Abstract
Characterizing Epstein–Barr virus (EBV) dynamics in asymptomatic immunocompetent persons provides a baseline for defining quantitative thresholds associated with EBV disease. Studying latent membrane protein (LMP)-1 sequence variation over time could establish the rates of reactivation and superinfection, and also trace transmission. Twelve asymptomatic adult subjects were evaluated prospectively nine times over 6 months. EBV serum antibodies were measured by enzyme immunoassay. EBV DNA in oral and whole-blood samples was quantitated by real-time (TaqMan) PCR and analyzed for LMP-1 sequence variability. All 11 antibody positive subjects had EBV DNA detected in their oral compartment at least once during the 6-month study. The quantities ranged from 1.70 to 4.91 log10 copies EBV per ml of oral cell pellet. One subject was continuously viremic for 79 days. Overall, EBV DNA was detected in 63 (24%) of 260 samples from 11 antibody-positive subjects and in 0/27 samples from an antibody-negative subject. The quantities in positive samples ranged from 1.7 to 4.9 log10 copies EBV per ml. EBV LMP-1 gene sequence variations in subjects were constant over time regardless of the compartment sampled. Subjects 18–30 years old had EBV DNA detected more frequently than subjects >30 years old (38/108 positive samples versus 25/152; P<0.001). In conclusion, EBV DNA shedding is common in asymptomatic adults. The younger adults shed more frequently, which may reflect a shorter time from their primary EBV infection to sampling. The LMP-1 sequence analysis method employed here could be used to trace person-to-person transmission because patterns remained almost identical over time.
Epstein–Barr virus (EBV), like all members of the human herpesvirus family, persists in the host after primary infection and may reactivate at any time. As EBV may replicate without causing apparent harm, it is important to be able to distinguish asymptomatic infection from impending EBV disease, which is troublesome in teenagers and young adults1 and is especially serious in the immunocompromised host.2 There are few data about EBV activity in immunocompetent, asymptomatic adults over time.3, 4 Characterizing viral–host interactions in healthy adults would provide a baseline for defining quantitative thresholds associated with EBV disease. Tracing the stability or variability of EBV strains over time could establish the rate of reactivation as compared with superinfection and could also be used to trace transmission. For these reasons, we performed a 6-month intensive prospective surveillance study in healthy immunocompetent adults.
Results
Characteristics of the study participants
Twelve consecutive volunteers were enrolled and followed for 6 months. Their characteristics on enrollment are given in Table 1. The subjects completed 97 (90%) of their 108 scheduled visits during which they provided 287 samples for EBV testing (median, 25 samples per subject; mean, 24 samples per subject) (Table 2). Eleven subjects had been previously infected by EBV and one volunteer was EBV naive. Three of the 11 antibody-positive subjects had positive viral capsid antigen (VCA) IgM enzyme immunoassay (EIA) indices (1.26–5.88). Their antibody profiles reflected recurrent rather than primary infection because all of them had robust EBV nuclear antigen 1 (EBNA-1) antibody levels (Table 3). Two of these subjects remained VCA IgM positive during the entire trial, whereas one subject became VCA IgM negative after 6 months.
Table 1. Characteristics of the 12 subjects on enrollment.
| Characteristic | |
|---|---|
| Age in years—median (mean, range) | 28.4 (33.7, 20–64) |
| Women—no. (%) | 8 (75) |
| History of infectious mononucleosis (%) | 2 (17)a |
| EBV infection status defined by profile of VCA IgG, VCA IgM, EBNA-1 IgG antibodies | |
| Past (positive for VCA and EBNA-1 IgG only) | 8 |
| Recurrent (positive for all three antibodies) | 3 |
| Naive (negative for all three antibodies) | 1 |
Abbreviations: EBV, Epstein–Barr virus; EBNA-1, EBV nuclear antigen 1; VCA, viral capsid antigen.
One of the two cases was laboratory confirmed.
Table 2. EBV DNA shedding pattern, proportion of positive samples and amount of EBV DNA detected by age group in antibody-positive subjects.
| Subjects 18–30 years old | Subjects >30 years old | All subjects | |
|---|---|---|---|
| Number of subjects | 5 | 6 | 11 |
| Mean age (range) | 22.5 (20–25) | 44.3 (30–64) | 34.4 (20–64) |
| Antibody profile | |||
| Past EBV infection | 3 | 5 | 8 |
| Recurrent EBV infection | 2 | 1 | 3 |
| Subjects positive at least once | 5 | 6 | 11 |
| Subjects viremic | 1 | 0 | 1 |
| Proportion of positive samples | |||
| Oral cells | 19/36 (53%)a | 15/51 (29%)a | 34/87 (39%) |
| Oral supernatant | 12/36 (33%) | 10/51 (20%) | 22/87 (25%) |
| Whole blood | 7/36 (19%)b | 0/50 (0%)b | 7/86 (8%) |
| All samples | 38/108 (35%)c | 25/152 (16%)c | 63/260 (24%) |
| Median log10copies EBV DNA in positive samples (mean, range) | |||
| Oral cells | 3.69 (3.49; 1.70–4.91) | 3.08 (2.92; 1.70–3.88) | 3.50 (3.24; 1.70–4.91) |
| Oral supernatant | 3.30 (3.47; 3.00–4.25) | 3.50 (3.51; 3.00–4.25) | 3.36 (3.49; 3.00–4.25) |
| Whole blood | 3.30 (3.39; 3.08–3.79) | — | 3.30 (3.39; 3.08–3.79) |
Abbreviation: EBV, Epstein–Barr virus.
P=0.04, Fisher's exact test.
P=0.002, Fisher's exact test.
P<0.001, Fisher's exact test.
Table 3. EBV antibody profile of subjects positive for VCA IgM antibody on enrollment.
| Subject (age and sex) |
Enrollment |
Final visita |
||||
|---|---|---|---|---|---|---|
| VCA IgM | VCA IgG | EBNA-1 IgG | VCA IgM | VCA IgG | EBNA-1 IgG | |
| 205 (25.1 F) | 1.26 | 5.36 | 3.99 | 0.64 | 5.18 | 4.93 |
| 208 (30.3 F) | 5.88 | 5.41 | 4.27 | 2.95 | 5.49 | 7.51 |
| 212 (20.7 F) | 2.32 | 2.89 | 6.45 | 1.81 | 3.47 | 3.45 |
Abbreviations: EBNA-1, EBV nuclear antigen 1; F, female; VCA, viral capsid antigen.
EIA index interpretation: negative, <0.90; equivocal, 0.90–1.09; and positive, ⩾1.10.
205, study day, 168; 208, study day 168; 212 study day, 90.
EBV DNA shedding
EBV DNA was present in 63 (24%) of 260 samples from the 11 antibody-positive adults and in 0 of 27 samples from the EBV naive subject. All antibody-positive participants had EBV DNA detected in at least one oral sample. One subject who was positive for VCA and EBNA IgG antibodies but negative for VCA IgM antibodies throughout the 6-month study had EBV DNA recovered from 17 of 24 samples including 7 of 8 whole-blood specimens. She had EBV DNAemia on seven consecutive visits spanning 79 days (quantitative range, 3.1–3.8 log10 copies EBV per ml of whole blood). Considering all subjects, the oral cells contained the highest proportion of detectable EBV DNA, followed by the oral supernatant and whole blood (Table 2). The subjects fit into two age groups: 18 to 30 years old (five subjects) and >30 years old (six subjects). The younger group was more likely to have EBV DNA recovered than the older group and the differences in the proportions of positive samples in the oral cell compartment and whole blood were statistically significant (Table 2). There were no significant differences between older and younger subjects in terms of the average, minimum or maximum quantities of EBV DNA.
EBV strain analysis
We chose to compare nucleotide differences in the C terminus of the EBV latent membrane protein (LMP)-1 gene to characterize our subjects' EBV strains because the region has been shown to be highly variable5 and this approach was used successfully by van Kooij et al.6 to define patterns of sequence variation in healthy individuals and patients with postransplant lymphoproliferative disorder.
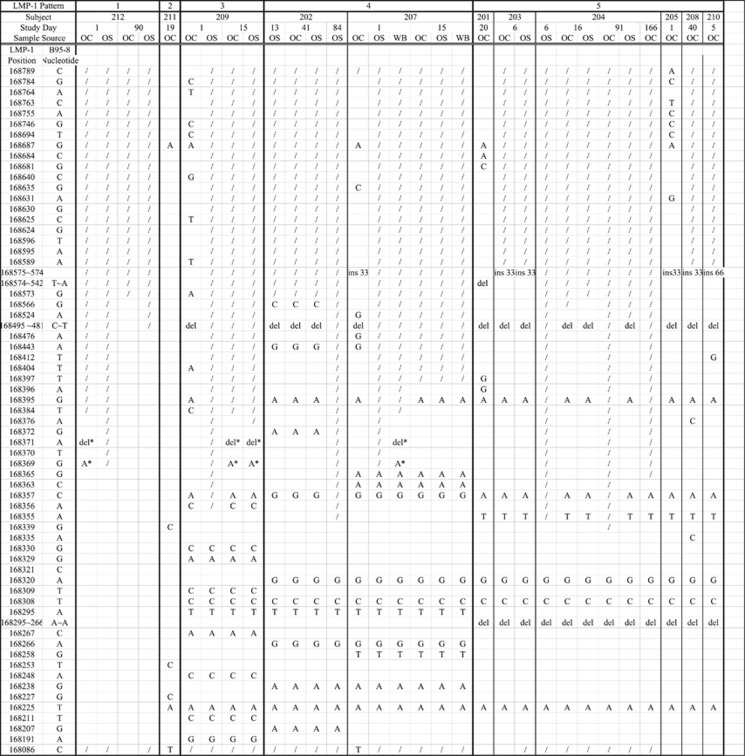
LMP-1 sequence variation analysis was performed on 31 selected samples from the 11 antibody-positive subjects (Figure 1). For subjects who shed EBV on more than one visit, samples were chosen as far apart in time as possible. Specimens containing as few as 160 copies per ml of sample could be sequenced. We were able to identify the LMP-1 sequences from all antibody-positive patients. The EBV LMP-1 sequences of the subjects fit into five patterns. Pattern 5 was the most common, being found in 6 of the 11 volunteers. Six subjects had LMP-1 sequences analyzed from more than one compartment or time point. Of the 26 samples sequenced from these subjects, 23 (88%) were identical and 3 contained only 2 positional mismatches. Ten subjects harbored EBV EBNA-2 type 1 virus and one (subject 210) had EBV EBNA-2 type 2 virus.
Figure 1.
Five LMP-1 sequence patterns of EBV strains from 11 subjects. The symbol / indicates regions where nucleotides could not be determined. The symbol * denotes positional mismatches from a subject's other samples. Letters indicate nucleotides that differ from the B95-8 sequence. OC, oral cell compartment sample; OS, oral supernatant sample; WB, whole-blood sample; ins, insertion; del, deletion.
Discussion
All 11 healthy adults previously infected by EBV had EBV DNA detected in at least one oral sample during the 6-month observation period. The five subjects in the 18–30-year-old group (age range, 20–25 years) had significantly more EBV-positive samples than the 6 subjects over 30 (age range, 30.3–64 years). Also, the only viremic subject was in the younger age group. Interestingly, she was negative for VCA IgM antibodies throughout the 6-month study. We speculate that the frequency of EBV DNA shedding in asymptomatic immunocompetent hosts steadily declines with time after primary infection. Thus, the greater EBV shedding among younger subjects may reflect a shorter time from their primary EBV infection to sampling. Although the quantity of EBV DNA in these asymptomatic subjects was generally less than that reported in patients with infectious mononucleosis,1 chronic active EBV infection7 or transplant recipients.8 However, 4.0 log10 copies EBV per ml were present in 6 (18%) of 34 oral cell pellet samples, and in 3 (14%) of 22 oral supernatant samples. The viremic subject had >3.0 log10 copies EBV per ml in seven consecutive blood samples over 79 days. This should be considered when attempting to associate the quantity of EBV DNA in certain compartments with its pathogenic potential and evaluating the need for a therapeutic intervention.
Our findings are consistent with two previous reports of EBV shedding in healthy antibody-positive subjects.3, 4 However, our study differs in that our subjects were sampled more frequently (nine times in 6 months), there was a significantly higher prevalence of samples positive for EBV DNA among younger subjects, and there was no difference in the proportion of positive samples during any of the 6 months studied (data not shown).
EBV strain identification based on the LMP-1 sequence patterns revealed a single EBV strain in every healthy antibody-positive individual. Of the 11 subjects tested, five unique patterns of LMP-1 were identified, ranging from variants similar to the B95-8 strain to strains that included the 30-bp deletion (Figure 1). For subjects with EBV DNA detected in multiple compartments, the LMP-1 sequences were nearly identical and these sequences did not change over time. We postulate that the LMP-1 sequence identified for each subject was the dominant strain.
Although we have not detected mixtures with our method, other groups have regularly reported multiple strain infections from asymptomatic carriers with a varying degree of prevalence. Walling et al.9 detected multiple strains in two of nine healthy adults using a direct LMP-1 amplification genotyping method. Within these two patients, they noted compartmental differences between the oral cavity and peripheral blood, as well as temporal changes. Using a highly sensitive heteroduplex tracking assay of the LMP-1 gene locus, Sitki-Green et al.10 found mixtures in 20 of 20 asymptomatic carriers. As many as six different variants were reported in some subjects. Sixteen of the 20 subjects exhibited differences between their oral wash and peripheral blood at a given time. However, other investigators such as Meijer et al.11 have not observed EBV strain differences in body compartments. Most likely, the differences are methodological.
The importance of minority variants in EBV epidemiology has yet to be defined.12 It could be that minority variants evolve from an individual's viral swarm over time and do not signify acquisition of exogenous strains. We believe that detecting the dominant strain is advantageous for providing information regarding the source of the subject's EBV infection. The similarity of the LMP-1 sequence pattern in different body compartments in the same subject and its apparent stability over time suggest that our method of LMP-1 sequencing could prove to be a powerful tool for investigating person-to-person transmission. As an example, we recently reported that this method confirmed donor to recipient transmission of EBV during pediatric kidney transplantation.13
Methods
Clinical trial design
Healthy consecutively acquired volunteers ⩾18 years old were seen nine times over a 6-month period (twice weekly for 3 weeks, and once at 6, 12 and 24 weeks). A brief medical history was obtained at the time of enrollment. At every visit, venous blood and oral washes were collected as previously described.14 This study was approved by the Research Subjects Protection Program of the University of Minnesota, and subjects gave written informed consent before participation.
Quantitation of EBV DNA in oral washes and blood
A real-time TaqMan PCR assay was used to quantitate EBV in whole blood, oral cells and oral supernatant samples collected at every visit.14 The amplicon for the quantitative EBV assay was a 71-bp portion of the EBNA-1 gene. The forward primer was: 5′-GAC TGT GTG CAG CTT TGA CGA T-3′ the reverse primer was: 5′-CGG CAG CCC CTT CCA-3′ and the probe was: 5′-(FAM) TAG ATT TGC CTC CCT GGT TTC CAC CTA TG-(TAMRA)-3′. Quantitative EBV data were expressed as viral copies per ml of oral wash or whole blood. The reliable limit of detection of the assay was 4 copies/reaction, which equates to 16 copies per ml for the oral wash fluid-derived cell pellet and 80 copies per ml for the oral wash fluid-derived supernatant and the whole blood.
EBV antibody tests and classification of EBV infection
EBV antibody assays were performed on serum samples collected on enrollment and at 6, 12 and 24 weeks using commercially available enzyme immunoassay (EIA) kits and a MAGO Plus Automated EIA Processor (Diamedix Corporation, Miami, FL, USA). Results were expressed as the index value, which was the absorbance of the patient's sample divided by the mean absorbance of three replicate dilutions of a weakly positive control supplied by the manufacturer. The results were classified according to their index value as: negative, <0.90; equivocal, 0.90–1.09; and positive, ⩾1.10.
The stage of EBV infection was defined by the following antibody profiles: past infection, positive for IgG antibodies against both EBV VCA and EBNA-1, and negative for IgM antibodies against VCA; recurrent infection, positive for VCA IgM, VCA IgG, and EBNA-1 IgG; and naive to EBV, negative for VCA IgM, VCA IgG, and EBNA-1 IgG.
LMP-1 sequence variation and EBNA-2 typing
The primers used for determining LMP-1 sequence variation and EBNA-2 typing are listed in Table 4. The LMP-1 primers were those of van Kooij et al.6 with modifications. The EBNA-2 primers were those described by Higa et al.15 with minor modifications. PCR was performed in an ABI 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA). The program consisted of 1 cycle at 95 °C for 10 min, 40 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 60 s, followed by 72 °C for 10 min after which the temperature was reduced to 4 °C and held. PCR products were run in a 1% agarose gel. The Namalwa cell line (ATCC CRL1422), which contains two integrated copies of EBV per cell,16 was used as the positive control and the negative control contained no template. EBNA-2 genotyping was based on the nested PCR product size: type 1 was 497 bp and type 2 was 162 bp. LMP-1 PCR products were purified with a QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA). Purified PCR products were sequenced by the University of Minnesota's Advanced Genetic Analysis Center using an ABI 3100 DNA sequencer and Big Dye chemistry (Applied Biosystems, Foster City, CA, USA). Chromatograms were compared with the EBV B95-8 reference strain using the Sequencher program (Gene Codes, Ann Arbor, MI, USA).
Table 4. Primers used for LMP-1 sequence analysis and EBNA-2 typing.
| Primer sequence | Nested PCR run | |
|---|---|---|
| LMP-1 primers | ||
| LMP-168843R | CTACAACAAAACTGGTGGACT | 1st PCR |
| LMP-168039F | AGACAGTGTGGCTAAGGGAGT | |
| LMP-168810R | CTCCTTTGGCTCCTCCTGTT | 2nd PCR |
| LMP-168039F | AGACAGTGTGGCTAAGGGAGT | |
| LMP-168689R or LMP-168390R | CTGGCCATGAATCTGACTCT AGCGACTCTGCTGGAAATGAT | 3rd PCR |
| LMP-168075F | TGATTAGCTAAGGCATTCCCA | |
| EBNA-2 primers | ||
| EBNA-2-48810F | AGGGATGCCTGGACACAAGA | 1st PCR |
| EBNA-2-49411R | TGTGCTGGTGCTGCTGGTGG | |
| EBNA-2-1-Fa | TCTTGATAGGGATCCGCTAGGATA | 2nd PCR |
| EBNA-2-1-Ra | ACCGTGGTTCTGGACTATCTGGAC | |
| EBNA-2-2-Fb | CATGGTAGCCTTAGGACATA | 2nd PCR |
| EBNA-2-2-Rb | AGACTTAGTTGATGCCCTAG | |
Abbreviations: EBV, Epstein–Barr virus; EBNA-1, EBV nuclear antigen 1; LMP, latent membrane protein.
EBNA-2 type 1 EBV yields a 497 bp product.
EBNA-2 type 2 EBV yields a 162 bp product.
Statistics
Differences in proportions of samples positive for EBV DNA were compared using the Fisher's exact test. Two-sided P-values <0.05 were considered significant. Differences in the subject average, minimum and maximum EBV DNA values between subjects 18–30 years of age versus those >30 years old were examined using an unpaired t-test. Similarly, differences between males and females were tested. Significance was determined using a two-tailed P-value <0.05. Statistics were performed using GraphPad InStat version 3.0, GraphPad Software, San Diego, CA, USA.
Acknowledgments
This study was approved by the Research Subjects Protection Program of the University of Minnesota, and subjects gave written informed consent before participation. The research was supported by grants from the University of Minnesota Foundation and the University of Minnesota International Center for Antiviral Research and Epidemiology (I CARE).
The authors declare no conflict of interest.
References
- Balfour HH Jr, Odumade OA, Schmeling DO, Mullan BD, Ed JA, Knight JA et al. Behavioral, virologic, and immunologic factors associated with acquisition and severity of primary Epstein-Barr virus infection in university studentsJ Infect Dis 2013; 207: 80–88. [DOI] [PMC free article] [PubMed]
- Odumade OA, Hogquist KA, Balfour HH Jr. Progress and problems in understanding and managing primary Epstein-Barr virus infections. Clin Microbiol Rev 2011; 24: 193–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao QY, Rickinson AB, Epstein MA. A re-examination of the Epstein-Barr virus carrier state in healthy seropositive individuals. Int J Cancer 1985; 35: 35–42. [DOI] [PubMed] [Google Scholar]
- Ling PD, Lednicky JA, Keitel WA, Poston DG, White ZS, Peng R et al. The dynamics of herpesvirus and polyomavirus reactivation and shedding in healthy adults: a 14-month longitudinal study. J Infect Dis 2003; 187: 1571–1580. [DOI] [PubMed] [Google Scholar]
- Miller WE, Edwards RH, Walling DM, Raab-Traub N. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J Gen Virol 1994; 75 (Pt 10): 2729–2740. [DOI] [PubMed] [Google Scholar]
- van Kooij B, Thijsen SF, Meijer E, Niesters HG, van Esser JW, Cornelissen JJ et al. Sequence analysis of EBV DNA isolated from mouth washings and PBMCs of healthy individuals and blood of EBV-LPD patients. J Clin Virol 2003; 28: 85–92. [DOI] [PubMed] [Google Scholar]
- Maeda A, Wakiguchi H, Yokoyama W, Hisakawa H, Tomoda T, Kurashige T. Persistently high Epstein-Barr virus (EBV) loads in peripheral blood lymphocytes from patients with chronic active EBV infection. J Infect Dis 1999; 179: 1012–1015. [DOI] [PubMed] [Google Scholar]
- Holman CJ, Karger AB, Mullan BD, Brundage RC, Balfour HH Jr. Quantitative Epstein-Barr virus shedding and its correlation with the risk of post-transplant lymphoproliferative disorder. Clin Transplant 2012; 26: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling DM, Brown AL, Etienne W, Keitel WA, Ling PD. Multiple Epstein-Barr virus infections in healthy individuals. J Virol 2003; 77: 6546–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitki-Green D, Covington M, Raab-Traub N. Compartmentalization and transmission of multiple Epstein-Barr virus strains in asymptomatic carriers. J Virol 2003; 77: 1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer E, Spijkers S, Moschatsis S, Boland GJ, Thijsen SF, van Loon AM et al. Active Epstein-Barr virus infection after allogeneic stem cell transplantation: re-infection or reactivation? Transpl Infect Dis 2005; 7: 4–10. [DOI] [PubMed] [Google Scholar]
- Puchhammer-Stockl E, Gorzer I. Cytomegalovirus and Epstein-Barr virus subtypes— the search for clinical significance. J Clin Virol 2006; 36: 239–248. [DOI] [PubMed] [Google Scholar]
- Verghese PS, Schmeling DO, Knight JA, Matas AJ, Balfour HH Jr. Valganciclovir administration to kidney donors to reduce the burden of cytomegalovirus and Epstein-Barr virus transmission during transplantation. Transplantation 2015; 99: 1186–1191. [DOI] [PubMed] [Google Scholar]
- Balfour HH Jr, Holman CJ, Hokanson KM, Lelonek MM, Giesbrecht JE, White DR et al. A prospective clinical study of Epstein-Barr virus and host interactions during acute infectious mononucleosis. J Infect Dis 2005; 192: 1505–1512. [DOI] [PubMed] [Google Scholar]
- Higa M, Kinjo T, Kamiyama K, Iwamasa T, Hamada T, Iyama K. Epstein-Barr virus (EBV) subtype in EBV related oral squamous cell carcinoma in Okinawa, a subtropical island in southern Japan, compared with Kitakyushu and Kumamoto in mainland Japan. J Clin Pathol 2002; 55: 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Villnave CA, Singer RH. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell 1988; 52: 51–61. [DOI] [PubMed] [Google Scholar]