Abstract
The transcriptomes of five pear cultivars, ‘Hosui’ (P. pyrifolia), ‘Yali’ (P. bretschneideri), ‘Kuerlexiangli’ (P. sinkiangensis), ‘Nanguoli’ (P. ussuriensis), and ‘Starkrimson’ (P. communis) were sequenced at seven key fruit developmental stages, from fruit setting to maturation and fruit senescence after harvesting. In total, 33,136 genes that could be mapped by reads, were analyzed. Most gene expression cluster models showed a steadily decreasing trend. Gene expression patterns had obvious differences according to maturity type, that is, post-ripening cultivars were still vigorous at maturity, and showed a higher proportion of up-regulated genes; non post-ripening cultivars had a gradually decreasing tendency during fruit maturation. Meanwhile, differentially expressed genes related to fruit quality and development, such as stone cells, sugar, acid and hormones, were identified. Co-expression analysis revealed that several ethylene synthesis genes and polyphenoloxidase-related genes interacted with each other directly, and an indirect relationship was reflected between ethylene synthesis genes and ethylene response genes. In addition, the highly diverse SNPs represented the great differences between oriental and occidental pears. Understanding how RNA-seq based gene-expression patterns and differential gene expression contribute to fruit quality allows us to build models for gene-expression for fruit development of Pyrus species.
Pear (Pyrus L.) belongs to the family of Rosaceae, with a history of cultivation dating back more than two thousand years in China1. It is well known that the pear originated in southwest of China, with diverse centers of evolution in China, Central Asia and Near Easter/Asia Minor2. Even though up to 22 pear species have been identified3, only five species are cultivated worldwide, four deriving from oriental countries, including P. pyrifolia, P. bretschneideri, P. sinkiangensis and P. ussuriensis, with only P. communis deriving from an occidental country. The geographical distribution and fruit genotype are diverse between the five pear species. In China, all five species have been cultivated around the country in appropriate cultivation areas. P. pyrifolia is cultivated in the south of China, P. bretschneideri in northern China, P. ussuriensis usually in the northeast region with extreme low temperatures, while P. sinkiangensis is only found in Xinjiang, and P. communis is cultivated in the north of China with cold climate, such as Beijing and Shandong province. For different cultivated species, typical maturity phenotypes exist between the five pear species. Generally speaking, fruit of P. communis and P. ussuriensis ripen after harvest, and their pears are usually eatable with good flavor and fruit quality after post harvest storage, while the other three cultivated species are eatable immediately when harvested at mature stage. In addition, different fruit traits are also found in the five species. The P. communis pears are gourd shaped, have soft and smooth flesh, with few stone cells4 and high sugar and acid content5, with sweet aroma. P. ussuriensis pears are usually high in volatile compounds6 and concentrated fruit flavor. On the contrary, the other oriental pears have crisp flesh, low aroma and flavor, and some of them have high sugar and low acid content5.
Until now, some physiological and molecular mechanism studies have been carried out to investigate different fruit development characteristics and fruit quality. For the fruit maturation process, fruit texture is affected by the ACO (1-aminocyclopropane-1-carboxylate oxidase) gene, and then XTH (Xyloglucan endotransglucosylase/hydrolase)-related genes can lead to cell wall disassembly and loosening7. ‘Bartlett’ occidental pears are prevented from ripening on the tree, but ethylene was able to stimulate fruit softening on the tree8 and endo-PG genes have an impact on many different maturation characteristics8,9. Microarrays were developed for pear to illustrate the absence of cupin family protein-related genes and two un-annotated genes in Japanese pear (Pyrus pyrifolia), which may induce post-ripening in pears10. Sugar is one of the most important factors for fruit quality, and provides the main energy for fruit metabolism. In Pyrus species, sucrose and fructose are the major soluble sugars, and overall sugar content varies greatly between different cultivars5. It was found that sucrose synthase and sucrose-phosphate synthase were highly correlated with sucrose levels in Japanese pear11. Soluble acid invertase accumulates hexoses and regulates sucrose-to-hexose ratio, and two invertase genes, PsS-AIV1 and PsS-AIV2, have been cloned in pears, but are expressed differently during fruit development12. Lignified stone cells are a special characteristic of pears, which severely affect fruit quality. The stone cells have a mosaic pattern in the flesh of pear fruits, with a higher concentration of larger stone cells near the core, and smaller ones around the pericarp13. The ratio of G-type and S-type lignin and polymerization degree have been evaluated among various pear species14. Highly expressed gene families of HCT, C3′H, and CCOMT contribute to high accumulation of both G-lignin and S-lignin in ‘Dansuansuli’ fruit. However, there are few reports on all five cultivated pear species toward understanding the different molecular regulation mechanisms of fruit development and quality.
Next-generation RNA sequencing can sensitively and rapidly reflect gene expression with an enormous amount of data, and is an efficient method to explore fruit development characteristics. Besides pear, many Rosaceae family fruits are popular, such as apple, peach, berry, strawberry, plum, loquat, and apricot. The whole genomes of apple15, strawberry16, plum17, pear18 and peach19 have been sequenced, making possible differentially expressed gene analysis based on the reference genome to reveal different genotypes. In a previous study, transcriptome sequencing of apple helped to find the Gypsy-like retrotransposon in the bud mutant, affecting internode length20. Peach21 and berry22 have also been analyzed by high-throughout sequencing technology to reveal internal mechanisms of fruit development and quality. The whole genome sequence of pear has also been published with high quality assembly. Therefore, a good platform is available to study the genetic characteristics of fruit development and quality conformation in pear. The transcriptome of Japanese pears has been analyzed to reveal the flower bud transitioning through endodormancy23, and occidental pear24 was analyzed to study fruit size. However, many fruit developmental characters and transcriptome-based differences in all five pear species are still unclear. RNA-seq technology can be an effective method to reveal gene expression patterns and difference during fruit development and maturation for distinguished cultivated species of pear.
In this research, we performed large-scale RNA sequencing from seven fruit developmental stages in the five cultivated species of pear to effectively reveal the main differences between fruit-related metabolism, especially fruit post- or non post-ripening. We used the ‘Dangshansuli’ (Pyrus bretschneideri Rehd.) genome as a reference to analyze various fruit developmental patterns and differentially expressed genes, which mainly regulate the pear growth and fruit quality. Results contribute to a better understanding of the dynamic metabolism and molecular mechanism of different pear species, which provides a significant basis to improve fruit quality of pear, and even other species in Rosaceae.
Results and Discussion
RNA sequencing of different developmental stages of pear fruit in five cultivars
In this study, the transcriptomes of five representative cultivars: ‘Hosui’ (P. pyrifolia), ‘Yali’ (P. bretschneideri), ‘Kuerlexiangli’ (P. sinkiangensis), ‘Nanguoli’ (P. ussuriensis) and ‘Starkrimson’ (P. communis), which are the main cultivated species worldwide, were selected to reveal gene expression patterns and explore relationships between different fruit traits. A total of seven periods (date details in materials), including six key fruit developmental stages (periods 1–6) and one fruit senescence stage after harvesting (period 7) were analyzed. Fruit developmental stages were: fruit setting (period 1), physiological fruit dropping (period 2), fruit rapid enlargement (period 3), a month after fruit enlargement (period 4), pre-mature (period 5) and mature (period 6). Each was sequenced by the Illumina2000 platform. There were more than 10 M raw reads for each sample analysis (Table 1). The original data are available on NCBI: project number PRJNA309745, SRA trace number SRP070620. The mean quality score was above 30, and after adapter removing and ribosomal RNA filtering, a total of 33,136 mapping genes were aligned to the reference pear (Pyrus bretschneideri Rehd.) genome18, and analyzed based on the 49 bp RNA-seq single end data. The mapping gene numbers were 28,621 (86.4%,‘Hosui’), 29,041 (87.6%, ‘Kuerlexiangli’), 28,925 (87.3%, ‘Nanguoli’), 29,294 (88.4%, ‘Starkrimson’) and 28,905 (87.2%, ‘Yali’). Transcript sets of five cultivars had no bias in mapping gene number, which reflected good sequencing distribution. Little difference was found in the percentage of aligned gene numbers accounting for clean reads during fruit developmental stages, however, it was relatively low for all cultivars at maturation or senescence (Table 1). One major reason might be that many genes have no expression or greatly decreased expression during fruit maturation. Another natural reason could be low sequencing depth, and the limited sequencing data, 10 M reads with 49 bp length for each sample. In tomato, as the cell wall decomposed during maturation, the expression of many genes involved in cell wall carbohydrates also decreased25. The same phenomenon was also present in watermelon, with CmXTH expression down during mature-fruit abscission26.
Table 1. Read number and unique gene mapping for RNA-seq.
| Samples | No. of raw reads | No. of clean reads | No. of mapping genes | Percentage of clean read mapping rates (%) |
|---|---|---|---|---|
| HS1 | 11,302,851 | 10,328,076 | 25,688 | 75.63 |
| HS2 | 10,154,316 | 9,197,441 | 24,765 | 75.11 |
| HS3 | 12,117,316 | 9,398,656 | 23,071 | 74.06 |
| HS4 | 11,213,396 | 10,109,015 | 23,635 | 74.63 |
| HS5 | 11,243,703 | 10,216,762 | 22,655 | 75.76 |
| HS6 | 11,950,723 | 11,000,668 | 22,663 | 75.42 |
| HS7 | 10,668,693 | 9,779,232 | 21,374 | 73.93 |
| KRLXL1 | 12,362,986 | 11,781,418 | 26,289 | 73.72 |
| KRLXL2 | 12,239,312 | 11,619,212 | 24,464 | 73.25 |
| KRLXL3 | 11,970,718 | 11,341,031 | 24,826 | 73.18 |
| KRLXL4 | 12,395,731 | 11,272,539 | 23,929 | 73.38 |
| KRLXL5 | 11,885,029 | 10,253,055 | 23,148 | 72.44 |
| KRLXL6 | 12,197,320 | 11,152,288 | 23,282 | 73.74 |
| KRLXL7 | 11,847,303 | 11,274,519 | 22,249 | 73.20 |
| NG1 | 12,516,088 | 12,006,578 | 26,173 | 74.42 |
| NG2 | 11,942,855 | 11,447,523 | 25,681 | 74.55 |
| NG3 | 11,548,878 | 9,242,452 | 23,346 | 75.35 |
| NG4 | 12,396,607 | 10,214,122 | 23,310 | 75.36 |
| NG5 | 12,533,756 | 8,731,570 | 21,469 | 75.09 |
| NG6 | 12,428,560 | 11,425,645 | 22,259 | 74.98 |
| NG7 | 12,461,123 | 11,408,415 | 21,801 | 74.97 |
| RC1 | 11,854,995 | 11,287,205 | 25,675 | 73.62 |
| RC2 | 11,695,092 | 11,085,589 | 25,046 | 73.10 |
| RC3 | 12,087,344 | 11,566,921 | 24,728 | 71.81 |
| RC4 | 11,734,817 | 11,298,495 | 24,419 | 73.45 |
| RC5 | 11,678,523 | 11,180,375 | 24,539 | 72.98 |
| RC6 | 11,693,653 | 11,288,167 | 22,323 | 74.25 |
| RC7 | 12,468,584 | 11,734,107 | 22,473 | 76.01 |
| YL1 | 12,098,295 | 11,396,714 | 26,062 | 75.83 |
| YL2 | 12,449,966 | 11,712,248 | 25,643 | 75.47 |
| YL3 | 12,564,888 | 11,788,331 | 24,220 | 73.63 |
| YL4 | 12,257,369 | 10,177,818 | 23,495 | 73.74 |
| YL5 | 12,304,901 | 10,475,211 | 23,431 | 74.09 |
| YL6 | 11,932,440 | 11,371,346 | 22,837 | 74.24 |
| YL7 | 11,123,512 | 10,582,639 | 21,394 | 75.92 |
Transcriptome sequencing revealed divergent gene expression patterns in pear fruit
Regular mRNA abundance was calculated by reads per exon kilobase per million mapped sequence reads (RPKM). Cluster 2.027 used clustering correlation matrixes to generate a tree. As Fig. 1 shows, there were two main groups in the tree, group I and group II. Most fruit setting stages (period 1) and fruit physiological dropping stages (period 2) were gathered together in group I by the Spearman correlation method. The occidental pear ‘Starkrimson’ made up a separate subgroup B, and other oriental pears made subgroup A. Group II also divided into two subgroups, C and D, with subgroup C clustering the stages of full development to mature stage of all oriental cultivars, while subgroup D included post-harvest stages of all five cultivars and three developmental stages of occidental pear ‘Starkrimson’. It was interesting to find that the most closely related stages of the same cultivar usually first clustered together, and then clustered with stages of other cultivars, indicating different gene expression patterns between the five cultivars, which represent the five cultivated species to some extent. In addition, occidental pear ‘Starkrimson’ showed a relatively distant relationship with other oriental cultivars, in both groups I and II.
Figure 1. Cluster tree of five pear cultivars during fruit development and maturation stages.
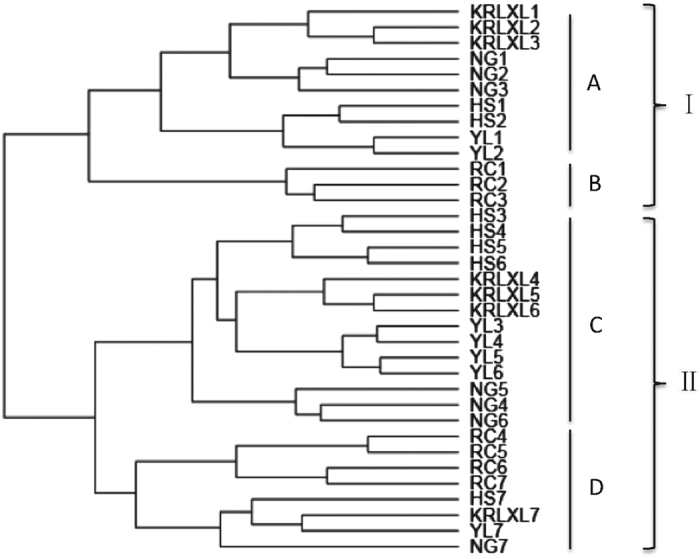
HS: Hosui, YL: Yali, KRLXL: Kuerlexiangli, NG: Nanguo, RC: Red Clapp’s Favorite mutant (‘Starkrimson’), 1–7: seven fruit developmental stages. Spearman correlation tree has two major groups, I and II and four subgroups, A–D.
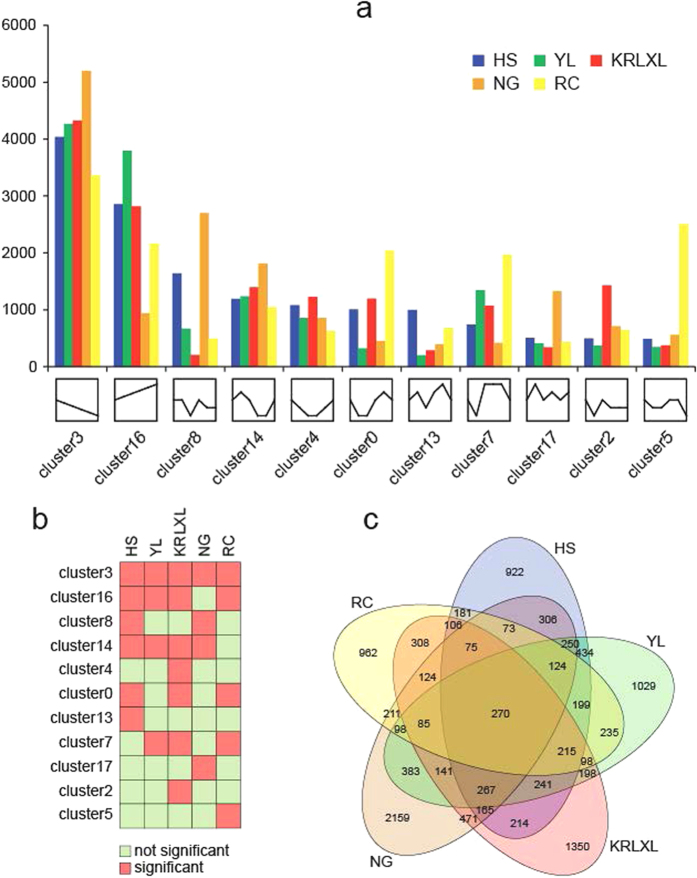
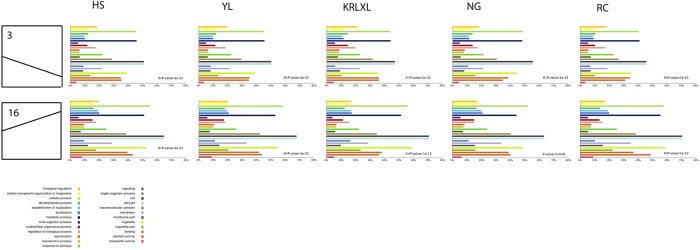
Models for genes with the same expression pattern were defined by STEM analysis28, and a total of 20 gene cluster modules were generated for each sample. We selected the top eleven with the same expression model, shown in Fig. 2a. In general, the gene expression models cluster 3–steady decrease–and cluster 16–steady increase–represented the most common gene expression models across all seven stages and different cultivars. Cluster 3 included the largest number of genes (more than 3,000 genes for each cultivar), indicating that all five cultivars had most genes expressed with the model of steady decrease. A total of 270 genes were identified in all five cultivars in cluster 3, demonstrated in the Venn diagram (Fig. 2c). Gene expression modules of five pear cultivars were significantly enriched (P-value < 1e-5, except for ‘Nanguo’ in cluster 16) in two main clusters, and presented different genetic functions by Gene Ontology (GO) (Fig. 3). Continuous increase cluster 16 had a relatively higher percentage of genes responsible for cellular component organization or biogenesis, single–organism process, cell part and transporter activity, compared with the steady decrease cluster 3 (Fig. 3). We used the 270 genes in cluster 3 for KEGG pathway enrichment, and the genes were most enriched in the photosynthesis pathway (map00195 with p-value of 4.15e-12 and map00196 with p-value of 5.68e-6). In previous gene expression pattern research, sweet orange showed the largest cluster (3,075 genes, accounting for 16.3%) expressed as continuous increase during four fruit development and ripening stages, and the genes in that cluster had functions such as cysteine proteinase sucrose phosphate synthase, and a sucrose transporter29. However, the steadily changing gene-expression patterns were not the main models22 in studies of the berry transcriptome, based on analysis of three stages of fruit development.
Figure 2. Gene expression clusters for different Pyrus species.
(a) Main clusters of gene expression pattern of five Pyrus species. X-axis: type of main clusters and Y-axis: number of genes in different clusters. (b) Significance level of gene number in different clusters for five species. (c) Venn diagram for gene number in the continuous decrease pattern (cluster 3).
Figure 3. Common and different gene expression clusters and their function in five cultivars.
Two main gene expression patterns are illustrated on the left, namely cluster 3 and cluster 16. Functional analysis based on the five cultivars is presented in different colors.
Specific gene expression patterns showed that occidental pears had a gradually decreasing tendency of gene expression in stage 6 of ‘Starkrimson’ – a typically European pear species, expressed genes mostly grouped in cluster 3, cluster 16, cluster 0, cluster 7, and cluster 5 (Fig. 2b). All the gene expression tendencies in these clusters were down-regulated at the mature stage, except for the common model of cluster 16. Cluster 2 represented a specific gene expression pattern of ‘Kuerlexiangli’, because the gene numbers in that pattern were significantly high (under the significance level of 0.05, Fig. 2b). In addition, ‘Nanguoli’ showed its own special fruit developmental model with cluster 17, and ‘Hosui’ had a special gene expression cluster 13. How these specific gene expression patterns regulate the particular fruit development of each cultivar still needs further exploration and study.
Specific gene expression characteristics for post-ripening and non post-ripening cultivars in pear
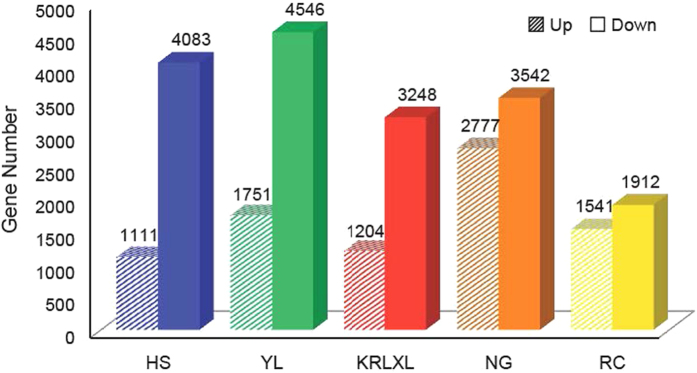
We sequenced samples from the stage of fruit maturity to senescence to detect maturation characteristics of pear. Of the five cultivated species, ‘Starkrimson’ and ‘Nanguoli’ are post-ripening pears, while other three cultivars, ‘Hosui’, ‘Yali’ and ‘Kuerlexiangli’ are eatable directly after harvest. Analysis revealed that post-ripening cultivars were still vigorous after harvest, furthermore we found that they had a higher proportion of up-regulated genes. ‘Nanguoli’ had 43.95% (Fig. 4) up-regulated genes from maturity to fruit senescence. The same tendency was followed by another post-ripening occidental pear, ‘Starkrimson’, with 44.63% (Fig. 4). However, up-expressed genes in ‘Hosui’, ‘Yali’, and ‘Kuerlexiangli’ only accounted for 21.39%, 24.33%, and 27.04% (Fig. 4), respectively. The ratio of up-expressed genes between post-ripening and non post-ripening pear cultivars had significant differences by t-test (p-value = 0.005032).
Figure 4. Up- and down-regulated genes from maturity to post harvest in pear species.
X-axis indicates five samples, and Y-axis is gene number from maturity to post harvest. Bars with diagonal lines are up-regulated gene numbers, and solid bars are down-regulated gene numbers.
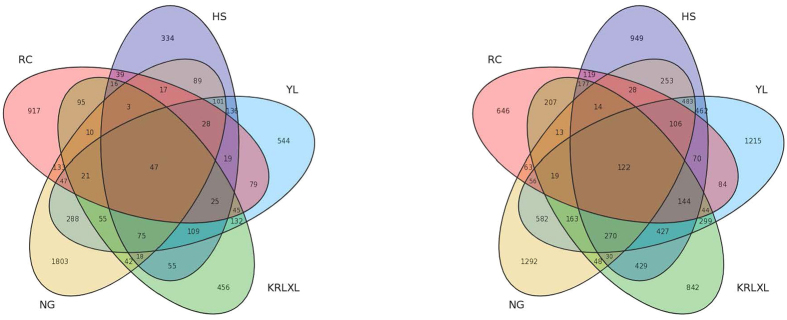
In order to distinguish the two extreme gene expression patterns in post-ripening and non post-ripening pears, we explored gene functions within the two patterns. The number of genes only up-expressed in post-ripening cultivars was 133, while the gene number of only down-expressed in non post-ripening cultivars was 47, as Venn diagrams in Fig. 5 show for the up-regulated and down-regulated genes. Previous studies proved that ethylene plays an important role in catalyzing ripening in European pears30. Ethylene catalyzed the maturity process through different methods such as with the involvment of one cupin family protein gene and two unannotated genes in occidental pears10. In this study, functional analysis showed that the only up-regulated genes in post-ripening cultivars were responsible for nucleosome assembly and protein-DNA complex assembly based on the gene ontology (GO) analysis. However, the only down-expressed genes in non post-ripening cultivars were in the categories metabolism and transportation.
Figure 5. Venn diagram of up- and down-regulated genes.
Gene expression differences reveal zeatin and ethylene regulating development and maturation of pear fruit
Zeatin is a plant hormone, first isolated from maize. It controls many aspects of plant development, such as cell division31 and regulation of environmental stress responses32. Based on the heat map of gene expression at later stages of fruit development, we found cytokinin oxidize (CKX) to be differentially expressed (with log2 fold change from 4 to 8) in all five species (Fig. S1). CKX is the key gene for zeatin synthesis, and is highly conserved in function and structure. Not only did the gene expression pattern of CKX seem to be the same among different pear cultivars (Fig. S1), but also the structure of CKX was conserved between families33.
Ethylene regulates fruit maturity and also responses to stress in plants34. We selected rate-limiting genes in ethylene biosynthesis and key genes for ethylene receptors, though maturity characteristics regulated by ethylene varied between different pear cultivars. Genes corresponding to ethylene biosynthesis had two expressed forms, one pattern was steady increase, and the other pattern steady decreased during fruit development. S-adomet synthetase (SAMS), 1-aminocyclo-propane-1-carboxylic acid synthase (ACS) and 1-aminocyclo-propane -1-carboxylic acid oxidase (ACO) were rate-limiting for ethylene synthesis. Expression of Pbr018549.1, Pbr037756.1, Pbr0001686.1, and Pbr008602.1 (Fig. S2) was continually high for SAMS, but low for Pbr020754.1, Pbr026061.1, Pbr006707.1 and Pbr018549.1 in five cultivars. The expression of two ACS related genes (Pbr030234.1 and Pbr029891.1) was especially high at the stage of fruit maturation. The oriental post-ripening cultivar ‘Nanguoli’ also showed a high expression of ACS at the mature stage (period 6) and fruit senescence stage (period 7), and the same result was verified in previous studies35. In Pyrus pyrifolia, ethylene receptors and ACS have also been identified as regulating pear maturation in the late developmental stages36. We found that most ethylene receptors had steady expression. Ethylene response (ETR), constitutive triple response (CTR) and ethylene insensitive (EIL) are the main ethylene receptors; a heat map of these gene expression was shown in Fig. S2. In addition, RAN (response to antagonist) is a copper transporter necessary for ethylene signaling37. In our study, RAN (Pbr002476.1) was shown to express especially highly at the early stage of all five pear cultivars (Fig. S2).
Gene expression reveals genetic differences related to fruit quality in pear
Stone cell
The stone cell is a special characteristic of pear fruit that can considerably influence fruit quality. Lignin has been found to be the main component of stone cells38. So, in our study, lignin synthesis-related genes were selected for gene expression analysis (Fig. S3). The results showed that several genes such as 4CL (4-coumarate CoA ligase), C3H (p-coumarate 3-hydroxylase), CA5H (coniferylaldehyde 5-hydroxylas), and CAD (cinnamyl alcohol dehydrogenase) had relatively high expression at the early fruit development for all five cultivars (Fig. S3), which was consistent with previous studies showing that stone cells could be detected from sixty days after full blooming39. Genes regulating hydroxycinnamoyl transferases (HCT), which reduce the H-lignin content40, were identified as expressed at the early stage of fruit development (Fig. S3). Evaluation of stone cell content in 304 pear cultivars from five species revealed that the mean stone cell content in P. ussuriensis was the highest4. In this study, it was found that Caffeoyl-CoA o-methyltransferase (CCOMT) related gene (Pbr034039.1) expressed at an especially low level during ‘Nanguoli’ development, with higher expression in the other four cultivars. Based on the lignin pathway analysis from previous study, CCOMT uses the lignin precursor to synthesize G-lignin and S-lignin41. Therefore, we speculated that ‘Nanguoli’ might have relatively high H-type lignin.
Sugar and acid biosynthesis
Sugar and acid are among the most important fruit quality indicators, and the ratio of sugar/acid is a large component of fruit flavor. Sugars are not only energy for metabolism, but also signaling materials for plants. Fructose accounts for the greatest sugar content at maturity for the five pear cultivars, from components of sucrose, glucose, fructose, and sorbitol (Fig. S4). Among these cultivars, ‘Starkrimson’ had high fructose content, exceeding others at the mature stage (Fig. S4). Meanwhile, we found that expression of one hexokinase (HK) related gene in ‘Starkrimson’ was not as high as in the other four species (Fig. S5, Pbr005841.1). Since hexokinase can transfer fructose into fructose-6-phosphate, we speculated that low HK expression in ‘Starkrimson’ induced higher fructose content. Similarly, HK was also able to induce fructose repression in yeast42.
Organic acid is an important factor that can affect fruit flavor. Malate and citrate are the main acids in pear fruit. From malate, citrate, quinate and shikimic acid content at the mature stage, citrate in ‘Starkrimson’ was the highest among all the acids of five species43. It was found that citrate synthase related genes (CS, Pbr004985.1) always had superior expression in ‘Starkrimson’ during fruit development, and could be an important candidate gene for controlling acid content. In a previous report, two Chinese pear accessions were analyzed to find that CS also responds for citrate accumulation44.
Gene networks reflect ethylene synthesis and ethylene receptor interaction
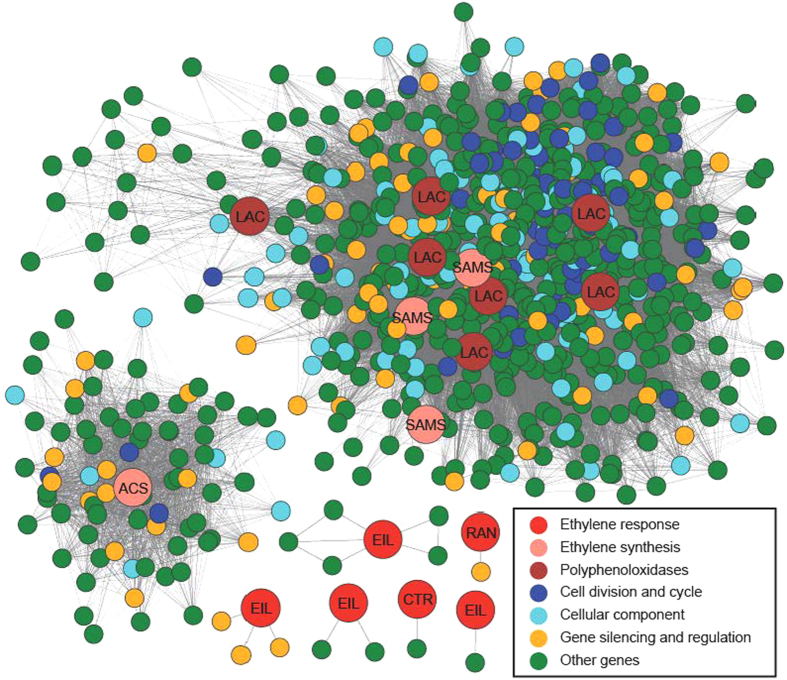
In order to understand which gene networks are active in pear fruit development, we explored the ethylene synthesis and ethylene receptor interaction with the R package. Based on the results presented in Fig. 6, we found that several ethylene synthesis genes and polyphenoloxidase-related genes interacted with each other directly, and indirect relation was reflected between some ethylene synthesis genes and ethylene response genes. Gene interaction analysis illustrated that SAMS (S-adomet synthetase) regulated ethylene synthesis together with LAC (laccase), which had a tight relationship during pear fruit development. In addition, genes controlling cell division, cellular components, gene silencing, and regulation functioned in a network with indirect complex interaction (Fig. 6). It was reported that S-adomet synthetase (SAMS) can synthesize S-adenosyl-methionine (S-AdoMet) from methionine with ATP45. The laccase-related gene, as an ethylene receptor, regulates the downstream ethylene signal transduction pathway46. Researchers found that a cDNA induced by ethylene was homologous to a LAC gene in Rosaceae species of miniature roses47, which was also the verification of the close connection of ethylene and LAC.
Figure 6. Network of ethylene-related genes for pears.
ACS: 1-aminocyclo-propane-1-carboxylic acid synthase; SAMS: S-adomet synthetase; LAC: laccase.
SNP mining for expressed genes in fruit from five different pear cultivars
SNP calling based on transcriptome
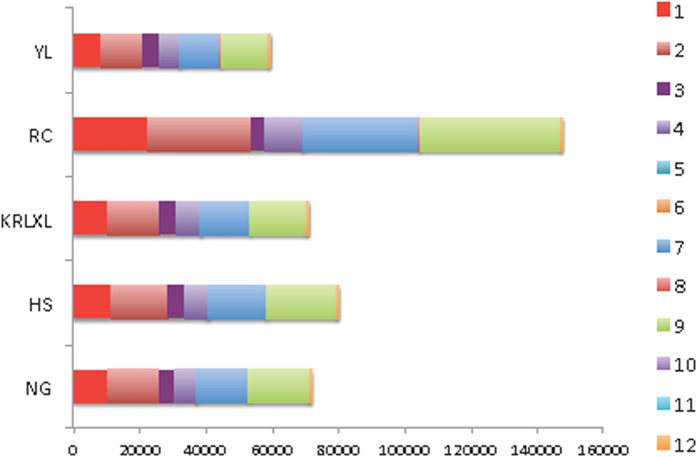
SNPs were detected in ‘Hosui’, ‘Yali’, ‘Kuerlexiangli’, ‘Nanguoli’, and ‘Starkrimson’, based on the RNA-seq data during fruit development in comparison with 'Dangshansuli’ (Pyrus bretschneideri) genome18. SNP calling based on the transcriptome is meaningful, because some SNPs can affect protein function. Oriental pears ‘Hosui’, ‘Yali’, ‘Kuerlexiangli’, and ‘Nanguoli’ had 83543, 62522, 74814 and 75463 filtered SNPs, respectively. We found the most polymorphic cultivar was ‘Starkrimson’, with 154,404 SNPs after filtering (Fig. 7). The nearly doubly high diversity rate represented the many differences between oriental pear ‘Dangshansuli’ and occidental pear of ‘Starkrimson’ on the transcriptome level.
Figure 7. SNP number and effectiveness in five pear species.
X-axis presents SNP effectiveness number, and Y-axis shows five pear species. SNP effectiveness types: 1 SNPs are present in the upstream region of 2 Kb. 2 SNPs are present in the downstream region of 2 Kb. 3 SNPs are present 2 bp around the boundaries of exon and intron. 4 SNPs are in introns caused by selective splicing. 5 SNPs appear in the initiation codon and change the function. 6 SNPs appear in the termination codon and change the function. 7 SNPs appear in the exon and cause synonymous mutation. 8 SNPs are present in the exon and cause premature stop codons. 9 SNPs appear in the exon and cause nonsynonymous mutation. 10 SNPs appear in the termination codon and do not change the function. 11 SNPs appear in the initiation codon and do not change the function. 12 SNPs meet the missing ‘N’ in the genome.
Effectiveness of non-synonymous SNPs
In previous reports, SNPs induced mutants for gene function, for example, peach genotype of flesh melting was affected by SNPs at the melting flesh (M) locus48, and SNP241 in watermelon resulted in a resistant gene mutation on exon49. Arrays of apple50 and peach51 have also been developed based on SNPs, and could be used for genetic mapping, genetic relatedness and marker assisted selection in breeding programs52. In our research, SNPs were classified by the location and effectiveness in genes. Effectiveness included synonymous mutations, non-synonymous mutations, start codon mutations and stop codon mutations, among others, the proportion varied in species (in Fig. 7). Synonymous mutations were overwhelmingly prevalent over other types in all five cultivars. Non-synonymous SNPs in the initiation codon (type a, eg: HS accounted for 0.015% in 79,724 SNPs), SNPs in the termination codon (type b, eg: HS: 0.039%), and SNPs changing protein function by early termination (type d, eg: HS: 0.05%) accounted for only a small portion in the whole SNPs (in Table 2 and Fig. 7), however, these SNPs sometimes can completely change gene function and effect the biology trait significantly.
Table 2. GO slim gene counts in five pear cultivars.
| GO Slim Term | HS | KRLXL | NG | RC | YL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | a | b | c | d | a | b | c | d | a | b | c | d | a | b | c | d | |
| metabolic process | 3 | 8 | 2427 | 8 | 3 | 11 | 2304 | 9 | 0 | 14 | 2422 | 9 | 4 | 17 | 3961 | 9 | 1 | 9 | 1928 | 7 |
| biological process | 3 | 10 | 2178 | 9 | 2 | 8 | 2004 | 5 | 3 | 10 | 2105 | 9 | 8 | 14 | 3342 | 20 | 2 | 11 | 1657 | 8 |
| cellular process | 4 | 8 | 2109 | 8 | 2 | 9 | 1992 | 6 | 2 | 13 | 2044 | 7 | 3 | 15 | 3404 | 7 | 1 | 10 | 1656 | 6 |
| protein metabolic process | 2 | 3 | 932 | 4 | 1 | 4 | 842 | 5 | 0 | 5 | 926 | 2 | 1 | 3 | 1437 | 4 | 1 | 4 | 729 | 3 |
| biosynthetic process | 1 | 4 | 730 | 2 | 1 | 5 | 711 | 2 | 0 | 5 | 725 | 3 | 2 | 6 | 1240 | 0 | 0 | 5 | 578 | 5 |
| nucleobase-containing compound metabolic process | 0 | 2 | 583 | 3 | 0 | 1 | 563 | 1 | 0 | 4 | 567 | 1 | 1 | 5 | 977 | 0 | 0 | 1 | 446 | 2 |
| cellular protein modification process | 1 | 1 | 557 | 2 | 0 | 2 | 495 | 1 | 0 | 3 | 554 | 0 | 0 | 2 | 866 | 3 | 0 | 2 | 426 | 0 |
| transport | 1 | 1 | 413 | 1 | 0 | 0 | 443 | 1 | 3 | 1 | 426 | 2 | 0 | 1 | 702 | 4 | 0 | 1 | 361 | 1 |
| carbohydrate metabolic process | 0 | 0 | 250 | 1 | 0 | 0 | 211 | 0 | 0 | 0 | 235 | 2 | 0 | 2 | 377 | 0 | 0 | 0 | 191 | 0 |
| response to stress | 1 | 3 | 222 | 2 | 0 | 4 | 209 | 0 | 0 | 1 | 213 | 1 | 0 | 3 | 322 | 1 | 0 | 2 | 200 | 0 |
| lipid metabolic process | 1 | 0 | 166 | 0 | 1 | 2 | 148 | 0 | 0 | 1 | 158 | 1 | 1 | 1 | 249 | 1 | 1 | 1 | 112 | 0 |
| catabolic process | 0 | 0 | 147 | 1 | 0 | 1 | 129 | 0 | 0 | 0 | 141 | 1 | 0 | 2 | 223 | 1 | 0 | 0 | 117 | 0 |
| cellular component organization | 0 | 0 | 144 | 1 | 0 | 0 | 134 | 0 | 0 | 1 | 130 | 1 | 0 | 0 | 208 | 1 | 0 | 0 | 111 | 0 |
| translation | 0 | 2 | 128 | 0 | 0 | 2 | 117 | 2 | 0 | 2 | 123 | 1 | 0 | 1 | 189 | 0 | 0 | 2 | 85 | 2 |
| cell communication | 1 | 1 | 122 | 1 | 0 | 1 | 122 | 1 | 0 | 1 | 125 | 0 | 0 | 3 | 216 | 1 | 0 | 2 | 109 | 0 |
| signal transduction | 1 | 1 | 108 | 0 | 0 | 1 | 110 | 0 | 0 | 1 | 105 | 0 | 0 | 3 | 184 | 0 | 0 | 2 | 93 | 0 |
| DNA metabolic process | 0 | 0 | 81 | 1 | 0 | 0 | 79 | 0 | 0 | 1 | 73 | 0 | 0 | 0 | 115 | 0 | 0 | 0 | 63 | 0 |
| generation of precursor metabolites and energy | 0 | 0 | 53 | 0 | 0 | 0 | 36 | 0 | 0 | 0 | 47 | 0 | 0 | 1 | 77 | 0 | 0 | 0 | 38 | 0 |
| response to biotic stimulus | 0 | 1 | 13 | 0 | 0 | 1 | 6 | 0 | 0 | 1 | 8 | 0 | 0 | 1 | 14 | 0 | 0 | 1 | 7 | 0 |
| pollination | 0 | 0 | 13 | 1 | 0 | 0 | 11 | 1 | 0 | 0 | 19 | 0 | 0 | 0 | 31 | 1 | 0 | 0 | 15 | 0 |
| pollen-pistil interaction | 0 | 0 | 13 | 1 | 0 | 0 | 11 | 1 | 0 | 0 | 19 | 0 | 0 | 0 | 31 | 1 | 0 | 0 | 15 | 0 |
a: SNPs appear in the initiation codon and change the amino acid function; b: SNPs appear in the termination codon and change the amino acid function; c: SNPs appear in the exon and cause amino acid non-synonymous mutation; d: SNPs appear before the termination codon and change the amino acid function by early termination.
Based on the gene ontology of the predicted SNP-induced varied genes in Table 2, we found that genes responsible for transport in ‘Nanguo’ had the largest number of mutations in the start codon regions, and GO slim term of response to stress was prone to mutate in the stop codons comparing with other GO terms. We demonstrated all the SNPs, which induced mutations in start codon, stop codon and premature stop codon in Table S1. Conserved function domains (https://www.ncbi.nlm.nih.gov/cdd) were analyzed based on these genes (Table S2). Many SNPs are related to fruit development. We found a SNP mutation in the start codon of Pbr038562.1, which had the functional domain of “ethylene responsive element binding protein (EREBP)” in cultivar of ‘Starkrimson’. In Arabidopsis, EREBPs can interact with the 11 bp GCC box in the promoter of target genes and activate downstream ethylene responses53. Furthermore, in the stop codon region, the SNP from ‘Kuerlexiangli’, ‘Nanguo’ and ‘Starkrimson’ might influence Pbr026622.1 with domains from the DUF647 superfamily that has functions in auxin transport. A study found that a member of the conserved DUF647 protein family plays an essential role in the regulation of polar auxin transport by maintaining the proper level of auxin transporters on the plasma membrane54. The predicted non-synonymous SNPs provided valuable information for candidate genes associated with fruit development, even though more gene function experiment should be performed to verify the SNP effectiveness derived gene mutations.
RNA expression verification by q-PCR on ‘Starkrimson’
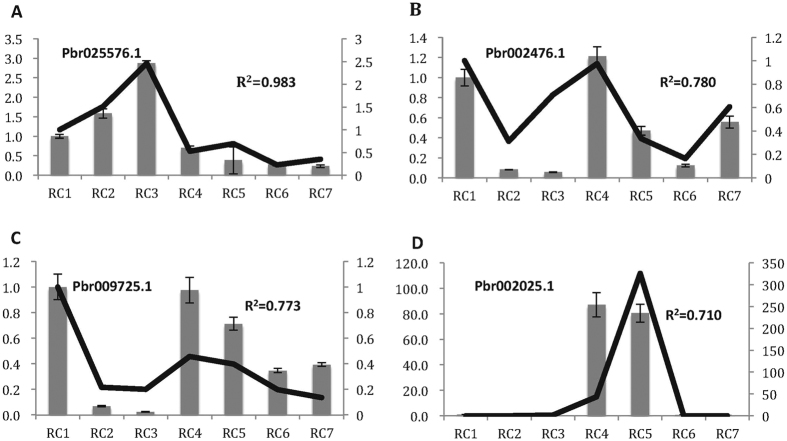
We selected five genes to verify the RNA-seq gene expression data from seven stages of fruit development for ‘Starkrimson’ by q-PCR. Each gene was repeated in three biological samples with the first period’s expression as a standard level. Data for the genes β-amyrin synthesis (Pbr025576.1), ethylene receptor (Pbr002476.1), ABA synthesis (Pbr009725.1), and zeatin synthesis (Pbr002025.1) are shown in Fig. 8. Correlations of the RPKM of four genes and the relative qPCR expression were 0.983 (Pbr025576.1), 0.780 (Pbr002476.1), 0.773 (Pbr009725.1), and 0.710 (Pbr002025.1), throughout fruit development, respectively. The ethylene receptor showed fluctuating expression. We found that the ABA synthesis related-gene had high expression at the early stage of fruit development, when xanthoxin is being converted to ABA55. The zeatin-related gene was found to have a peak in the middle stage of fruit metabolism in ‘Starkrimson’, but was highly expressed in the early stage of fruit development in blueberry56.
Figure 8. Comparison of RNA-seq based gene expression and q-PCR verification.
Figures represent expression of four genes for seven fruit developmental stages of ‘Red Clapp’s Favorite’. X-axis represents the fruit developmental stages. Y-axis on the left shows the relative gene expression levels of q-PCR by bar charts, and the Y-axis on right illustrated the relative values of RPKM with line charts. The RPKM of Y-axis on the right were unified by dividing the first stage’s gene expression value in (A–C).
Methods
Pear Cultivar selection
Cultivars of ‘Hosui’, ‘Yali’, ‘Kuerlexiangli’, ‘Nanguoli’ and ‘Starkrimson’ originate from different species, and have a range of maturity characteristics. We collected pear fruits from the Jiangpu Farm of Nanjing Agricultural University. A total of seven developmental stages were collected from the fruit-setting period, which was 15 days after full blooming (15 DAB, period 1). Pear fruits were selected from the physiological fruit dropping stage (30 DAB, period 2), fruit rapid enlargement stage (55 DAB, period 3), a month after fruit enlargement stage (85 DAB, period 4), pre-mature stage (115 DAB, period 5), mature stage (varied according to species, period 6) and fruit senescence stage (period 7). Then pear fruit flesh was cut into small pieces and stored in the freezer at −80 °C.
RNA sequencing and sequence mapping
A total of 35 samples’ (five species X seven stages) RNA was extracted from fruit flesh. Illumina standard mRNA-Seq Prep Kit (TruSeq RNA and DNA Sample Preparation Kits version 2) was used for construction of RNA sequencing libraries. Single end RNA-seq data were generated with length of 49 bp. Reads were filtered and trimmed, and then they were mapped onto ‘Dangshansuli’ (Pyrus bretschneideri) CDS sequences by software of SOAPaligner57.
Gene expression and clustering analysis
The gene expression profile was presented by in-house perl scripts using RPKM algorithm58. The Java-based program STEM28 was used for clustering to build gene expression model. All of the mapping units with at least five expression sets were used to map onto the gene model.
Gene co-expression analysis
WGCNA (weighted gene co-expression network analysis) package59 was used to find clusters (modules) of highly correlated genes and construct co-expression networks. Then Cytoscape (http://www.cytoscape.org) was used to visualize the correlation relationships between specified genes.
SNP calling
Burrows-Wheeler Alignment (BWA)60 was used for RNA-seq read mapping (-n 1 -o 1 -e 0). After files were aligned for all 35 samples, bam format files were merged together from seven stages of the same cultivar by samtools61. SNP calling was performed with the module ‘mpileup’ in samtools. Effectiveness of the SNPs was analyzed with an in-house perl script (https://github.com/Sunhh/NGS_data_processing).
Sugar content measurements
Pear fruit sugars including sucrose, glucose, fructose and sorbitol were measured using High Performance Liquid Chromatography (HPLC). Sugars were extracted from pear flesh by grounding, and then were dissolved and filtered through a SEP-C18 cartridge (Waters, WAT021515) and Sep-Pak filter. Waters 1525 system (Waters, Shanghai, China) was used to measure sugars. The column was 6.5 mm × 300 mm, Inner Diameter, 10 um particle size (Waters), with a guard column cartridge of Sugar-pak 1 Guard-Pak Holder and Insert (Waters). Column temperature was 85 °C, and the reference cell temperature was 35 °C.
RT-PCR validation
Several expressed genes were selected for verification by quantitative RT-PCR (qRT-PCR) on ‘Starkrimson’. Coding sequences of the genes were acquired from the pear genome project (http://peargenome.njau.edu.cn). Primers for the candidate genes were designed (Table S1) with the online software Primer 3 (http://simgene.com/Primer3). The LightCycler 480 SYBR GREEN Master (Roche, USA) system was used, with GAPDH as the house-keeping gene. Reactions of target genes and reference genes were performed in triplicate in a total volume of 20 μl. PCR reactions were carried out via an initial incubation at 95 °C for 10 min and at 95 °C for 15 s, and then cycled at 60 °C for 15 s, and 72 °C for 20 s for 40 cycles, and final extension at 72 °C for 30 min.
Additional Information
How to cite this article: Zhang, M.-Y. et al. Distinct transcriptome profiles reveal gene expression patterns during fruit development and maturation in five main cultivated species of pear (Pyrus L.). Sci. Rep. 6, 28130; doi: 10.1038/srep28130 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31171928), the earmarked fund for the China Agriculture Research System (CARS-29), the Science Foundation for Distinguished Young Scientists in Jiangsu Province (BK20150025) and the Ministry of Education Program for New Century Excellent Talents in University (NCET-13-0864).
Footnotes
Author Contributions Experiment design: J.W. Data analyzed: M.Y.Z. and H.H.S. Experiment performed: C.X. and M.F.Q. Material preparation: S.L.Z. and L.L.X. Wrote the paper: M.Y.Z. and J.W.
References
- Rom R. C. & Carlson R. F. Rootstocks for fruit crops. (Wiley, 1987). [Google Scholar]
- Vavilov N. I. The origin, variation, immunity and breeding of cultivated plants. Soil Science 72, 482 (1951). [Google Scholar]
- Kimura T. et al. Identification of Asian pear varieties by SSR analysis. Breeding Science 52, 115–121 (2002). [Google Scholar]
- CAO Y.-f., Tian L.-m., LI L.-l. & GAO Y. Comparison Studies on the Stone Cell Content in Flesh of Pear Cultivars. Acta Horticulturae Sinica 37, 1220–1226 (2010). [Google Scholar]
- YAO G.-f. et al. Characteristics of Components and Contents of Soluble Sugars in Pear Fruits from Different Species. Scientia Agricultura Sinica 43, 4229–4237 (2010). [Google Scholar]
- Qin G. et al. Evaluation of the volatile profile of 33 Pyrus ussuriensis cultivars by HS-SPME with GC-MS. Food Chem 134, 2367–2382 (2012). [DOI] [PubMed] [Google Scholar]
- Fonseca S., Monteiro L., Barreiro M. G. & Pais M. S. Expression of genes encoding cell wall modifying enzymes is induced by cold storage and reflects changes in pear fruit texture. J Exp Bot 56, 2029–2036 (2005). [DOI] [PubMed] [Google Scholar]
- Murayama H. et al. Effect of girdling above the abscission zone of fruit on ‘Bartlett’ pear ripening on the tree. J Exp Bot 57, 3679–3686 (2006). [DOI] [PubMed] [Google Scholar]
- Hiwasa K. et al. European, Chinese and Japanese pear fruits exhibit differential softening characteristics during ripening. J Exp Bot 55, 2281–2290 (2004). [DOI] [PubMed] [Google Scholar]
- Nashima K. et al. Microarray analysis of gene expression patterns during fruit development in European pear (Pyrus communis). Sci Hortic-Amsterdam 164, 466–473 (2013). [Google Scholar]
- Zhang H. P. et al. The role of sucrose-metabolizing enzymes in pear fruit that differ in sucrose accumulation. Acta Physiol Plant 36, 71–77 (2014). [Google Scholar]
- Yamada K. et al. Cloning of two isoforms of soluble acid invertase of Japanese pear and their expression during fruit development. J Plant Physiol 164, 746–755 (2007). [DOI] [PubMed] [Google Scholar]
- Tao S. T., Khanizadeh S., Zhang H. & Zhang S. L. Anatomy, ultrastructure and lignin distribution of stone cells in two Pyrus species. Plant Sci 176, 413–419 (2009). [Google Scholar]
- Yan C. C. et al. Stone cell distribution and lignin structure in various pear varieties. Sci Hortic-Amsterdam 174, 142–150 (2014). [Google Scholar]
- Velasco R. et al. The genome of the domesticated apple (Malus x domestica Borkh.). Nature genetics 42, 833–839 (2010). [DOI] [PubMed] [Google Scholar]
- Shulaev V. et al. The genome of woodland strawberry (Fragaria vesca). Nature genetics 43, 109–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. et al. The genome of Prunus mume. Nature Communications 3, 1318 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Research 23, 396–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Peach Genome, I. et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nature genetics 45, 487–494 (2013). [DOI] [PubMed] [Google Scholar]
- Otto D. et al. The columnar mutation (“Co gene”) of apple (Malus x domestica) is associated with an integration of a Gypsy-like retrotransposon. Mol Breeding 33, 863–880 (2014). [Google Scholar]
- Lombardo V. A. et al. Metabolic Profiling during Peach Fruit Development and Ripening Reveals the Metabolic Networks That Underpin Each Developmental Stage. Plant Physiol 157, 1696–1710 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S. et al. Characterization of Transcriptional Complexity during Berry Development in Vitis vinifera Using RNA-Seq. Plant Physiol 152, 1787–1795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S. L. et al. Transcriptome Analysis of Japanese Pear (Pyrus pyrifolia Nakai) Flower Buds Transitioning Through Endodormancy. Plant and Cell Physiology 54, 1132–1151 (2013). [DOI] [PubMed] [Google Scholar]
- Nashima K. et al. Transcriptome analysis of flower receptacles of the European pear (Pyrus communis L.) ‘La France’ and its giant fruit sport using next-generation sequencing technology. J Hortic Sci Biotech 89, 293–300 (2014). [Google Scholar]
- Zouari I. et al. From root to fruit: RNA-Seq analysis shows that arbuscular mycorrhizal symbiosis may affect tomato fruit metabolism. Bmc Genomics 15, 221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbacho J. et al. Transcriptomic Events Involved in Melon Mature-Fruit Abscission Comprise the Sequential Induction of Cell-Wall Degrading Genes Coupled to a Stimulation of Endo and Exocytosis. Plos One 8, e58363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O. & Botstein D. Cluster analysis and display of genome-wide expression patterns. P Natl Acad Sci USA 95, 14863–14868 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J. & Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. Bmc Bioinformatics 7, 191 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K. Q. et al. Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). Bmc Genomics 13, 10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiwasa K. et al. Ethylene is required for both the initiation and progression of softening in pear (Pyrus communis L.) fruit. J Exp Bot 54, 771–779 (2003). [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Huntley R., Jacqmard A. & Murray J. A. H. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544 (1999). [DOI] [PubMed] [Google Scholar]
- Ha S. et al. Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci 17, 172–179 (2012). [DOI] [PubMed] [Google Scholar]
- Immanen J. et al. Characterization of cytokinin signaling and homeostasis gene families in two hardwood tree species: Populus trichocarpa and Prunus persica. Bmc Genomics 14, 885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey R. F. & Binder B. M. How plants sense ethylene gas - The ethylene receptors. J Inorg Biochem 133, 58–62 (2014). [DOI] [PubMed] [Google Scholar]
- Huang G. H. et al. Comparative Transcriptome Analysis of Climacteric Fruit of Chinese Pear (Pyrus ussuriensis) Reveals New Insights into Fruit Ripening. PLos One 9, e107562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H. Y. et al. PpACS1b, a pear gene encoding ACC synthase, is regulated during fruit late development and involved in response to salicylic acid. Sci Hortic-Amsterdam 164, 602–609 (2013). [Google Scholar]
- Binder B. M., Rodriguez F. I. & Bleecker A. B. The Copper Transporter RAN1 Is Essential for Biogenesis of Ethylene Receptors in Arabidopsis. Journal of Biological Chemistry 285, 37263–37270 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. W. The course of stone cell formation in pear fruits[J]. Plant physiology. Plant Physiol 10, 587 (1935). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. H. & Lee S. H. Distribution of stone cell in Asian, Chinese, and European pear fruit and its morphological changes. Journal of Applied Botany and Food Quality 86, 185–189 (2013). [Google Scholar]
- Besseau S. et al. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19, 148–162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K. et al. Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol 117, 761–770 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWinde J. H. et al. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur J Biochem 241, 633–643 (1996). [DOI] [PubMed] [Google Scholar]
- YAO G.-f. et al. Characteristics of Components and Contents of Organic Acid in Pear Fruits from Different Cultivated Species Acta Horticulturae Sinica 41, 755–764 (2014). [Google Scholar]
- Lu X. P. et al. Identification of organic acid-related genes and their expression profiles in two pear (Pyrus pyrifolia) cultivars with difference in predominant acid type at fruit ripening stage. Sci Hortic-Amsterdam 129, 680–687 (2011). [Google Scholar]
- McManus M. T. The plant hormone ethylene. (Wiley-Blackwell, 2012). [Google Scholar]
- Nergi M. A. D. & Ahmadi N. Effects of 1-MCP and ethylene on postharvest quality and expression of senescence-associated genes in cut rose cv. Sparkle. Sci Hortic-Amsterdam 166, 78–83 (2014). [Google Scholar]
- Ahmadi N., Mibus H. & Serek M. Characterization of ethylene-induced organ abscission in F1 breeding lines of miniature roses (Rosa hybrida L.). Postharvest Biol Tec 52, 260–266 (2009). [Google Scholar]
- Morgutti S. et al. Changes in endopolygalacturonase levels and characterization of a putative endo-PG gene during fruit softening in peach genotypes with nonmelting and melting flesh fruit phenotypes. New Phytol 171, 315–328 (2006). [DOI] [PubMed] [Google Scholar]
- Ling K. S. et al. Non-synonymous single nucleotide polymorphisms in the watermelon eIF4E gene are closely associated with resistance to Zucchini yellow mosaic virus. Theoretical and Applied Genetics 120, 191–200 (2009). [DOI] [PubMed] [Google Scholar]
- Kumar S. et al. Genomic Selection for Fruit Quality Traits in Apple (Malus x domestica Borkh.). Plos One 7, e36674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I. et al. Development and Evaluation of a 9K SNP Array for Peach by Internationally Coordinated SNP Detection and Validation in Breeding Germplasm. Plos One 7, e35668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco L. et al. Development and Validation of a 20 K Single Nucleotide Polymorphism (SNP) Whole Genome Genotyping Array for Apple (Malus x domestica Borkh). Plos One 9, e110377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. L. C., Li H. & Ecker J. R. Ethylene biosynthesis and signaling networks. Plant Cell 14, S131–S151 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. et al. Root Ultraviolet B-Sensitive1/Weak Auxin Response3 Is Essential for Polar Auxin Transport in Arabidopsis. Plant Physiol 162, 965–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J., Flors V. & Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends Plant Sci 14, 310–317 (2009). [DOI] [PubMed] [Google Scholar]
- Zifkin M. et al. Gene Expression and Metabolite Profiling of Developing Highbush Blueberry Fruit Indicates Transcriptional Regulation of Flavonoid Metabolism and Activation of Abscisic Acid Metabolism. Plant Physiol 158, 200–224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Q. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009). [DOI] [PubMed] [Google Scholar]
- Mortazavi A. et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Langfelder P. & Horvath S. WGCNA: an R package for weighted correlation network analysis. Bmc Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.