Abstract
Background
Magnetic resonance imaging (MRI) is a well-developed technique in neuroscience. Limitations in applying MRI to rodent models of neuropsychiatric disorders include the large number of animals required to achieve statistical significance, and the paucity of automation tools for the critical early step in processing, brain extraction, which prepares brain images for alignment and voxel-wise statistics.
New method
This novel timesaving automation of template-based brain extraction (“skull-stripping”) is capable of quickly and reliably extracting the brain from large numbers of whole head images in a single step. The method is simple to install and requires minimal user interaction.
Results
This method is equally applicable to different types of MR images. Results were evaluated with Dice and Jacquard similarity indices and compared in 3D surface projections with other stripping approaches. Statistical comparisons demonstrate that individual variation of brain volumes are preserved.
Comparison with existing methods
A downloadable software package not otherwise available for extraction of brains from whole head images is included here. This software tool increases speed, can be used with an atlas or a template from within the dataset, and produces masks that need little further refinement.
Conclusions
Our new automation can be applied to any MR dataset, since the starting point is a template mask generated specifically for that dataset. The method reliably and rapidly extracts brain images from whole head images, rendering them useable for subsequent analytical processing. This software tool will accelerate the exploitation of mouse models for the investigation of human brain disorders by MRI
Keywords: Mouse brain extraction, Automated skull stripping, Magnetic resonance imaging, Freeware, Software tools
Graphical abstract

1. Introduction
Automated computational image analysis drives discovery of brain networks involved in either task-oriented or task-free mental activity. Human fMRI has spurred great strides in developing computational tools, including automated brain extraction, otherwise referred to as “skull-stripping” or “brain extraction”, a process that masks non-brain tissues present in MR images of the head when studying living human subjects. For rodents, which can provide robust models of human neuropsychopathology (Neumann et al., 2011), automation software has not been as easy to develop nor as readily available. Automation tools for expedient processing and analysis are integral in allowing researchers to investigate mechanisms of these disorders in rodent models quickly and reliably, thus containing cost.
Skull-stripping, a first step in image processing, involves masking extra-meningeal tissues, including skull, mouth, nares, ears, etc. Programs that use edge detection methods, like BrainSuite (Shattuck et al, 2002; Sandor and Leahy, 1997) or Brain Extraction Toolkit (BET) (Smith, 2002), are fast, effective and reliable when applied to MR images of human brain because the spaces occupied by cerebral spinal fluid and relevant boundaries are wider, and thus more distinct in human than in rodent images. For rodents, the space between brain and skull is narrow, and the brain less spherical than the human brain. Both of these attributes complicate direct application of human tools to mouse or rat MR images, which require manual refinement to acquire precise segmentation of brain from non-brain tissue. Thus, manual refinement of images produced by these automated stripping routines has been essential for subsequent alignment of rodent brain images and for statistical parametric mapping to identify differences between images from large cohorts of individual animals over time, such as we and others have previously reported (Bearer et al, 2007, 2009; Gallagher et al., 2012, 2013; Zhang et al., 2010; Chen et al, 2006; Spring et al., 2007; van Eede et al, 2013; Vousden et al., 2014).
Manual segmentation with hand-drawn masks to separate brain from non-brain tissue in whole-head images is a common and laborious solution used both to create a mask or to refine a mask generated using human brain extraction tools. Hand-drawn masks are made by working through a single image, slice by slice in each of the three coordinates, drawing the mask by hand on an interactive screen or with a cursor. This process can be expedited somewhat by using a tool such as BrainSuite’s Brain Surface Extractor (BSE) (Shattuck et al., 2002) to produce an initial mask that then requires refinement by hand. Drawbacks of this semi-manual segmentation process include the time involved, ranging from 1 to 4 h per image depending on how effectively BSE estimates the initial mask; and consistency, since masks created or refined manually differ slightly from each other. Other methods to mask non-brain structures include warping the new target image to an atlas (Ma et al., 2008, 2014; Benveniste et al., 2007). To date there are 7 atlases available online, many of which lack the original whole head images (see Ma et al., 2014 for a list of atlases). While these are helpful for atlas-based segmentation of structures within the brain (Ma et al., 2014; Hayes et al., 2014; Scheenstra et al, 2009), they are not easily applied for brain extraction from whole head images. Online publicly available digital atlases may also sometimes be useful as a template for alignment of brain images that have been masked, such as the ones we reported (http://www.loni.usc.edu/Software/MBAT) (Mackenzie-Graham et al., 2007), and labs also prepare in-house atlases. Best subsequent alignments and analysis are likely achieved when using a template image or atlas that was acquired with the same imaging parameters as the dataset to be extracted—not always available from pre-existing sources.
To overcome these issues and simplify skull stripping, we developed a new software tool based on the software package, NiftyReg (Modat et al, 2010; Modat, in press), in which a new mask for a new image is derived from a template mask made for a template image of the whole head captured with the same MR parameters as the images to be stripped. The template image thus has similar or identical experimental conditions, MR parameters and image properties, enabling alignment of whole head images from the entire experimental cohort. Creation of this dataset-specific template mask is the only time-consuming step in this extraction approach. Once the template mask is created, automated skull-stripping of whole-head images for the rest of the cohort takes only minutes to perform and results in uniform, cleanly extracted brain images. Our protocol aligns the whole head image of the template to the target, thus no alteration of target images subsequently used for brain alignments and analysis occurs. Because this method uses a template from within the dataset to be stripped, it allows application of automated skull-stripping to nonstandard images, such as manganese-enhanced MR images (MEMRI) where other semi-automated methods require significant time for manual correction due to the hyper-intense T1 weighted signal from the Mn2+ (Bearer et al, 2009; Gallagher et al., 2012, 2013; Zhang et al., 2010).
2. Method
2.1. Acquisition of MR Images
Three sets of mouse head images collected with different MR imaging modalities were tested with this approach: T1-weighted images of living animals captured with either a RARE (rapid acquisition with relaxation enhancement, or fast spin echo) (Bearer et al., 2007; Gallagher et al, 2012), or a FLASH (fast low angle shot, or spoiled gradient echo) protocol (Malkova et al., 2014); and T2-weighted images of fixed heads using a conventional pulsed-gradient spin echo, with isotropic diffusion-weighting (Bearer et al., 2009).
For RARE imaging, living mice (n = 9) were anesthetized and injected in the right CA3 of the hippocampus (coordinates x=−3.2mm (midline), y=−4.1mm (Bregma), z = 3.4mm (down) with 5 nL of a 200 mM MnCl2 aqueous solution and imaged at 30min, 6h, and 24 h post-injection in a 11.7T Bruker Avance Biospin vertical bore MR scanner. Images were captured with a RARE factor of 4 and the following parameters: 4 averages, TR/TE = 300ms/10.18ms; matrix size of 256 × 160 × 128; FOV 23.04 14.4, 11.52 mm; yielding 90 μm isotropic voxels with a 46 min scan time.
For FLASH images, intraperitoneal injection of aqueous MnCl2 was performed in living mice (n = 12) (Malkova et al., 2014; Yu et al., 2005) and images captured at 24 h post-injection in the MR scanner. Images from these mice were obtained with a 3D FLASH sequence (Frahm et al., 1986) with RARE factor of 4 and the following parameters: no averages, TR/TE = 25 ms/5 ms, matrix size 200 × 124 × 82, FOV 20 × 12.4×8.2 mm, for a resolution of 100 μm isotropic voxels and a 15 min scan time.
Finally, we imaged fixed brains (n = 9) in a Teflon holder immersed in perfluoropolyether (Fomblin®, Solvay Solexis, Inc, Thorofare, NJ) with a conventional pulsed-gradient spin echo (PGSE) sequence (TR/TE = 300 ms/11.9 ms, 256× 150× 120 matrix, with a field of view of 25.6 × 15 × 12 mm, giving a resolution of 100 μm isotropic voxel size as described (Bearer et al, 2009; Tyszka et al., 2006).
Thus here we show application of our software in 3 different MR imaging modalities of large cohorts of 9–12 images each (33 different individuals) with different FOV and resolution. In addition to the examples shown here, we have applied our stripping program to over a hundred images of different animals collected with different imaging parameters in two different labs and acquired rapidly generated consistent results.
2.2. Manual extraction and preparation of the template mask
A neuropathologist-trained research assistant manually created masks for 3D MR images of whole mouse heads, one template image from each of the datasets tested here, using a combination of BrainSuite’s (Version 13a) Brain Surface Extractor (BSE) and manual refinement according to our usual procedure, which was the gold-standard for brain extractions in our group (Bearer et al., 2007, 2009; Gallagher et al., 2012, 2013; Zhang et al., 2010). Each mask was further refined by senior authors and then considered to be the “template mask” for our study purposes. We found that while template images and masks from datasets with different imaging parameters were sometimes applicable to images captured with different parameters, the best output was when the template image and its mask were from within the dataset to be stripped. The template whole head image and its mask are then used in the automation to produce masks for the other images in the cohort.
2.3. Software development
Our strategy for automation is shown in a cartoon diagram in Fig. 1. The program starts with two template images, a whole head image and its mask, and one or more target images for which masks are needed. In the diagram, the whole head image is shown with its mask overlay—masked non-brain in gray and extracted brain in yellow. Real images produced by the automation as diagrammed in Fig. 1 are shown in Fig. 2 and are color-coded with the same titles so that the steps in the diagram and its results on images can be readily compared between the two figures. Three intermediate steps diagrammed in Fig. 1 do not produce images while running the program, the affine transform matrix, the control point grid, and the align-warped template mask. We included image diagrams for these steps to help the reader understand what the effects of the process would be on an image.
Fig. 1. Diagram of the automation strategy.
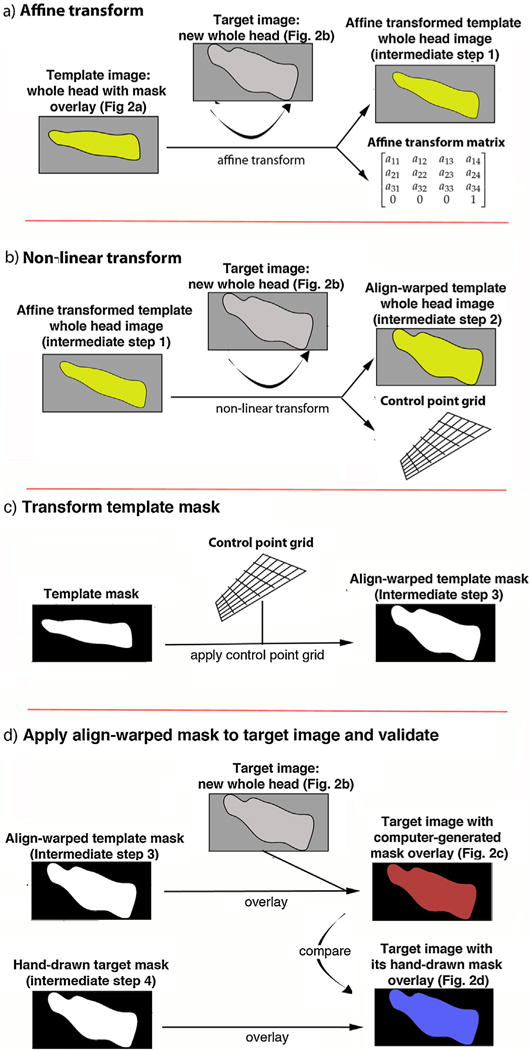
(a) Step 1: Affine transform. The template is a whole head image for which a hand drawn mask has been made (whole head non-brain tissue in gray with mask overlay leaving extracted brain pseudo-colored yellow). Although we diagram the mask overlay on the template head image here, the mask is not used in these steps, and only becomes necessary for the final transformation shown in (c). The target image, a new whole head image for which a mask is needed, is shown in two shades of gray: brain light gray, non-brain darker gray. Linear alignment of the template head image to the target head image creates an affine transformation matrix. The effect of this transformation on the template head image is also diagramed (Intermediate step 1), although the software does not produce an image from this step. See Fig. 2a and b for examples of MR images for these steps. (b) Step 2: Non-linear transform. Using the affine transformed template image/matrix as the starting point, a non-linear transform is performed between the template whole head image and the same target whole head image as in (a), which results in a control point grid containing both the affine transform and the non-linear warp field. This step further transforms the template head image to align with the target head image (Intermediate step 2). (c) Step 3: Transform template mask: The control-point grid, which includes information from the affine transformation matrix, is then applied to the template mask, resulting in a new align-warped mask fitting the target head image (Intermediate step 3). (d) Step 4: Apply align-warped mask to target image and validate: The mask produced in (c) is applied to the target head image, with extracted brain diagramed in red. For examples of images resulting from this procedure, see Fig. 2c. Once the mask is applied, the non-brain tissue is masked in the image, shown in black in the diagram. To validate our processing, we compared hand-drawn mask overlays with computer-generated masks in a variety of ways. Shown in the diagram is a visual comparison, with an example in Fig. 2d.
Fig. 2. Examples of automated and manual brain extraction.
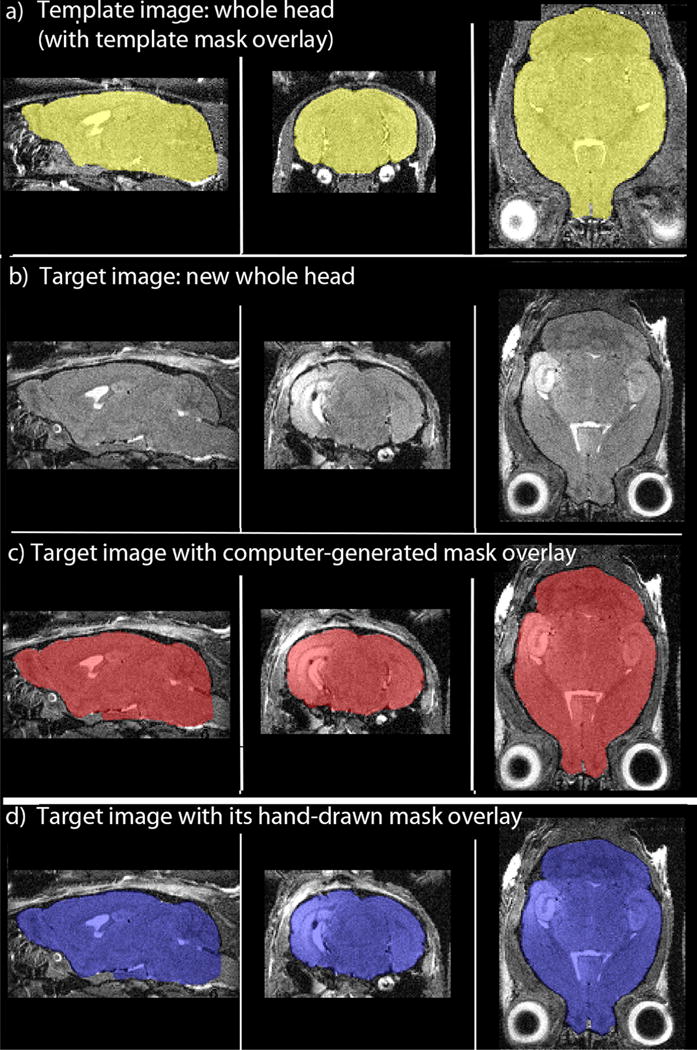
Examples of a template image and a target image with their respective masks. Orthogonal slices in sagittal, coronal and axial projections of the 3D datasets for each condition are shown, with the whole head MR image in grayscale with mask overlays leaving the extracted brain in pseudo-color. Images are color-coded to correlate with steps shown in Fig. 1. (a) The template whole head image overlaid with its hand-drawn mask from a cohort of mice imaged with a RARE pulse sequence protocol. The extracted brain is colored yellow. This is a pre-injection image with no hyper-intense Mn2+ signal. (b) The target image, anew whole head image for which a mask is needed, from the same cohort, imaged by the same protocol, but with a hyper-intense signal in CA3 of the hippocampus from a Mn2+ injection (Bearer et al., 2007). (c) Same target image as in (b) with its computer-generated mask derived from the template of a different brain, as shown in (a). Extracted brain in red. Note the differences in orientation and intensities between the two brains in (a), template, and (c), target, and compare the morphology of the masks between (c), computer-generated, and (d), hand drawn overlays on the same target head image. (d) Same target image as in (b) with its hand-drawn mask overlay. Brain is colored blue. A Jacquard similarity index comparing the mask in Image (c) with the one in (d) gives a similarity of 0.92, and a Dice index of 0.96, indicating highly precise correspondence between manually stripping, the gold standard, which takes several hours, and our new automated stripping procedure, which takes less than a few minutes.
First, as shown in Fig. 1a, our program performs a rigid body rotation of the template whole head image to the new target whole head image orientation, using reg_aladin from NiftyReg (Modat, in press). Reg_aladin works by using a block-matching approach to find key points and normalized cross-correlations to find correspondence, and computes the affine transform using a trimmed least-squares scheme. An affine transformation preserves the points, lines and planes of the original image, and sets of parallel lines remain parallel after affine transformation. This step provides an affine transform matrix (Fig. 1a).
We then align-warp the affine transform matrix of template whole brain image (for which we already have a mask) to the target, new whole head Image (Ourselin et al., 2001, 2002) (Fig. 1b). For this the affine transform matrix is used as a starting point by another NiftyReg algorithm, regf3d, which performs a nonlinear registration of the two Images (Rueckert et al., 1999). The deformation field (control point grid) is generated using cubic B-splines. This warp field includes the affine transform matrix and produces a control point grid that maps the deformation of the source (template whole head image) to the target whole head image.
Next, the control point grid is applied to the template mask (Fig. 1c), which produces a mask that is align-warped to fit the new target whole head image.
The program concludes with a final step to apply the mask (Fig. 1d). Application of the new mask to the target image, either within the SkullStrip program or by other means, yields the desired skull-stripped image of the brain (Figs. 1d and 2c, masked brain in red, non-brain in gray). Our program sets a threshold for the masks at 0.5 and applies the mask to the whole head images as the final output. Masks may also be applied using fslmaths, a program in FMRIB Software Library (FSL) (Jenkinson et al., 2012; Woolrich et al., 2009), which is a freeware software library of imaging analysis tools (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) that relies on MATLAB. Finally, here we compare the computer-generated masked image to the gold-standard hand-drawn masked image visually or with additional computational approaches (Figs. 1d and 2d, masked brain shown in blue).
The program can be run on a single target whole head image or as a loop on a dataset of multiple target images. All images must have identical headers, including the mode, bit size, dimensions and resolution. We typically use 16 bit integer unsigned, and perform N3 correction and modal scaling of the whole head images prior to running them through the program, which improves output. The processing environment is written in Python and runs in Linux on either Mac, Windows-Linux, or virtual Linux PC machines. Typically our machines have 8 Gb of RAM and plenty of hard drive space to store and deposit images as they move through the processing steps. We utilize shell scripting in this program to perform skull-stripping in a single step that automates transitions between all of the processing required to segment the brain from the whole head image. A README and Instructions are included with the software in Supplemental Materials.
We refer to our new program as “SkullStrip”.
2.4. Validation
2.4.1. Visual inspection
Masks created either manually or with our program were overlaid on their respective whole head image and the image details reviewed in 3D displays using FSLView, ImageJ and other 3D viewers. Visual inspection may seem informal and unreliable, but has been shown to detect small differences when comparing MR and CT images accurately (Fitzpatrick et al., 1998).
Surface projections of the same mouse brain stripped with three different methods (manual stripping, automation with BrainSuite, and our new SkullStrip) were visualized with AMIRA (FEI, Hillsboro, Oregon).
2.4.2. Statistical comparisons
We used statistical comparison methods to validate the automated stripping protocol. Both Dice (Dice, 1945) and Jacquard (Zijdenbos et al., 1994) similarity indices were applied to compare the results of our automated stripping to our expert, manual stripping of the same whole brain image. The Dice similarity index D is derived from the equation:
And the Jacquard similarity index J is obtained from the following equation: where Mh, Mc are ahuman-generated mask and a computer-generated mask, respectively. The values for M are the binary value (0,1) at each position in the mask matrix. Masks were also visually assessed for consistency.
2.4.3. Alignment success
We skull-stripped a large dataset (n = 9 mice) by both processing approaches—manually and computationally with our new program. Masks were applied to their respective image, and each set of 9 images, either hand-stripped or computer-stripped, were aligned and resliced to a template image using FSL’s FLIRT (Spring et al., 2007; Ellegood et al., 2015) (resulting in 0.08 mm isotropic voxels). Individual images were bias-field corrected using MIPAV (Sled et al., 1998; Nieman et al., 2005), scaled to the mode of the template image’s histogram using a custom MATLAB routine, and smoothed in SPM8. After processing, the two groups of images (hand-stripped and computer-stripped) were averaged using FSL (Woolrich et al., 2009) and the averaged images visualized with ImageJ.
2.4.4. Volumetric comparisons
A dataset of 9 brain images from a cohort of individuals imaged under identical MR parameters were manually stripped and also stripped by our new program. The volume of each stripped brain image was measured with the volume function (−V) in the fslstats program in FSL from the Oxford Centre for Functional Magnetic Resonance Imaging of the Brain (FMRIB) (Jenkinson et al., 2012; Woolrich et al., 2009), and the extracted brain volumes produced by the two stripping methods were quantitatively compared using Excel formulas, including paired and homoscedastic T.TESTs. The average difference between the volumes of brain with hand-drawn versus computer-generated masks was determined by calculating the difference for each pair and dividing by the average volumes of all the brains.
3. Results
We applied our stripping program to images collected from three different MR imaging modalities with differing resolution (Figs. 2 and 3). For each case we selected examples from larger 3D datasets and show mask overlays in orthogonal slices in the sagittal, coronal and axial planes of whole brain images, with the whole head image in gray scale and the segmented brain in color.
Fig. 3. Examples of application of computer-stripping to two other imaging modalities.
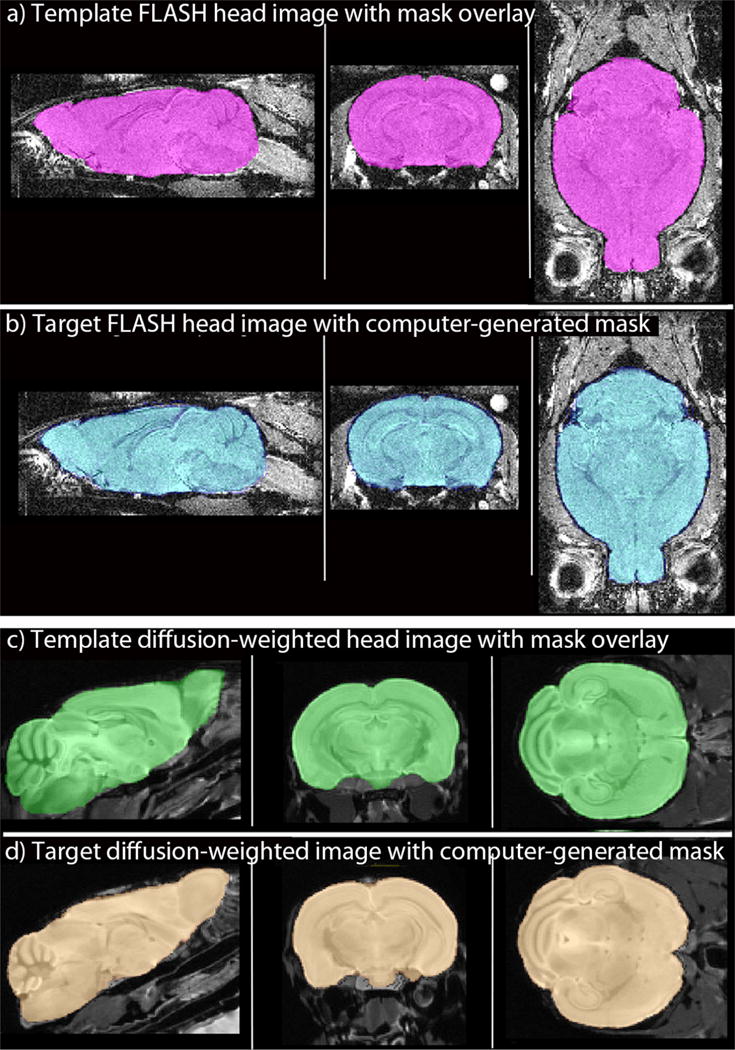
(a and b) 3D MR images acquired with a FLASH protocol are equally well stripped by automation, as shown in selected sagittal, coronal and axial slices: (a) Template whole head image with its hand-drawn mask overlay, extracted brain shown in pink. (b) Target image and its computer-generated mask, created from the hand-drawn mask in (a) overlaid on a different image from the same cohort of mice imaged with the same imaging parameters. Extracted brain shown in blue. (c and d) Slices from 3D images acquired with a diffusion-weighted protocol (Bearer et al., 2009): (c) Template image with its hand-drawn mask used to generate the mask for the target image in (d). Extracted brain in green. (d) Target image with computer-generated mask. Extracted brain in copper.
Examples of extracted brains from whole head images obtained from 3D T1 RARE imaging with masks created by either laborious hand-drawing or by our stripping program demonstrate that our stripping protocol segments the brain at least as precisely as an expert anatomist (Fig. 2). We show mask overlays on the whole head image to reveal to the reader how precisely the masks distinguish brain from non-brain tissue. The template whole head image is shown with its template mask overlay (Fig. 2a). The whole head image (the gray scale image in the figure) was aligned to the target image, a new whole head image of a different animal imaged with the same parameters (Fig. 2b). To demonstrate the power of this processing procedure, we chose here to show a template image captured before Mn2+ injection (Fig. 2a) and a target image from a different animal captured after Mn2+ injection which has a bright signal in the hippocampus not present in the template image (Fig. 2b). Visual inspection comparing the computer-generated mask created by our program using the template whole head image and its hand-drawn mask as reference images (Fig. 2c) with a hand-drawn mask for the same target image (Fig. 2d) finds no obvious differences. Thus, neither differing orientation between the target and template images nor the bright signal present only in the target image interfered with the whole head alignment producing the control point grid that yielded the new computer-generated mask.
To quantify similarity between expert hand-drawn masks and our computer-generated masks we applied statistical methods. The Dice and Jacquard similarity coefficients (Zijdenbos et al., 1994) comparing computer-generated masks to our gold standard, hand-drawn masks, were 0.96 and 0.92, respectively, demonstrating a very high similarity between the two types of masks. This result confirms the visual impression that our automated stripping protocol performs at least as well as manual stripping in extracting the brain from whole-head images, even when a region of hyper-intensity is present in the target but not the template whole head image.
We then applied our stripping algorithm to two other MR imaging modalities: T1-weighted FLASH and isotropic diffusion-weighted imaging (Fig. 3). A template whole head image with its mask overlay (Fig. 3a) was used to generate new masks for a set of different whole head images using our program (Fig. 3b). Thus the different grayscale patterns of the whole head images did not preclude alignments between template and target images captured with the same imaging parameters. We found that images captured with different parameters were sometimes useable across other imaging protocols, although often this introduced stripping artifacts. Hence best practice was to hand-draw a mask on one image from a larger cohort of individuals and use that whole head and mask as the template for the program to apply to the rest of the images in the experimental cohort.
Application of our program to fixed heads imaged by isotropic-diffusion-weighting also produced excellent brain extraction (Fig. 3c and d). Despite low contrast in the non-brain structures and pronounced differences in orientation of the brains, our program produced excellent masks.
We then created surface projections of the same brain (Fig. 4), shown above in Fig. 2c and d, extracted by three different methods (Fig. 4a–c): Manual extraction (Fig. 4a); BrainSuite Brain Surface Extractor (BSE) (Shattuck et al., 2002; Sandor and Leahy, 1997) (Fig. 4b); and our new automated skull stripping program, SkullStrip (Fig. 4c). Manual stripping is the gold standard of mouse brain extraction, while BrainSuite automation almost always leaves surface areas in rodent brains requiring further manual refinement (notice the rough patches on the surface projection, Fig. 4b). In contrast, our skull-stripper program produces an image as good as expert manual stripping (compare Fig. 4a with c)—and takes only minutes to perform rather than the hours needed to manually extract the brain from a mouse whole head image.
Fig. 4. Surface renderings of different stripping procedures.
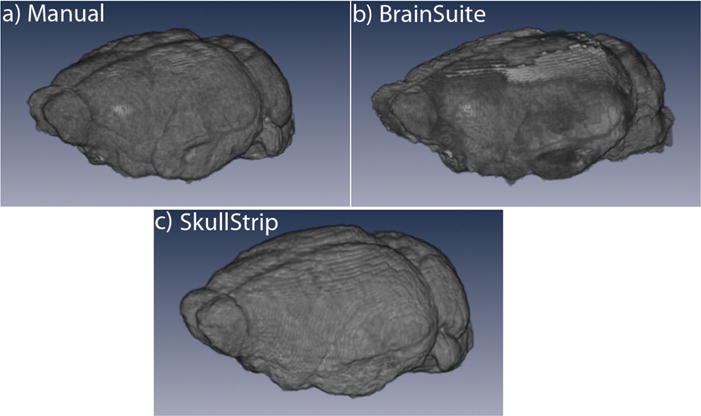
Surface renderings of the same brain extracted by three different approaches: (a) manual refinement in BrainSuite, a gold standard or ground truth; (b) preliminary BrainSuite BSE automated stripping. Shown is a best-case result, not all are this good; (c) application of a mask created by our SkullStrip program.
Next, we reviewed the impact of our automation on alignments (Fig. 5a and b). Nine living mice were imaged with a FLASH protocol, and brain extractions performed either manually (Fig. 5a) or with our software, producing two datasets of nine images each, one for each type of brain extraction (Fig. 5b). After application of the masks, each set of images, hand-stripped or generated by the Skull-Strip program, was aligned and inspected with SPM “check regs” and an averaged image produced. Visual inspection, an accepted method to validate alignments (Woods, 2000), of axial projections of the nine averaged images demonstrate that automated stripping appears to have resulted in improved alignments, as witnessed by increased detail in deeper brain structures and in the cerebellum (Fig. 5c and d). We speculate that our skull stripping program produces more consistent brain extraction throughout the dataset, and thus improves alignments between images. Such improved alignment would result in increased detail of anatomic structures through more precise co-registration of contrast in the MR images.
Fig. 5. Automated processing of brain extraction improves alignments and preserves brain volumes.
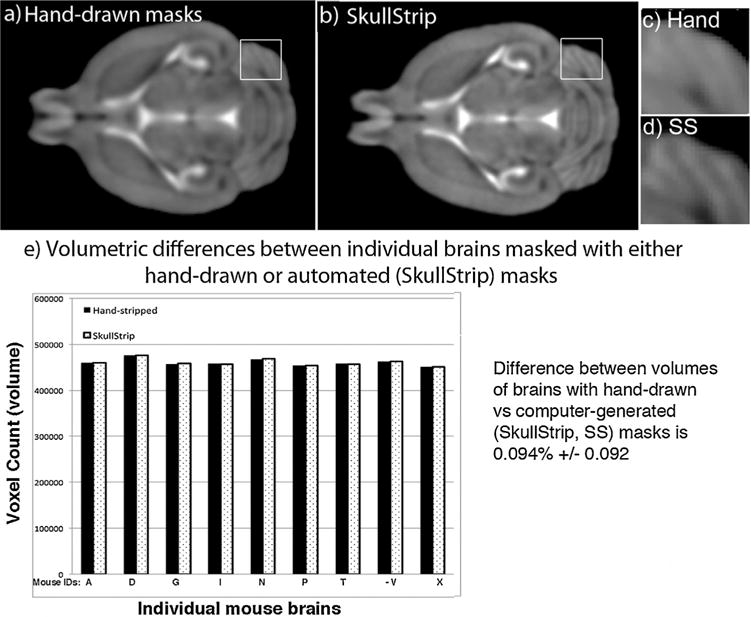
(a and b) Shown are axial slices from aligned and averaged images (n = 9) that were either stripped manually by an expert (a) or stripped by automation with SkullStrip (b). The increased definition of the cerebellar cortical surface folds in (b) shows that alignment of brains stripped by our new automation are at least as good as those manually extracted by a trained person (a), our previous gold standard. (c and d) Enlarged images from areas identified by white squares in a and b.(c) Hand drawn masks; (d) Skullstrip (SS) automated masks. (e) Comparison of the brain volumes after either manual or automated stripping. Volumes (numbers of voxels) were determined for each brain image in the dataset after extraction. The average difference between hand-drawn or computer-stripped images was 0.094% ± 0.092. The average brain volume was 460,809 voxels, with an average difference of 434 voxels ±458 between hand-and computer-stripping. A 1.6% difference was found between brains within each dataset regardless of stripping approach, demonstrating that automation preserved individual variations at least in this range. Comparison of the volumes by homoscedastic t-test gave a probability of P= 0.998 that volumes by either extraction technique were the same.
As a final validation step, we compared the volumes of all nine images in the hand-stripped and in the computer-stripped datasets (Fig. 5e). Preservation of volume is critical for some types of data analysis, particularly for study of neurodegenerative, neuropsychiatric and vascular diseases affecting the brain. We found that the average difference in volume between a brain that was manually extracted with a hand-drawn mask and the same brain masked by our program was 0.093% ± 0.92. This was 15-fold smaller than the average differences in volumes between brains within both datasets regardless of stripping approach, which was 1.6%, consistent with previous studies (Chen et al., 2006; Kovacevic et al., 2005; Nieman et al., 2007). The similar variance in volume for both datasets prompted us to apply a homoscedastic Student’s t-test, which compares two samples assuming equal variance, to test the null hypothesis that there was no difference. This between-group statistical comparison gave a probability of 0.998 that volumes produced by either approach, hand-drawn masks or our computer automation program, were the same, i.e. accepting the null hypothesis.
The source code and documentation for our program, Skull-Strip, can be downloaded from our Center website under the Tools and Data tab at http://stmc.health.unm.edu/tools-and-data/index.html. Please also see Supplemental Materials for instructions, README and downloadable software program.
4. Discussion
Our automated method both saves time and results in precise, uniform masks for rodent whole head images. It is straightforward to use for any type of rodent brain MR, as it relies on a template image generated from within the dataset to be analyzed. Uniformity of brain extraction significantly improves post-alignment image analysis, avoiding most stripping artifacts and the variability introduced by manual processing, yet preserving biological variation between individuals. In our work, for a single experiment, it is not uncommon that to obtain statistical power a hundred or more images must be analyzed (Bearer et al., 2007, 2009; Gallagher et al., 2012, 2013; Zhang et al., 2010). This analysis begins with brain extraction, which until now has been a time-consuming, error-prone process. The methodology reported here greatly reduces the time and effort needed to produce masks yielding brain-only images for subsequent analysis. Automated brain extraction also yields highly consistent results, which improves alignments. Our volumetric analysis demonstrates that automated stripping with our program preserves normal variation in brain volume. While many labs may already have in-house semi-automated stripping approaches, this new automated method, which is freely accessible, will facilitate standardization of brain extraction through a rapid and easily applied computational approach.
The advantage of aligning the template whole brain into the new raw image is that the details within the whole image provide many points for alignments. Masks, which are binary, provide less detail for accurate alignments. Additionally our approach has no effect on the new whole brain image, which is not altered by the processing.
Our automation can be applied to any type of MR imaging, including T1 and T2 weighted images such as those acquired by RARE, FLASH and diffusion-weighted images. Resolution has no impact, as long as all images have the same “header” details as to image dimensions, resolution, and mode (bit dimension and integer, signed/unsigned, float, etc.). While it is time-consuming to create the initial template mask, usually by manual refinement of a mask generated by other automation programs, this is a minor time investment compared to manual stripping of dozens to hundreds of images in a large dataset. Since one template mask is applied to all target images, the target masks will have few random, manually introduced, differences. This uniformity improves alignments, as demonstrated by increased detail in aligned images. While the particular resolution of any image dataset does not interfere with the processing, all the images within a dataset to be stripped must have the same resolution. This can readily be done if necessary by processing through a variety of different freeware programs, such as MIPAV (http://mipav.cit.nih.gov/documentation.php) from the NIH.
A few publications specifically describe automation approaches for brain extraction, although most reports of MR mouse brain images give little detail of how the brain was extracted from non-brain tissue of the head, a critical step in pre-processing of whole head images. One of these reports uses a template-based approach (Lin et al., 2013) that may be analogous to the one we describe here, but the details are not well described and the software tools not available. A first step in segmentation of sub-brain regions sometimes involves brain extraction, and in at least one report this involves alignment of the whole head target images to atlas images of unspecified provenance. The transform matrix generated by this alignment is then inverted and used to propagate the atlas brain masks onto the target Images (Ma et al., 2014). Utilization of this approach requires that there be a set of whole head atlas images for which masks have been generated, that the imaging parameters are similar between atlas and targets, and that software tools to perform processing steps are available. Our software reported here would be applicable to automate this approach also, and to standardize across investigators.
Other approaches extract the brain using more complicated image analysis procedures, including constraint level sets (Uberti et al., 2009). RATS, a novel automated tissue segmentation program relies on LOGISMOS-based graph segmentation based on grayscale mathematical morphology creating a new mask specifically for each whole head Image (Oguz et al., 2014). De novo extraction of the brain from non-brain tissue when not based on a template is more complicated computationally, and may lack the ability to improve alignments of datasets if applied individually to each image in the dataset separately.
Our analysis of mouse brain images demands brain extraction from a large number of whole head images, which would be very time consuming to perform manually without this automation. While other approaches have been described in most cases the details are not explicit and the software tools used not reported and not freely available.
Our method described here overcomes these drawbacks in a straightforward and simple manner, and is freely available from our website (unm.stmc.edu). We welcome others to the application of the power of MR imaging to mouse brain analysis in pursuit of answers to the critical biomedical questions: How do mental illnesses evolve, and what are the temporal processes that lead to cognitive or emotional disability.
Supplementary Material
HIGHLIGHTS.
We present a new software tool for automated mouse brain extraction.
The new software tool is rapid and applies to any MR dataset.
We validate the output by comparing with manual extraction (the gold standard).
Brain extraction with this tool preserves individual volume, and improves alignments.
Acknowledgments
The authors gratefully acknowledge Vince Calhoun for review of this work, Art Toga for inspiring us to pursue creation of this new processing tool, and technical assistance from Xiaowei Zhang, Sharon Lin, Kevin P. Reagan, Kathleen Kilpatrick, Amber Zimmerman and Frances Chaves. This work is supported by The Harvey Family Endowment (ELB), and the NIH: R01MH087660 and P5OGM08273 (ELB).
Appendix A. Supplementary data
Supplementary data including the instructions, README and software program, associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/jjneumeth.2015.09.031.
This article should be cited whenever this software is used.
References
- Bearer EL, Zhang X, Jacobs RE. Live imaging of neuronal connections by magnetic resonance: robust transport in the hippocampal-septal memory circuit in a mouse model of Down syndrome. NeuroImage. 2007;37:230–42. doi: 10.1016/j.neuroimage.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer EL, Zhang X, Janvelyan D, Boulat B, Jacobs RE. Reward circuitry is perturbed in the absence of the serotonin transporter. NeuroImage. 2009;46:1091–104. doi: 10.1016/j.neuroimage.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste H, et al. Anatomical and functional phenotyping of mice models of Alzheimer’s disease by MR microscopy. Ann N Y Acad Sci. 2007;1097:12–29. doi: 10.1196/annals.1379.006. [DOI] [PubMed] [Google Scholar]
- Chen XJ, et al. Neuroanatomical differences between mouse strains as shown by high-resolution 3D MRI. NeuroImage. 2006;29:99–105. doi: 10.1016/j.neuroimage.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Dice L. Measure of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- Ellegood J, et al. Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Mol Psychiatry. 2015;20:118–25. doi: 10.1038/mp.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick JM, et al. Visual assessment of the accuracy of retrospective registration of MR and CT images of the brain. IEEE Trans Med Imaging. 1998;17:571–85. doi: 10.1109/42.730402. [DOI] [PubMed] [Google Scholar]
- Frahm J, Haase A, Matthaei D. Rapid three-dimensional MR imaging using the FLASH technique. J Comput Assist Tomogr. 1986;10:363–8. doi: 10.1097/00004728-198603000-00046. [DOI] [PubMed] [Google Scholar]
- Gallagher JJ, Zhang X, Ziomeck GJ, Jacobs RE, Bearer EL. Deficits in axonal transport in hippocampal-based circuitry and the visual pathway in APP knock-out animals witnessed by manganese enhanced MRI. NeuroImage. 2012;60(3):1856–66. doi: 10.1016/j.neuroimage.2012.01.132. http://dx.doi.org/10.1016/j.neuroimage.2012.01.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JJ, et al. Altered reward circuitry in the norepinephrine transporter knockout mouse. PLoS ONE. 2013;8:e57597. doi: 10.1371/journal.pone.0057597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes K, et al. Comparison of manual and semi-automated segmentation methods to evaluate hippocampus volume in APP and PS1 transgenic mice obtained via in vivo magnetic resonance imaging. J Neurosci Methods. 2014;221:103–11. doi: 10.1016/j.jneumeth.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. NeuroImage. 2012;62:782–90. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Kovacevic N, et al. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb Cortex. 2005;15:639–45. doi: 10.1093/cercor/bhh165. [DOI] [PubMed] [Google Scholar]
- Lin L, Wu S, Yang C. A template-based automatic skull-stripping approach for mouse brain MR microscopy. Microsc Res Tech. 2013;76:7–11. doi: 10.1002/jemt.22128. [DOI] [PubMed] [Google Scholar]
- Ma Y, et al. In vivo 3D digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Front Neuroanat. 2008;2:1. doi: 10.3389/neuro.05.001.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, et al. Automatic structural parcellation of mouse brain MRI using multi-atlas label fusion. PLoS ONE. 2014;9:e86576. doi: 10.1371/journal.pone.0086576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie-Graham AJ, et al. Multimodal, multidimensional models of mouse brain. Epilepsia. 2007;48(Suppl 4):75–81. doi: 10.1111/j.1528-1167.2007.01244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova NV, Gallagher JJ, Yu CZ, Jacobs RE, Patterson PH. Manganese-enhanced magnetic resonance imaging reveals increased DOI-induced brain activity in a mouse model of schizophrenia. Proc Natl Acad Sci USA. 2014;111:E2492–500. doi: 10.1073/pnas.1323287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modat M, et al. Fast free-form deformation using graphics processing units. Comput Methods Programs Biomed. 2010;98:278–84. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Modat, M. NiftyReg (Aladin and F3D); from http://cmictig.cs.ucl.ac.uk/research/software.
- Neumann ID, et al. Animal models of depression and anxiety: what do they tell us about human condition? Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1357–75. doi: 10.1016/j.pnpbp.2010.11.028. [DOI] [PubMed] [Google Scholar]
- Nieman BJ, et al. Magnetic resonance imaging for detection and analysis of mouse phenotypes. NMR Biomed. 2005;18:447–68. doi: 10.1002/nbm.981. [DOI] [PubMed] [Google Scholar]
- Nieman BJ, et al. Mouse behavioral mutants have neuroimaging abnormalities. Hum Brain Mapp. 2007;28:567–75. doi: 10.1002/hbm.20408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguz I, Zhang H, Rumple A, Sonka M. RATS: rapid automatic tissue segmentation in rodent brain MRI. J Neurosci Methods. 2014;221:175–82. doi: 10.1016/j.jneumeth.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourselin S, Roche A, Subsol G, Pennec X, Ayache N. Reconstructing a 3D structure from serial histological sections. Image Vis Comput. 2001;19:25–31. [Google Scholar]
- Ourselin S, et al. Robust registration of multi-modal images: towards real-time clinical applications. In: Dohi T, Kikinis R, editors. MICCAI 2002. Berlin Heidelberg: Springer-Verlag; 2002. pp. 140–7. [Google Scholar]
- Rueckert D, et al. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–21. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Sandor S, Leahy R. Surface-based labeling of cortical anatomy using a deformable atlas. IEEE Trans Med Imaging. 1997;16:41–54. doi: 10.1109/42.552054. [DOI] [PubMed] [Google Scholar]
- Scheenstra AE, et al. Automated segmentation of in vivo and ex vivo mouse brain magnetic resonance images. Mol Imaging. 2009;8:35–44. [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002;6:129–42. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–55. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring S, Lerch JP, Henkelman RM. Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. NeuroImage. 2007;35:1424–33. doi: 10.1016/j.neuroimage.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Tyszka JM, Readhead C, Bearer EL, Pautler RG, Jacobs RE. Statistical diffusion tensor histology reveals regional dysmyelination effects in the shiverer mouse mutant. NeuroImage. 2006;29:1058–65. doi: 10.1016/j.neuroimage.2005.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti MG, Boska MD, Liu Y. A semi-automatic image segmentation method for extraction of brain volume from in vivo mouse head magnetic resonance imaging using Constraint Level Sets. J Neurosci Methods. 2009;179:338–44. doi: 10.1016/j.jneumeth.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eede MC, Scholz J, Chakravarty MM, Henkelman RM, Lerch JP. Mapping registration sensitivity in MR mouse brain images. Neuroimage. 2013;82:226–36. doi: 10.1016/j.neuroimage.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Vousden DA, et al. Whole-brain mapping of behaviourally induced neural activation in mice. Brain Struct Funct. 2015;220(4):2043–57. doi: 10.1007/s00429-014-0774-0. http://dx.doi.org/10.1007/s00429-014-0774-0, Epub 2014 Apr 24. [DOI] [PubMed] [Google Scholar]
- Woods R. Validation of registration and accuracy. In: Isaac N. Bankman John Hopkins Univ., L., MD, editor. Handbook of medical imaging. Orlando, FL, USA: Academic Press; 2000. pp. 491–7. [Google Scholar]
- Woolrich MW, et al. Bayesian analysis of neuroimaging data in FSL. NeuroImage. 2009;45:S173–86. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- Yu X, Wadghiri YZ, Sanes DH, Turnbull DH. In vivo auditory brain mapping in mice with Mn-enhanced MRI. Nat Neurosci. 2005;8:961–8. doi: 10.1038/nn1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Altered neurocircuitry in the dopamine transporter knockout mouse brain. PLoS ONE. 2010;5:e11506. doi: 10.1371/journal.pone.0011506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC. Morphometric analysis of white matter lesions in MR images: method and validation. IEEE Trans Med Imaging. 1994;13:724–6. doi: 10.1109/42.363096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


