Abstract
Background
The thermophilic, anaerobic bacterium Thermoanaerobacterium saccharolyticum digests hemicellulose and utilizes the major sugars present in biomass. It was previously engineered to produce ethanol at yields equivalent to yeast. While saccharolytic anaerobes have been long studied as potential biomass-fermenting organisms, development efforts for commercial ethanol production have not been reported.
Results
Here, we describe the highest ethanol titers achieved from T. saccharolyticum during a 4-year project to develop it for industrial production of ethanol from pre-treated hardwood at 51–55 °C. We describe organism and bioprocess development efforts undertaken to improve ethanol production. The final strain M2886 was generated by removing genes for exopolysaccharide synthesis, the regulator perR, and re-introduction of phosphotransacetylase and acetate kinase into the methyglyoxal synthase gene. It was also subject to multiple rounds of adaptation and selection, resulting in mutations later identified by resequencing. The highest ethanol titer achieved was 70 g/L in batch culture with a mixture of cellobiose and maltodextrin. In a “mock hydrolysate” Simultaneous Saccharification and Fermentation (SSF) with Sigmacell-20, glucose, xylose, and acetic acid, an ethanol titer of 61 g/L was achieved, at 92 % of theoretical yield. Fungal cellulases were rapidly inactivated under these conditions and had to be supplemented with cellulosomes from C. thermocellum. Ethanol titers of 31 g/L were reached in a 100 L SSF of pre-treated hardwood and 26 g/L in a fermentation of a hardwood hemicellulose extract.
Conclusions
This study demonstrates that thermophilic anaerobes are capable of producing ethanol at high yield and at titers greater than 60 g/L from purified substrates, but additional work is needed to produce the same ethanol titers from pre-treated hardwood.
Keywords: Cellulosic ethanol, Consolidated bioprocessing, Organism development, Metabolic engineering, Bioprocess development, Thermophilic bacteria
Background
Biotechnology for the conversion of biomass to fuels has the potential to reduce the need for carbon-intensive fossil fuels, but must be cost-competitive to be commercialized. Ethanol is the first commercial cellulosic biofuel and the logical proving ground for innovations aimed at reducing production costs. To be cost-competitive, an improved process must generate ethanol at high yield. Sufficiently high ethanol titers, generally at or above 40 g/L [1, 2], are also required to avoid high costs for fermentation and distillation. The upper limit of ethanol titer that can be achieved with lignocellulosic feedstocks is considerably lower than can be achieved from starch due to the lower fraction of fermentable sugar and materials handling issues [2]. As a result, both near-term and futuristic designs for cellulosic ethanol plants often involve ethanol titers in the range of 50–60 g/L [3, 4].
Thermophilic, anaerobic bacteria exhibit distinctively high rates of cellulose and plant cell wall solubilization [2, 5], with fermentation of cellulose and hemicellulose usually carried out by different species. Thermoanaerobacterium saccharolyticum ferments xylan, the main polymer in hemicellulose, and also utilizes all other major biomass sugars, including cellobiose, glucose, mannose, xylose, galactose, and arabinose. This microorganism does not, however, ferment cellulose to any significant degree. Organic fermentation products from wild-type strains of T. saccharolyticum strains include ethanol, acetic acid, and lactic acid. By deleting the genes encoding lactate dehydrogenase, phosphotransacetylase, and acetate kinase, an engineered strain was developed that produces ethanol at greater than 90 % of theoretical yield, equivalent to yeast and other homoethanologens [6]. T. saccharolyticum is naturally competent and recombinogenic, making genetic manipulation relatively easy [7]. The genome sequence and other genomic resources have been recently published [8]. Beginning with a homoethanologenic strain of T. saccharolyticum, Shaw et al. [9] achieved an ethanol titer of 54 g/L by introducing genes encoding urease and using urea as the nitrogen source. To our knowledge, this is the highest titer of produced ethanol reported for a thermophilic bacterium.
The US Department of Energy Biomass Program and Mascoma Corporation funded a 4-year project to develop T. saccharolyticum as a biocatalyst for the production of ethanol from pre-treated hardwood [10]. The two main components of the project were organism and bioprocess development activities. Organism development efforts were aimed at generating strains to produce high ethanol titers in the presence of inhibitors found in pre-treated biomass, using a combination of rational genetic engineering, classical mutagenesis/selection, and genome-scale resources. Bioprocess development efforts were aimed at meeting specific performance targets using optimization of media, enzyme addition, growth on hardwood substrates, and process integration. The two activities were pursued in parallel and subsequently brought together to achieve high ethanol titers, first with purchased model substrates, nutrients and inhibitors, and then progressing to pre-treated hardwood.
The original vision was to use T. saccharolyticum in a simultaneous saccharification and fermentation (SSF) process configuration. Since the fermentation temperature of T. saccharolyticum matches the optimal temperature for many fungal cellulases, we expected to add less cellulase than would otherwise be necessary. However, we discovered mid way through the project that commercial fungal cellulases are reversibly inactivated by the low-redox fermentation conditions [11]. A related project aimed to express cellulases in T. saccharolyticum [12], but the maximal expression and secretion levels were insufficient. Ultimately, cellulosome preparations from C. thermocellum were used to overcome the limitations of fungal cellulase, as described below. We also describe the rationale for directed strain modifications and the sequence-level effects of selections and adaptations. Finally, we present performance data for both model substrates and conditions more representative of an industrial process.
Results and discussion
Strain development
We previously described a method to perform markerless genetic manipulations in T. saccharolyticum. It is “markerless” in so far as it allows the removal of the antibiotic resistance genes (i.e., markers) after they are used [13]. The method is based on negative selection against the presence of the pta and ack genes with chloroacetate. It was used to eliminate lactate and acetate production in wild-type strain JW/SL-YS485 (DSM 8691), creating homoethanologen strain M355 [13]. This strain was then subjected to multiple rounds of nitrosoguanidine mutagenesis and screening for high ethanol titers in the presence of an enzymatic hydrolysate from pre-treated hardwood by Panlabs Biologics in Taiwan.
The 14 top-performing strains from that effort (M796–M809) were mixed and used as inoculum into a cytostat containing a mixture of inhibitory chemicals found in pre-treated hardwood and 20 g/L ethanol. A cytostat is a cell density-regulated continuous culture that uses a highly sensitive flow cytometer to measure cell density, allowing the culture to be maintained continuously at low cell density and fast growth rates [14]. A single clone was isolated from the cytostat and designated M863 (Table 1).
Table 1.
Strains used in present study
| Strain # | Description of genetic manipulation | Genotype |
|---|---|---|
| M355 | Markerless KO of genes for acetate and lactate production | pta/ack(−) L-ldh(−) |
| M795–M809 | Mix of strains generated by Panlabs using NTG mutagenesis and selection on wood hydrolysate | pta/ack(−) L-ldh(−) |
| M863 | Selection in Cytostat with synthetic mix of inhibitors and 20 g/L ethanol | pta/ack(−) L-ldh(−) |
| M1151 | Addition of urease, fix of metE and markerless KO of Tsac_0795 | pta/ack(−) L-ldh(−) Tsac_0795(−) urease(+) metE(+) |
| M1291 | Markerless KO of putative EPS operon (phosphoglucomutase, UDP-G1P transferase, transmembrane protein, near gene Tsac_1471) | pta/ack(−) L-ldh(−) Tsac_0795(−) urease(+) metE(+) EPSoperon(−) |
| M1442a | Selection in auxostat for fast growth in glucose, xylose, arabinose, and acetic acid | pta/ack(−) L-ldh(−) Tsac_0795(−) urease(+) metE(+) EPSoperon(−) |
| M2476 | Markerless KO of perR | pta/ack(−) L-ldh(−) Tsac_0795(−) urease(+) metE(+) EPSoperon(−) perR(−) |
| M2886 | Insertion of pta/ack-KanR into methylglyoxal synthase mgs | pta/ack(−) L-ldh(−) Tsac_0795(−) urease(+) metE(+) EPSoperon(−) perR(−) mgs::pta/ack-KanR |
aStrain M1442 is also known as LL1049
Using an approach as described previously [15], a library of clones was created that positioned random pieces of T. saccharolyticum DNA down-stream from a strong promoter integrated into the T. saccharolyticum chromosome, with the expectation that overexpression of some genes would lead to improved inhibitor tolerance. The library was selected on solid or liquid media containing extracts from pre-treated hardwood. Sequencing the inserts showed that 19 out of 23 selected clones had the pta/ack gene pair inserted. This was surprising, since the strain had been engineered to eliminate acetate production by the removal of these genes. Also intriguing, the library-selected strains did not produce wild-type levels of acetate and the pta/ack genes confer inhibitor tolerance even without net acetate production. An investigation of this result is published elsewhere [16].
A related cloning strategy was used to create a random deletion library in T. saccharolyticum which was subjected to selection in the cytostat with mixed inhibitors and in auxostat cultures with extracts of pre-treated hardwood. An auxostat is a continuous culture in which the feed rate is indirectly coupled to growth rate. In this case, growth caused a drop in pH from the uptake of ammonia, which was countered by automatic addition of a base solution to maintain a constant pH mixed with growth-inhibitory extract. The dilution rates of both cytostats and auxostats are proportional to growth, but in practice, the auxostat has a higher cell density and slower growth rate. The deletion library yielded a wider assortment of genotypes than the overexpression library, but both cytostat and auxostat selected for clones with a deletion in the gene Tsac_0795, encoding a possible helicase or protein kinase. Further strain improvement consisted of a knockout of Tsac_0795, while simultaneously adding beneficial genes. The urease genes from C. thermocellum were inserted in place of Tsac_0795 to allow the use of urea as nitrogen source, which was shown to result in higher ethanol titers [9]. Also inserted at the same locus was the metE gene from Caldicellulosiruptor kristjanssonii to restore vitamin B-12-independent methionine synthesis, compensating for the disrupted native metE gene in T. saccharolyticum.
We next deleted a 4-gene putative operon that appeared to be related to exopolysaccharide synthesis: genes Tsac_1474-Tsac_1477, annotated as phosphoglucomutase, NGN domain-containing protein, UTP-glucose-1-phosphate uridylyltransferase, and lipopolysaccharide biosynthesis protein. The resulting strain M1291 produced more ethanol than its parent strain M1151 (Table 2), possibly due to diversion of intracellular glucose from anabolism (polymerization) to catabolism (glycolysis). This strain was then selected for rapid growth on mixed sugars by growing it for 425 h in a pH-controlled auxostat containing xylose, glucose, arabinose, and acetic acid, at growth rates from 0.09 to 0.37 h−1.
Table 2.
Production of ethanol from 60 g/L cellobiose, 90 g/L maltodextrin by strains M1151, M1291, and M1442 in bottles
| Medium | Strain | Final ethanol (g/L) | SD (g/L) |
|---|---|---|---|
| TSC-3 | M1151 | 61.0 | 1.9 |
| TSC-3 | M1291 | 65.1 | 2.0 |
| TSC-3 | M1442 | 70.1 | 1.0 |
| TSC-4 | M1442 | 60.0 | 0.4 |
The next modification consisted of a markerless deletion of the regulatory gene perR to generate strain M2476. PerR is a repressor of oxidative stress response genes, and its deletion has been shown to increase aerotolerance in C. acetobutylicum [17]. Microarray studies with T. saccharolyticum looking at the response to inhibitors in pre-treated hardwood suggested an oxidative challenge [8], and we reasoned that overexpression of the perR regulon would increase tolerance to these inhibitors. Indeed, knockout mutants of perR in T. saccharolyticum (gene Tsac_2491) produced more ethanol than their parent from inhibitory concentrations of pre-treated hardwood hemicellulose extract (data not shown). The bacterium was also able to survive up to 4 h of air exposure on a pertri plate without an observable drop in viability. In contrast, the parent began to lose viability after 1 h under the same conditions.
Finally, the gene-encoding methylglyoxal synthase (mgs, Tsac_2114) was deleted by insertion of the kanamycin resistance marker and the pta/ack genes, creating strain M2886. While T. saccharolyticum grows well in high levels of starch and cellobiose, it is inhibited by monosaccharides at concentrations greater than 40 g/L. Glucose toxicity has been shown to correlate with the production of methylglyoxal [18]. The strain M2886 grew at 100 g/L glucose and produced more ethanol from pre-treated hardwood hydrolysate than other candidate strains.
It should be noted that many other approaches, both rational and selection-based, were tested in addition to those that were used to generate strain M2886. Strain benchmark tests were performed throughout its development with up to 30 strains at a time in standardized conditions to identify the best-performing strains and eliminate less-beneficial approaches. The benchmark tests comprised bottle cultures with high sugars (e.g., Table 2), SSFs on purified cellulose or challenges with inhibitory levels of pre-treated hardwood extracts, with maximum ethanol titer being the key metric. The strain lineage described here represents the top-performing modifications from each round of strain evaluation.
Resequencing results
Strains M863, M1442, and M2886 were resequenced by Illumina sequencing, and compared to the wild-type JW/SL-YS485 genome sequence. Strain LL1025, which is another clone of JW/SL-YS485, was also sequenced as a control. Small-scale sequence variations are shown in Table 3. Seven sequence differences were found in all four strains, including LL1025 (rows 1–7), indicating possible errors in the Genbank genome sequence. Rows 8–10 show differences detected only in strain M863. Since the later strains were descended from M863, they should also contain these differences yet do not, suggesting that they are artifacts. A total of 16 small variations were detected in strain M863 and the later strains, likely arising during the extensive selections that took place to generate M863. These include mutations in the genes for the bifunctional acetaldehyde/alcohol dehydrogenase gene adhE, and in the hfs hydrogenase cluster, whose effect on ethanol production has been described elsewhere [16]. Selection in continuous culture preceding the isolation of M1442 resulted in nine mutations compared to the parent strain. Five additional small mutations arose in generating strain M2886.
Table 3.
Genomic resequencing results from strains in the present study: small-scale sequence variations and their occurrence in each strain
| Row | Gene | Nucleotide | Type | Reference | Change | Non-synonymous | Amino acid change | Percent of sequencing reads | Description | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YS485 (WT) | M863 | M1442 | M2886 | |||||||||
| 1 | Tsac_0902 | 944418 | Deletion | G | – | Intergenic | 99 | 95 | 98 | 99 | Hypothetical protein | |
| 2 | Tsac_1176 | 1217778 | Insertion | – | A | Intergenic | 100 | 100 | 99 | 100 | Histidinol dehydrogenase, prokaryotic-type | |
| 3 | Tsac_1332 | 1388136 | Insertion | – | A | Intergenic | 100 | 100 | 99 | 100 | Hypothetical protein | |
| 4 | Tsac_1641 | 1717467 | Insertion | – | T | Yes | Tyr 18 FS | 100 | 100 | 100 | 100 | MerR family transcriptional regulator |
| 5 | Tsac_2098 | 2143046 | Insertion | – | A | Intergenic | 100 | 100 | 100 | 100 | Peptidase S11, d-alanyl-d-alanine carboxypeptidase A | |
| 6 | Tsac_2308 | 2368691 | Deletion | A | – | Intergenic | 100 | 100 | 100 | 100 | HEPN domain protein | |
| 7 | Tsac_2828 | 21 (megaplasmid) | MNV | GA | AG | Yes | Lys 104 Glu | 96 | 88 | 97 | 95 | Hypothetical protein, on megaplasmid CP003185 |
| 8 | Tsac_0096 | 92464 | Insertion | – | T | Intergenic | 100 | PWT | 99 | 100 | LuxR family transcriptional regulator | |
| 9 | Tsac_0303 | 322547 | Insertion | – | A | Intergenic | 99 | 40 | 99 | 92 | Fibronectin type III domain protein | |
| 10 | Tsac_0902 | 944612 | SNV | A | T | No | PWT | 49 | PWT | PWT | Putative transposase YhgA family protein | |
| 11 | Tsac_0032 | 30909 | Insertion | – | A | Yes | Gly 177 FS | PWT | 100 | 99 | 100 | PTS system, N-acetylglucosamine-specific IIBC subunit |
| 12 | Tsac_0079 | 79429 | Deletion | A | – | Yes | Asn 86 FS | PWT | 74 | 99 | 100 | Uncharacterised conserved protein UCP018688 |
| 13 | Tsac_0144 | 150053 | SNV | C | T | Intergenic | PWT | 98 | 99 | 100 | 4-Deoxy-l-threo-5-hexosulose-uronate ketol-isomerase | |
| 14 | Tsac_0163 | 177035 | SNV | A | C | Yes | Phe 104 Val | PWT | 100 | 100 | 100 | Altronate oxidoreductase |
| 15 | adhE Tsac_0416 | 448858 | SNV | G | A | Yes | Gly 544 Asp | PWT | 98 | 100 | 100 | Bifunctional alcohol/aldehyde dehydrogenase |
| 16 | Tsac_0838 | 884294 | Deletion | T | – | Yes | Ser 102 FS | PWT | 85 | 99 | 100 | Protein of unknown function DUF324 |
| 17 | Tsac_0948 | 993551 | SNV | A | T | Yes | Phe 361 Tyr | PWT | 100 | 100 | 100 | Spore germination protein |
| 18 | hfsB Tsac_1551 | 1627813 | Deletion | A | – | Yes | Lys 146 FS | PWT | 100 | 99 | 100 | Hydrogenase large subunit domain protein |
| 19 | hfsD Tsac_1553 | 1630591 | SNV | A | T | Yes | Arg 107 Ser | PWT | 98 | 100 | 100 | Iron hydrogenase, large subunit, C-terminal |
| 20 | Tsac_1726 | 1800969 | Insertion | – | A | Yes | Tyr 454 FS | PWT | 100 | 100 | 100 | Serine/threonine protein kinase with PASTA sensor(s) |
| 21 | pta Tsac_1744a,b | 1814403 | Insertion | – | G | Yes | Ile 3 FS | PWT | 100 | 97 | PWT | Phosphate acetyltransferase |
| 22 | Tsac_1782 | 1848944 | Deletion | G | – | Yes | Glu 180 FS | PWT | 100 | 99 | 100 | Flagellar motor switch protein FliG |
| 23 | Tsac_2119 | 2160615 | SNV | C | T | Yes | Gly 488 Glu | PWT | 97 | 100 | 100 | Penicillin-binding protein 2 |
| 24 | Tsac_2196 | 2239931 | SNV | G | A | No | PWT | 100 | 99 | 100 | Phenylalanyl-tRNA synthetase, class IIc, alpha subunit | |
| 25 | Tsac_2229 | 2273080 | SNV | A | G | Yes | Ser 283 Pro | PWT | 82 | 99 | 100 | Bacterial inner-membrane translocator |
| 26 | Tsac_2476 | 2526425 | SNV | A | G | No | PWT | 94 | 100 | 100 | RNA polymerase, sigma 70 subunit, RpoD subfamily | |
| 27 | Tsac_0535 | 559915 | SNV | C | T | Yes | Gly 257 Asp | PWT | PWT | 100 | 100 | Cyanophycin synthetase |
| 28 | Tsac_0795a | 835013 | Deletion | A | – | Yes | Ile 1000 FS | PWT | PWT | 100 | 100 | SNF2-related protein |
| 29 | Tsac_1054 | 1089642 | SNV | T | C | Intergenic | PWT | PWT | 100 | 100 | Thioredoxin reductase | |
| 30 | Tsac_1296a | 1356848 | SNV | A | G | Yes | Glu 105 Gly | PWT | PWT | 100 | 100 | UDP-glucose 4-epimerase |
| 31 | Tsac_1419 | 1491395 | SNV | C | A | Yes | Ala 187 Asp | PWT | PWT | 99 | 100 | ATPase, F0 complex, subunit A |
| 32 | Tsac_1477 | 1557984 | Insertion | – | G | Yes | Ile 267 FS | PWT | PWT | 100 | 100 | Lipopolysaccharide biosynthesis |
| 33 | Tsac_1712 | 1787698 | SNV | C | A | Yes | Pro 161 Thr | PWT | PWT | 100 | 100 | Diaminopimelate epimerase |
| 34 | Tsac_2390 | 2460517 | SNV | C | T | Yes | Leu 311 Phe | PWT | PWT | 99 | 100 | Hypothetical protein |
| 35 | Tsac_2507 | 2555785 | SNV | G | A | Yes | Gln 840 stop | PWT | PWT | 100 | 100 | PTS system transcriptional activator |
| 36 | Tsac_1486 | 1566717 | MNV | AA | TT | Intergenic | PWT | PWT | PWT | 100 | Flavodoxin/nitric oxide synthase | |
| 37 | Tsac_1725 | 1799018 | SNV | G | A | Yes | Ala 50 Thr | PWT | PWT | PWT | 100 | Protein serine/threonine phosphatase |
| 38 | Tsac_2189 | 2229111 | Insertion | – | T | Yes | Asp 144 FS | PWT | PWT | PWT | 100 | Two component transcriptional regulator, LuxR family |
| 39 | perR Tsac_2491a | 2541836 | Deletion | C | – | Yes | Ser 113 FS | PWT | PWT | PWT | 100 | Ferric uptake regulator, Fur family |
| 40 | perR Tsac_2491a | 2541840 | Deletion | T | – | Yes | Tyr 115 FS | PWT | PWT | PWT | 100 | Ferric uptake regulator, Fur family |
| 41 | Tsac_0622 | 659800 | SNV | T | G | Intergenic | PWT | PWT | PWT | 45 | Hypothetical protein | |
| 42 | Tsac_0634 | 677092 | SNV | T | G | Intergenic | PWT | PWT | PWT | 41 | Glucokinase, ROK family | |
Where the sequence analysis software detected the mutation in greater than 20 % of reads, the percent of reads with the mutation is given. Otherwise, the percent of sequencing reads is not calculated
SNV single nucleotide variation, FS frame shift, PWT presumptively wild type, MNV multiple nucleotide variation
aThese variations occurred in the small residual sequences that were not removed when the genes were knocked out
bThe pta/ack genes were re-introduced into strain M2886
Table 4 shows nine larger-scale variations that were identified in the resequencing data. Six of these were the engineered deletions, but the others appear to be spontaneous. Two deletions occurred in intergenic repeat regions, one of which is CRISPR-associated. In the promoter region of gene Tsac_2564 encoding a phosphotransferase subunit, there is a possible transposon insertion. No sequencing reads span the insertion site, but they contain the duplicated sequence ATTTTTAATTATTTT and additional sequence that matches part of the gene Tsac_0046-encoding pyruvate-ferredoxin oxidoreductate (PFOR), a critical gene for ethanol production [19].
Table 4.
Genomic resequencing results from strains in the present study: large-scale sequence variations and their occurrence in each strain
| Locus | Nucleotides | Description | LL1025 (WT) | M863 | M1442 | M2886 |
|---|---|---|---|---|---|---|
| Tsac_0179 | Engineered ldh deletion | WT | Deletion | Deletion | Deletion | |
| Tsac_0389 | 424,393–424,493 | Small deletion in CRISPR repeat region | WT | Deletion | Deletion | Deletion |
| Tsac_0832 | 875,581–875,753 | Small deletion in intergenic repeat region | WT | Deletion | Deletion | Deletion |
| Tsac_1744–1745 | Engineered pta/ack deletion | WT | Deletion | Deletion | Deletion | |
| Tsac_2564 | 2618,783–2618,797 | Transposon insertion in putative promoter of gene for PTS IIA subunit | WT | Putative transposon | Putative transposon | Putative transposon |
| Tsac_0795 | Engineered deletion | WT | WT | Deletion | Deletion | |
| Tsac_1474–1477 | Engineered deletion of EPS gene cluster | WT | WT | Deletion | Deletion | |
| Tsac_2114 | Engineered mgs deletion/insertion | WT | WT | WT | Breakpoints in 23 % of reads b | |
| Tsac_2491 | Engineered perR deletion | WT | WT | WT | Deletion |
aThe pta/ack genes were re-introduced elsewhere in the genome
bThe fraction of the reads supporting the mutation (left and right breakpoints averaged). This value was >90 % for all other breakpoints
For most of the spontaneous mutations in Table 3, it is unknown whether they conferred adaptive phenotypes. Although creation of isogenic strains for each allele is required to rigorously establish genotype:phenotype relationships, inferences about the importance of various mutations may be made based on their recurrence in multiple lineages. Table 5 shows recurrent mutations from all strains resequenced under this project. We observed independent occurrence of mutations in the adhE and hfs cluster genes as reported previously, along with 11 others. Of particular interest, two sets of mutations occurred in PTS-related transcriptional regulators encoded by Tsac_1263 and Tsac_2568, and another in a PTS IIBC subunit encoded by Tsac_0032. Recurrent mutations in Tsac_0825-encoding inorganic diphosphatase and Tsac_1419-encoding ATPase are also noteworthy for their potential impact on ethanol production. The mutations in Tsac_0361 are also interesting, because the protein encoded by this gene is one of the most abundant secreted proteins and a primary component of the S-layer [20].
Table 5.
Genomic resequencing results from all strains sequenced in this project: recurrent mutations
| Gene | Description | Independent alleles | Present in this lineage |
|---|---|---|---|
| Tsac_0032 | PTS system, N-acetylglucosamine-specific IIBC subunit | 2 | Yes |
| Tsac_0079 | Uncharacterised conserved protein UCP018688 | 2 | Yes |
| Tsac_0361 | S-layer domain-containing protein | 4 | |
| adhE Tsac_0416 | Bifunctional alcohol/aldehyde dehydrogenase | 4 | Yes |
| Tsac_0644 | Hypothetical protein | 2 | |
| Tsac_0653 | Methionyl-tRNA synthetase | 2 | |
| Tsac_0825 | Inorganic diphosphatase | 2 | |
| Tsac_0838 | Protein of unknown function DUF324 | 3 | Yes |
| Tsac_1263 | PTS system transcriptional activator | 3 | |
| Tsac_1419 | ATPase, F0 complex, subunit A | 2 | Yes |
| Tsac_1520 | ATP:corrinoid adenosyltransferase BtuR/CobO/CobP | 2 | |
| hfs Tsac_1550-1553 | Hydrogenase large subunit domain protein | 8 | Yes |
| Tsac_2568 | PTS modulated transcriptional regulator, MtlR family | 2 |
Fermentations
Fermentation conditions were developed to reach the highest possible ethanol titer with T. saccharolyticum in batch format, at 20 mL liquid volume in anaerobic 125 mL serum bottles. These conditions were used to benchmark different strains for ethanol production. We found that cellobiose and starch were readily fermented and well-tolerated at relatively high concentrations. A mixture of 60 g/L cellobiose and 90 g/L maltodextrin in TSC3 rich medium yielded a maximum of 70 g/L ethanol (Table 2). An excess of calcium carbonate provided excellent buffering at a pH of 5.5, which is close to the pH optimum for T. saccharolyticum. For reasons we do not fully understand, the same growth media in 1 L fermenters yielded 5–10 g/L less ethanol (Fig. 1).
Fig. 1.
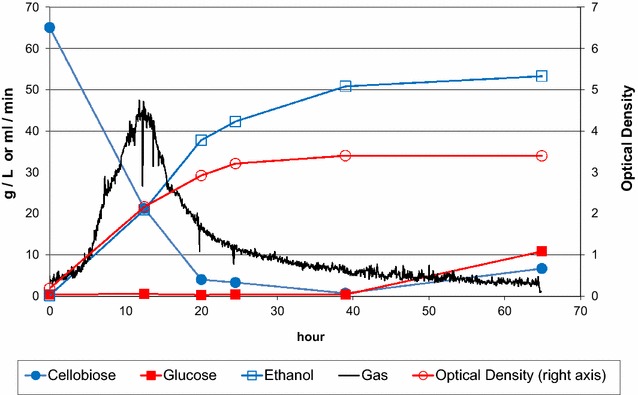
Fermentation of cellobiose and maltodextrin. Strain M1151 was grown in TSC3 medium containing 90 g/L maltodextrin and 60 g/L cellobiose at 1 L scale. Cellobiose, glucose, and ethanol levels are shown in units of g/L on the left axis
Fermentation conditions were then developed to reach the highest possible ethanol titer in a Simultaneous Saccharification and Co-Fermentation (SSCF) configuration with substrates approximating the conditions we expected from pre-treated hardwood (i.e., a “mock hydrolysate”). The fermentation contained 100 g/L purified cellulose (Sigmacell-20) and 10 g/L acetic acid, and was fed with 35 g/L xylose and 20 g/L of glucose. We had found that commercially available cellulases were inactivated by low redox and ethanol [11], so we added a mixture of fungal and bacterial cellulase from C.thermocellum (see “Methods” section). The T. saccharolyticum inoculum was drawn from a chemostat, so that it was active and had a consistently high optical density (5–10 OD units). The results of this fermentation are shown in Table 6, comparing the previously published strain ALK2 to the improved strain M1442. An ethanol titer of 61 g/L was reached in 93 h by strain M1442 while strain ALK2 produced 46 g/L, leaving some residual xylose. The metabolic yield for both strains was greater than 90 % of the theoretical maximum, while the cellulose conversion by the enzyme mix was 71–75 %. Scaled up to 8 L, strain M1442 produced 55 g/L ethanol.
Table 6.
Fermentation data comparing strains ALK2, M1442, and M2886 in pH-controlled bioreactors in SSCF or SHF process configurations
| Substrate | Mock hydrolysate | Mock hydrolysate | Mock hydrolysate | Pre-treated hardwood | Pre-treated hardwood | Pre-treated hardwood | Hardwood hydrolysate |
|---|---|---|---|---|---|---|---|
| Process type | SSCF | SSCF | SSCF | SSCF | SSCF | SSCF | SHF |
| Fermentation volume (liters) | 1 | 1 | 8 | 1 | 1 | 100 | 1 |
| Strain | ALK2 | M1442 | M1442 | ALK2 | M2886 | M2886 | M2886 |
| Initial concentrations | |||||||
| Solids (%) | 16.5 | 16.5 | 16.5 | 12.0 | 12.0 | 12.0 | 0.0 |
| Cellulose (g/L) | 100 | 100 | 100 | 64.5 | 64.5 | 64.5 | 0.0 |
| Glucose (g/L) | 20 | 20 | 20 | 1.0 | 1.0 | 1.2 | 88.6 |
| Xylose (g/L) | 35 | 35 | 35 | 13.9 | 13.9 | 16.6 | 24.3 |
| Other sugars (g/L) | 3.8 | 3.8 | 3.2 | 5.2 | |||
| Acetic acid and other inhibitors (g/L) | 10.5 | 10.5 | 10.0 | 6.3 | 6.3 | 6.3 | 4.5 |
| Fermentation performance | |||||||
| Fermentation time (hours) | 97 | 93 | 90 | 60 | 60 | 60 | 60 |
| Final ethanol titer (g/L) | 45.7 | 61.4 | 54.7 | 32.6 | 32.0 | 30.8 | 49.5 |
| Cellulose conversion (%) | 71.0 | 75.1 | 83.4 | 83.6 | 80.4 | 77.2 | n/a |
| Glucose utilization (%) | 79.5 | 93.6 | 85.9 | 97.2 | 94.7 | 99.5 | 89.3 |
| Xylose utilization (%) | 58.1 | 99.6 | 80.9 | 82.3 | 86.7 | 100.0 | 91.6 |
| Metabolic yield (%) | 90.5 | 91.5 | 85.8 | 75.9 | 81.0 | 78.4 | 90.1 |
An SSCF was also performed with pre-treated hardwood at 12 % solids concentration, comparing two strains in duplicate. A concentrated, polymeric hemicellulose extract was fed, and activated carbon was used to reduce the toxicity of both the solids and the liquid feed. Again, a mixture of fungal and C. thermocellum cellulases was used, and cellulose conversion was 80–84 %. Strain M2886 produced 32 g/L ethanol in 60 h, while ALK2 produced 33 g/L ethanol, at 81 and 76 % of theoretical metabolic yield, respectively. Scaled up to 100 L, strain M2886 produced 31 g/L ethanol. Other fermentations at 22 % solids loading performed poorly (not shown), likely due to the presence of inhibitors at levels higher than the cells could tolerate. At 12 % solids, there was a little difference in performance between the project’s starting and final strains (ALK2 and M2886, respectively, Fig. 2), while at 22 % solids, both strains were inhibited. We can speculate that at some intermediate level of solids loading, inhibition would be enough to better distinguish the performance of the two strains, but not too much for M2886 to grow. Figure 2 shows that at approximately 40 h, the glucose levels in all fermentations were below 1 g/L and ethanol was greater than 30 g/L, suggesting that the cultures were limited by the availability of glucose (i.e. the activity of the cellulases) at that time. Some glucose accumulated by 60 h, suggesting that cellulase-mediated solubilization rates exceeded the rate of fermentation.
Fig. 2.
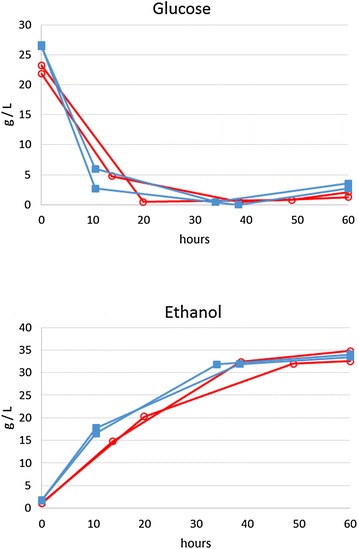
Fermentation of 12 % solids pre-treated hardwood, fed with hemicellulose extract. Duplicate 1 L fermentations with strain ALK2 are shown in red with open circles and with strain M2886 in blue filled squares
To demonstrate the ability of T. saccharolyticum to produce high ethanol titers when cellulase activity is not limiting, a separate hydrolysis and fermentation (SHF) was performed with pre-treated hardwood hydrolysate and hemicellulose extract (last column of Table 6). After 60 h of fermentation, the ethanol titer reached 50 g/L, while sugar utilization and metabolic yield were 90 %.
Thermoanaerobacterium saccharolyticum is distinct from other homoethanologens in its native ability to digest polymeric hemicellulose and to co-ferment all the resulting sugars at high ethanol yield. Commercial bioprocessing configurations can be considered where hemicellulose is separated from biomass by hot water extraction and fermented separately. T. saccharolyticum would be a good choice of organism for such fermentations, because it can mediate hydrolysis of the polymeric hemicellulose without added enzymes or acid, though it needs to be able to handle the acetic acid and other inhibitors that normally accompany it. Some level of detoxification can be considered, but the cost must be kept very low.
A number of strains were evaluated at varying levels of hemicellulose extract, as shown in Fig. 3. At low concentrations of extract (13 g/L total sugar), the ethanol yields exceeded 90 %, but the yields declined rapidly at higher concentrations of extract. Lime treatment and nanofiltration were used to detoxify the extract, which was fermented in fed-batch at 1 L scale (Fig. 4). After 47 h, 25 g/L of ethanol was produced, and increased to 26 g/L by 73 h. Xylose, the main sugar component, was low throughout the fermentation, and arabinose was undetectable by 23 h. The final metabolic ethanol yield was 78 % of theoretical.
Fig. 3.
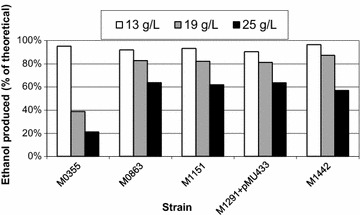
Strain comparison in varying levels of hemicellulose extract. Bars indicate ethanol produced in batch fermentations as a percent of the maximum theoretical ethanol possible for the amount of sugar provided by the extract, indicated by bars: 13 g/L (white), 19 g/L (gray), and 25 g/L (black)
Fig. 4.
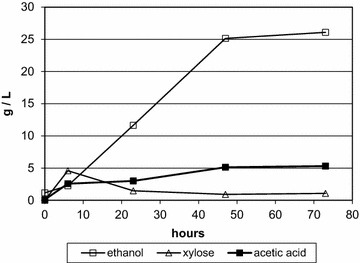
Fermentation of detoxified hemicellulose extract. Strain M1732 was grown in TSC7 medium containing hemicellulose extract at 1 L volume at 51 °C at pH 5.8. The fermentor contained 42 g/L available sugar (76 % xylose, 11 % mannose, 6 % glucose, 5 % galactose, and 2 % arabinose, as polymeric hemicellulose) at the start, and was fed an additional 25 g/L over two feedings at 24 and 47 h. The hemicellulose was detoxified by lime treatment and nanodiafiltration
It has been noted in the literature that tolerance to added ethanol is often higher than the maximum titers of ethanol that are produced, but this ‘gap’ can be eliminated by strain adaption and engineering [21]. The maximum titer of produced ethanol reported here (70 g/L) is consistent with reports for the maximum concentrations of added ethanol at which thermophilic anaerobes will grow after selection for ethanol tolerance—generally in the range of 50–70 g/L [22]. Thus, the strain and pathway reported here represent a new example of success in closing the titer gap among thermophilic ethanol producers. Production of ethanol beyond the maximum at which growth occurs is possible based on uncoupled metabolism, although this has received relatively little study in thermophiles to date. The ethanol tolerance of thermophilic strains selected for growth in the presence of ethanol is similar to that described for engineered strains of E. coli, but not as high as either the bacterium Zymomonas mobilis or Saccharomyces cerevisiae. Higher ethanol titers can be achieved for a given species or strain at lower temperatures within its growth range [23], but we have no reason to think that an interspecies comparison between thermophiles and mesophiles would show the same trend. It should be noted, however, that beyond approximately 40 g/L, ethanol titer has a diminishing effect on distillation costs, and lignocellulosic materials are difficult to convert to ethanol at much more than 50 g/L due to inherent limitations such as mixability and the fraction of fermentable sugar [1, 2].
Conclusions
Production of ethanol at greater than 90 % yield and at titers greater than 60 g/L from model cellulosic substrates were demonstrated using T. saccharolyticum in an SSCF configuration in the presence of 10 g/L acetate. However, maximum ethanol titers were lower using steam pre-treated hardwood or hemicellulose extract. The complex inhibitors present in pre-treated wood are problematic for T. saccharolyticum above moderate concentrations. Random and directed strain modifications, along with detoxification steps, have made improvements in increasing substrate tolerance, but not enough to fully overcome the problem. Further work will be needed to analyze what compounds or combinations of compounds are actually inhibitory, or to more fully detoxify the material in a cost-effective way. Alternately, these inhibitors could be simply avoided by elimination of pre-treatment from the bioprocess. The provision of sufficient cellulase activity for T. saccharolyticum to be used in SSF has proved to be problematic with existing technology. Development of a bacterial lignocellulose solubilization system and/or an understanding of the limitations of fungal cellulases at low-redox levels are necessary for the further development of T. saccharolyticum as biocatalyst for SSF of pre-treated hardwood. However, the high titers and yields we observed support the feasibility of using engineered thermophiles for industrial ethanol production if challenges associated with pre-treatment inhibitors can be avoided.
Methods
Plasmids, primers, and genetic engineering
All markerless gene knockouts were performed as described earlier [13]. The chromosomal flanking regions were PCR amplified with primers listed in Table 7. These PCR products were fused to plasmid pMU433 to create the following gene knockout plasmids: pMU1546 targeting the EPS cluster, including gene Tsac_1474-Tsac_1477; pMU1301 targeting the perR gene Tsac_2491; and pMU3014 targeting the mgs gene Tsac_2114.
Table 7.
Oligonucleotide primers
| Primer | Description | Sequence |
|---|---|---|
| X04986 | perR up-stream forward primer | tttcgactgagcctttcgttttatttgatgcctggTTTGTAATAAAGTCTGCCGT |
| X04987 | perR up-stream reverse primer | AATTGTAGAATACAATCCACTTCACAATGGGCACGTTTTCTTTCAGGATTGACGA |
| X04989 | perR down-stream reverse primer | CCGTCAGTAGCTGAACAGGAGGGACAGCTGATAGAGGCGATAAAGACTATGTAGA |
| X05122 | perR down-stream forward primer | aggggtcccgagcgcctacgaggaatttgtatcgCACAGATTACCTTTTGATGG |
| X07562 | EPS up-stream forward primer | tttcgactgagcctttcgttttatttgatgcctggccgaaaggataagagagcttgc |
| X07563 | EPS up-stream reverse primer | AATTGTAGAATACAATCCACTTCACAATGGGCACGGCATGATGAGGCGATACCTTGATG |
| X07564 | EPS down-stream forward primer | aggggtcccgagcgcctacgaggaatttgtatcggttcctgataaacctgtatcgccc |
| X07565 | EPS down-stream reverse primer | CCGTCAGTAGCTGAACAGGAGGGACAGCTGATAGACTGCCAGCGATGTAAAGCATAG |
| X07568 | EPS external primer 1 | acttggatacaggcagtggaggaa |
| X07569 | EPS external primer 2 | TCCAGCATAGCCTGCAACTGGATA |
| X13281 | perR external primer 1 | agctatgctttctacccttgccca |
| X13282 | perR external primer 2 | AACGACAAGCAGTTTGTGCTTCCG |
| X15225 | mgs up-stream forward primer | agcttgatatcgaattcctgcagcccgggggatctCAGTGCGTCACACGCAGTTG |
| X15226 | mgs up-stream reverse primer | agaatacaatccacttcacaatgggcacgGGATCCGATCTTTTGCCTTCGCATCCC |
| X15227 | mgs down-stream forward primer | gtcccgagcgcctacgaggaatttgtatcgGATCCGGATTTTTGGAATGGAGAGATG |
| X15228 | mgs down-stream reverse primer | accgcggtggcggccgctctagaactagtGGATCTGGTCCTGCTAATGCGATGATG |
| X15767 | mgs external primer 1 | TGCACATTCAGTGCCGTTGTC |
| X15768 | mgs external primer 2 | GTAATCCAACTGAGTGCCGATG |
Classical mutagenesis and selection
An enzymatic hydrolysate was prepared to serve as substrate for mutagenized cultures. Pre-treated hardwood was hydrolyzed with 30 mg/g Accellerase (DuPont) cellulase in a 10 L bioreactor at 10 % initial solids and subsequently fed additional solids up to 20 %. The bioreactor temperature was 50 °C and the pH was 4.8. After 5 days of hydrolysis, the enzymes were heat inactivated at 80 C for 1 h, and the liquids were filtered with Whatman Shark Skin filter paper to remove solids, and then filter sterilized. T. saccharolyticum was mutagenized with 100–160 ppm nitrosoguanidine for 30–60 min at Panlabs Biologics (Taiwan), then diluted and cultured on petri plates in an anaerobic chamber to isolate clones. The clones were screened by culturing in tubes containing BA medium, 1–19 g/L each of xylose, glucose, and/or cellobiose, and up to 25 % volume of enzymatic hydrolysate. HPLC was used to measure ethanol production and substrate utilization, and the best clones were chosen for additional rounds of mutagenesis and screening.
Library construction
A Gateway Cloning (Life Technologies, Carlsbad, CA) destination vector called pMU1035 was constructed with the cellobiose phosphorylase promoter from C. thermocellum positioned up-stream from a cloning site and a ccdB cassette for negative selection. Adjacent to these were sequences flanking the T. saccharolyticumldh gene, chosen as the site for chromosomal integration. It was constructed by inserting the cellobiose phosphorylase promoter between the up-stream ldh flanking region and the kanamycin resistance gene in plasmid pMU433 [13] using yeast-mediated ligation [24]. The resulting plasmid was digested with the enzyme SnaBI and a PCR product containing the ccdB gene was ligated. A library of randomly cleaved genomic DNA from T. saccharolyticum was cloned first into the pCR8/GW/Topo entry plasmid and then transferred into pMU1035 by a clonase LR reaction. The reaction mix was transformed into E. coli strain Mach1 (Life Technologies) and selected for kanamycin resistance, generating the overexpression library. Plasmid DNA from this library was used to transform T. saccharolyticum and selected for kanamycin resistance before being used in growth selection experiments.
The T. saccharolyticum knockout library was generated by modifying the previously created overexpression library. Briefly, the overexpression library was digested with a set of three restriction enzymes that frequently cut T. saccharolyticum genomic DNA but do not cut anywhere on the cloning vector backbone. The kanamycin resistance gene was ligated into the digested library, transformed into E. coli, and 2000–6000 kanamycin-resistant colonies were collected for each of the enzymes used. This produced a large number of plasmids containing the kanamycin resistance marker flanked by T. saccharolyticum genomic DNA on either side, which were transformed and integrated into the T. saccharolyticum genome. These transformants were selected for kanamycin resistance, then screened or selected for inhibitor tolerance. To identify the overexpressed or knockout gene, genomic DNA was isolated and cloned into an E. coli plasmid vector and selected for kanamycin resistance. The resulting colonies were then sequenced.
Resequencing
Raw data for strain M863 were generated at the National Center for Genome Resources (Santa Fe, NM) using an Illumina Solexa Genome Analyzer. The data comprised single 36 bp reads (non-paired).
Raw data for strains M1442 and wild-type JW/SL-YS485 were generated by the Joint Genome Institute (JGI) with an Illumina MiSeq instrument as described by Zhou and coworkers [19]. Unamplified libraries were generated using a modified version of Illumina’s standard protocol. 100 ng of DNA was sheared to 500 bp using a focused ultrasonicator (Covaris). The sheared DNA fragments were size selected using SPRI beads (Beckman Coulter). The selected fragments were then end repaired, A tailed, and ligated to Illumina compatible adapters (IDT Inc.) using KAPA- Illumina library creation kit (KAPA biosystems). Libraries were quantified using KAPA Biosystem’s next-generation sequencing library qPCR kit and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were then multiplexed into pools for sequencing. The pools were loaded and sequenced on the Illumina MiSeq sequencing platform utilizing a MiSeq Reagent Kit v2 (300 cycle), following a 2 × 150 indexed run recipe. Paired-end reads were generated, with an average read length of 150 bp and paired distance of 500 bp.
Raw data for strain M2886 were generated at the Oak Ridge National Laboratory. Illumina TruSeq libraries were prepared as described in the manufacturer’s methods (Part# 15005180 RevA) following the low throughput protocol. In short, 3 ug of DNA was sheared to a size between 200 bp and 1000 bp by nebulization using nitrogen gas for 1 min at 30 psi. Sheared DNA was purified on a Qiagen Qiaquick Spin column (Qiagen). The sheared material was assessed for quantity with a Qubit broad range double stranded DNA assay (Life Technologies) and quality by visualization on an Agilent Bioanalyzer DNA 7500 chip (Agilent). One microgram of sheared DNA was used for library preparation following the manufacturer’s protocol. Libraries were validated by Qubit (Life Technologies) and Agilent Bioanalyzer for appearance and size determination. Samples were normalized using Illumina’s Library dilution calculator to a 10 nM stock and diluted further for sequencing. Clustering was completed on an Illumina CBot, and paired-end sequencing was completed on an Illumina HiSeq instrument (101 bp for each end and 7 bp for the index) using TruSeq sequencing-by-synthesis chemistry.
Data analysis was performed using CLC Genomics Workbench, version 8.5 (Qiagen, USA). Reads were mapped to the reference genome (NC_017992). Mapping was improved by two rounds of local realignment. The CLC probabilistic variant detection algorithm was used to determine small mutations (single and multiple nucleotide polymorphisms, short insertions, and short deletions). Variants occurring in less than 90 % of the reads and variants that were identical to those of the wild-type strain (i.e., due to errors in the reference sequence) were filtered out. The fraction of the reads containing the mutation is shown in Table 3. To determine larger mutations, the CLC InDel and Structural Variant algorithm was run. This tool analyzes unaligned ends of reads and annotates regions where a structural variation may have occurred, which are called breakpoints. Since the read length averaged 150 bp and the minimum mapping fraction was 0.5, a breakpoint can have up to 75 bp of sequence data. The resulting breakpoints were filtered to eliminate those with fewer than ten reads or less than 20 % “not perfectly matched.” The breakpoint sequence was searched with the Basic Local Alignment Search Tool (BLAST) algorithm for similarity to known sequences [25]. Pairs of matching left and right breakpoints were considered evidence for structural variations, such as transposon insertions and gene deletions.
Media and bottle cultures
Growth media were prepared as 10× concentrates and filter sterilized, then immediately added to fermenters or stored in sterile, nitrogen-flushed serum bottles. Chemicals were from Sigma-Aldrich (St. Louis, MO, USA). The medium TSC3 at 1× concentration contained: 8.5 g/L yeast extract, 4 g/L trisodium citrate dihydrate, 2.0 g/L monobasic potassium phosphate, 2.0 g/L magnesium sulfate heptahydrate, 5 g/L urea, 0.2 g/L calcium chloride dihydrate, 0.1 g/L iron sulfate heptahydrate, 0.12 g/L l-methionine, and 0.5 l-cysteine hydrochloride. Medium TSC6 at 1× concentration contained: 8.5 g/L yeast extract, 0.5 g/L trisodium citrate dihydrate, 2.0 g/L monobasic potassium phosphate, 2.0 g/L magnesium sulfate heptahydrate, 5 g/L urea, 0.2 g/L calcium chloride dihydrate, 0.2 g/L iron sulfate heptahydrate, 0.12 g/L l-methionine, and 0.5 l-cysteine hydrochloride. Medium TSC7 at 1× concentration contained: 8.5 g/L yeast extract, 1.0 g/L trisodium citrate dihydrate, 1.0 g/L monobasic potassium phosphate, 2.0 g/L magnesium sulfate heptahydrate, 1.85 g/L ammonium sulfate, 0.2 g/L calcium chloride dihydrate, 0.2 g/L iron sulfate heptahydrate, 0.12 g/L l-methionine, and 0.5 l-cysteine hydrochloride. The medium BA at 1× concentration contained: 3 g/L trisodium citrate dihydrate, 1.5 g/L monobasic potassium phosphate, 2.4 g/L magnesium sulfate heptahydrate, 2 g/L ammonium sulfate, 0.2 g/L calcium chloride dihydrate, 0.1 g/L iron sulfate heptahydrate, 0.015 g/L l-methionine, 0.02 g/L para-amino benzoic acid, 0.02 g/L thiamine, and 0.0001 g/L vitamin B12.
Bottle cultures were performed in 125 ml serum bottles sealed with blue butyl rubber stoppers and crimp seals. Culture volumes were 20 or 50 ml in 125 ml bottles, and those with high sugar concentrations were vented periodically to prevent hazardous pressure build-up. Sugars were dissolved in de-ionized water, and calcium carbonate was added to a final concentration of 10 g/L. The bottles were sealed and then flushed with a 5 % carbon dioxide, 95 % nitrogen gas mixture. They were incubated at 51–55 °C in an incubator shaking at 125–150 rpm. In Fig. 3, cultures were performed in anaerobic tubes with 5 ml liquid volume, using TSC6 medium with 15 g/L calcium carbonate and 1.85 g/L ammonium sulfate in place of urea as nitrogen source. The hemicellulose extract was concentrated by evaporation and analyzed by quantitative saccharification analysis. Inoculations for Fig. 3 were 10 % of the total volume.
Cytostat
To adapt T. saccharolyticum to rapid growth in a mixture of inhibitors found in pre-treated hardwood, a cytostat was constructed and operated as per [14]. The medium used for continuous cultivation of T. saccharolyticum contained (per liter): 20 g ethanol, 24 mg gallic acid, 395 mg hydroxymethylfurfural, 405 mg furfural, 95 mg 3,4-dihydroxybenzoic acid, 19 mg syringic acid, 37 mg vanillin, and 61 mg syringaldehyde.
Fermentations
Fermentations were conducted in 2 L Biostat A reactors (Sartorius AG, Goettingen, Germany) at 1 L working volume. Sugars or pre-treated hardwood along with 10 g/L calcium carbonate and 10 g/L Norit PAC200 activated carbon were added to de-ionized water, and the fermenters were autoclaved. They were sparged with a 5 % carbon dioxide, 95 % nitrogen gas mixture while cooling to fermentation temperature of 51–55 °C. Medium TSC7, prepared at 10× concentration, was filter sterilized and added to the reactors. The pH was set to 5.5 with ammonium hydroxide. Before inoculation of SSFs, cellulase was added for 3–5 h of prehydrolysis. An inoculum of 100 ml was added from a chemostat maintained at a dilution rate of 0.1 h−1 with TSC7 medium with 38 g/L glucose plus 11 g/L total sugars in extract from pre-treated hardwood, at pH 5.8 and 55 °C. For the SSCF fermentations shown in Fig. 2, a feed of 80 mL of activated carbon-treated and dialyzed hemicellulose extract was started after inoculation and 90 mL of C. thermocellum cellulase was added.
SHF fermentations were performed as fed-batch in duplicate, feeding a mixture of liquid solutions prepared from pre-treated hardwood. Polymeric hemicellulose (mostly 5-carbon sugars) was extracted from pre-treated hardwood, treated with lime and activated carbon, and concentrated with nanofiltration. The water-washed solid pre-treated hardwood (mostly 6-carbon sugars) was enzymatically digested with fungal cellulase, concentrated, and treated with activated carbon. The two preparations were mixed in proportion to the abundance of sugars in unfractionated pre-treated hardwood. Glucose levels in the fermentation were monitored carefully and feed rate adjusted to keep the glucose levels less than 0.5 g/L, which we had determined to be important for optimizing ethanol production.
Cellulases
The SSCF of Sigmacell-20 (a purified cellulose sold by Sigma-Aldrich, St. Louis, MO) shown in Table 6 was conducted with 10 mg enzyme per gram of dry solids using a 3:1 mixture of monocomponent CBHI and Endoglucanase from AB Enzymes (Darmstadt, Germany). The SSCF of pre-treated hardwood shown in Fig. 2 was conducted with 20 mg/g CTec3 from Novozymes (Bagsvaerd, Denmark). To supplement fungal cellulases, bacterial cellulase was prepared by growing C. thermocellum strain ATCC 27405 on 5 g/L avicel until early stationary phase. The culture broth was left to settle overnight at 4 °C, and then decanted. The supernatant was concentrated 5- to 10-fold using a 500 kDa filter in tangential flow filtration, then frozen until needed. Before use, cellulosome preparations were centrifuged briefly then filter sterilized. Fungal cellulases were stored at 4 °C and bacterial cellulase was stored at −20 °C.
HPLC
Fermentation products and residual sugars were acidified with sulfuric acid and analyzed using an Aminex HPX-87H (300 × 7.8 mm) column (Bio-Rad Laboratories, Hercules, CA, USA), protected by an in-line frit (0.2um) and Cation-H guard column. Analytes were detected by refractive index and optional UV detector. Eluent was 5 mM sulfuric acid diluted in de-ionized water and the flow rate was 0.7 mL/min at 65 °C.
Authors’ contributions
CDH wrote the manuscript, designed, and conducted experiments and co-directed the project; WRK and AJS designed and conducted experiments and co-directed the project; SFC, JZ, WRS, VT, JSB, SRR, PGT, JPJ, AF, and IDS designed and conducted experiments; DGO analyzed genome sequence data; DMK and SDB generated genome sequence data; BHD and LRL supervised research; and DAH supervised research and co-directed the project. All authors read and approved the final manuscript.
Acknowledgements
The authors wish to acknowledge valued intellectual contributions from Hans van Dijken and from Simon Shih and Panlabs Biologics Inc.
Availability of data
Genome sequence data are available from the Joint Genome Institute under accession numbers SRA234880, SRA233073, and SRP070696.
Competing interests
Most of the authors are former employees of Mascoma Corporation, which has held a financial interest and intellectual property in the technology described herein. There are no non-financial conflicts of interest for any of the authors.
Funding
This work was supported by Mascoma Corporation and the Department of Energy (DOE) under Award Number DE-FC36-07G017057. Manuscript preparation was supported in part by the BioEnergy Science Center, a US DOE Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. The work conducted by the U.S. DOE Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. ORNL is managed by UT-Battelle, LLC, Oak Ridge, TN, USA, for the DOE under contract DE-AC05-00OR22725. United States Government retains and the publisher, by accepting the article for publication, acknowledges that the US Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for US Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE-Public Access Plan (http://energy.gov/downloads/doe-public-access-plan). Disclaimer “This report was prepared as an account of work sponsored by an agency of the US Government. Neither the US Government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the US Government or any agency thereof. The views and opinions of authors expressed herein do not necessarily state or reflect those of the US Government or any agency thereof.”
Abbreviations
- SSF
simultaneous saccharification and fermentation
- SSCF
simultaneous saccharification and co-fermentation
- SHF
separate hydrolysis and fermentation
- DNA
deoxyribonucleic acid
- PCR
polymerase chain reaction
- EPS
exopolysaccharide
- HPLC
high-performance liquid chromotography
- UV
ultraviolet
Contributor Information
Christopher D. Herring, Email: chrisherringfish@gmail.com
William R. Kenealy, Email: wrkenealy@gmail.com
A. Joe Shaw, Email: joeshawiv@gmail.com
Sean F. Covalla, Email: scovalla@lallemand.com
Daniel G. Olson, Email: dan268@gmail.com
Jiayi Zhang, Email: jiayizhang0313@gmail.com.
W. Ryan Sillers, Email: ryansillers@gmail.com
Vasiliki Tsakraklides, Email: vt@aya.yale.edu.
John S. Bardsley, Email: jbardsley@lallemand.com
Stephen R. Rogers, Email: Stephen.r.rogers@icloud.com
Philip G. Thorne, Email: pthorne@lallemand.com
Jessica P. Johnson, Email: jessica92116@yahoo.com
Abigail Foster, Email: abigailfoster123@gmail.com.
Indraneel D. Shikhare, Email: Indraneel.shikhare@gmail.com
Dawn M. Klingeman, Email: klingemandm@ornl.gov
Steven D. Brown, Email: brownsd@ornl.gov
Brian H. Davison, Email: davisonbh@ornl.gov
Lee R. Lynd, Email: lee.r.lynd@dartmouth.edu
David A. Hogsett, Email: dhogsett@xymurgy.com
References
- 1.Zacchi G, Axelsson A. Economic evaluation of preconcentration in production of ethanol from dilute sugar solutions. Biotechnol Bioeng. 1989;34:223–233. doi: 10.1002/bit.260340211. [DOI] [PubMed] [Google Scholar]
- 2.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humbird D, Davis R, Tao L, Kinchin C, Hsu D, Aden A, Schoen P, Lukas J, Olthof B, Worley M, Sexton D, Dudgeon D. Process design and economics for conversion of lignocellulosic biomass to ethanol. NREL technical report NREL/TP-5100–51400. 2011.
- 4.Laser M, Jin H, Jayawardhana K, Dale BE, Lynd LR. Projected mature technology scenarios for conversion of cellulosic biomass to ethanol with coproduction thermochemical fuels, power, and/or animal feed protein. Biofuels Bioprod Biorefin. 2009;3:231–246. doi: 10.1002/bbb.131. [DOI] [Google Scholar]
- 5.Paye J, Guseva A, Hammer S, Gjersing E, Davis M, Davison B, Olstad J, Donohoe B, Nguyen T, Wyman C, Pattathil S, Hahn M, Lynd L. Biological lignocellulose solubilization: comparative evaluation of biocatalysts and enhancement via cotreatment. Biotechnol Biofuels. 2016;9:8. doi: 10.1186/s13068-015-0412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw AJ, Podkaminer KK, Desai SG, Bardsley JS, Rogers SR, Thorne PG, Hogsett D, Lynd LR. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci USA. 2008;105:13769–13774. doi: 10.1073/pnas.0801266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw AJ, Hogsett DA, Lynd LR. Natural competence in thermoanaerobacter and thermoanaerobacterium species. Appl Environ Microbiol. 2010;76:4713–4719. doi: 10.1128/AEM.00402-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Currie DH, Raman B, Gowen CM, Tschaplinski TJ, Land ML, Brown SD, Covalla SF, Klingeman DM, Yang ZK, Engle NL, Johnson CM, Rodriguez M, Shaw a J, Kenealy WR, Lynd LR, Fong SS, Mielenz JR, Davison BH, Hogsett D, Herring CD. Genome-scale resources for Thermoanaerobacterium saccharolyticum. BMC Syst Biol. 2015;9:30. doi: 10.1186/s12918-015-0159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw AJ, Covalla SF, Miller BB, Firliet BT, Hogsett DA, Herring CD. Urease expression in a Thermoanaerobacterium saccharolyticum ethanologen allows high titer ethanol production. Metab Eng. 2012;14:528–532. doi: 10.1016/j.ymben.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Herring C, Kenealy W, Shaw A, Raman B, Tschaplinski T, Brown S, Davison B, Covalla S, Sillers W, Xu H, Tsakraklides V, Hogsett D. Final report on development of Thermoanaerobacterium saccharolyticum for the conversion of lignocellulose to ethanol. US Dep Energy Tech Rep DOE/GO/017057-1 2012.
- 11.Podkaminer KK, Kenealy WR, Herring CD, Hogsett DA, Lynd LR. Ethanol and anaerobic conditions reversibly inhibit commercial cellulase activity in thermophilic simultaneous saccharification and fermentation (tSSF) Biotechnol Biofuels. 2012;5:43. doi: 10.1186/1754-6834-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Currie DH, Herring CD, Guss AM, Olson DG, Hogsett DA, Lynd LR. Hogsett DA, Lynd LR: Functional heterologous expression of an engineered full length CipA from Clostridium thermocellum in Thermoanaerobacterium saccharolyticum. Biotechnol Biofuels. 2013;6:32. doi: 10.1186/1754-6834-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw AJ, Covalla SF, Hogsett DA, Herring CD. Marker removal system for Thermoanaerobacterium saccharolyticum and development of a markerless ethanologen. Appl Environ Microbiol. 2011;77:2534–2536. doi: 10.1128/AEM.01731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kacmar J, Gilbert A, Cockrell J, Srienc F. The cytostat: a new way to study cell physiology in a precisely defined environment. J Biotechnol. 2006;126:163–172. doi: 10.1016/j.jbiotec.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 15.Borden JR, Papoutsakis ET. Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl Environ Microbiol. 2007;73:3061–3068. doi: 10.1128/AEM.02296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw AJ, Miller BB, Rogers SR, Kenealy WR, Meola A, Bhandiwad A, Sillers WR, Shikhare I, Hogsett DA, Herring CD. Anaerobic detoxification of acetic acid in a thermophilic ethanologen. Biotechnol Biofuels. 2015;8:75. doi: 10.1186/s13068-015-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillmann F, Fischer R-J, Saint-Prix F, Girbal L, Bahl H. PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol Microbiol. 2008;68:848–860. doi: 10.1111/j.1365-2958.2008.06192.x. [DOI] [PubMed] [Google Scholar]
- 18.Russell JB. Glucose toxicity in Prevotella ruminicola: methylglyoxal accumulation and its effect on membrane physiology. Appl Environ Microbiol. 1993;59:2844–2850. doi: 10.1128/aem.59.9.2844-2850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Olson DG, Lanahan AA, Tian L, Murphy SJL, Lo J, Lynd LR. Physiological roles of pyruvate ferredoxin oxidoreductase and pyruvate formate-lyase in Thermoanaerobacterium saccharolyticum JW/SL-YS485. Biotechnol Biofuels. 2015;8:138. doi: 10.1186/s13068-015-0304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Currie DH, Guss AM, Herring CD, Giannone RJ, Johnson CM, Lankford PK, Brown SD, Hettich RL, Lynd LR. Profile of secreted hydrolases, associated proteins, and SlpA in Thermoanaerobacterium saccharolyticum during the degradation of hemicellulose. Appl Environ Microbiol. 2014;80:5001–5011. doi: 10.1128/AEM.00998-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23(3):396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 22.Lynd LR, Guss AM, Beri D, Herring C, Holwerda EK, Murphy SJ, Olson DG, Paye J, Rydzak T, Shao X, Tian L, Worthen R. Advances in consolidated bioprocessing using Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. In: Liao J, Wittmann C, editors. Industrial biotechnology. Weinheim: Wiley-VCH; 2016. [Google Scholar]
- 23.Cray JA, Stevenson A, Ball P, Bankar SB, Eleutherio ECA, Ezeji TC, Singhal RS, Thevelein JM, Timson DJ, Hallsworth JE. Chaotropicity: a key factor in product tolerance of biofuel-producing microorganisms. Curr Opin Biotechnol. 2015;49:228–259. doi: 10.1016/j.copbio.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O’Toole GA. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol. 2006;72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;13:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]


