysis-induced reaction trajectory in Figure 6A. This suggests that there is no inherent kinetic difference between the mixing and photolysis methods.
Figure 6.
Photolysis-induced electron transfer from 1.7 µM CO-bound 1 to 3.4 µM cyt c at pH 7.9 at several ionic strengths. A: Traces follow the change at the His rebinding isosbestic point of 432 nm and are normalized to the completed value at 100 ms. Concentrations of NaCl and Na2HPO4 respectively are: 11 mM & 5 mM; 33 mM & 15 mM; 55 mM & 25 mM; 70 mM & 32 mM; 90 mM & 41 mM; 110 mM & 50 mM. B: Ionic strength dependence of log10 of electron-transfer half-times from Figure 6A, referenced to the half time at 11 mM NaCl, 5 mM Na2HPO4. Curve represents a Debye-Hückel model with a charge product of −150 charge units squared and protein radii of 17 Å.
Figure 6B shows the dependence of the reaction half-time on the square root of ionic strength, based on a Debye-Hückel model [37] assuming a radius of 17 Å for both cyt c [38] and maquette 1. This estimate for the effective radius of 1 matches the measured radius of gyration from small angle X-ray scattering [Nathan Ennist, personal communication]. The model predicts a charge product of −150; thus, maquette 1 acts similar to a sphere of net charge −19 given the cyt c net charge of +8.
4. Discussion
Creating novel proteins with functions designed for human, as opposed to cellular, needs and integrating them into metabolism to produce useful products is a key goal of synthetic biology. Maquette proteins have already demonstrated that a wide range of light capture, charge separating and redox functions can be established in these helical scaffolds. Moreover, maquettes can be expressed and fitted with cofactors in vivo [17,39]. Maquettes’ uncomplicated helical bundle frame supports selection of cofactor type and position, allowing the designer to integrate redox and energetic properties for single or multi-cofactor function [17]. The next step for in vivo integration is a practical understanding of design principles for electron-transfer connections with appropriate natural redox protein partners.
It has long been understood that successful electron-transfer contact between mobile redox protein partners can be guided by complementary surface electrostatics [1,38,40] and that within the cell, multiple electron-transfer proteins may exploit similar charge patterning to create branching electron-transfer networks [7]. Soluble c-type cytochromes and other small diffusing redox proteins serve as electron transporters, connecting larger catalytic redox complexes in a network that often tolerates replacement of one diffusing transporter with another [7]. As maquettes are about the same size as natural electron transporters and tolerate dramatic sequence and charge changes without compromising redox function, they are excellent prospects for insertion into natural electron-transfer networks to engineer novel inter-protein connections. Already, a choice of maquettes with a variety of redox cofactor types, including iron-sulfur cluster [19], flavin [21], quinone [22], copper [41] and irons [42], are available for external patterning.
By comparing the electron-transfer activity of oppositely charged maquettes 2 and 1, Figure 2 shows that we can indeed create an electrostatically modulated connection between heme maquettes and the physiologically common redox carrier cyt c. Even at relatively high ionic strength conditions where significant charge screening is expected, there is a 50-fold increase in the estimated second-order rate constant induced by changing the net charge from roughly +7 to −20. Assuming typical protein reorganization energies of 1 to 1.2 eV [35], the increase in electron-transfer driving force from −0.41 eV in 2 to −0.55 eV in 1 would lead a modest 3-fold acceleration. The much larger magnitude of the observed acceleration is consistent with an electrostatic interaction between maquette 1 and cyt c.
1 reacts with cyt c sufficiently quickly for effective function in vivo. Indeed, it is fast enough that simple stopped-flow measurements provide inadequate time resolution and the faster method of CO photolysis-initiated electron transfer is needed to describe the ionic strength dependence of the reaction. The photoreduction of cyt c by CO-bound maquette 1 is analogous to the natural diffusive photooxidation of cyt c by bacterial photosynthetic reaction centers [43]. Photo-excited reaction centers of Rb. sphaeroides oxidize cyt c with a second-order rate constant of 1.4 × 108 M−1s−1 in 30 mM NaCl, 10 mM Tris [43], remarkably similar to the 1.8 × 108 M−1s−1 observed between 1 and cyt c at 33 mM NaCl, 15 mM K2HPO4 pH 7.9 (Figure 6A, blue trace). This rate approaches that expected for a diffusion-limited reaction. Using the Smulochowski expression [44] with similar radii of 17Å and similar diffusion constants for both 1 and cyt c of 1.1×10−8 cm2s−1, the expected second-order rate constants for uniformly reacting spheres is about 4.5 × 108 M−1s−1. Because not every contact between 1 and cyt c will have appropriate distances for rapid electron transfer, we expect electrostatic attraction to increase the effective collisional frequency and significantly enhance the electron-transfer rate.
As expected for an electrostatically-mediated reaction, electron-transfer rate between 1 and cyt c increases with falling ionic strength (Figure 6). A simple Debye-Hückel model for diffusing spheres using uniform surface charges of radius 17 Å for cyt c [37] and the SAXS derived radius of gyration for maquette 1 also of 17 Å (Nathan Ennist, personal communication), goes far in explaining the ionic strength dependence of the rate if the product of the charges is around −150 charge units squared (Figure 6B). Employing this model using a net charge for cyt c of +8 [1], the experimentally indicated charge on 1 is −19, quite close to the expected value of −20 for 1 with two reduced hemes and all surface exposed amino acids and heme propionates ionized. Because there is no strong decrease in the rate above 100 mM ionic strength, there does not appear to be a significant dipole interaction controlling the electron-transfer event [37]. Indeed, we made no attempt in the design of 1 to introduce a strong dipole asymmetry, although other maquettes (not tested here) have been designed with marked dipolar character [45]. Although the maquette system displays very fast electron transfer under low salt conditions, the approximately linear dependence on cyt c concentration shown in Figure 5b argues against a simple first-order reaction from a 1:1 maquette-cyt c complex.
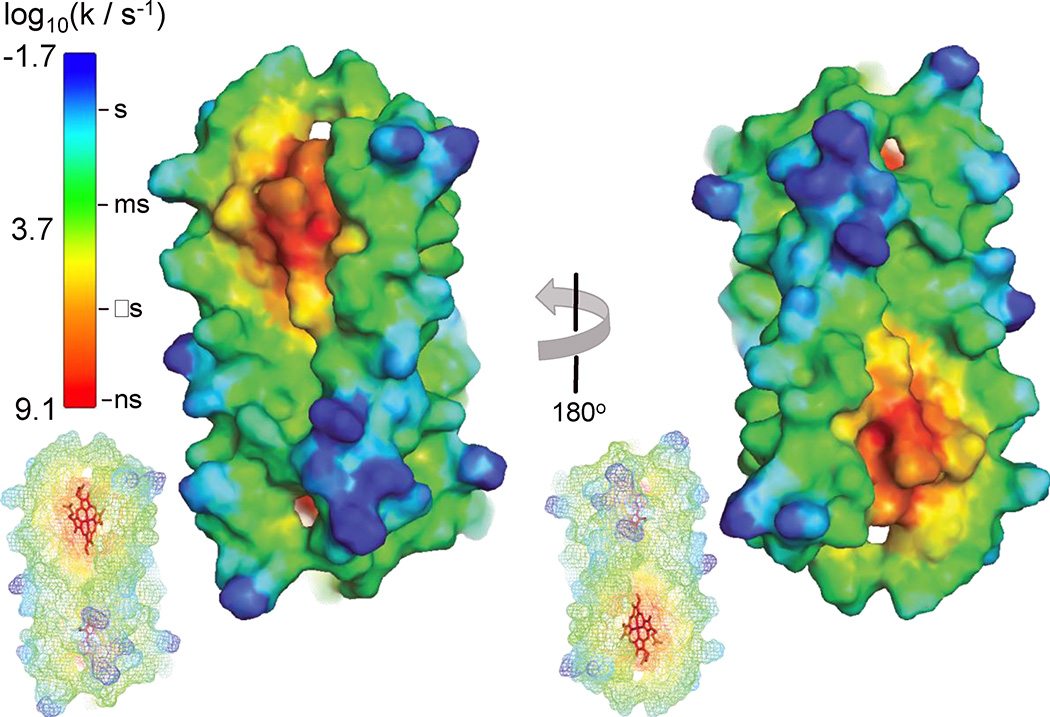
There are different views on the precision of the electrostatic interactions required by biological electron-transfer networks, ranging from specific arrangements of inter-protein amino acid salt bridges [40] to a broader delocalized charge complementarity [46]. We propose that the maquette-cyt c electron-transfer system supports the latter view. Figure 7 displays the expected first-order electron-tunneling rates from 1 to an optimally-oriented cyt c located at points along the maquette surface. From this result, we may infer that productive electron transfer at low salt conditions is limited to cyt c heme edge contacting a maximum of 15%–20% of the maquette surface; these are the yellow through red regions with a calculated half-time less than 200 µs as observed in the low-salt experiments. As this is a significant fraction of the maquette surface, there was no need to design a highly specific docking site for successful electron transfer beyond a coarse scale electrostatic attraction.
Figure 7.
Surface representation of maquette 1 with 2 bound hemes. Colors represent estimated electron tunneling rates to a cyt c redox partner. Calculation assumes the cyt c heme cleft is in VdW contact with given atom on maquette surface. Insets show maquette heme orientation (red sticks). Contact must be within the yellow to red region to support the observed reaction rates at low ionic strength.
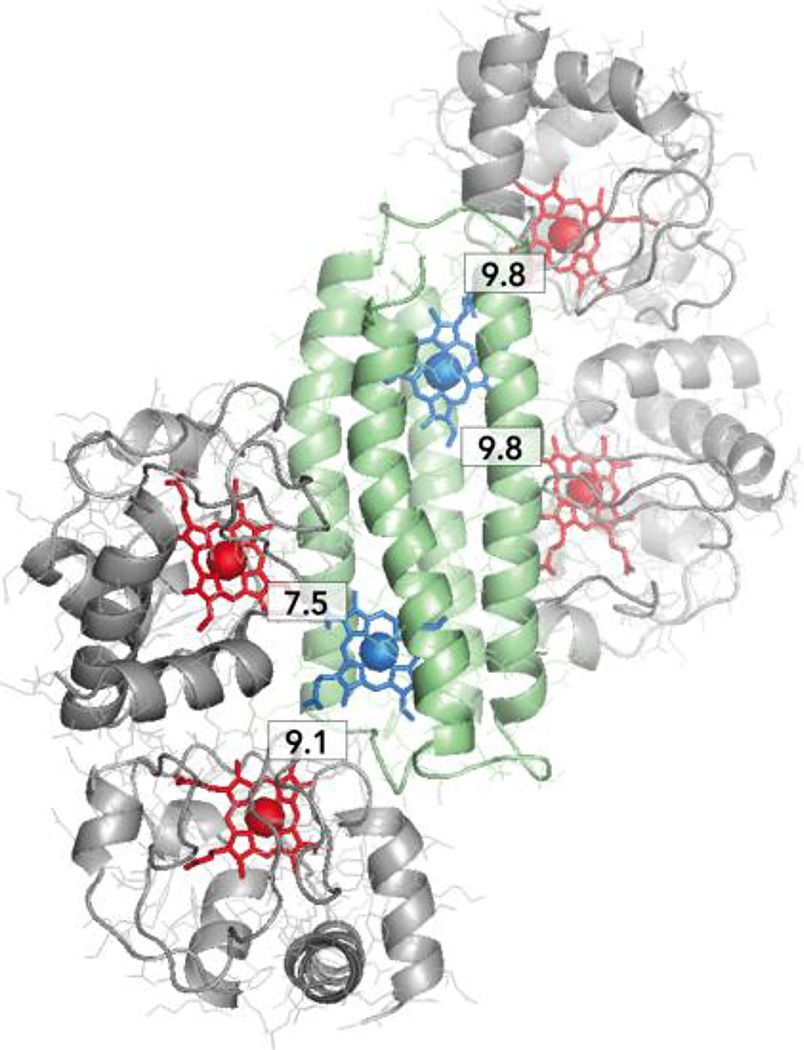
More extensive measurements will be needed to exclude the possibility that at the lowest ionic strengths, there may be loose complexes between 1 and multiple cytochromes c, leading to increased electron-transfer rates as the cyt c stoichiometry is increased. There is room on the maquette surface for such interactions (Figure 8). To better resolve the stoichiometry and electron-transfer dynamics of active natural protein-maquette complexes, maquettes are now being designed with two additional features: 1) asymmetric clusters of negative and positive charges around single heme sites and 2) site-specific single His Zn-porphyrin binding. This will enable light activated electron transfer between maquette and cytochrome c analogous to those previous work establishing binding and stoichiometry of photosynthetic reaction centers with c type cytochromes [43]. Such constructs will further clarify the practical rules controlling interactions with natural redox partners and the effect of enhancing binding interactions between maquettes and natural proteins.
Figure 8.
Cartoon representation of hypothetical complex of maquette 1 (green, blue heme) with 4 cyt c molecules (grey, red heme). Numerical labels show electron transfer distances, as measured between porphyrin ring edges in 1 and cyt c.
In vivo integration of maquettes provides a powerful new tool in the synthetic biology toolbox. So far, maquettes show little or no toxicity in E. coli [17] even when over-expressed; they also demonstrate successful interaction with cellular cofactor maturation machinery [39]. Tolerance to extensive sequence modification and ability to modulate interprotein interactions via elementary electrostatics, combined with the evident flexibility and promiscuity of many natural interprotein electron-transport systems, suggest that maquettes are a versatile means to establish in vivo electrical connections with bioenergetic pathways.
Supplementary Material
Highlights.
Man-designed protein reduces cyt c at natural rates
Electrostatic interaction revealed by charge patterning and ionic strength
Atomistic precision not required for inter-protein electron-transfer control
Acknowledgments
This work support by NIH GM48130, and NSF AIR ENG-1312202 and the Nano/Bio Interface Center NSF NSEC DMR08-32802.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koppenol WH, Vroonland CAJ, Braams R. The electric potential field around cytochrome c and the effect of ionic strength on reaction rates of horse cytochrome c. Biochim. Biophy. Acta. 1978;503:499–508. doi: 10.1016/0005-2728(78)90149-4. [DOI] [PubMed] [Google Scholar]
- 2.McLendon G. Control of biological electron transport via molecular recognition and binding: the “velcro” model. Structure and Bonding. 1991;75:159–174. [Google Scholar]
- 3.Volkov AN, van Nuland NAJ. Electron Transfer Interactome of Cytochrome c. PLoS Comput Biol. 2012;8:e1002807. doi: 10.1371/journal.pcbi.1002807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schilder J, Ubbink M. Formation of transient protein complexes. Current Opinion in Structural Biology. 2013;23:911–918. doi: 10.1016/j.sbi.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 6.Keilin D. On cytochrome, a respiratory pigment, common to animals, yeast, and higher plants. Proceedings of the Royal Society of London Series B. 1925 [Google Scholar]
- 7.Meschi F, Wiertz F, Klauss L, Blok A, Ludwig B, Merli A, Heering HA, Rossi GL, Ubbink M. Efficient Electron Transfer in a Protein Network Lacking Specific Interactions. J. Am. Chem. Soc. 2011;133:16861–16867. doi: 10.1021/ja205043f. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier H, Kraut J. Crystal Structure of a Complex Between Electron Transfer Partners, Cytochrome c Peroxidase and Cytochrome c. Science. 1992;258:1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 9.Hall J, Zha X, Durham B, O'Brien P, Vieira B, Davis D, Okamura M, Millett F. Reaction of cytochrome c and c2 with the Rhodobacter sphaeroides reaction center involves the heme crevice domain. Biochem. 2002 doi: 10.1021/bi00388a048. [DOI] [PubMed] [Google Scholar]
- 10.Witthuhn VC, Gao J, Hong S, Halls S, Rott MA, Wraight CA, Crofts AR, Donohue TJ. Reactions of Isocytochrome c2in the Photosynthetic Electron Transfer Chain of Rhodobacter sphaeroides. Biochem. 1997;36:903–911. doi: 10.1021/bi961648k. [DOI] [PubMed] [Google Scholar]
- 11.Page CC, Moser CC, Dutton PL. Mechanism for electron transfer within and between proteins. Curr Opin Chem Biol. 2003;7:1–6. doi: 10.1016/j.cbpa.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhen YJ, Hoganson CW, Babcock GT, Ferguson-Miller S. Definition of the interaction domain for cytochrome c on cytochrome c oxidase - I Biochemical, spectral, and kinetic characterization of surface mutants in subunit II of Rhodobacter sphaeroides cytochrome aa(3) J. Biol. Chem. 1999;274:38032–38041. doi: 10.1074/jbc.274.53.38032. [DOI] [PubMed] [Google Scholar]
- 13.Axelrod HL, Abresch EC, Okamura MY, Yeh AP, Rees DC, Feher G. X-ray Structure Determination of the Cytochrome c2: Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides. J. Mol. Biol. 2002;319:501–515. doi: 10.1016/S0022-2836(02)00168-7. [DOI] [PubMed] [Google Scholar]
- 14.Nicholls P, Mochan E. Formation of a Stable “Active” Complex between Cytochrome c and Yeast Peroxidase. Nature. 1971;230:276–277. doi: 10.1038/newbio230276a0. [DOI] [PubMed] [Google Scholar]
- 15.Tian H, Sadoski R, Zhang L, Yu CA, Yu L, Durham B, Millett F. Definition of the interaction domain for cytochrome c on the cytochrome bc(1) complex Steady-state and rapid kinetic analysis of electron transfer between cytochrome c and Rhodobacter sphaeroides cytochrome bc(1) surface mutants. J. Biol. Chem. 2000;275:9587–9595. doi: 10.1074/jbc.275.13.9587. [DOI] [PubMed] [Google Scholar]
- 16.Robertson DE, Farid RS, Moser CC, Urbauer JL, Mulholland SE, Mulholland SE, Pidikiti R, Lear JD, Wand AJ, Degrado WF, Dutton PL. Design and Synthesis of Multi-Heme Proteins. Nature. 1994;368:425–431. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 17.Farid TA, Kodali G, Solomon LA, Lichtenstein BR, Sheehan MM, Fry BA, Bialas C, Ennist NM, Siedlecki JA, Zhao Z, Stetz MA, Valentine KG, Anderson JLR, Wand AJ, Discher BM, Moser CC, Dutton PL. Elementary tetrahelical protein design for diverse oxidoreductase functions. Nat. Chem. Biol. 2013;9:826–833. doi: 10.1038/nchembio.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solomon LA, Kodali G, Moser CC, Dutton PL. Engineering the Assembly of Heme Cofactors in Man-Made Proteins. J. Am. Chem. Soc. 2014;136:3192–3199. doi: 10.1021/ja411845f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibney BR, Mulholland SE, Mulholland SE, Rabanal F, Dutton PL. Ferredoxin and ferredoxin-heme maquettes. Proc. Natl. Acad. Sci. U.S.a. 1996;93:15041–15046. doi: 10.1073/pnas.93.26.15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grzyb J, Xu F, Weiner L, Reijerse EJ, Lubitz W, Nanda V, Noy D. De novo design of a non-natural fold for an iron-sulfur protein: alpha-helical coiled-coil with a four-iron four-sulfur cluster binding site in its central core. Biochim. Biophys. Acta. 2010;1797:406–413. doi: 10.1016/j.bbabio.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Design, synthesis, and characterization of a photoactivatable flavocytochrome molecular maquette. 1998;95:10465–10470. doi: 10.1073/pnas.95.18.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichtenstein BR, Moorman VR, Cerda JF, Wand AJ, Dutton PL. Electrochemical and structural coupling of the naphthoquinone amino acid. Chem. Commun. 2012;48:1997–1999. doi: 10.1039/c2cc16968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koder RL, Anderson JLR, Solomon LA, Reddy KS, Moser CC, Dutton PL. Design and engineering of an O2 transport protein. Nature. 2009;458:305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibney BR, Isogai Y, Rabanal F, Reddy KS, Grosset AM, Moser CC, Dutton PL. Self-assembly of heme A and heme B in a designed four-helix bundle: implications for a cytochrome c oxidase maquette. Biochem. 2000;39:11041–11049. doi: 10.1021/bi000925r. [DOI] [PubMed] [Google Scholar]
- 25.Roy A, Sarrou I, Vaughn MD, Astashkin AV, Ghirlanda G. De novo design of an artificial bis[4Fe-4S] binding protein. Biochem. 2013;52:7586–7594. doi: 10.1021/bi401199s. [DOI] [PubMed] [Google Scholar]
- 26.Solomon LA. Thesis. University of Pennsylvania; 2013. The physical chemistry underlying the assembly and midpoint potential control in a series of designed protein-maquettes. [Google Scholar]
- 27.Chance B, Graham N, Legallais V. Low temperature trapping method for cytochrome oxidase oxygen intermediates. Analytical Biochemistry. 1975;67:552–579. doi: 10.1016/0003-2697(75)90331-0. [DOI] [PubMed] [Google Scholar]
- 28.Eleff S, Kennaway NG, Buist NR, Darley-Usmar VM, Capaldi RA, Bank WJ, Chance B. 31P NMR study of improvement in oxidative phosphorylation by vitamins K3 and C in a patient with a defect in electron transport at complex III in skeletal muscle. Proc. Natl. Acad. Sci. U.S.a. 1984;81:3529–3533. doi: 10.1073/pnas.81.11.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Analytical Biochemistry. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 30.Wildes PD, Pacifici JG, Irick G, Whitten DG. Solvent and substituent on the thermal isomerization of substituted azobenzenes Flash spectroscopic study. J. Am. Chem. Soc. 1971;93:2004–2008. [Google Scholar]
- 31.The PyMOL Molecular Graphics System, Version 13r1. Schrodinger: LLC; 2010. [Google Scholar]
- 32.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communications. 1995;91:43–56. [Google Scholar]
- 33.Šali A, Blundell TL. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.a. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. Nature of Biological Electron-Transfer. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 36.Sheehan MM. Thesis. University of Pennsylvania; 2014. Engineering oxygen reactivity in heme-protein maquettes. [Google Scholar]
- 37.Van Leeuwen JW. The ionic strength dependence of the rate of a reaction between two large proteins with a dipole moment. Biochim. Biophys. Acta. 1983;743:408–421. doi: 10.1016/0167-4838(83)90400-4. [DOI] [PubMed] [Google Scholar]
- 38.Dixon DW, Hong X, Woehler SE. Electrostatic and steric control of electron self-exchange in cytochromes c, c551, and b5. Biophys. J. 1989 doi: 10.1016/S0006-3495(89)82680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kodali G, Lichtenstein BR, Mancini JA, Farid TA, Moser CC, Dutton PL. Constructing a man-made c-type cytochrome maquette in vivo: electron transfer, oxygen transport and conversion to a photoactive light harvesting maquette. 2014;5:507–514. doi: 10.1039/C3SC52019F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margoliash E, Bosshard HR. Guided by electrostatics, a textbook protein comes of age. Trends Biochem. Sci. 1971;8:316–320. [Google Scholar]
- 41.Tegoni M. De Novo Designed Copper α-Helical Peptides: From Design to Function. Eur. J. Inorg. Chem. 2014;2014:2177–2193. [Google Scholar]
- 42.Faiella M, Andreozzi C, de Rosales RTM, Pavone V, Maglio O, Nastri F, DeGrado WF, Lombardi A. An artificial di-iron oxo-protein with phenol oxidase activity. Nat. Chem. Biol. 2009;5:882–884. doi: 10.1038/nchembio.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moser CC, Dutton PL, Dutton PL. Cytochrome c and c2 binding dynamics and electron transfer with photosynthetic reaction center protein and other integral membrane redox proteins. Biochem. 1988;27:2450–2461. doi: 10.1021/bi00407a031. [DOI] [PubMed] [Google Scholar]
- 44.von Smoluchowski M. Investigation of a Mathematical Theory on the Coagulation of Colloidal Suspensions. Z. Physik. Chem.(Ger.) 1917 [Google Scholar]
- 45.Chen X, Moser CC, Pilloud DL, Gibney BR, Dutton PL. Engineering Oriented Heme Protein Maquette Monolayers through Surface Residue Charge Distribution Patterns. J. Phys. Chem. B. 1999;103:9029–9037. [Google Scholar]
- 46.Tiede DM, Vashishta AC, Gunner MR. Electron-transfer kinetics and electrostatic properties of the Rhodobacter sphaeroides reaction center and soluble c-cytochromes. Biochem. 1993;32:4515–4531. doi: 10.1021/bi00068a006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.