ABSTRACT
Beyond their mere presence, the distribution pattern of inflammatory cells is of special interest. Our hypothesis was that random distribution may be a clear indicator of being non-functional as a consequence of lack of interaction. Here, we have assessed the implication of cell-to-cell distances among inflammatory cells in anal squamous cell carcinoma and a possible association with survival data. Thirty-eight patients suffering from anal carcinoma were studied using tissue microarrays, double staining immunohistochemistry, whole slide scanning and image analysis software. Therapy consisted of concurrent radiochemotherapy. Numbers of stromal and intraepithelial tumor-infiltrating inflammatory cells (TIC) and the distances between cells were quantified. Double-staining of FoxP3+ cells with either CD8+, CD1a+ or CD20+ cells was performed. Measured cell-to-cell distances were compared to computer simulated cell-to-cell distances leading to the assumption of non-randomly distributed and therefore functional immune cells. Intraepithelial CD1a+ and CD20+ cells were randomly distributed and therefore regarded as non-functional. In contrary, stromal CD20+ cells had a non-random distribution pattern. A non-random distance between CD20+ and FoxP3+ cells was associated with a clearly unfavorable outcome. Measured distances between FoxP3+ cells were distinctly shorter than expected and indicate a functional active state of the regulatory T cells (Treg). Analysis of cell-to-cell distances between TIC has the potential to distinguish between suppressed non-functional and functionally active inflammatory cells. We conclude that in this tumor model most of the CD1a+ cells are non-functional as are the intraepithelial CD20+ cells, while stromal CD20+ cells and FoxP3+ cells are functional cells.
KEYWORDS: Anal squamous cell carcinoma, B cells, cell-to-cell distances, cell distribution pattern, dendritic cells, FoxP3, image analysis software, regulatory T cells, radiochemotherapy, short distances, tumor-infiltrating inflammatory cells
Introduction
Inflammatory cells in the microenvironment of solid tumors have become a crucial point in cancer research.1 The so-called TIC are now considered an important part of the host–tumor interaction representing a key player in both host antitumor immunity and tumorous immunoevasion.2,3 In an effort to elucidate the complex cell interactions, research has been strongly focused on quantitative assessments of TIC in the core tumor and the invasive margins. Recent data suggested to include an “immunoscore” in the TNM classification of malignant tumors (TNM).4 Yet, the interaction of inflammatory cells might not only depend on the frequency of immune cells, but also on the inter-individual distances as cells might interact via soluble factors but also via cell-cell contact. Accordingly, the mapping of the tumor microenvironment has recently gained importance.5 Especially Treg interact and regulate the activation, proliferation and functions of different inflammatory cells.5,6 Tregs can modulate the functions of at least CD4+ and CD8+ T cells, natural killer and natural killer T cells, B cells and antigen-presenting cells (APCs).7 Thus, many cancers induce Treg proliferation or generation from naive T cells resulting in an immunoevasion by active immunoediting.8,9 To exert their immunosuppressive function, Tregs need either a direct cell-to-cell contact or a spatial proximity to the target cells for an effective secretion of immunomodulatory cytokines.10 Therefore, a non-random distribution of cells or short cell-to-cell distances could be expected. New methods exists to map the spatial and morphological patterns of cancer and normal cells and can contribute to a more comprehensive understanding of the highly heterogeneous tumor microenvironment.11 The following prerequisites are necessary for the analysis of the distribution pattern of inflammatory cells. First, digital slide scanners enable the acquisition of whole tissue images for analyzing the distribution pattern of a representative amount of cells. Additional imaging analysis software is needed which can identify both tissue structures and individual cells and is able to compute cell-to-cell distances.11,12
Therefore, in the present study we set out to explore whether not only the mere number of TIL in anal cancer biopsies, but also the proximity of individual cell types might have an influence on the outcome of the patients. We used double staining methods to study the interaction of FoxP3+ cells to CD8+, CD1a+ and CD20+ cells and compared the measured cell-to-cell distances with computer simulation-predicted distances.
Results
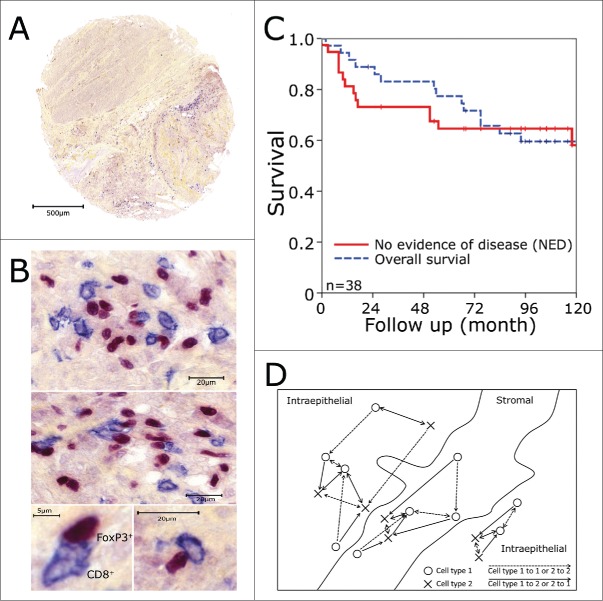
Overall- and no evidence of disease (NED)-survival rates of all patients were 77% and 65% at 5 y and 59% and 58% at 10 y (Fig. 1C), respectively, as published earlier.13,14 Median follow up time was 8 y (range, 2 to 147 mo). In the previous study, only single subsets of TIC in the tumor epithelium were evaluated. Here, we performed double stainings for FoxP3+ (Treg) and CD8 (cytotoxic T cells, CTL) (Fig. 1A, B), CD1a (immature dendritic cells, iDC) or CD20 (B cells), respectively. Additionally, we quantified the numbers of inflammatory cells in both the stromal and intraepithelial compartment and estimated the shortest distance between two cells of the same subset or of different subsets using an image analysis system. Distances were calculated separately for stromal and intraepithelial cells (Fig. 1D). We measured the shortest distances between different cell subsets in the two respective directions, as they may differ distinctly. This is the case when different numbers of the cell subsets are present and they are unevenly distributed. For example, one cell of one type and two cells of the other type are nearest together. So for the single cell, only the shortest distance to the nearest cell of the other subset is calculated, while each of the two other cells has a an individual shortest distance to the single cell (Fig. 1D).
Figure 1.
Tissue microarrays and immunohistochemistry. Biopsy specimens were processed into tissue microarrays using a core diameter of 1.6 mm. Tissue microarray tumor punches (A) were stained for FoxP3+ (red nucleic staining) and CD8+ (blue predominantly membranous staining) as shown in (B). (C) Kaplan–Meier plots for overall survival and no evidence of disease over the whole patients collective. (D) Schematic view on the counting strategy used with the image analysis system. The shortest distance to the next cell was calculated from cell type I to cell type I (dashed line) and from cell type I to cell type II and vice versa (continuous line). Only the cells marked with an arrow were calculated in reference to the other cell. Cells in stromal or intraepithelial compartment were analyzed separately.
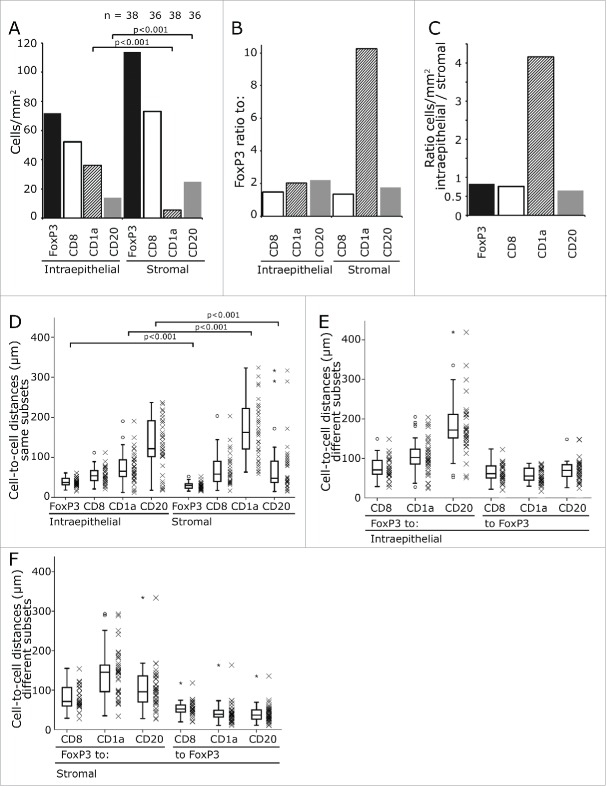
In both compartments, FoxP3+ Tregs were the predominant inflammatory cell subtype followed by the CD8+ CTL (Fig. 2A). FoxP3+ cells outnumbered the other cell types by a factor of 1.5 to 2.2 with the exception of the very rare stromal CD1a+ iDC which they exceeded by a factor of 10 (Fig. 2B). CD1a+ iDC were four times more frequent in the epithelial than in the stromal compartment while all other cell subtypes predominated in the stroma with a factor ranging between 1.2- and 1.5-fold (Fig. 2C).
Figure 2.
Frequency of inflammatory cells and their cell-to-cell distances. Mean values of infiltrating cells in the stromal and intraepithelial compartment. The counts are given per mm2. The numbers on top of the graph give the patients included in the analysis (A). The ratio between FoxP3+ cells and the other cell subsets are displayed (B) and the ratio of the different subsets between intraepithelial and stromal compartment (C). The distances between same cell subsets including FoxP3+ cells (D) and FoxP3+ and the different cell subsets in the intraepithelial (E) and stromal compartment (F) are shown. The distances from FoxP3+ to the other cell subsets and from the other cell subset to FoxP3+ was calculated separately (E, F). The significance values are marked by square brackets and the p value is given on top of it.
The distances of FoxP3+ Treg between each other or other inflammatory cell subtypes varied greatly from a mean distance of about 30 µm to more than 200 µm (Fig. 2D - F). As lymphocytic cells in general have a diameter of about 8–10 µm a measured distance of 10 µm to 14 µm is approximately the distance between two adjacent cells. In our stainings, the surface stained inflammatory cells had a diameter of about 12 µm and the nuclei of the FoxP3 stained cells 6 to 8 µm each in the long axis (Fig. 1B).
We compared the distribution density of inflammatory cells with the cell-to-cell distances (Fig. 3). One would expect that the distances between randomly distributed cells inside an area would be the inverse square root of the cell number per area. The expected cell-to-cell distances in dependence of cell number per mm2 were computer simulated by generating randomly distributed cell-coordinates and calculating the cell-to-cell distances. The expected cell-to-cell distances for randomly distributed cells are included in the graphs. Additionally, the 5% and 95% confidential interval of 200 simulation repetitions were included. The observed experimental data were fitted by Equation 3 (see Materials and Methods). The function takes into account that cells have a spatial extension. Therefore, an offset value in the X and Y direction is included in the used function. The data fits of the simulated values of randomly distributed coordinates can be identified by a curve shape starting with low cell density and a high cell-to-cell distance and a steep decrease of the cell-to-cell distance and an asymptotic shape in the X direction (Fig. 3A–D). Additionally, we used the number of cells per area of each patient and simulated the expected cell-to-cell distance. The computer simulated distances of the 38 patients was averaged and compared to the mean of the observed distances in our collective (Table 1).
Figure 3.
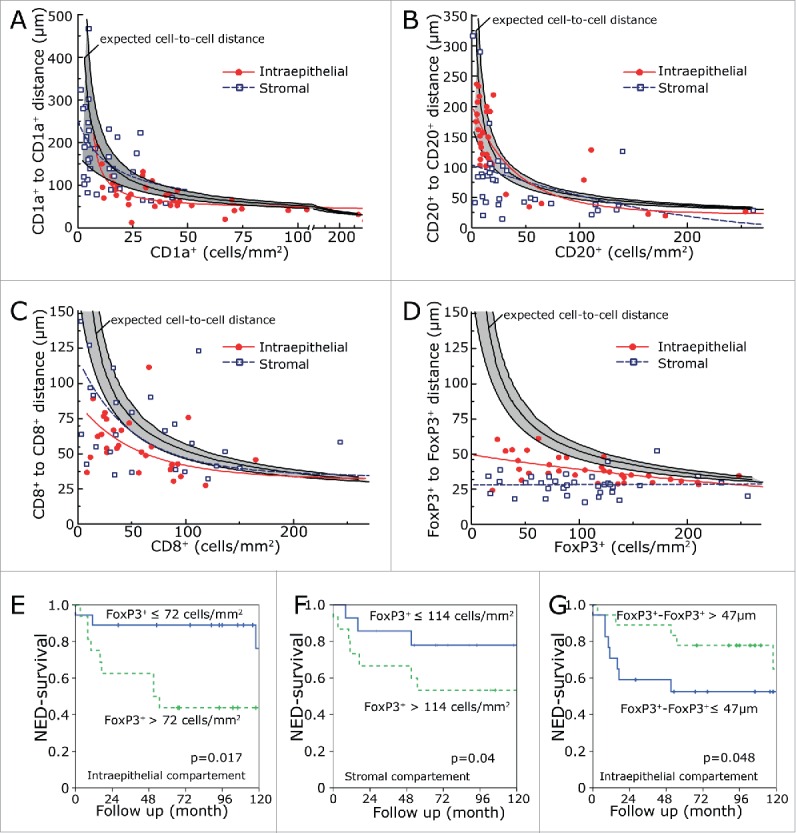
Inflammatory cell-to-cell distances in comparison to inflammatory cell numbers per area. CD1a+-to-CD1a+ distances (A), CD20+-to-CD20+ distances (B), CD8+-to-CD8+ distances (C) and FoxP3+-to-FoxP3+ distances (D) were compared to the numbers of corresponding cells per square mm2. The solid line in the middle of the gray region is the simulated cell-to-cell distance derived from randomly distributed cells. The black lines delimiting the gray region are the 5% and 95% confidential interval of 200 simulation repetitions. The values were fitted by equation 3. Solid red lines represent intraepithelial inflammatory cells and the dashed blue lines the stromal inflammatory cells. Data sets which could not be fitted by equation 3 were fitted by a linear regression. NED survival rates in relation to the intraepithelial (E) and stromal (F) numbers of FoxP3+ cells per mm2. NED survival rates in dependence of the shortest distance between FoxP3+-to-FoxP3+ (G) in the intraepithelial compartment.
Table 1.
Average frequency of inflammatory FoxP3+, CD1a+, CD20+ and CD8+ cells in the intraepithelial and stromal areas and simulated (expected) and observed distances between the lymphocytes.
| Same cell subsets | Different cell subsets |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell-to-cell |
FoxP3-to- |
–to-FoxP3 |
|||||||||
| Cells (mm2) | Expected (µm) | Observed (µm) | Factor | Expected (µm) | Observed (µm) | Factor | Expected (µm) | Observed (µm) | Factor | ||
| FoxP3+ | Intraepithelial | 105.1 | 71.1 | 46 | 0.65 | — | — | — | — | — | — |
| Stromal | 126.9 | 59.8 | 32 | 0.54 | — | — | — | — | — | — | |
| CD1a+ | Intraepithelial | 51.3 | 73.2 | 74 | 1.01 | 76.6 | 105.1 | 1.37 | 56.2 | 55.6 | 0.99 |
| Stromal | 12.4 | 160.5 | 172 | 1.07 | 196.0 | 142.8 | 0.73 | 58.3 | 40.6 | 0.70 | |
| CD20+ | Intraepithelial | 47.4 | 153.2 | 150 | 0.98 | 167.1 | 168.3 | 1.01 | 75.2 | 71.8 | 0.96 |
| Stromal | 71.9 | 116.7 | 76 | 0.65 | 130.3 | 105.2 | 0.81 | 53.2 | 40.5 | 0.76 | |
| CD8+ | Intraepithelial | 71.0 | 83.3 | 55 | 0.66 | 86.1 | 74.6 | 0.87 | 78.0 | 64.3 | 0.82 |
| Stromal | 92.9 | 96.0 | 70 | 0.73 | 99.3 | 81.48 | 0.82 | 66.8 | 53.7 | 0.80 | |
Observed distance is the estimated shortest distance between two cells of the same subset or of different subsets by the use of an image analysis system. Expected distance is computer simulated by the use of the cell numbers of the 38 samples to simulate the expected cell-to-cell distances and to compare it to the calculated distances in the tissue samples. The observed values were divided by the expected values to yield the portion (factor). The same was done for the distances between the FoxP3 cells and CD1a, CD20+ and CD8+ cells and from the CD1a, CD20+ and CD8+ to the FoxP3 cells.
Cell-to-cell distances of intraepithelial and stromal CD1a, CD20+, CD8+ and FoxP3+ cells were each compared to the computer simulated values (Fig. 3A–D). For CD1a, cells cell-to-cell distances were quite similar to the expected values (Fig. 3A). The same is true for intraepithelial CD20 cells (Fig. 3B). Stromal CD20 cell-to-cell distances were shorter than expected. The same is true for CD8+ cells and FoxP3 cells (Fig. 3C, D). Especially, FoxP3-to-FoxP3 distances were distinctly shorter than expected both for intraepithelial and stromal location by a factor of 0.65 and 0.54, respectively (Fig. 3D, Table 1).
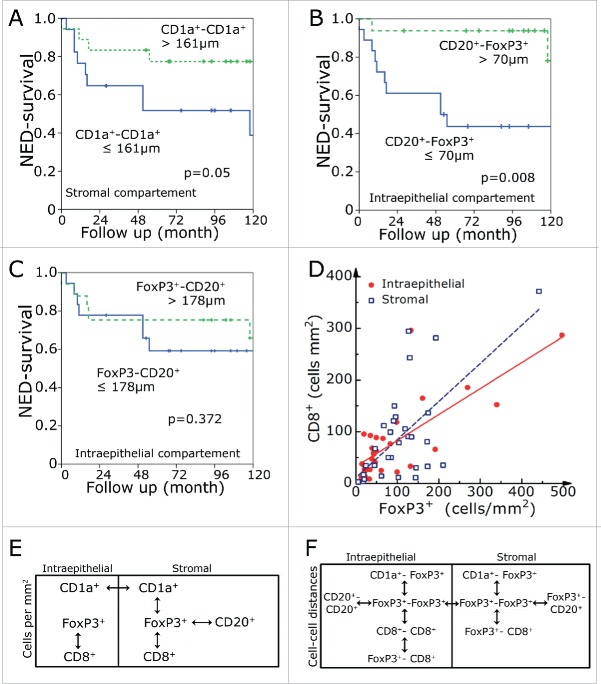
In our previous study, only intraepithelial inflammatory cell numbers were counted and we could not find a relationship between FoxP3 and prognosis.14 In the present investigation, however, we found an association of low FoxP3+ cell counts with a favorable prognosis in both the intraepithelial (p = 0.017) and stromal (p = 0.04) compartment (Fig. 3E, F). In line with this, long distances between intraepithelial FoxP3+ lymphocytes were prognostically beneficial (Fig. 3G). None of intraepithelial or stromal counts of CD8+ CTL, CD1a+ iDC or CD20+ B cell reached a significant prognostic value (Table S1).
CD1a+-to-CD1a+ cell distances corresponded much more to the computer simulated cell-to-cell distances than FoxP3+-to-FoxP3+. This is true for both the intraepithelial and stromal compartment. Patients with CD1a+-to-CD1a+ cell distances longer than 161 µm in the intraepithelial compartment had an improved prognosis (p = 0.05) (Fig. 4A). Additionally, we determined the distances between the different lymphocyte subsets. As stated above, we measured the shortest distance of a cell subset to the respective other surrounding cell subset. The CD20+-FoxP3+ distances in the intraepithelial compartment reached a clear prognostic value (Fig. 4B). Interestingly, the FoxP3+-CD20+ distances had no significant prognostic value (Fig. 4B). None of the other distances between different lymphocyte subsets had a clear prognostic value. All of the prognostic relevant markers for NED had at least a trend for a better overall survival (Fig. S1).
Figure 4.
NED survival rates in dependence of the shortest distance between stromal CD1a +-to-CD1a+ cells (A) and distances between intraepithelial CD20+-to-FoxP3+ (B) and FoxP3+-to-CD20+ (C) cells. Correlation of FoxP3+ and CD8+ cells numbers in the intraepithelial and stromal compartment (D). Interrelations within the intraepithelial and stromal regions and between the two regions for frequency of cells (E) and cell-to-cell distance (F). The double-headed arrows indicate a significance level of p < 0.001.
Additionally, we were interested in the correlations between cell counts and cell-to-cell distances. We studied the correlations and found that FoxP3+ numbers correlate with CD8+ numbers in the stromal and intraepithelial compartment. In between the two compartments only CD1a+ cells had a positive correlation (Fig. 4E). The distances of FoxP3+-to-FoxP3+ correlated between stromal and intraepithelial compartment and to the CD1a+-to-CD1a+ distances. Intraepithelial FoxP3-distances correlated to CD20+ and CD8+ distances. Stromal FoxP3+ distances correlated to the FoxP3+-CD20+ and FoxP3+-CD8+ distances (Fig. 4F).
Discussion
Here, we used double staining of FoxP3+ cells with CD1a+, CD20+ or CD8+, whole slide scanning and image analysis software to determine the distances between the same cell subsets and between FoxP3+ cells and the other cell subsets. Additionally, we computer simulated the expected cell-to-cell distances based on the cell numbers of the different cell subsets and compared it to the observed cell-to-cell distances.
We hypothesize that active inflammatory cells are not randomly distributed in the tissue, because lymphocytes interact with antigens and therefore have a target driven, non-random distribution. In reverse, we speculate that suppressed inflammatory cells have a more random distribution pattern and have cell-to-cell distances in accordance with our computer simulated cell-to-cell distances. We observed that tumor infiltrating lymphocytes have different distribution patterns. Dendritic CD1a+ cells and intraepithelial CD20+ cells are randomly distributed. Contrary to this finding, stromal CD20+ cells have a clearly non-random distribution. So we might interpret that all CD1a cells and the intraepithelial CD20+ cells in the tumor core are suppressed by the tumor microenvironment, while stromal CD20+ cells located in the invasive margin are not exposed to this suppressive influence. Further evidence for this assumption is that short distances between CD20+ cells and FoxP3+ Tregs were associated with an unfavorable prognosis regarding NED- and overall survival, indicating that Tregs provoke this effect by suppressing CD20+ cells.
Intraepithelial and stromal CD8+ cells have a similar distribution pattern and a somehow shorter cell-to-cell distance than CD8+-to-FoxP3+ distance as expected. However, CD8+ is not prognostically relevant and it is difficult to judge whether the CD8+ cells are suppressed by FoxP3+ cells or not.
Especially, FoxP3+ cells have short cell-to-cell distances regardless of the number of cells per area. FoxP3+ cells are closer to other FoxP3+ cells in the stromal compartment than in the intraepithelial compartment and stromal FoxP3+ cells have shorter distances to the different other cell subsets than expected. This indicates that FoxP3+ cells are functionally active cells possibly having the ability to suppress other cells. This is supported by the finding that low numbers of FoxP3+ cells are prognostically favorable. It is in line with the general assumption that high numbers of Tregs suppress the host immune response against cancer cells. However, in several tumor entities an inverse effect was found with high numbers of Tregs associated with a good prognosis.15 Using a similar approach in a collective of patients with gastric cancer, we found that high numbers of FoxP3+ cells and short CD8+-to-FoxP3 distances have a favorable metastasis-free survival. We argued that Treg could control cancer-associated inflammation and thus control inflammatory mediators and therefore tumor growth.16 Here, long FoxP3+-to-FoxP3+ distances are associated with a favorable prognosis. This is in line with high FoxP3 cell numbers per square mm2 having an unfavorable prognosis. Long distances could be a consequence of low FoxP3+ numbers or a result of more randomly distributed FoxP3+ cells indicating a less active FoxP3+ subset. Yet, there are some limitations to our study: given the retrospective nature of our histologic approach, no in vivo characteristics such as chemokine microenvironment or dynamic cells interactions could be investigated. Further functional in vivo studies are necessary to elucidate this aspect. Moreover, we selectively investigated one tumor entity, namely anal carcinoma. Following studies will have to focus on the validity of our findings for other tumor entities.
Conclusions
The analysis of cell-to-cell distances between TIC has the potential to distinguish between non-functional suppressed and functional active inflammatory cells. We conclude that in our anal cancer cohort most of the CD1a+ cells are non-functional as well as the intraepithelial CD20+ cells, while stromal CD20+ cells and FoxP3+ cells are functional cells.
Materials and methods
Patient selection
A total of 112 patients with anal squamous carcinoma were treated by combined radiochemotherapy at the Erlangen University Hospital between 1985 and 2001. For this study, a subgroup of 38 patients was selected according to the following criteria as presented in a previous study.13 Only HIV negative patients with anal carcinoma >2 cm without evidence of distant metastases treated by radiochemotherapy and with available paraffin embedded biopsy specimen were included. For a detailed resume of patient pathologic characteristics and treatment protocols, see the study by Grabenbauer et al.14 The use of the tissue sections and the patients’ survival data following patient consent was approved by the Friedrich-Alexander University Erlangen-Nuremberg ethics committee.
Treatment protocol
The primary tumor region as well as inguinal, perirectal and internal iliac/presacral nodes were treated by radiation using a four-field box technique with 10 to 15 MV photons with single fractions of 1.8 Gy and a total dose of 50.4 Gy. All patients received a simultaneous chemotherapy with two cycles of 5-fluorouracil and a single i.v. bolus of mitomycin C. Regular clinical follow-ups including rectoscopy, CT scans of the abdominal pelvis and chest X-ray were performed as previously described.13,14
Tissue microarray and immunohistochemistry
Paraffin-embedded biopsy samples obtained from 38 patients with anal squamous cell carcinoma before radiochemotherapy were processed into a tissue microarray (Biocat, Heidelberg, Germany) using a core diameter of 1.6 mm (Fig. 1A). Between 2 and 7 cores were taken per patient (mean 2.8), resulting in a total of 120 cores. Analyzed intraepithelial and stromal area and counted cell numbers are given in Table S2. Immunohistochemistry of paraffin sections was carried out using an immunohistochemical double staining method including streptavidin biotinylated alkaline phosphatase (ABC-AP, DakoCytomation, Hamburg, Germany).17,18 Each paraffin section was double stained with the following primary antibodies FoxP3–CD1, (Abcam, Cambridge, United Kingdom–DakoCytomation, Hamburg, Germany), FoxP3–CD8+ (Abcam–DakoCytomation, Fig. 1B), FoxP3–CD20 (Abcam–DakoCytomation) followed by secondary antibody. The detailed procedure of the immunohistochemical double staining method is as follows.
The paraffin sections were dewaxed using first Xylol and then descending alcohol series. After rehydration and treatment with 3% H2O2 for 10 min at room temperature to block endogenous peroxidase paraffin sections were cooked in a steam cooker (Biocarta Europe, Hamburg, Germany) for 5 min with Target retrieval solution (DakoCytomation, Hamburg, Germany), pH 6.0. Overnight sections were incubated with diluted primary antibody for FoxP3 (polyclonal goat-anti-FoxP3; 1:100) and then the biotin-labeled secondary antibody (biotin rabbit-anti-goat; 1:100) was added for 30 min at RT. Afterwards sections were incubated with streptavidin-biotinylated horse-radish-peroxidase-complex for 30 min (ABC-PO, DakoCytomation, Hamburg, Germany) followed by tyramide signal amplification (TSA) for 10 min to enhance the signal. Biotin was visualized using streptavidin-biotinylated horse-radish-peroxidase-complex (ABC-PO, DakoCytomation, Hamburg, Germany). Aminoethyl carbazole (AEC, Zytomed, Berlin, Germany) was used as chromogen.
All slides stained in a second step using a double staining enhancer (Zymed, San Francisco, California, USA) for 30 min at RT followed by an avidin and biotin block (Avidin/Biotin Blocking Kit, Vector Laboratories, Inc.. Burlingame, CA) for 15 min each at RT to hinder unspecific bindings. Thereafter, slides were incubated with primary antibodies against CD1a (DakoCytomation, Hamburg, Germany; mouse-anti-CD1a; 1:2), CD8+ (DakoCytomation, Hamburg, Germany; mouse-anti-CD8+; 1:100) or CD20 (DakoCytomation, Hamburg, Germany; mouse-anti-CD20; 1:500) for 1 h at RT. After applying a post-Block-solution, secondary antibodies connected covalently with an AP-Polymer (ZytoChem-Plus, Berlin, Germany). Finally, Fast Blue was used as chromogen.
Stained slides were scanned with a high throughput scanner (Zeiss, Mirax MIDI Scan, Göttingen, Germany) at a magnification of 1:200 and transferred to a PC (resolution 0.2308 µm/pixel). TIC were counted using the semiautomatic image analysis program COUNT (Biomas, Erlangen, Germany).14,19 The software automatically identifies cells with a high specificity. With the subsequent visual adjustment, a high sensitivity is reached. We used a three-color thresholding approach to detect the two different colors of the double staining. Numbers of labeled tumor-infiltrating cells were determined per 1 mm2 and the shortest mean distance between cells of the same subset and of the different subsets was calculated (Fig. 1D). According to histopathology, two distinct compartments were investigated: on the one hand cells were counted intraepithelially in the tumor core, on the other hand cells counts were determined in the surrounding stromal compartment.
Cell-to-cell distances
When measuring the cell-to-cell distance some points have to be considered: There is a relationship between the distribution density of cells and the distances between the cells. For uniformly distributed cells inside an area the distances between cells would be expected to be the inverse square root of the lymphocytes per area:
where n is the number of cells per area and p0 is the matching coefficient. However, this equation is only true for theoretical cells without any extension and with a uniform spatial distribution. Therefore, to fit the data points we used a similar equation and introduced a constant for the Y offset due to the diameter of the cells and the non-uniform distribution of the cells to fit the data points:
where p2 is an offset in the Y direction (Fig. 1 SB). Additionally, this function is not defined for zero. Toward zero the function’s value goes to infinity. As there are no infinite distances between cells, another parameter is introduced which moves the function to the left:
where p1 is an offset in the X direction (Fig. S1C). The constants p0, p1 and p2 are calculated by the computer algebra system Techplot (Techplot7, SFTek, Braunschweig, Germany).
Simulation of the cell-to-cell distances
Mean cell-to-cell distances of randomly distributed cell-coordinates were simulated for certain cell frequencies per mm2. The random positioning of the cells is carried out with the aid of random numbers that are generated by visual basic software of the spreadsheet program Excel. The x and y coordinates of the desired quantity of “cells” were generated and the shortest distance of each cell to the nearest cell was calculated and then the shortest distances were averaged. This procedure was repeated for 200 times and the mean and 5% and 95% confidential interval was calculated.
Statistical methods
Overall survival and NED survival rates were calculated according to Kaplan–Meier20 and the log-rank test was used to compare survival rates 21 between patients with good and poor prognosis. Therefore, the median value was used as cut-off. All methods were analyzed by the statistical software SPSS (SPSS Inc., Chicago, IL). The interval between end of radiochemotherapy and last follow-up or death was regarded as follow-up time. Death from anal squamous carcinoma was defined as the end point in overall survival, whereas the appearance of local, nodal, or distant recurrence was defined as the end point in disease-free survival (NED survival).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Birgit Meyer and Christa Winkelmann for excellent technical assistance. We thank the Tumor Center at the Friedrich-Alexander University Erlangen-Nürnberg, Erlangen, Germany for providing us with patient data. The present work was performed in fulfillment of the requirements for obtaining the degree “Dr. med..”
References
- 1. Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 2013; 39:11-26; PMID:23890060; http://dx.doi.org/ 10.1016/j.immuni.2013.07.008 [DOI] [PubMed] [Google Scholar]
- 2. Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity 2004; 21:137-48; PMID:15308095; http://dx.doi.org/ 10.1016/j.immuni.2004.07.017 [DOI] [PubMed] [Google Scholar]
- 3. Yu P, Fu YX. Tumor-infiltrating T lymphocytes: friends or foes? Lab Invest 2006; 86:231-45; PMID:16446705; http://dx.doi.org/ 10.1038/labinvest.3700389 [DOI] [PubMed] [Google Scholar]
- 4. Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, Bifulco C, et al. Towards the introduction of the 'Immunoscore' in the classification of malignant tumours. J Pathol 2014; 232:199-209; PMID:24122236; http://dx.doi.org/ 10.1002/path.4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Piccirillo CA, Thornton AM. Cornerstone of peripheral tolerance: naturally occurring CD4+CD25+ regulatory T cells. Trends Immunol 2004; 25:374-80; PMID:15207505; http://dx.doi.org/ 10.1016/j.it.2004.04.009 [DOI] [PubMed] [Google Scholar]
- 6. Zhang X, Kelaria S, Kerstetter J, Wang J. The functional and prognostic implications of regulatory T cells in colorectal carcinoma. J Gastrointestinal Oncol 2015; 6:307-13; PMID:26029458; http://dx.doi.org/20559327 10.3978/j.issn.2078-6891.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol 2010; 10:490-500; PMID:20559327; http://dx.doi.org/ 10.1038/nri2785 [DOI] [PubMed] [Google Scholar]
- 8. Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol 2004; 22:329-60; PMID:15032581; http://dx.doi.org/ 10.1146/annurev.immunol.22.012703.104803 [DOI] [PubMed] [Google Scholar]
- 9. Wang Q, Feng M, Yu T, Liu X, Zhang P. Intratumoral regulatory T cells are associated with suppression of colorectal carcinoma metastasis after resection through overcoming IL-17 producing T cells. Cell Immunol 2014; 287:100-5; PMID:24487033; http://dx.doi.org/ 10.1016/j.cellimm.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 10. Askenasy N, Kaminitz A, Yarkoni S. Mechanisms of T regulatory cell function. Auto Immunity Rev 2008; 7:370-5; PMID:25599534; http://dx.doi.org/25599534 10.1016/j.autrev.2008.03.001 [DOI] [PubMed] [Google Scholar]
- 11. Heindl A, Nawaz S, Yuan Y. Mapping spatial heterogeneity in the tumor microenvironment: a new era for digital pathology. Lab Invest 2015; 95:377-84; PMID:25599534; http://dx.doi.org/ 10.1038/labinvest.2014.155 [DOI] [PubMed] [Google Scholar]
- 12. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013; 39:782-95; PMID:24138885; http://dx.doi.org/ 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 13. Grabenbauer GG, Kessler H, Matzel KE, Sauer R, Hohenberger W, Schneider IH. Tumor site predicts outcome after radiochemotherapy in squamous-cell carcinoma of the anal region: long-term results of 101 patients. Dis Colon Rectum 2005; 48:1742-51; PMID:15991058; http://dx.doi.org/ 10.1007/s10350-005-0098-5 [DOI] [PubMed] [Google Scholar]
- 14. Grabenbauer GG, Lahmer G, Distel L, Niedobitek G. Tumor-infiltrating cytotoxic T cells but not regulatory T cells predict outcome in anal squamous cell carcinoma. Clin Cancer Res 2006; 12:3355-60; PMID:16740757; http://dx.doi.org/ 10.1158/1078-0432.CCR-05-2434 [DOI] [PubMed] [Google Scholar]
- 15. Haas M, Dimmler A, Hohenberger W, Grabenbauer GG, Niedobitek G, Distel LV. Stromal regulatory T-cells are associated with a favourable prognosis in gastric cancer of the cardia. BMC Gastroenterol 2009; 9:65; PMID:19732435; http://dx.doi.org/ 10.1186/1471-230X-9-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feichtenbeiner A, Haas M, Buttner M, Grabenbauer GG, Fietkau R, Distel LV. Critical role of spatial interaction between CD8(+) and Foxp3(+) cells in human gastric cancer: the distance matters. Cancer Immunol Immunother 2014; 63:111-9; PMID:24170095; http://dx.doi.org/ 10.1007/s00262-013-1491-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rave-Frank M, Tehrany N, Kitz J, Leu M, Weber HE, Burfeind P, Schliephake H, Canis M, Beissbarth T, Reichardt HM, et al. Prognostic value of CXCL12 and CXCR4 in inoperable head and neck squamous cell carcinoma. Strahlenther Onkol 2016; 192:47-54; PMID:26374452; http://dx.doi.org/25575976 10.1007/s00066-015-0892-5 [DOI] [PubMed] [Google Scholar]
- 18. Tehrany N, Kitz J, Rave-Frank M, Lorenzen S, Li L, Kuffer S, Hess CF, Burfeind P, Reichardt HM, Canis M, et al. High-grade acute organ toxicity and p16(INK4A) expression as positive prognostic factors in primary radio(chemo)therapy for patients with head and neck squamous cell carcinoma. Strahlenther Onkol 2015; 191:566-72; PMID:25575976; http://dx.doi.org/ 10.1007/s00066-014-0801-3 [DOI] [PubMed] [Google Scholar]
- 19. Distel L, Fickenscher R, Dietel K, Hung A, Iro H, Zenk J, Nkenke E, Büttner M, Niedobitek G, Grabenbauer G. Tumour infiltrating lymphocytes in squamous cell carcinoma of the oro-and hypopharynx: Prognostic impact may depend on type of treatment and stage of disease. Oral Oncol 2009; 45(10):e167-74; PMID:19576838; http://dx.doi.org/ 10.1016/j.oraloncology.2009.05.640 [DOI] [PubMed] [Google Scholar]
- 20. Kaplan EL, Meier P. Non parametric estimation from incomplete observations. J Am Stat Assoc 1958; 53:457-81; http://dx.doi.org/ 10.1080/01621459.1958.10501452 [DOI] [Google Scholar]
- 21. Bland JM, Altman DG. The logrank test. BMJ 2004; 328:1073; PMID:15117797; http://dx.doi.org/ 10.1136/bmj.328.7447.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.