ABSTRACT
This study aims to investigate the clinical significance of serum autoantibodies against MDM2 and c-Myc and evaluate their feasibility in the immunodiagnosis of lung cancer. 50 sera samples with 43 available paired lung cancer tissue and adjacent normal tissue slides with follow up information and 44 sera from normal human controls (NHC) were used in the research group. Another 62 lung cancer sera and 43 NHC sera were used in the validation group. The results of IHC showed that MDM2 and c-Myc protein were overexpressed in lung cancer tissues compared to adjacent normal tissues (p < 0.001). Likewise, significantly higher levels of serum autoantibodies against MDM2 and c-Myc were found in lung cancer compared to NHC both in research and validation groups. Further analysis on IHC and ELISA results showed that serum level of autoantibodies against these two TAAs were positively associated with tissue staining scores (both p < 0.05). The area under curve (AUC) values of anti-MDM2 and anti-cMyc autoantibodies for discriminating lung cancers from NHC were 0.698 and 0.636 in research group, 0.777 and 0.815 in the validation group, respectively. Both anti-MDM2 and anti-c-Myc autoantibodies can discriminate stage I lung cancer patients from NHC with AUC values of 0.703 and 0.662. Kaplan–Meier analysis showed that higher level of serum anti-c-Myc autoantibodies was significantly related to shortened disease-free survival (DFS) (p = 0.041). In conclusion, our finding suggested that serum MDM2 and c-Myc autoantibodies may have the potential to serve as non-invasive diagnostic biomarkers in patients with lung cancer.
KEYWORDS: Autoantibody, biomarker, c-Myc, immunodiagnosis, lung cancer, MDM2, tumor-associated antigen
Introduction
Lung cancer is the most common cause of cancer death both in men and women in the United States.1 Main symptoms of lung cancer include cough, chest pain, hoarseness, weight loss and loss of appetite. However, these symptoms are not specific to lung cancer. Due to the atypical symptoms of early stage lung cancer, most patients are diagnosed with advanced lung cancer when surgical removal of the cancer are less curable because metastasis may have already occurred.2 Overall, eighty percent of lung cancer patients were diagnosed with late stage disease owing to the paucity of early diagnostic methods.3 Five-year survival rate following removal of Stage IA/B lung cancer is approximately 70% and for Stage IIA/B disease is in the range of 50% while overall five-year survival for lung cancer is only 18%.4,5 So it would be of great significance to find effective and applicable clinical screening methods or biomarkers. In the past few decades, the National Lung Screening Trial (NLST) reported that low-dose CT (LDCT) can greatly improve the likelihood of detection of lung cancer at an earlier and potentially more curable stage in people at high risk.6,7 However, the low specificity of LDCT scan has limited its clinical implementation as a screening tool. As early as in the 1980s, serum carcinoembryonic antigen (CEA) and tissue polypeptide antigen (TPA) were introduced as early detection biomarkers for lung cancer.8 After that, several trials have provided more serum biomarkers with definitive clinical relevance and applicability, including serum protein and circulating microRNA.9-13 Although current serum biomarkers are convenient, they are not feasible to serve as screening tool due to the lack of adequate diagnostic values. Previous studies have shown that tumor-associated antigen (TAA) that aberrantly expressed in cancers could be sensed as foreign antigens and autoantibodies were subsequently produced.14 Autoantibodies against TAAs in the serum of cancer patients are more stable and last longer than other potential markers, including TAAs themselves.15 In recent years, the utilization of TAAs and anti-TAA antibody systems as early cancer biomarkers to monitor therapeutic outcomes or as indicators of disease prognosis have been extensively explored.15 Our previous studies showed that anti-MDM2 and anti-c-Myc autoantibodies could serve as diagnostic biomarkers in solid tumors, including hepatocellular carcinoma and esophageal squamous cell carcinoma.16,17
MDM2, an oncogene, was firstly discovered in a locus amplified on double minute chromosomes in a tumorigenic mouse cell line (3T3-DM),18 which can attenuate p53 function, increase cancer risk, accelerate tumor formation and progression,19,20 Accumulating data indicate that MDM2/MDM2 is involved in a variety of tumors including invasive ductal breast cancer, gastric cancer, urothelial neoplasia and hepatocellular carcinoma.21-25 Martin et al.24 reported that MDM2 is a potential predictive biomarker for the neo-adjuvant chemotherapy response in pediatric osteosarcoma. As an oncogenic transcription factor, c-Myc plays a pivotal role in function of cellular proliferation. Brian et al.26 showed that MDM2-binding protein (MTBP) could increase Myc-mediated transcription, proliferation and transformation, and it was a novel regulator of MYC. C-Myc has been identified as a driver of gene signatures associated with a poor prognosis in different cancers including lung cancer, and it is a necessary component of metastatic cell behavior.27,28
However, their feasibility as biomarkers in lung cancer remains largely unknown. The present study aims to evaluate the clinical significance of the expression of MDM2 and c-Myc in tissues from lung cancer patients and their corresponding autoantibodies in serum samples from lung cancer patients.
Materials and methods
General information of sera and tissue samples
A total of 50 sera from lung cancer patients with detailed clinical information and 3-y-follow-up information, with 43 pairs of corresponding lung cancer tissue and adjacent normal tissue slides (corresponding tissue slides of 7 among these 50 patients were unavailable) were provided by Lung Cancer Biospecimen Resource Network (University of Virginia, USA). These sera were collected before the patients received lung cancer surgery. The patients' clinical characteristics are summarized in the results section. Forty-four normal human sera were obtained from the serum bank of Cancer Autoimmunity and Epidemiology Research Laboratory at the University of Texas, El Paso (UTEP). These sera were collected during annual health examinations. Another 62 lung cancer sera and 43 normal human sera were also obtained from the same serum bank to serve as validation group. All patients with lung cancer were confirmed by histopathological diagnosis. This study was approved by the Institutional Review Board of University of Texas at El Paso and the Institutional Review Board of University of Virginia.
Immunohistochemistry (IHC) with tissue slides
IHC analysis was performed using mouse monoclonal anti-MDM2 antibody (ab3110, Abcam Inc., Cambridge, MA, 1:1000 dilution) and rabbit monoclonal antibody against c-Myc (ab32072, Abcam Inc., Cambridge, MA, 1:200 dilution) according to the manufacturers' instructions. All the results were analyzed by two independent pathologists. Intensity as well as the area stained were considered in the IHC scoring process.29 A four-level scoring system was used to evaluate the staining intensity as follows: “−,” negative, score 0; “+,” low expression, score 1; “++,” moderate expression, score 2; and “+++,” high expression, score 3. The percentage of positive cells were divided into four levels and scored accordingly, as were 1 (up to 25%), 2 (25%–50%), 3 (50%–75%) and 4 (75%–100%). Final results were scored by multiplying the intensity by the percentage of positivity cells scores. Final scores greater than 2 were considered as positive results.
Purification of recombinant proteins
Recombinant proteins of MDM2, c-Myc were derived from our previous studies. The purification of MDM2 and c-Myc was described previously.30-32
Enzyme-linked immunosorbent assay (ELISA)
Autoantibodies against MDM2 and c-Myc in human sera were tested by ELISA, which was described in detail in our previous study.31 The optical density (OD) value of each well was read at 405 nm, and the cut-off value for determining a positive reaction was designated as the mean OD value of control sera plus 3 standard deviations (mean + 3SD). All samples were tested in duplicate to confirm the results.
Western blotting and serum absorption
Serum samples that were determined to contain autoantibodies by ELISA were further tested by immunoblotting to confirm the immunoreactivity of the sera. The purified recombinant proteins were transferred to a nitrocellulose membrane and then cut into 15 to 20 stripes and incubated with selected sera diluted at 1:200 respectively, and finally incubated with HRP-conjugated goat anti-human IgG diluted 1:10000 (Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA). Positive signals were captured by autoradiography using chemiluminescence (Piece Biotechnology, Rockford, IL), according to the manufacturer's instructions.
Statistical analysis
All data are presented as mean ± standard deviation (SD). Independent t test, Fisher's exact test, One-way ANOVA or χ2 test was used to check whether there was statistic difference where applicable. Spearman rank correlation analysis was used to check the association between serum levels of autoantibodies against MDM2 or c-Myc and IHC scores in lung cancer patients. Multivariable or univariate Cox proportional hazards regression model was used to discover predictors for lung cancer overall survival (OS) and disease-free survival (DFS). Receiver operating characteristic (ROC) was curved to illustrate the diagnostic performance of serum anti-MDM2 and anti-c-Myc autoantibodies. DFS curves were yielded by using the Kaplan–Meier method, and differences were examined using log-rank tests. All p values were two sided, and those less than 0.05 were considered statistically significant. All statistical analysis was carried out using the IBM SPSS Statistics 21.0 (IBM Corp. Armonk, NY).
Results
MDM2 and c-Myc protein expression were higher in lung cancer tissues than in adjacent normal tissues
As shown in Table 1, 39 out of 43 lung cancer tissues (90.7%) were positively stained with MDM2, and 42 out of 43 lung cancer tissues (97.7%) were positive with c-Myc. Out of the 43 adjacent normal tissues, 16 (37.2%) were positively stained with MDM2, and 14 (32.6%) were positively stained with c-Myc. The frequency of MDM2 and c-Myc expression in lung cancer tissues was significantly higher than that in the adjacent normal tissues (both p < 0.001). IHC staining patterns and cellular localizations of MDM2 and c-Myc protein in the lung cancer tissues are shown in Fig. 1. Due to the limited number of tissue specimens in this study, the analysis on the frequency of MDM2, c-Myc positive staining in the lung cancer tissues with different clinical parameters such as tumor stages, gender and age showed no significant correlation (data not shown).
Table 1.
MDM2 and c-Myc Protein Expression in 43 pair-wise lung cancer tissues.
| Type of tissues | Cases | MDM2 positive | p | c-Myc positive | p |
|---|---|---|---|---|---|
| Adjacent normal tissue | 43 | 16 (37.2%) | 0.000** | 14 (32.6%) | 0.000** |
| Lung cancer tissue | 43 | 39 (90.7%) | 42 (97.7%) | ||
| Adenocarcinoma | 22 | 20 (90.9%) | 21 (95.5%) | ||
| Squamous cell carcinoma | 16 | 15 (93.8%) | 16 (100%) | ||
| Others | 5 | 4 (80%) | 5 (100%) |
p < 0.01 (χ2-test)
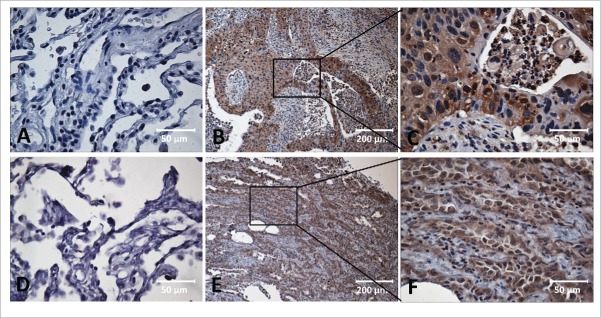
Figure 1.
Immunohistochemical staining of MDM2, c-Myc in Lung cancer tissue and adjacent normal tissue slides. Tissue slides were stained with monoclonal anti-MDM2 antibody (A, B, C) and anti-c-Myc antibody (D, E, F), respectively. (A) Negative stain pattern of MDM2 in representative adjacent normal tissue (×400 magnification). (B and C) Positive stain pattern of MDM2 in representative lung cancer tissue (×100 and ×400 magnification). (D) Negative stain pattern of c-Myc in representative adjacent normal tissue (×400 magnification). (E and F) Positive stain pattern of c-Myc in representative lung cancer tissue (×100 and ×400 magnification).
Frequency of autoantibodies against MDM2 and c-Myc in sera from patients with lung cancer
The frequencies of serum autoantibodies against MDM2 and c-Myc in lung cancer sera are shown in Table 2. When the cut-off value for determining a positive reaction was set as mean plus 3SD, 18% (9/50) anti-MDM2 and 16% (8/50) anti-c-Myc autoantibodies were detected in sera from 50 patients with lung cancer in the research group. However, the reactivity of normal human sera was relatively low, the frequencies were 2.3% (1/44, anti-MDM2), 2.3% (1/44, anti-c-Myc), respectively. Combinational utilization of anti-MDM2 and anti-c-Myc autoantibodies can increase the positive rate to 26% (13/50). In the validation group, autoantibodies against these two TAAs also showed high positive rate (Table 2).
Table 2.
Frequency of autoantibody against MDM2 and c-Myc in human sera by ELISA.
| Sera | anti-MDM2 | p | anti-c-Myc | p | anti-MDM2 or anti-c-Myc | p | |
|---|---|---|---|---|---|---|---|
| Research group | Lung cancer (n = 50) | 9 (18.0% ) | 8 (16.0%) | 13 (26.0%) | |||
| NHC (n = 44) | 1 (2.3% ) | 0.018* | 1 (2.3% ) | 0.034* | 2 (4.5%) | 0.005** | |
| Validation group | Lung cancer (n = 62) | 23 (37.1%) | 22 (35.5%) | 37 (59.7%) | |||
| NHC (n = 43) | 1 (2.3%) | 0.000** | 1 (2.3%) | 0.000** | 2 (4.7%) | 0.000** |
p < 0.05
p < 0.01 (χ2-test or Fisher's exact test)
IHC scores of MDM2 and c-Myc were positively associated with serum autoantibodies against these two TAAs in lung cancer patients
Spearman's rank correlation analysis was performed to explore the link between MDM2 and c-Myc IHC scores and serum autoantibodies against these two TAAs in 43 lung cancer cases with paired tissues and sera samples. The results showed that MDM2 and c-Myc protein expression in lung cancer tissues were positively associated with serum autoantibodies against these two TAAs (rs= 0.360, p = 0.018 and rs = 0.526, p < 0.001, respectively. Fig. 2). The results indicated that higher serum autoantibodies against these two TAAs were positively correlated with their protein expression levels in lung cancer tissues respectively. So we further checked whether serum autoantibodies against these two TAAs could serve as diagnostic biomarkers for lung cancer.
Figure 2.
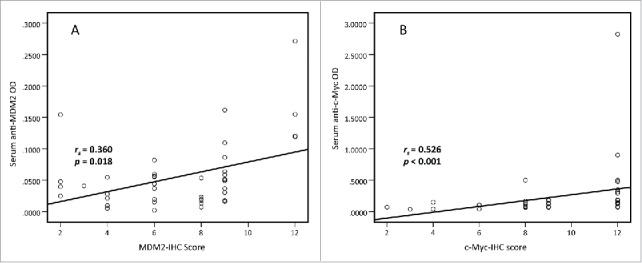
Spearman correlation of MDM2 and c-Myc IHC scores and serum levels of autoantibodies against MDM2 and c-Myc. Spearman correlation analysis was used to explore the link between MDM2 and c-Myc IHC scores and serum autoantibodies against these two TAAs in 43 lung cancer cases with paired tissues and sera samples. The results show that MDM2 and c-Myc protein expression in lung cancer tissues are positively associated with serum autoantibodies against these two TAAs respectively. (rs = 0.360, p = 0.018 and rs = 0.526, p < 0.001, respectively).
Autoantibodies against MDM2 and c-Myc were relatively higher in lung cancer than in normal controls
The presence of circulating anti-MDM2 and anti-c-Myc autoantibodies in sera from 50 lung cancer patients (including 13 patients with grade 1 lung cancer, 37 patients with grade 2 or 3 lung cancer) and 44 NHC were detected by ELISA. Compared to NHC, serum autoantibodies against these two TAAs were significantly higher in patients with lung cancer (Fig. 3A, p = 0.001; Fig. 4A, p = 0.049). Subsequently, we generated ROC curves to assess the potential significance of serum anti-MDM2 and anti-c-Myc antibodies as a noninvasive biomarker for the immunodiagnosis of lung cancer. ROC curve analyses revealed that serum anti-MDM2 and anti-c-Myc autoantibodies were robust in discriminating lung cancer patients from NHC with AUC values of 0.698 (95% CI: 0.592–0.805) and 0.636 (95% CI: 0.524–0.747), respectively (Fig. 3B, Fig. 4B). Moreover, one-way ANOVA analysis showed that serum anti-MDM2 autoantibodies significantly increased in patients with stage I and III lung cancer (p = 0.006 and p = 0.001 respectively, Fig 3E). However, serum anti-c-Myc autoantibodies showed no significantly difference between NHC and patients with stage I, II and III lung cancer (p = 0.093, p = 0.295 and p = 0.281 respectively; Fig. 4E). We further conducted ROC curve analyses to check whether anti-MDM2 or anti-c-Myc autoantibodies could discriminate stage-specific lung cancer from NHC. Serum anti-MDM2 autoantibodies could discriminate stage I and III lung cancer from NHC with AUC values of 0.703 and 0.866 respectively (both p < 0.01; Fig. 3F, 3H), and anti-c-Myc autoantibodies could discriminate stage I lung cancer from NHC with AUC values of 0.662 (p = 0.020; Fig. 4F). However, anti-MDM2 autoantibodies failed to discriminate stage II lung cancer from NHC (p = 0.161; Fig. 3G) and anti-c-Myc autoantibodies failed to discriminate stage II and III lung cancer from NHC (p = 0.444 and p = 0.152, respectively; Fig. 4G, 4H).
Figure 3.
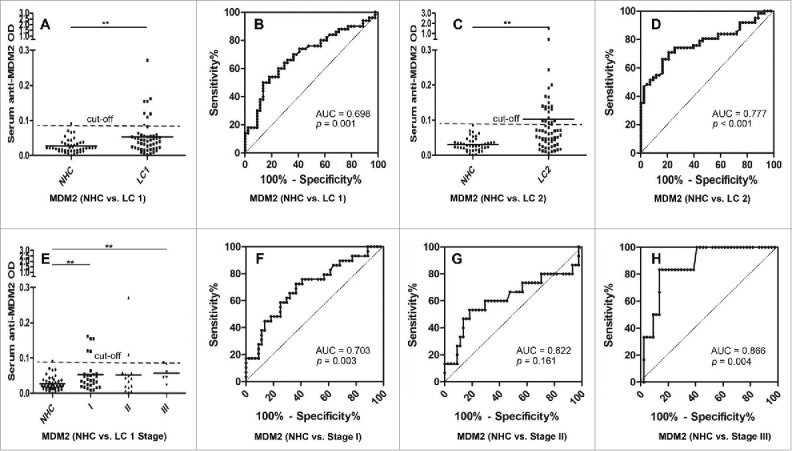
Serum anti-MDM2 autoantibodies in lung cancer and NHC. In Fig. 3A, 3C and 3E, Y-axis represents OD value and X-axis represents serum samples (*p < 0.05; **p < 0.01). (A) Serum anti-MDM2 autoantibodies distribution in NHC (n = 44) and lung cancer patients (n = 50) (p = 0.001). (B) ROC curve analysis using serum anti-MDM2 autoantibodies for discriminating lung cancer patients from NHC in research group (AUC: 0.698, 95% CI: 0.592–0.805, p = 0.001). (C) Serum anti-MDM2 autoantibodies distribution in the validation group including NHC (n = 43) and lung cancer patients (n = 62) (p = 0.005). (D) ROC curve yielded by OD values of serum anti-MDM2 autoantibodies for discriminating lung cancer patients from NHC in the validation group (AUC: 0.777, 95% CI: 0.689–0.865, p < 0.001). (E) Serum anti-MDM2 autoantibodies distribution and in NHC and lung cancer patients with different TNM stage. (F) ROC curve yielded by OD values of anti-MDM2 autoantibodies in discriminating stage I lung cancer patients from NHC (AUC: 0.703; 95% CI: 0.5800–0.827, p = 0.003). (G) ROC curve yielded by OD values of anti-MDM2 autoantibodies in discriminating stage II lung cancer patients from NHC (AUC: 0.622; 95% CI: 0.432–0.812, p = 0.161). (H) ROC curve yielded by OD values of anti-MDM2 autoantibodies in discriminating stage I lung cancer patients from NHC (AUC: 0.866; 95% CI: 0.741–0.991, p = 0.004). Note: LC1: 50 lung cancer patients from research group. LC2: 62 lung cancer patients from validation group.
Figure 4.
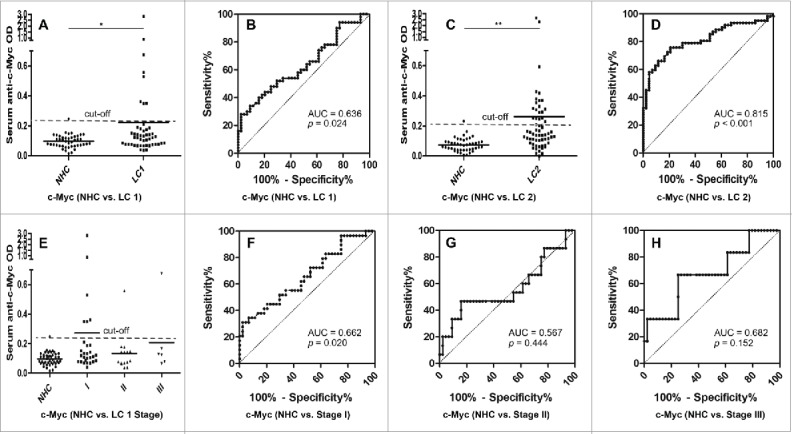
Serum anti-c-Myc autoantibodies in lung cancer patients and NHC. In Fig 4A, 4C and 4E, Y-axis represents OD value and X-axis represents serum samples (*p < 0.05; **p < 0.01). (A) Serum anti-c-Myc autoantibodies distribution in NHC (n = 44) and lung cancer patients (n = 50) (p = 0.049). (B) ROC curve analysis using serum anti-c-Myc autoantibodies for discriminating lung cancer patients from NHC in research group (AUC: 0.636, 95% CI: 0.524–0.747, p = 0.024). (C) Serum anti-c-Myc autoantibodies distribution in the validation group including NHC (n = 43) and lung cancer patients (n = 62) (p = 0.001). (D) ROC curve yielded by OD values of serum anti-c-Myc autoantibodies for discriminating lung cancer patients from NHC in the validation group (AUC: 0.815, 95% CI: 0.733–0.896, p < 0.001). (E) Serum anti-c-Myc autoantibodies distribution and in NHC and lung cancer patients with different TNM stage. (F) ROC curve yielded by OD values of anti-c-Myc autoantibodies in discriminating stage I lung cancer patients from NHC (AUC: 0.662; 95% CI: 0.532–0.791, p = 0.020). (G) ROC curve yielded by OD values of anti-c-Myc autoantibodies in discriminating stage II lung cancer patients from NHC (AUC: 0.567; 95% CI: 0.377–0.756, p = 0.444). (H) ROC curve yielded by OD values of anti-c-Myc autoantibodies in discriminating stage I lung cancer patients from NHC (AUC: 0.682; 95% CI: 0.439–0.924, p = 0.152). Note: LC1: 50 lung cancer patients from research group. LC2: 62 lung cancer patients from validation group.
To further confirm the reproducibility of serum anti-MDM2 and anti-c-Myc autoantibodies as diagnostic markers in lung cancer, we have tested these two biomarkers in a validation group, including patients with lung cancer (n = 62) and NHC (n = 43). Interestingly, autoantibodies against these two TAAs were also significantly higher in patients with lung cancer (both p < 0.01; Fig. 3C, Fig. 4C). ROC curves analyses further confirmed that anti-MDM2 and anti-c-Myc autoantibodies were robust for discriminating lung cancer patients from NHC with AUC of 0.777 (95% CI: 0.689–0.865, p < 0.001) and 0.815 (95% CI: 0.733–0.896, p < 0.001) respectively (Fig 3D, Fig 4D). Due to the lack of clinical characteristics in the validation group, we were not able to do further analyze on other clinical parameters.
Anti-MDM2 autoantibodies were significantly lower in lung cancer patients with history of malignancies in 50 lung cancer patients
We have summarized the characteristics of 50 lung cancer patients in the research group in Table 3. Furthermore, we divided lung cancer cases from the research group into different subgroups according to different clinical parameters and tried to explore their relations. Table 3 illustrates that serum level anti-MDM2 autoantibodies is significantly lower in lung cancer patients with history of malignancies (some primary lung cancer patients accompanied with other malignancies when enrolled, such as basal cell carcinoma, multiple myeloma and melanoma.) (p = 0.005). For other clinicopathologic features, the results showed no significant difference.
Table 3.
Relationship between clinicopathological factors and autoantibodies against MDM2 and c-Myc in lung cancer patients.
| anti-MDM2 |
anti-c-Myc |
||||
|---|---|---|---|---|---|
| Mean ± SD | p | Mean ± SD | p | ||
| Mean age ± SD | 66.0±9.9 | ||||
| Age range | 45-86 | ||||
| 21 | 0.0641 ± 0.0600 | 0.1434 ± 0.1079 | |||
| ≥65 | 29 | 0.0448 ± 0.0400 | 0.178 | 0.2643 ± 0.5240 | 0.305 |
| Gender | |||||
| Male | 28 | 0.0544 ± 0.0608 | 0.2195 ± 0.5155 | ||
| Female | 22 | 0.0510 ± 0.0320 | 0.812 | 0.2059 ± 0.2097 | 0.908 |
| TMN stages | |||||
| 1 (IA+ IB) | 29 | 0.0525 ± 0.0448 | 0.2656 ± 0.5214 | ||
| 2 (≥II A) | 21 | 0.0536 ± 0.0570 | 0.939 | 0.1416 ± 0.1230 | 0.292 |
| Depth of tumor invasion | |||||
| 1(T1) | 15 | 0.0687 ± 0.0752 | 0.1522 ± 0.1249 | ||
| 2(≥T2) | 35 | 0.0462 ± 0.0329 | 0.282 | 0.2398 ± 0.4790 | 0.491 |
| Lymph node Metastasis | |||||
| Negative | 34 | 0.0493 ± 0.0427 | 0.460 | 0.2527 ± 0.4856 | 0.326 |
| Positive | 16 | 0.0606 ± 0.0632 | 0.1303 ± 0.1022 | ||
| Histological grade | |||||
| I | 13 | 0.0457 ± 0.0455 | 0.1263 ± 0.1200 | ||
| II | 19 | 0.0447 ± 0.0255 | 0.342 | 0.1651 ± 0.1948 | 0.326 |
| III | 18 | 0.0668 ± 0.0683 | 0.3276 ± 0.6356 | ||
| Histology | |||||
| Adenocarcinoma | 26 | 0.0468 ± 0.0398 | 0.1633 ± 0.1779 | ||
| Squamous cell | 16 | 0.0587 ± 0.0622 | 0.2963+0.6777 | ||
| carcinoma | |||||
| Others | 8 | 0.0614 ± 0.0554 | 0.663 | 0.2111 ± 0.1873 | 0.598 |
| Post-op chemo | |||||
| None | 33 | 0.0487 ± 0.0439 | 0.2439 ± 0.4938 | ||
| yes | 17 | 0.0612 ± 0.0602 | 0.407 | 0.1545 ± 0.1139 | 0.467 |
| Tumor size | |||||
| < 4cm | 21 | 0.0563 ± 0.0661 | 0.1395 ± 0.1071 | ||
| ≥4cm | 29 | 0.0505 ± 0.0346 | 0.719 | 0.2670 ± 0.5235 | 0.278 |
| Chest wall/Mediastinal invasion | |||||
| None | 45 | 0.0520 ± 0.0517 | 0.1544 ± 0.1526 | ||
| yes | 5 | 0.0615 ± 0.0286 | 0.689 | 0.7453 ± 1.1732 | 0.323 |
| Metastasis | |||||
| None | 36 | 0.0612 ± 0.0546 | 0.2326 ± 0.4703 | ||
| Yes | 14 | 0.0317 ± 0.0250 | 0.059 | 0.1642 ± 0.1541 | 0.598 |
| History of Malignancies | |||||
| None | 34 | 0.0629 ± 0.0569 | 0.005* | 0.2483 ± 0.4864 | 0.382 |
| Yes | 16 | 0.0318 ± 0.0165 | 0.1393 ± 0.1042 | ||
p < 0.05
p < 0.01 (t-test or One-way ANOVA)
Cox proportional hazards regression model analysis and survival analysis
In order to determine whether serum anti-MDM2 and anti-c-Myc autoantibodies could serve as predictors for lung cancer patients' outcome, we divided 50 lung cancer patients from the research group into two subgroups by using median OD values of anti-MDM2 and anti-c-Myc autoantibodies as cutoff values, respectively. Kaplan–Meier analysis revealed that patients with higher level of serum anti-c-Myc autoantibodies showed a significantly shorter DFS (p = 0.041, log-rank test; Fig. 5). Univariate and multivariate Cox proportional hazards regression model was further used in this study to identify important prognostic factors for OS and DFS (Table 4). Univariate analysis showed that chest wall/mediastinal invasion and metastasis were significantly related to shortened DFS (p = 0.017 and p = 0.037, respectively). However, no significant findings were found for the relations of these factors and OS. Multivariate analysis revealed that TNM stage was independent predictor for OS (p = 0.046), chest wall/mediastinal invasion and metastasis were independent predictors for DFS (p = 0.008 and p = 0.019, respectively).
Figure 5.
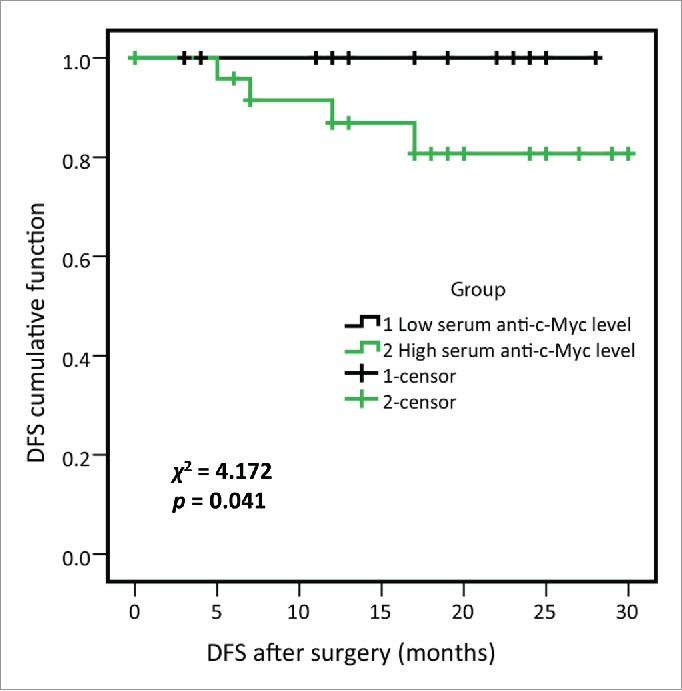
Disease-free survival (DFS) of high- and low-serum anti-c-Myc autoantibodies levels in lung cancer patients from research group. 50 lung cancer patients from research group were divided into two subgroups by using the median OD value of serum anti-c-Myc autoantibodies, as were high- and low-serum anti-c-Myc autoantibodies expression subgroups respectively, and 25 lung cancer patients were included in each subgroup. Log-rank test showed that lung cancer patients with higher anti-c-Myc autoantibodies were associated with shortened DFS (χ2 = 4.172, p = 0.041).
Table 4.
Univariate and Multivariate analyses of OS/DFS (Cox proportional hazards regression model).
| Factors | OS univariate analysis |
DFS univariate analysis |
||
|---|---|---|---|---|
| HR(95% CI) | p | HR(95% CI) | p | |
| Pathology | 1.271 (0.281–5.740) | 0.755 | 1.416 (0.329–6.182) | 0.635 |
| T | 0.983 (0.098–9.861) | 0.988 | 1.544(0.159–14.980) | 0.708 |
| N | 6.523 (0.556–76.475) | 0.135 | 2.468 (0.346–17.617) | 0.368 |
| Stage | 6.740 (0.673–67.458) | 0.104 | 5.106 (0.527–49.475) | 0.159 |
| Grade | 2.492 (0.373–16.675) | 0.346 | 0.292 (0.030–2.807) | 0.286 |
| History of malignancies | 1.787 (0.248–12.856) | 0.564 | 2.484 (0.350–17.641) | 0.363 |
| Family Hx of lung cancer | 2.026 (0.180–22.743) | 0.567 | 1.074 (0.111–10.355) | 0.951 |
| Largest tumor on CT/PET scan (cm) | 0.792 (0.111–5.605) | 0.817 | 0.922 (0.129–6.592) | 0.935 |
| Chest wall/Mediastinal invasion | 7.101 (0.960–52.519) | 0.055 | 11.090 (1.584–79.437) | 0.017* |
| Post-op chemo | 2.584 (0.352–18.431) | 0.354 | 2.043 (0.287–14.529) | 0.476 |
| Metastasis after surgery | 8.770 (0.905–85.020) | 0.061 | 11.920 (1.165–121.938) | 0.037* |
| Serum c-Myc level (high/low) | 54.131 (0.002–1414979.961) | 0.442 | 68.224 (0.022–212584.586) | 0.304 |
| Serum MDM2 level (high/low) | 0.670 (0.087–5.131) | 0.700 | 1.091 (0.153–7.749) | 0.931 |
| Factors | OS multivariate analysis |
DFS multivariate analysis |
||
| |
HR(95% CI) |
p |
HR(95% CI) |
p |
| Stage | 5.398 (1.028–28.339) | 0.046* | – | – |
| Chest wall/Mediastinal invasion | – | – | 28.261 (2.364–337.872) | 0.008** |
| Metastasis after surgery | – | – | 27.377 (1.721–435.490) | 0.019* |
p < 0.05
p < 0.01
Western blot confirmed the ELISA results
Serum samples were also analyzed by Western blot to confirm the results from ELISA. Sixteen sera from research group and validation group were chosen to do Western Blot as described previously. Negative control and positive control were also set as quality control. The results of Western Blot further confirmed the results from ELISA. All the results are shown in Fig. 6.
Figure 6.
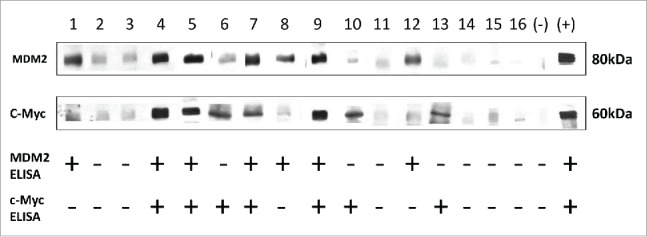
Western Blot results validated the results from ELISA. Western Blot analysis with representative sera from lung cancer patients and NHC. Lanes 1–10 (research group), Lanes 11, 12 (validation group), sera from lung cancer patients; Lanes 13–16, sera from NHC. Lanes 4, 5, 7 and 9 show immunoreactivity with both MDM2 and c-Myc recombinant proteins; Lanes 6, 10 and 13 show strong reactivity with c-Myc recombinant protein, while they show weak reactivity with MDM2; Lanes 1, 8 and 12 show strong reactivity with MDM2 recombinant protein, while they show weak reactivity with c-Myc; Lanes 2, 3, 11, 14, 15 and 16 show no reactions with MDM2 or c-Myc. The last two lanes are negative control and positive control of MDM2 and c-Myc, respectively. Note: MDM2 and c-Myc proteins transferred to the NC membrane were purified his-tag proteins.
Discussion
Lung cancer remains the most common cancer worldwide, both in term of new cases and cancer deaths.33 Due to the lack of ideal biomarkers, it is difficult to diagnose lung cancer while in its early stage.34-36 Up to now, only four non-small cell lung cancer biomarkers have been considered as “emerging” markers to clinical molecular diagnostics: EGFR gene expression, ERCC gene expression, RRM1 gene expression, K-ras gene mutation.37 Predictive (diagnostic) and prognostic biomarkers are helpful in matching targeted therapies with patients and in preventing toxicity of standard (systemic) therapies.38 It has long been known that the sera from cancer patients contain antibodies that can react with a unique group of autologous cellular antigens called TAAs.14,15 Many studies have demonstrated that MDM2 is a key negative regulator of the tumor suppressor protein p53, which is mutated in a variety of cancers, including breast carcinoma, sarcoma, leukemia, melanoma and glioblastoma.39,40 Accumulating evidences have also shown that MDM2 may inhibit p53 bioactivity by blocking its transcriptional activity and promoting p53 protein degradation.41-43 c-Myc is an oncogene that has overexpressed/dysregulated expression in 70% of human malignancies,28 and its activation appears to be associated with cell growth, division, and differentiation.44 It is demonstrated by Zhang et al. that Nilotinib can treat patients whose cancer cells overexpress MDM2.45 Therefore, the strategy of targeting MDM2 and c-Myc via utilization of antagonists has been indicated as a potential approach for anticancer therapy.26,46
In the present study, we found that the expression of MDM2 and c-Myc protein in lung cancer tissues were significantly higher than that in adjacent normal lung tissues using IHC analysis (both p < 0.001). These data are consistent with reports from other groups and further confirm that MDM2 and c-Myc may play an important role in the tumorigenesis of lung cancer.47,48 Moreover, we analyzed the presence of anti-MDM2 and anti-c-Myc autoantibodies in sera from lung cancer patients. It was worth noting that serum autoantibodies against these two TAAs were positively associated with IHC scores in 43 available lung cancer tissues. This indicates that serum levels of autoantibodies against these two TAAs might be useful in discriminating lung cancers from normal controls. To further demonstrate the potential feasibility of serum autoantibodies against-MDM2 and c-Myc in the immunodiagnosis of lung cancer, we further analyzed the diagnostic value of these two autoantibodies in the research group and validation group. Current study shows a significant increase of serum anti-MDM2 and anti- c-Myc autoantibodies in lung cancer patients compared to controls, and the results were further confirmed in the validation group. Some studies found that MTBP not only can regulate MDM2, but also can serve as a potential transcriptional regulator of Myc to enhance the activation of Myc-dependent genes, which are critical for proliferation and transformation.26,49 These evidences further demonstrated that mutant MDM2 can induce c-Myc imbalance and accelerate carcinogenic process. ROC curve analyses were further used to assess the diagnostic performance of anti-MDM2 and anti-c-Myc autoantibodies in lung cancer. Evidence is provided here that these two biomarkers have good diagnostic performance both in research group and validation group. The reason why sensitivity is higher in the validation group needs to be further explored since clinical information of lung cancer patients from the validation group is unavailable. The higher frequency and higher titer of anti-MDM2 and anti-c-Myc autoantibodies in sera from patients with lung cancer also support the possibility of using these anti-TAA autoantibodies as biomarkers in lung cancer detection. ROC curve analyses in stage-specific lung cancer patients and NHC indicated that these two biomarkers have good diagnostic value in early stage lung cancer.
Upon the availability of clinical features in lung cancer patients from the research group, we have analyzed these features to check which can serve as predictors for OS and DFS. Univariate Cox proportional hazards regression model revealed that chest wall/mediastinal invasion and tumor metastasis were independent predictors for DFS. Multivariate Cox proportional hazards regression model revealed that TNM stage was independent predictor of OS, and further confirmed that chest wall/mediastinal invasion and tumor metastasis were independent predictors for DFS. Kaplan–Meier survival analysis illustrated that higher serum level of anti-c-Myc autoantibodies was related with shortened DFS. This is partially consistent with similar finding in breast cancer that increased MYC gene signature was associated with shortened DFS in breast cancer.50 It indicates that higher level of serum anti-c-Myc autoantibodies might be an indicator for tumor recurrence. One of the interesting findings is that serum anti-MDM2 antibody level was significantly lower in lung cancer patients with history of malignancies (p = 0.005). We hypothesize that for some patients with history of malignancies, standard cancer treatments which were used may trigger immune responses and further affect tumor control which might influence autoantibody responses. However, more studies are warranted to further confirm these findings.
In conclusion, MDM2 and c-Myc may play a key role in the development and progression various cancers including lung cancer, and autoantibodies against MDM2 and c-Myc are presented in lung cancer sera during the malignant transformation. From a clinical perspective, our data provide novel evidence that serum anti-MDM2 and/or anti-c-Myc antibodies can serve as potential biomarkers for the early diagnosis and indicators for tumor recurrence in lung cancer patients. However, more studies with larger sample size should be performed in the future.
Abbreviations
- AUC,
area under curve;
- CEA,
carcinoembryonic antigen;
- DFS,
disease-free survival;
- ELISA,
enzyme-linked immunosorbent assay;
- IHC,
immunohistochemistry;
- LDCT,
low-dose computed tomography;
- MTBP,
MDM2-binding protein;
- NHC,
normal human control;
- NLST,
the National Lung Screening Trial;
- OD,
optical density;
- OS,
overall survival;
- ROC,
receiver operating characteristic;
- SD,
standard deviation;
- TAA,
tumor-associated antigen;
- TNM,
the TNM classification of malignant tumors;
- TPA,
tissue polypeptide antigen.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
The following authors conceived and designed the experiments: PL JXS LPD YRC HFZ MK PK BFY JYZ. The following authors performed the experiments: PL JXS LPD YRC HFZ MK PK JYZ. The following authors analyzed the data: PL JXS LPD HFZ. The following authors contributed reagents/materials/analysis tools: PL JXS LPD YRC HFZ MK PK JYZ. The following authors contributed to the writing of the manuscript: PL JXS LPD JYZ.
Funding
This work was supported by a grant (SC1CA166016) from the National Institutes of Health (NIH), Program for New Century Excellent Talents in University, Ministry of Education, China (NCET-11-0949), a grant from National Science Foundation of China (NSFC, 81372371), and a grant from Greater East Cancer Center through MDHonors/WorldOne, London, UK. We would also like to thank the Border Biomedical Research Center (BBRC) Core facilities at The University of Texas at El Paso (UTEP) for their help, which were funded by NIH grant (5G12MD007592).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65:5–29; PMID:25559415; http://dx.doi.org/ 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 2.Midthun DE. Early diagnosis of lung cancer. F1000 Prime Rep 2013; 5:12; PMID:23585930; http://dx.doi.org/ 10.12703/P5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arkin DM, Ferlay J, Curado MP, Bray F, Edwards B, Shin HR, Forman D. Fifty years of cancer incidence: CI5 I-IX. Int J Cancer 2010; 127:2918–27; PMID:21351270; http://dx.doi.org/ 10.1002/ijc.25517 [DOI] [PubMed] [Google Scholar]
- 4.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L, International Association for the Study of Lung Cancer International Staging C et al.. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007; 2:706–14; PMID:17762336; http://dx.doi.org/ 10.1097/JTO.0b013e31812f3c1a [DOI] [PubMed] [Google Scholar]
- 5.Howlader N, Noone A, Krapcho M, Garshell J, Miller D, Altekruse S, Kosary C, Yu M, Ruhl J, Tatalovich Z et al.. SEER cancer statistics review, 1975–2012. National Cancer Institute; Bethesda, MD: http://seercancergov/csr/1975_2012/2015 [Google Scholar]
- 6.Henschke CI, McCauley DI, Yankelevitz DF, Naidich DP, McGuinness G, Miettinen OS, Libby DM, Pasmantier MW, Koizumi J, Altorki NK et al.. Early Lung Cancer Action Project: overall design and findings from baseline screening. Lancet 1999; 354:99–105; PMID:10408484; http://dx.doi.org/ 10.1016/S0140-6736(99)06093-6 [DOI] [PubMed] [Google Scholar]
- 7.National Lung Screening Trial Research T, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM et al.. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011; 365:395–409; PMID:21714641; http://dx.doi.org/ 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salvati F, Antilli A, Cruciani AR, Pau F, Flore F, Munno R, De Angelis G, Cipri A, Pigorini F. Plasma carcinoembryonic antigen and tissue polypeptide antigen levels in lung cancer: correlation with cell types and stage. Cancer Detect Prev 1985; 8:111–4; PMID:4064030 [PubMed] [Google Scholar]
- 9.Huang HL, Wu YC, Su LJ, Huang YJ, Charoenkwan P, Chen WL, Lee HC, Chu WC, Ho SY. Discovery of prognostic biomarkers for predicting lung cancer metastasis using microarray and survival data. BMC Bioinformatics 2015; 16:54; PMID:25881029; http://dx.doi.org/ 10.1186/s12859-015-0463-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiura K, Shiraishi A, Suzuki C, Takamura K, Yamamoto M, Komori H, Watanabe Y, Iwaki-Egawa S. MMP-9/ANC score as a predictive biomarker for efficacy of bevacizumab plus platinum doublet chemotherapy in patients with advanced or recurrent non-squamous non-small cell lung cancer. Cancer Biomark 2015; 15:433–40; PMID:25835177; http://dx.doi.org/ 10.3233/CBM-150483 [DOI] [PubMed] [Google Scholar]
- 11.Wang RJ, Zheng YH, Wang P, Zhang JZ. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol 2015; 8:765–71; PMID:25755772 [PMC free article] [PubMed] [Google Scholar]
- 12.Thunnissen E, van der Oord K, den Bakker M. Prognostic and predictive biomarkers in lung cancer. A review. Virchows Arch 2014; 464:347–58; PMID:24420742; http://dx.doi.org/ 10.1007/s00428-014-1535-4 [DOI] [PubMed] [Google Scholar]
- 13.Ahn J, Cho J. Current serum lung cancer biomarkers. J Mol Biomark Diagn 2013; 4:2; http://dx.doi.org/10.4172/2155-9929.S4-001 [Google Scholar]
- 14.Tan EM, Zhang JY. Autoantibodies to tumor-associated antigens: reporters from the immune system. Immunol Rev 2008; 222:328–40; PMID:18364012; http://dx.doi.org/ 10.1111/j.1600-065X.2008.00611.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang JY, Tan EM. Autoantibodies to tumor-associated antigens as diagnostic biomarkers in hepatocellular carcinoma and other solid tumors. Expert Rev Mol Diagn 2010; 10:321–8; PMID:20370589; http://dx.doi.org/ 10.1586/erm.10.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Zheng SJ, Chen Y, Li N, Ren PF, Dai LP, Duan ZP, Zhang JY. Autoantibody response to murine double minute 2 protein in immunodiagnosis of hepatocellular carcinoma. J Immunol Res 2014; 2014:906532; PMID:24955377; http://dx.doi.org/10.1155/2014/906532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chai Y, Peng B, Dai L, Qian W, Zhang Y, Zhang JY. Autoantibodies response to MDM2 and p53 in the immunodiagnosis of esophageal squamous cell carcinoma. Scand J Immunol 2014; 80:362–8; PMID:24965442; http://dx.doi.org/ 10.1111/sji.12202 [DOI] [PubMed] [Google Scholar]
- 18.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM Jr, Hofmainster C, Alder H et al.. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010; 18:367–81; PMID:20951946; http://dx.doi.org/ 10.1016/j.ccr.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biomed Res 2013; 27:254–71; PMID:23885265; http://dx.doi.org/ 10.7555/JBR.27.20130030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 2010; 20:299–309; PMID:20172729; http://dx.doi.org/ 10.1016/j.tcb.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Q, Lao X, Chen Z, Lai H, Deng Y, Wang J, Mo C, Sui J, Wu J, Zhai L et al.. TP53 and MDM2 gene polymorphisms, gene-gene interaction, and hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLoS One 2013; 8:e82773; PMID:24376578; http://dx.doi.org/ 10.1371/journal.pone.0082773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park HS, Park JM, Park S, Cho J, Kim SI, Park BW. Subcellular localization of Mdm2 expression and prognosis of breast cancer. Int J Clin Oncol 2014; 19:842–51; PMID:24292333; http://dx.doi.org/ 10.1007/s10147-013-0639-1 [DOI] [PubMed] [Google Scholar]
- 23.Gunther T, Schneider-Stock R, Hackel C, Kasper HU, Pross M, Hackelsberger A, Lippert H, Roessner A. Mdm2 gene amplification in gastric cancer correlation with expression of Mdm2 protein and p53 alterations. Mod Pathol 2000; 13:621–6; PMID:10874665; http://dx.doi.org/ 10.1038/modpathol.3880107 [DOI] [PubMed] [Google Scholar]
- 24.Martin JW, Chilton-MacNeill S, Koti M, van Wijnen AJ, Squire JA, Zielenska M. Digital expression profiling identifies RUNX2, CDC5L, MDM2, RECQL4, and CDK4 as potential predictive biomarkers for neo-adjuvant chemotherapy response in paediatric osteosarcoma. PLoS One 2014; 9:e95843; PMID:24835790; http://dx.doi.org/ 10.1371/journal.pone.0095843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lianes P, Orlow I, Zhang ZF, Oliva MR, Sarkis AS, Reuter VE, Cordon-Cardo C. Altered patterns of MDM2 and TP53 expression in human bladder cancer. J Natl Cancer Inst 1994; 86:1325–30; PMID:8064890; http://dx.doi.org/ 10.1093/jnci/86.17.1325 [DOI] [PubMed] [Google Scholar]
- 26.Grieb BC, Gramling MW, Arrate MP, Chen X, Beauparlant SL, Haines DS, Xiao H, Eischen CM. Oncogenic protein MTBP interacts with MYC to promote tumorigenesis. Cancer Res 2014; 74:3591–602; PMID:24786788; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volm M, Koomagi R. Prognostic relevance of c-Myc and caspase-3 for patients with non-small cell lung cancer. Oncol Rep 2000; 7:95–8; PMID:10601599; http://dx.doi.org/10.3892/or.7.1.95 [DOI] [PubMed] [Google Scholar]
- 28.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8:976–90; PMID:19029958; http://dx.doi.org/ 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- 29.van Diest PJ, van Dam P, Henzen-Logmans SC, Berns E, Van der Burg M, Green J, Vergote I. A scoring system for immunohistochemical staining: consensus report of the task force for basic research of the EORTC-GCCG. European Organization for Research and Treatment of Cancer-Gynaecological Cancer Cooperative Group. J Clin Pathol 1997; 50:801; PMID:9462258; http://dx.doi.org/ 10.1136/jcp.50.10.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JY, Megliorino R, Peng XX, Tan EM, Chen Y, Chan EKL. Antibody detection using tumor-associated antigen mini-array in immunodiagnosing human hepatocellular carcinoma. J Hepatol 2007; 46:107–14; PMID:17067715; http://dx.doi.org/ 10.1016/j.jhep.2006.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang JY, Casiano CA, Peng XX, Koziol JA, Chan EK, Tan EM. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol Biomarkers Prev 2003; 12:136–43; PMID:12582023 [PubMed] [Google Scholar]
- 32.Dai L, Li J, Ortega R, Qian W, Casiano CA, Zhang JY. Preferential autoimmune response in prostate cancer to cyclin B1 in a panel of tumor-associated antigens. J Immunol Res 2014; 2014:827827; PMID:24860838; http://dx.doi.org/ 10.1155/2014/827827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136:E359–E86; PMID:25220842; http://dx.doi.org/ 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 34.Ardizzoni A, Cafferata MA, Tiseo M, Filiberti R, Marroni P, Grossi F, Paganuzzi M. Decline in serum carcinoembryonic antigen and cytokeratin 19 fragment during chemotherapy predicts objective response and survival in patients with advanced nonsmall cell lung cancer. Cancer 2006; 107:2842–9; PMID:17103443; http://dx.doi.org/ 10.1002/cncr.22330 [DOI] [PubMed] [Google Scholar]
- 35.Wang WJ, Tao Z, Gu W, Sun LH. Clinical observations on the association between diagnosis of lung cancer and serum tumor markers in combination. Asian Pac J Cancer Prev 2013; 14:4369–71; PMID:23992005; http://dx.doi.org/ 10.7314/APJCP.2013.14.7.4369 [DOI] [PubMed] [Google Scholar]
- 36.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13:83–96; PMID:23303139; http://dx.doi.org/ 10.1038/nrc3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 2013; 31:1039–49; PMID:23401433; http://dx.doi.org/ 10.1200/JCO.2012.45.3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalia M. Biomarkers for personalized oncology: recent advances and future challenges. Metab Clin Exp 2015; 64:s16-21; PMID:25468140; http://dx.doi.org/ 10.1016/j.metabol.2014.10.027 [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y, Yu H, Hu W. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim Biophys Sin (Shanghai) 2014; 46:180–9; PMID:24389645; http://dx.doi.org/ 10.1093/abbs/gmt147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol 2003; 13:49–58; PMID:12507556; http://dx.doi.org/ 10.1016/S1044-579X(02)00099-8 [DOI] [PubMed] [Google Scholar]
- 41.Nag S, Zhang X, Srivenugopal KS, Wang MH, Wang W, Zhang R. Targeting MDM2-p53 interaction for cancer therapy: are we there yet? Curr Med Chem 2014; 21:553–74; PMID:24180275; http://dx.doi.org/ 10.2174/09298673113206660325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res 2008; 14:5318–24; PMID:18765522; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toledo F, Wahl GM. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int J Biochem Cell Biol 2007; 39:1476–82; PMID:17499002; http://dx.doi.org/ 10.1016/j.biocel.2007.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan S, Sikora K. The potential of oncogene products as tumour markers. Cancer Surv 1987; 6:185–207; PMID:3322544 [PubMed] [Google Scholar]
- 45.Zhang H, Gu L, Liu T, Chiang KY, Zhou M. Inhibition of MDM2 by nilotinib contributes to cytotoxicity in both Philadelphia-positive and negative acute lymphoblastic leukemia. PLoS One 2014; 9:e100960; PMID:24968304; http://dx.doi.org/ 10.1371/journal.pone.0100960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res 2004; 2:1–8; PMID:14757840 [PubMed] [Google Scholar]
- 47.Chen X, Qiu J, Yang D, Lu J, Yan C, Zha X, Yin Y. MDM2 promotes invasion and metastasis in invasive ductal breast carcinoma by inducing matrix metalloproteinase-9. PLoS One 2013; 8:e78794; PMID:24236052; http://dx.doi.org/ 10.1371/journal.pone.0078794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee KB, Ye S, Park MH, Park BH, Lee JS, Kim SM. p63-Mediated activation of the β-catenin/c-Myc signaling pathway stimulates esophageal squamous carcinoma cell invasion and metastasis. Cancer Lett 2014; 353:124–32; PMID:25045846; http://dx.doi.org/ 10.1016/j.canlet.2014.07.016 [DOI] [PubMed] [Google Scholar]
- 49.Boyd MT, Vlatkovic N, Haines DS. A novel cellular protein (MTBP) binds to MDM2 and induces a G1 arrest that is suppressed by MDM2. J Biol Chem 2000; 275:31883–90; PMID:10906133; http://dx.doi.org/ 10.1074/jbc.M004252200 [DOI] [PubMed] [Google Scholar]
- 50.Horiuchi D, Kusdra L, Huskey NE, Chandriani S, Lenburg ME, Gonzalez-Angulo AM, Creasman KJ, Bazarov AV, Smyth JW, Davis SE et al.. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med 2012; 209:679–96; PMID:22430491; http://dx.doi.org/ 10.1084/jem.20111512 [DOI] [PMC free article] [PubMed] [Google Scholar]