ABSTRACT
Toll-like receptors (TLRs) are immunological receptors recognizing various microbial and endogenous ligands, such as DNA, RNA, and other microbial and host components thus activating immunological responses. The expression of TLRs in esophageal adenocarcinoma is not well known. The aim of this study was to evaluate expression patterns of those TLRs that sense nucleic acids in Barrett’s esophagus with and without dysplasia and in esophageal adenocarcinoma. TLRs 3, 7 and 8 were stained immunohistochemically and evaluated in a cohort of patients with esophageal adenocarcinoma or dysplasia. Specimens with normal esophagus (n = 88), gastric (n = 67) or intestinal metaplasia (n = 51) without dysplasia, and low-grade (n = 42) or high-grade dysplasia (n = 37) and esophageal adenocarcinoma (n = 99) were studied. We used immunofluorescence to confirm the subcellular localization of TLRs. We found abundant expression of TLR3, 7 and 8 in esophageal squamous epithelium, columnar metaplasia, dysplasia and adenocarcinoma. Cytoplasmic expression of TLR3, TLR7 or TLR8 did not associate to clinicopathological parameters or prognosis in esophageal cancer. High nuclear expression of TLR8, confirmed with immunofluorescence, in cancer cells was observed in tumors of high T-stage (p < 0.01) and in tumors with organ metastasis (p < 0.001). High nuclear TLR8 expression was associated with poor prognosis (p < 0.001). The expression of TLR3, TLR7 and TLR8 increased toward dysplasia and adenocarcinoma. We demonstrated nuclear localization of TLR8, which associates with metastasis and poor prognosis. TLR3 and TLR7 do not seem to have prognostic significance in esophageal adenocarcinoma.
KEYWORDS: Barrett’s esophagus, esophageal cancer, esophageal adenocarcinoma, toll-like receptor 3, toll-like receptor 7, toll-like receptor 8
Introduction
TLRs are innate immune receptors, which have unique antigen-recognition domains. TLR3 and the members of the TLR9 subfamily (TLRs 7, 8 and 9) specifically recognize different types DNA and RNA.1
Chronic inflammation and infection can affect carcinogenesis by altering cytokine and chemokine expression which regulate for example angiogenesis and metastasis.2 In addition to immune cells, TLRs are found in epithelial cells and fibroblasts. Epithelial cells can thus regulate the inflammatory response to luminal microbes and endogenous danger signals by TLR-mediated activation.3 The common adapter protein for TLRs, MyD88, has been shown to mediate inflammation via TLRs, but also affect Ras-MAPK signaling, cell-cycle control and malignant transformation.4
TLR9, which recognizes CpG-oligodeoxynucleotides, has been associated with survival in esophageal squamous cell carcinoma and adenocarcinoma.5,6 Stimulation of TLR9 induced invasion of esophageal cancer cells and its expression increases in metaplasia–dysplasia sequence.7,8 No published information, however, on TLRs 3, 7 or 8 in esophageal adenocarcinoma could be found. In an esophageal squamous cell carcinoma study by Sheydihin et al., increased TLR7 expression was associated with invasion to adventitia and worse histological grade. In the same study TLR3 correlated with cancer invasion to adventitia and lymph node metastasis.9 The current knowledge on TLRs in esophageal epithelium and esophageal cancer is summarized in a recent review.10
The aim of this study was to assess the expression of TLRs 3, 7 and 8 in different stages of esophageal metaplasia–dysplasia–adenocarcinoma sequence as well as associations between TLR 3, 7 and 8 expression and clinicopathological variables or survival in patients with esophageal adenocarcinoma.
Results
Expression of TLR3, TLR7 and TLR8 in esophageal epithelium, Barrett’s esophagus, dysplasia and cancer
TLR3, TLR7 and TLR8 were expressed epithelial cells in normal and metaplastic esophagus (Fig. 1; Table 1). In all of the lesions, the pattern of TLR expression was mainly cytoplasmic, with occasional nuclear expression of TLR3 and TLR8. Nuclear staining of TLR3 was found in 76 of 88 (86%) samples of normal epithelium and 81 of 99 (82%) cancers, and of TLR8 in 34 of 88 (39%) normal epithelium and 32 of 99 (32%) cancers. Nuclear localization of TLR8 was further verified with confocal microscopy of immunofluorescence staining, as shown in Fig. 2. For TLR3, the nuclear localization could not be verified (data not shown).
Figure 1.
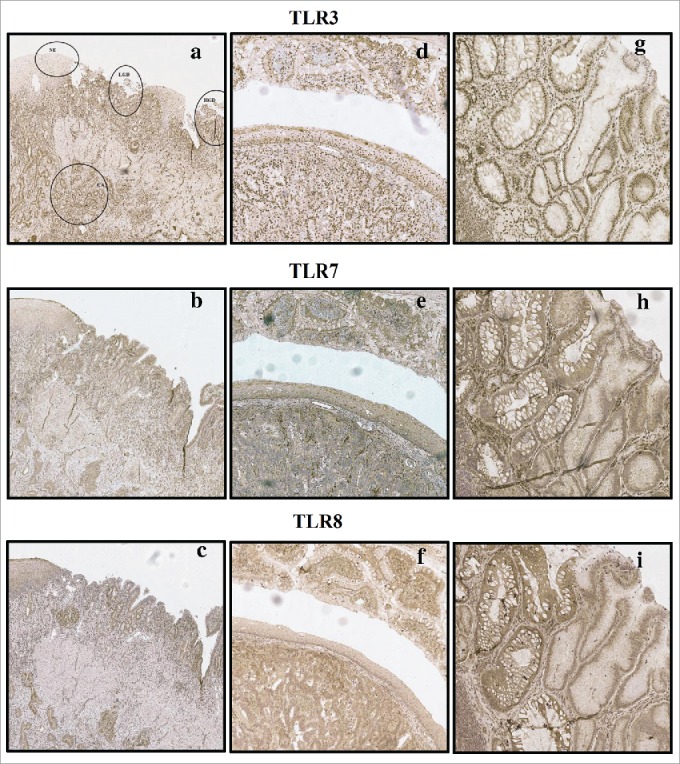
Examples of typical expression patterns of TLR3, TLR7 and TLR8. (A)–(C) representing the same sample with normal epithelium (NE), low-grade dysplasia (LGD), high grade dysplasia (HGD) and adenocarcinoma (CA) marked in (A). Gradual increase is found through normal epithelium–metaplasia–dysplasia sequence. (D)–(F) show intestinal type metaplasia (top), normal epithelium (middle) and adenocarcinoma (bottom). Intestinal metaplasia in TLR3, 7 show basal polarization, whereas TLR8 is expressed more diffusely. Expression pattern of all studied TLRs in adenocarcinoma is diffuse extending homogenously throughout the cell cytoplasm with no apparent basal polarization. (G)–(I) show intestinal metaplasia (left) and gastric type metaplasia (right). Gastric metaplasia presented a strong polarized staining to the basal cytoplasm in all studied TLRs. Magnifications 6× and 20× were used.
Table 1.
Baseline characteristics of TLR3, 7 and 8 expression in normal esophageal squamous epithelium and in different esophageal lesions. Intensity was assessed with a 4-point scale from negative (0) to strong intensity (3). The extent of the staining was expressed as percentage of positive cells and positive cell nuclei (0–100%). Histoscore is counted by multiplying intensity with the percentage of positive cells (0–300) Values are presented as mean, median and interquartile range (IQR). Statistically significant differences are shown for histoscores and nuclear TLR expression. Letters are placed to indicate the lesion with higher TLR expression in each comparison.
| Histoscore | Histoscore | Histoscore | Statistical | Nuclei | Statistical | |||
|---|---|---|---|---|---|---|---|---|
| TLR3 | mean | median | IQR | significance | mean | median | IQR | significance |
| Normal epithelium | 65 | 80 | 45 | 68 | 80 | 54 | b | |
| Gastric metaplasia | 85 | 80 | 35 | a | 59 | 70 | 55 | |
| Intestinal metaplasia | 110 | 90 | 68 | ab | 65 | 75 | 40 | |
| Low-grade dysplasia | 133 | 100 | 120 | ab | 72 | 85 | 30 | b |
| High-grade dysplasia | 119 | 100 | 78 | ab | 61 | 80 | 80 | |
| Adenocarcinoma | 139 | 100 | 100 | abc | 70 | 90 | 45 | bc |
| TLR7 | ||||||||
| Normal epithelium | 105 | 100 | 5 | — | ||||
| Gastric metaplasia | 139 | 135 | 105 | a | — | |||
| Intestinal metaplasia | 205 | 200 | 100 | ab | — | |||
| Low-grade dysplasia | 253 | 268 | 100 | abc | — | |||
| High-grade dysplasia | 266 | 300 | 50 | abce | — | |||
| Adenocarcinoma | 224 | 250 | 150 | abc | — | |||
| TLR8 | ||||||||
| Normal epithelium | 112 | 100 | 5 | 19 | 0.0 | 29 | ||
| Gastric metaplasia | 129 | 100 | 55 | 19 | 10 | 35 | ||
| Intestinal metaplasia | 183 | 200 | 138 | ab | 26 | 10 | 60 | |
| Low-grade dysplasia | 227 | 244 | 100 | abce | 31 | 0.0 | 60 | |
| High-grade dysplasia | 225 | 250 | 150 | abce | 23 | 0.0 | 60 | |
| Adenocarcinoma | 189 | 200 | 150 | ab | 23 | 0.0 | 55 |
compared to normal epithelium, p < 0.05
compared to gastric metaplasia, p < 0.05
compared to intestinal metaplasia, p < 0.05
compared to low-grade dysplasia, p < 0.05
compared to adenocarcinoma, p < 0.05
Figure 2.
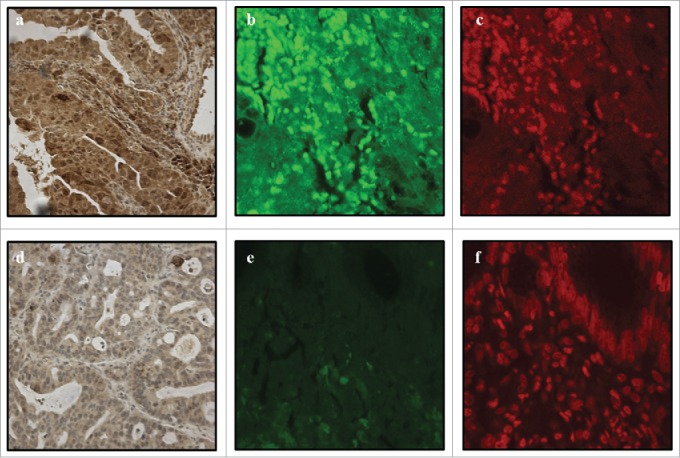
Positive TLR8 nuclear expression in immnohistochemistry (A), immunofluorescence photomicrograph obtained by confocal microscopy (B) and DNA-specific TO-PRO2 as a positive control (C). Negative TLR8 nuclear expression in immunohistochemistry (D) and immunofluorescence (E). Nuclei are visible with DNA-specific TO-PRO2 staining (F). Magnification of 40x was used.
TLR3 was expressed in 70/88 (79.5%) of normal epithelia, 63/67 (94%) of gastric metaplasia, 46/51 (90.2%) of intestinal metaplasia, 37/42 (88.1%) of low-grade dysplasia, 32/37 (86.5%) of high-grade dysplasia and 92/99 (92.9%) of adenocarcinomas. As we can see from Table 1, TLR3 histoscore increased from normal epithelium toward low-grade dysplasia and was significantly higher in all other lesions when compared to normal squamous epithelium. TLR3 histoscore was the highest in low-grade dysplasia and cancer, followed by high-grade dysplasia. TLR3 histoscore was also significantly higher in intestinal metaplasia, dysplasia and cancer, when compared to gastric-type metaplasia. The results are summarized in Table 1 and Fig. 1.
TLR7 was expressed in all (100%) of the lesions assessed, usually in all cells, however with varying intensity. The TLR7 histoscore was significantly higher in all other lesions compared to normal esophageal epithelium. Low- and high-grade dysplasia expressed the highest amount of TLR7, difference in comparison with non-dysplastic epithelial types being significant (Fig. 1; Table 1).
Similar to TLR7, TLR8 was also expressed in all (100%) of the lesions assessed with percentage of around 100% for all lesions. TLR8 histoscore increased from normal epithelium and gastric metaplasia toward intestinal metaplasia and dysplasia, which had the highest TLR8 histoscore. Nuclear expression of TLR8 varied highly among the lesions, with the lowest mean expression in normal esophagus and the highest mean expression in low-grade dysplasia. The expression patterns are summarized in Fig. 1 and Table 1.
TLR3, TLR7 and TLR8 expression, clinicopathological variables and survival in esophageal adenocarcinoma
TLR3 or TLR7 expression in cancer tissue was not associated to clinicopathological parameters (Table 2.) or prognosis (data not shown). TLR8 histoscore was significantly higher in inoperable tumors when compared to operatively treated tumors (Table 2). Nuclear expression of TLR8 associated strongly to higher T stage and presence of distant metastases (Table 3) and thus, to short survival (p < 0.01) in univariate analysis, but not in multivariate analysis (Fig. 3). TLR3 nuclear expression did not show any clinical associations (data not shown).
Table 2.
The relationships between TLR3, 7 and 8 histoscores compared to clinicopathological variables in esophageal adenocarcinoma.
| TLR3 histoscore n (%) |
TLR7 histoscore n (%) |
TLR8 histoscore n (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | n/N (%) | Weak | Strong | p | Weak | Strong | p | Weak | Strong | p |
| pT | ||||||||||
| T1–2 | 29/94 (31) | 5 (29) | 24 (31) | 0.887 | 9 (22) | 20 (38) | 0.100 | 12 (27) | 17 (35) | 0.400 |
| T3–4 | 65/94 (69) | 12 (71) | 53 (69) | 32 (78) | 33 (62) | 33 (73) | 32 (65) | |||
| Lymph nodes | ||||||||||
| Negative | 35/94 (37) | 7 (41) | 28 (36) | 0.710 | 14 (34) | 21 (40) | 0.586 | 14 (31) | 21 (43) | 0.239 |
| Positive | 59/94 (63) | 10 (59) | 49 (64) | 27 (66) | 32 (60) | 31 (69) | 28 (57) | |||
| Organ metastases | ||||||||||
| Negative | 63/94 (67) | 13 (77) | 50 (65) | 0.360 | 27 (66) | 36 (68) | 0.832 | 33 (73) | 30 (61) | 0.212 |
| Positive | 31/94 (33) | 4 (23) | 27 (35) | 14 (34) | 17 (32) | 12 (27) | 19 (39) | |||
| Grade | ||||||||||
| 1 | 29/93 (31) | 2 (12) | 27 (36) | 0.209 | 10 (24) | 19 (37) | 0.385 | 9 (20) | 20 (42) | 0.088 |
| 2 | 22/93 (24) | 4 (24) | 18 (24) | 10 (24) | 12 (24) | 11 (24) | 11 (23) | |||
| 3 | 42/93 (45) | 11 (65) | 31 (41) | 22 (52) | 20 (39) | 25 (56) | 17 (35) | |||
| Stage | ||||||||||
| I | 13/94 (14) | 3 (18) | 10 (13) | 0.895 | 4 (10) | 9 (17) | 0.672 | 4 (9) | 9 (18) | 0.194 |
| II | 37/94 (39) | 6 (35) | 31 (40) | 17 (42) | 20 (38) | 21 (47) | 16 (33) | |||
| III | 13/94 (14) | 3 (18) | 10 (13) | 7 (17) | 6 (11) | 8 (18) | 5 (10) | |||
| IV | 31/94 (33) | 5 (29) | 26 (34) | 13 (32) | 18 (34) | 12 (27) | 19 (39) | |||
| Tumor resection | ||||||||||
| Inoperable | 28/99 (28) | 6 (33) | 22 (27) | 0.599 | 11 (26) | 17 (30) | 0.601 | 5 (11) | 23 (43) | <0.001 |
| Operable | 71/99 (72) | 12 (67) | 59 (73) | 32 (74) | 39 (70) | 41 (89) | 30 (57) | |||
| Tumor size | ||||||||||
| Small (<40mm) | 38/92 (41) | 6 (38) | 38 (42) | 0.734 | 13 (33) | 25 (48) | 0.133 | 15 (33) | 23 (49) | 0.129 |
| Large (≥40mm) | 54/92 (59) | 10 (63) | 44 (58) | 27 (68) | 27 (52) | 30 (67) | 24 (51) | |||
Table 3.
Association of presence of nuclear TLR8 expression and clinicopathological variables in esophageal adenocarcinoma. Significant p values are shown in bold.
| Nuclear TLR8 expression |
||||||
|---|---|---|---|---|---|---|
| Variable pT | n/N (%) | Absent N (%) | Present N (%) | p | ||
| T1–2 | 29/94 (31) | 25 (40) | 4 (13) | 0.008 | ||
| T3–4 | 65/94 (69) | 38 (60) | 27 (87) | |||
| Lymph nodes | ||||||
| Negative | 35/94 (37) | 27 (43) | 8 (26) | 0.108 | ||
| Positive | 59/94 (63) | 36 (57) | 23 (74) | |||
| Organ metastases | ||||||
| Negative | 63/94 (67) | 51 (81) | 12 (39) | <0.001 | ||
| Positive | 31/94 (33) | 12 (19) | 19 (61) | |||
| Grade | ||||||
| 1 | 29/93 (31) | 20 (32) | 9 (29) | 0.448 | ||
| 2 | 22/93 (24) | 17 (27) | 5 (16) | |||
| 3 | 42/93 (45) | 25 (40) | 17 (55) | |||
| Stage | ||||||
| I | 13/94 (14) | 13 (21) | 0 (0) | <0.001 | ||
| II | 37/94 (39) | 29 (46) | 8 (26) | |||
| III | 13/94 (14) | 9 (14) | 4 (13) | |||
| IV | 31/94 (33) | 12 (19) | 19 (61) | |||
| Tumor resection | ||||||
| Inoperable | 28/99 (28) | 5 (8) | 23 (66) | <0.001 | ||
| Operable | 71/99 (72) | 59 (92) | 12 (34) | |||
| Tumor size | ||||||
| Small (<40mm) | 38/92 (41) | 26 (41) | 12 (41) | 0.992 | ||
| Large (≥40mm) | 54/92 (59) | 37 (59) | 17 (59) | |||
Figure 3.
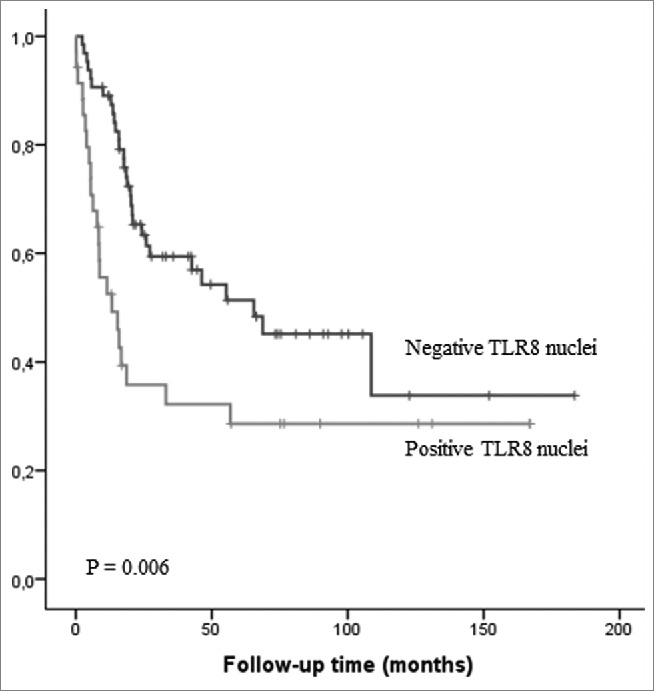
Kaplan–Meier survival curve in esophageal adenocarcinoma stratified by nuclear expression of TLR8 in the cancer cells.
Discussion
In this study, we demonstrate widespread expression of TLR3, TLR7 and TLR8 in normal, metaplastic and dysplastic esophageal epithelium as well as esophageal adenocarcinoma. Expression of TLR3, TLR7 and TLR8 showed variable patterns of increase during progression form normal squamous epithelium to columnar metaplasia, dysplasia and adenocarcinoma. Nuclear expression of TLR8 associated to distant metastases and poor cancer prognosis. These findings indicate that in addition to flagellin sensing TLR511 and bacterial DNA sensing TLR9,7 also other nucleotide sensing TLRs are involved in pathogenesis and progression of Barrett’s esophagus.
TLR3 recognizes viral RNA and nucleotides from necrotic tissues.12 We could confirm the presence of TLR3 expression in normal esophageal squamous epithelium.13 Novel finding is the increased expression of TLR3 along with development of columnar metaplasia and dysplasia in Barrett’s esophagus. However, we could not see any significant difference between non-dysplastic intestinal metaplasia and dysplasia suggesting that TLR3 expression does not serve as a marker of dysplasia.
Pathogenetic significance of increased TLR3 expression in Barrett’s dysplasia remains speculative. It has been shown, that in esophageal cell cultures necrotic epithelial cells induce a TLR3-mediated proinflammatory effect involving upregulation of interleukin-8 and NFkB,14 both of which have been suggested to play role in development and progression of Barrett’s esophagus.15 Role of TLR3 in gastrointestinal malignancies is largely unexplored. In our study of esophageal adenocarcinoma, TLR3 expression showed no association with prognosis or clinicopathological variables. In gastric cancer, high expression of TLR3 has been shown to associate with poor prognosis.16 Similarly, in esophageal squamous cell carcinoma increased TLR3 expression associated with occurrence of lymph node metastases.9 Accordingly, the role of TLR3 as a prognostic factor in different types of carcinoma seems to be inconstant, and could be related both with characteristics of the local microbiome and the epithelial cell type.
Single-stranded RNAs of viral origin are the best characterized ligands of TLR7.17 Based on our results, TLR7 is expressed in normal squamous epithelium as reported by Sheydihin et al.9 Throughout the esophageal adenocarcinoma carcinogenetic sequence, expression increased toward high-grade dysplasia. Due to overlap of histoscores between non-dysplastic columnar metaplasia and dysplasia, TLR7 seems not to be an optimal diagnostic tool, however. We found increased TLR7 expression in esophageal adenocarcinoma as compared with normal mucosa, but no association with characteristics or prognosis of cancer was seen. Oncological significance of TLR7 is controversial. Sheydihin et al. showed that TLR7 associated with worse histological grade in esophageal squamous cell carcinoma.9 In cervical squamous carcinoma cells, activation of TLR7 increased expression of a variety of inflammatory mediators.18 In summary, role of TLR7 expression and activation in esophageal adenocarcinoma remains unclear.
TLR8 binds viral single-stranded RNA or small interfering RNA and recruits NF-kappaB via MyD88 and is mainly involved in viral infections.19,20 In our study, cytoplasmic TLR8 was present in normal squamous epithelium and expression increased along with progression of Barrett’s esophagus to dysplasia. Highest expression of TLR8 was present in high-grade dysplasia. In esophageal adenocarcinoma, we demonstrated higher TLR8 expression in inoperable tumors. These findings suggest that cytoplasmic TLR8 plays a role in carcinogenesis of Barrett’s esophagus.
Interestingly, in addition to widespread cytoplasmic expression consistent with document endosomal localization of TLR8, we found TLR8 expression localized to cell nuclei in about one third of the cases. Using the freely available NucPred tool, which predicts the nuclear localization of proteins, we obtained a score of 0.88.21 This suggests that it is highly likely that TLR8 protein translocates to the nucleus. The finding was confirmed by immunofluorescence analysis with confocal microscopy (Fig. 2). Nuclear expression was seen all epithelial types including both normal squamous epithelium and different esophageal lesions, highest expression rates being seen in low-grade dysplasia. Interestingly, abundance of nuclear TLR8 expression was associated with advanced stage, presence of organ metastasis and shorter survival, suggesting that such subcellular localization has a pathogenetic role and prognostic significance.
Intracellular trafficking of TLR ligands and endosomal TLRs are poorly understood and there are no previous reports on nuclear expression of TLR8. We speculate, that viral infections may possibly affect normal cellular homeostasis and activate aberrant trafficking of TLR8 to the nucleus.22 Another link with both nuclear localization and role of such localization in the progression and prognosis of cancer might be related with the role of microRNAs as endogenous ligands of human TLR8. It has been suggested that interaction of TLR8 and such ligands modifies tumor microenvironment and is involved in the formation of metastasis.23 Although most miRNAs are present in both nucleus and cytoplasm, some mainly colocalize with nuclear structures such as transcription factors.24 Of the miRNA ligands of TLR823 miR-21 and let-7b can locate both in nucleus and cytoplasm, while miR-29a is mainly cytoplasmic.24 Clearly, more studies are needed to assess mechanisms linking nuclear TLR8 expression with the presence of distant metastases and the role of TLR8 endogenous ligands in such effect.
In conclusion, the upregulation of expression of TLRs 3, 7 and 8 seems to be an early event in esophageal carcinogenesis. This likely reflects the increased inflammatory activity in the premalignant stages of esophageal adenocarcinoma. Our findings suggest that TLRs 3 and 7 do not have major role in esophageal adenocarcinoma, but warrant further elucidation of the role of TLRs 3, 7 and 8 in transition of normal esophagus to metaplasia and dysplasia. TLRs 3 and 7 do not have prognostic significance in esophageal adenocarcinoma. However, cytoplasmic TLR8 expression is higher in inoperable esophageal adenocarcinomas. Interestingly, we demonstrated the presence of nuclear TLR8 expression, which indicates present distant organ metastasis and poor prognosis in esophageal adenocarcinoma.
Materials and methods
Patients
Paraffin-embedded, archival specimens of esophageal adenocarcinoma or esophageal dysplasia were obtained from the Department of Pathology, Oulu University Hospital, between the years 1987 and 2013. The final series consisted of 99 patients with esophageal adenocarcinoma, 10 with high-grade dysplasia, and 20 with low-grade dysplasia as the most advanced lesion. This resulted in evaluation of 88 normal esophageal epithelia, 67 gastric and 51 intestinal metaplasias without dysplasia, 42 low-grade and 37 high-grade dysplasias and 99 esophageal adenocarcinomas. The material has been earlier described elsewhere.11 The median age of the cancer patients was 64 y (range 43–90). The median follow-up time was 36 mo (range 0–288 mo). The patient survival data was acquired from Statistics Finland, and the other relevant data was acquired from the patient records. We could not retrieve treatment data from two of the patients, survival data from five and tumor size from seven of the patients.
The use of patient samples and the data inquiry were approved by the Oulu University Hospital Ethics Committee. The need to obtain a written or oral consent from the patients for using the samples in research was waived by the Finnish National Authority for Medicolegal Affairs (VALVIRA, Dnro 10832/06.01.03.01/2014).
Immunohistochemistry
Immunohistochemistry was performed on the tissue block sections, which were first selected by an expert gastrointestinal pathologist, on the basis of haematoxylin and eosin-staining, to be representative for the tumor mass in the resected specimen. TLR immunostaining was performed with commercial monoclonal antibodies (IMG-315A, mouse IgG1, Clone 40C1285.6 for TLR3, IMG-540, rabbit IgG, Clone N/A for TLR7 and IMG-321A, mouse IgG1, Clone 44C143 for TLR8 Imgenex) at a dilution of 1:25 (TLR3), 1:750 (TLR7) and 1:850 (TLR8). For immunohistochemical detection of the antibody reaction, we used the Dako Envision kit (Dako) with high temperature antigen retrieval in Tris-EDTA (for Ki67, TLR9) buffer for 15 min. Diaminiobenzidine (Dako basic DAB-kit) was used as a chromogen. All staining was done with Dako Autostainer (Dako).
We validated the immunohistochemical analysis through positive and two series of negative controls (omitting the primary antibody and by replacing primary antibody with the mouse primary antibody isotype control). Lymphocytes of the lymph nodes in the sample material were used as an internal positive control for TLR stainings.
Immunofluorescence
Formalin-fixed and paraffin-embedded esophageal sections were deparaffinized followed by treatment with 1% Triton X-100 in PBS for 5 min. Nonspecific staining was blocked by treatment with 1% bovine serum albumin for 20 min. Incubation with primary antibodies for 30 min at 37°C or 120 min at room temperature was then performed. The primary antibodies used are described in the immunohistochemistry section. After several washes, Alexa Fluor 488 or Alexa Fluor 568 conjugated to goat anti-mouse IgG (Life Technologies) was applied at appropriate dilutions and incubated for 60 min at 37°C. DNA specific TO-PRO2 staining was applied. Samples were mounted with Immumount (Thermo Scientific) and examined by using a Zeiss LSM510 confocal microscope. Images were analyzed with LSM510 Pascal software (Carl Zeiss).
Assessment of toll-like receptor expression
The histological hematoxylin-eosin stained sample slides were digitized using Aperio AT2 Console, Leica Biosystems Imaging Inc., Nussloch, Germany. The different epithelial lesions were pointed out and marked by expert gastrointestinal pathologist (T. J. K.). The immunohistochemical evaluation was done using regular light microscopy relating the sample material to the annotations in the H-E-stainings. The intensity and extent of different TLR expression in all epithelial cells in the each type of lesion, including normal squamous epithelium, columnar metaplasia and dysplasia, and carcinoma, was assessed semiquantitatively by two independent investigators (O. H and H. H.), as described previously.7,11 For interobserver calibration, a training set of samples were jointly evaluated and scored. The investigators were blinded to the clinical data and each other’s assessments. Assessment of the staining intensity was done using a 4-point scale from 0 (negative) to 1 (weak), 2 (moderate) and 3 (strong) according to most prevalent positive expression score. The extent of the staining was expressed as a percentage (0–100%) of the stained epithelial cells of lesion type. Percentage of the nuclei of the epithelial cells in the samples was similarly determined. Immune cells and stromal cells were not included in these analyses.
Before the beginning of the assessment, it was determined that if the values assessed by the investigators differed by one step in intensity or 30% in percentage, mean value of these two values would be used.7,11 In cases of more extensive differences between the investigators, a consensus would be reached by discussion with a third investigator (T. J. K.). None of the differences between the assessments exceeded these predetermined values. Thus, all of the values are means of intensities and percentages from two assessors.
Histoscore was then counted for each sample by multiplying the staining intensity by the percentage, resulting a number between 0 and 300. Expression levels were further dichotomized by dividing the histoscore by its median (95 for TLR3, 200 for TLR7 and 150 for TLR8) to weak and strong expression categories for further analysis. Nuclear staining was dichotomized to be either “absent” (percentage of 0) or “present” (percentage of >0).
Statistical analysis
We used IBM SPSS Statistics 22.0 (IBM corp.) for statistical analyses. To compare TLRs expression between different lesions we used Kruskall–Wallis due to skewed distributions. The chi-square test was used to calculate statistically significant differences between prognostic and clinicopathologic variables. Life tables were calculated according to the Kaplan–Meier method, and the survival curves were compared using the log-rank test. Cox proportional hazards model with backward selection was used for multivariate analysis with following covariates: Age, gender, T-stage, N-stage, M-stage and grade of differentiation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Erja Tomperi and Riitta Vuento for their excellent technical assistance, as well as statistician Pasi Ohtonen for his help with statistical problems.
Funding
This work was supported by grants from the Päivikki and Sakari Sohlberg Foundation (H. H.), the Emil Aaltonen Foundation (H. H), Georg C. and Mary Ehrnroot Foundation (H. H., J. H. K.), and the Finnish Medical Foundation (H. H., J. H. K.).
References
- 1. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol 2003; 21:335–76; PMID:12524386; http://dx.doi.org/ 10.1146/annurev.immunol.21.120601.141126 [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646–74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 3. Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A 2011; 108 Suppl 1:4607–14; PMID:20826446; http://dx.doi.org/ 10.1073/pnas.1000092107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coste I, Le Corf K, Kfoury A, Hmitou I, Druillennec S, Hainaut P, Eychene A, Lebecque S, Renno T. Dual function of MyD88 in RAS signaling and inflammation, leading to mouse and human cell transformation. J Clin Invest 2010; 120:3663–7; PMID:20941850; http://dx.doi.org/ 10.1172/JCI42771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takala H, Kauppila JH, Soini Y, Selander KS, Vuopala KS, Lehenkari PP, Saarnio J, Karttunen TJ. Toll-like receptor 9 is a novel biomarker for esophageal squamous cell dysplasia and squamous cell carcinoma progression. J Innate Immun 2011; 3:631–8; PMID:21876325; http://dx.doi.org/ 10.1159/000329115 [DOI] [PubMed] [Google Scholar]
- 6. Kauppila JH, Takala H, Selander KS, Lehenkari PP, Saarnio J, Karttunen TJ. Increased Toll-like receptor 9 expression indicates adverse prognosis in oesophageal adenocarcinoma. Histopathology 2011; 59:643–9; PMID:22014045; http://dx.doi.org/ 10.1111/j.1365-2559.2011.03991.x [DOI] [PubMed] [Google Scholar]
- 7. Huhta H, Helminen O, Kauppila JH, Takala H, Metsikko K, Lehenkari P, Saarnio J, Karttunen T. Toll-like receptor 9 expression in the natural history of Barrett mucosa. Virchows Arch 2015; 467:9–18; PMID:25838081; http://dx.doi.org/ 10.1007/s00428-015-1770-3 [DOI] [PubMed] [Google Scholar]
- 8. Kauppila JH, Karttunen TJ, Saarnio J, Nyberg P, Salo T, Graves DE, Lehenkari PP, Selander KS. Short DNA sequences and bacterial DNA induce esophageal, gastric, and colorectal cancer cell invasion. APMIS 2013; 121:511–22; PMID:23082743; http://dx.doi.org/ 10.1111/apm.12016 [DOI] [PubMed] [Google Scholar]
- 9. Sheyhidin I, Nabi G, Hasim A, Zhang RP, Ainiwaer J, Ma H, Wang H. Overexpression of TLR3, TLR4, TLR7 and TLR9 in esophageal squamous cell carcinoma. World J Gastroenterol 2011; 17:3745–51; PMID:21990957; http://dx.doi.org/ 10.3748/wjg.v17.i32.3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kauppila JH, Selander KS. Toll-like receptors in esophageal cancer. Front Immunol 2014; 5:200; PMID:24847326; http://dx.doi.org/ 10.3389/fimmu.2014.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Helminen O, Huhta H, Takala H, Lehenkari PP, Saarnio J, Kauppila JH, Karttunen TJ. Increased Toll-like receptor 5 expression indicates esophageal columnar dysplasia. Virchows Arch 2014; 464:11–8; PMID:24221343; http://dx.doi.org/ 10.1007/s00428-013-1505-2 [DOI] [PubMed] [Google Scholar]
- 12. Cavassani KA, Ishii M, Wen H, Schaller MA, Lincoln PM, Lukacs NW, Hogaboam CM, Kunkel SL. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. J Exp Med 2008; 205:2609–21; PMID:18838547; http://dx.doi.org/ 10.1084/jem.20081370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol 2007; 44:3100–11; PMID:17403538; http://dx.doi.org/ 10.1016/j.molimm.2007.02.007 [DOI] [PubMed] [Google Scholar]
- 14. Lim DM, Wang ML. Toll-like receptor 3 signaling enables human esophageal epithelial cells to sense endogenous danger signals released by necrotic cells. Am J Physiol Gastrointest Liver Physiol 2011; 301:G91–9; PMID:21474651; http://dx.doi.org/ 10.1152/ajpgi.00471.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Verbeek RE, Siersema PD, Ten Kate FJ, Fluiter K, Souza RF, Vleggaar FP, Bus P, van Baal JW. Toll-like receptor 4 activation in Barrett's esophagus results in a strong increase in COX-2 expression. J Gastroenterol 2014; 49:1121–34; PMID:23955118; http://dx.doi.org/ 10.1007/s00535-013-0862-6 [DOI] [PubMed] [Google Scholar]
- 16. Fernandez-Garcia B, Eiro N, Gonzalez-Reyes S, Gonzalez L, Aguirre A, Gonzalez LO, Del Casar JM, García-Muñiz JL, Vizoso FJ. Clinical significance of toll-like receptor 3, 4, and 9 in gastric cancer. J Immunother 2014; 37:77–83; PMID:24509170; http://dx.doi.org/ 10.1097/CJI.0000000000000016 [DOI] [PubMed] [Google Scholar]
- 17. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004; 303:1529–31; PMID:14976261; http://dx.doi.org/ 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- 18. Li L, Cheng FW, Wang F, Jia B, Luo X, Zhang SQ. The activation of TLR7 regulates the expression of VEGF, TIMP1, MMP2, IL-6, and IL-15 in Hela cells. Mol Cell Biochem 2014; 389:43–9; PMID:24347177; http://dx.doi.org/ 10.1007/s11010-013-1925-y [DOI] [PubMed] [Google Scholar]
- 19. Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. J Biochem 2007; 141:137–45; PMID:17190786; http://dx.doi.org/ 10.1093/jb/mvm032 [DOI] [PubMed] [Google Scholar]
- 20. Yu H, Wang Z, Sun G, Yu Y. Recognition of nucleic acid ligands by toll-like receptors 7/8: importance of chemical modification. Curr Med Chem 2012; 19:1365–77; PMID:22204348; http://dx.doi.org/ 10.2174/092986712799462603 [DOI] [PubMed] [Google Scholar]
- 21. Brameier M, Krings A, MacCallum RM. NucPred–predicting nuclear localization of proteins. Bioinformatics 2007; 23:1159–60; PMID:17332022; http://dx.doi.org/ 10.1093/bioinformatics/btm066 [DOI] [PubMed] [Google Scholar]
- 22. Lee BL, Barton GM. Trafficking of endosomal Toll-like receptors. Trends Cell Biol 2014; 24:360–9; PMID:24439965; http://dx.doi.org/ 10.1016/j.tcb.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A 2012; 109:E2110–6; PMID:22753494; http://dx.doi.org/ 10.1073/pnas.1209414109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roberts TC. The MicroRNA biology of the mammalian nucleus. Mol Ther Nucleic Acids 2014; 3:e188; PMID:25137140; http://dx.doi.org/ 10.1038/mtna.2014.40 [DOI] [PMC free article] [PubMed] [Google Scholar]


