Abstract
We analysed whole genome sequences of 560 breast cancers to advance understanding of the driver mutations conferring clonal advantage and the mutational processes generating somatic mutations. 93 protein-coding cancer genes carried likely driver mutations. Some non-coding regions exhibited high mutation frequencies but most have distinctive structural features probably causing elevated mutation rates and do not harbour driver mutations. Mutational signature analysis was extended to genome rearrangements and revealed 12 base substitution and six rearrangement signatures. Three rearrangement signatures, characterised by tandem duplications or deletions, appear associated with defective homologous recombination based DNA repair: one with deficient BRCA1 function; another with deficient BRCA1 or BRCA2 function; the cause of the third is unknown. This analysis of all classes of somatic mutation across exons, introns and intergenic regions highlights the repertoire of cancer genes and mutational processes operative, and progresses towards a comprehensive account of the somatic genetic basis of breast cancer.
Introduction
The mutational theory of cancer proposes that changes in DNA sequence, termed “driver” mutations, confer proliferative advantage upon a cell, leading to outgrowth of a neoplastic clone1. Some driver mutations are inherited in the germline, but most arise in somatic cells during the lifetime of the cancer patient, together with many “passenger” mutations not implicated in cancer development1. Multiple mutational processes, including endogenous and exogenous mutagen exposures, aberrant DNA editing, replication errors and defective DNA maintenance, are responsible for generating these mutations1–3.
Over the past five decades, several waves of technology have advanced the characterisation of mutations in cancer genomes. Karyotype analysis revealed rearranged chromosomes and copy number alterations. Subsequently, loss of heterozygosity analysis, hybridisation of cancer-derived DNA to microarrays and other approaches provided higher resolution insights into copy number changes4–8. Recently, DNA sequencing has enabled systematic characterisation of the full repertoire of mutation types including base substitutions, small insertions/deletions, rearrangements and copy number changes9–13, yielding substantial insights into the mutated cancer genes and mutational processes operative in human cancer.
As for many cancer classes, most currently available breast cancer genome sequences target protein-coding exons8,11–15. Consequently, there has been limited consideration of mutations in untranslated, intronic and intergenic regions, leaving central questions pertaining to the molecular pathogenesis of the disease unresolved. First, the role of activating driver rearrangements16–18 forming chimeric (fusion) genes/proteins or relocating genes adjacent to new regulatory regions as observed in haematological and other malignancies19. Second, the role of driver substitutions and indels in non-coding regions of the genome20,21. Common inherited variants conferring susceptibility to human disease are generally in non-coding regulatory regions and the possibility that similar mechanisms operate somatically in cancer was highlighted by the discovery of somatic driver substitutions in the TERT gene promoter22,23. Third, which mutational processes generate the somatic mutations found in breast cancer2,24. Addressing this question has been constrained because exome sequences do not inform on genome rearrangements and capture relatively few base substitution mutations, thus limiting statistical power to extract the mutational signatures imprinted on the genome by these processes24,25.
Here we analyse whole genome sequences of 560 cases in order to address these and other questions and to pave the way to a comprehensive understanding of the origins and consequences of somatic mutations in breast cancer.
Results
Cancer genes and driver mutations
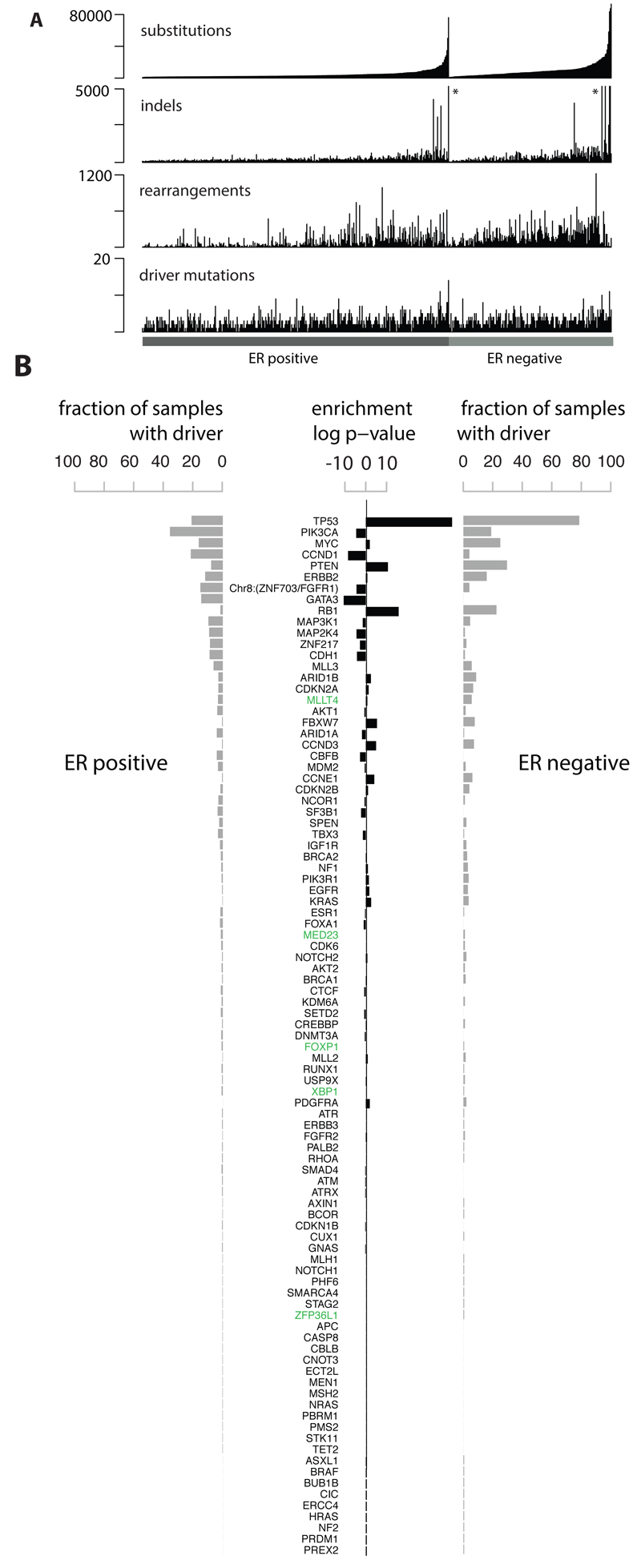
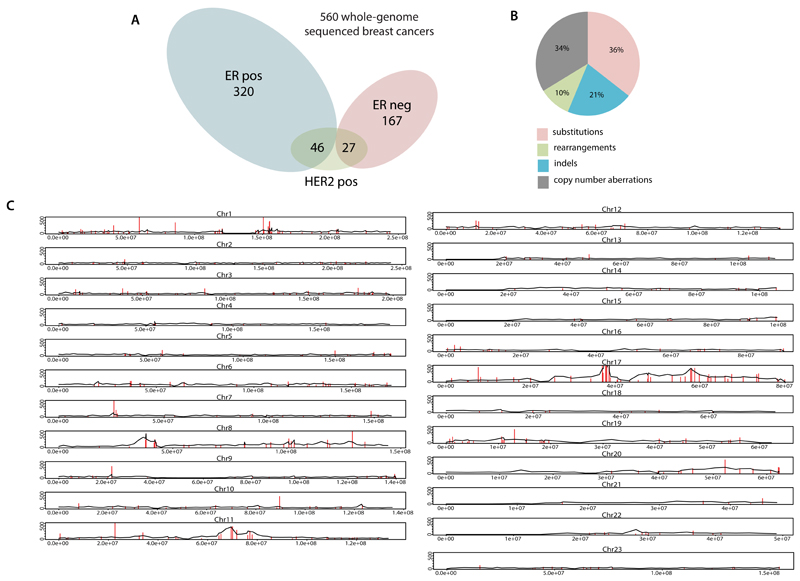
The whole genomes of 560 breast cancers and non-neoplastic tissue from each individual (556 female and four male) were sequenced (Fig.S1, Supplementary Table 1). 3,479,652 somatic base substitutions, 371,993 small indels and 77,695 rearrangements were detected, with substantial variation in the number of each between individual samples (Fig.1A, Supplementary Table 3). Transcriptome sequence, microRNA expression, array based copy number and DNA methylation data were obtained from subsets of cases.
Figure 1. Cohort and catalogue of somatic mutations in 560 breast cancers.
(A) Catalogue of base substitutions, insertions/deletions, rearrangements and driver mutations in 560 breast cancers (sorted by total substitution burden). Indel axis limited to 5,000(*).
(B) Complete list of curated driver genes sorted by frequency (descending). Fraction of ER positive (left, total 366) and ER negative (right, total 194) samples carrying a mutation in the relevant driver gene presented in grey. Log10 p-value of enrichment of each driver gene towards the ER positive or ER negative cohort is provided in black. Highlighted in green are genes for which there is new or further evidence supporting these as novel breast cancer genes.
To identify new cancer genes, we combined somatic substitutions and indels in protein-coding exons with data from other series12–15,26, constituting a total of 1,332 breast cancers, and searched for mutation clustering in each gene beyond that expected by chance. Five cancer genes were found for which evidence was previously absent or equivocal (MED23, FOXP1, MLLT4, XBP1, ZFP36L1), or for which the mutations indicate the gene acts in breast cancer in a recessive rather than in a dominant fashion, as previously reported in other cancer types (Supplementary Methods section 7.4 for detailed descriptions). From published reports on all cancer types (http://cancer.sanger.ac.uk/census), we then compiled a list of 727 human cancer genes (Supplementary Table 12). Based on driver mutations found previously, we defined conservative rules for somatic driver base substitutions and indel mutations in each gene and sought mutations conforming to these rules in the 560 breast cancers. 916 likely driver mutations of these classes were identified (Fig.1B, Supplementary Table 14, Extended Data Figure 1).
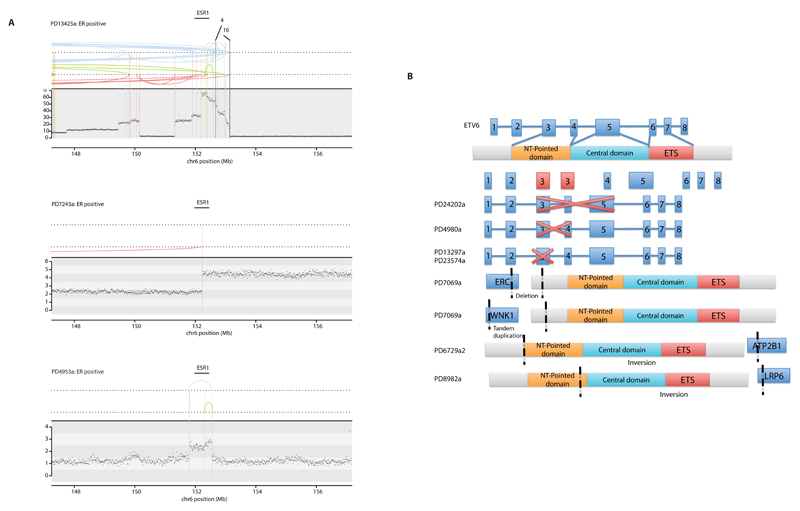
To explore the role of genomic rearrangements as driver mutations16,18,19,27, we sought predicted in-frame fusion genes that might create activated, dominant cancer genes. 1,278 unique and 39 infrequently recurrent in-frame gene fusions were identified (Supplementary Table 15). Many of the latter, however, were in regions of high rearrangement density, including amplicons28 and fragile sites, and their recurrence is likely attributable to chance27. Furthermore, transcriptome sequences from 260 cancers did not show expression of these fusions and generally confirmed the rarity of recurrent in-frame fusion genes. By contrast, recurrent rearrangements interrupting the gene footprints of CDKN2A, RB1, MAP3K1, PTEN, MAP2K4, ARID1B, FBXW7, MLLT4 and TP53 were found beyond the numbers expected from local background rearrangement rates, indicating that they contribute to the driver mutation burden of recessive cancer genes. Several other recurrently rearranged genomic regions were observed, including dominantly-acting cancer genes ETV6 and ESR1 without consistent elevation in expression levels, L1-retrotransposition sites29 and fragile sites. The significance of these recurrently rearranged regions remain unclear (Extended Data Figure 2).
Incorporation of recurrent copy number changes, including homozygous deletions and amplifications, generated a final tally of 1,628 likely driver mutations in 93 cancer genes (Fig.1B). At least one driver was identifiable in 95% of cancers. The 10 most frequently mutated genes were TP53, PIK3CA, MYC, CCND1, PTEN, ERBB2, chr8:ZNF703/FGFR1 locus, GATA3, RB1 and MAP3K1 (Fig.1B, Extended Data Figure 1) and accounted for 62% of drivers.
Recurrent somatic mutations in non-coding genomic regions
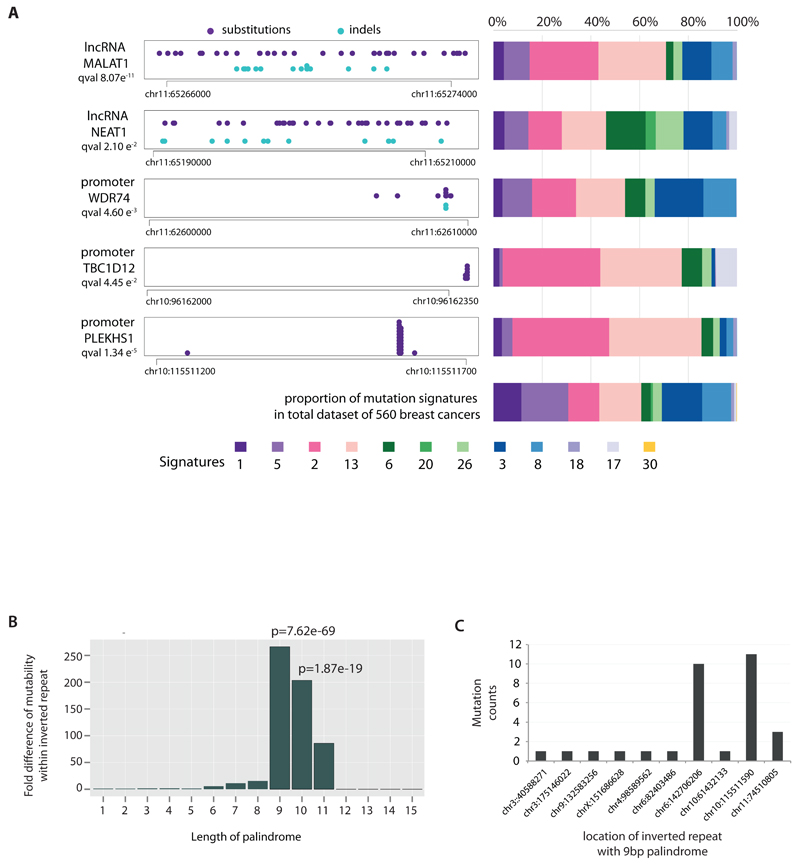
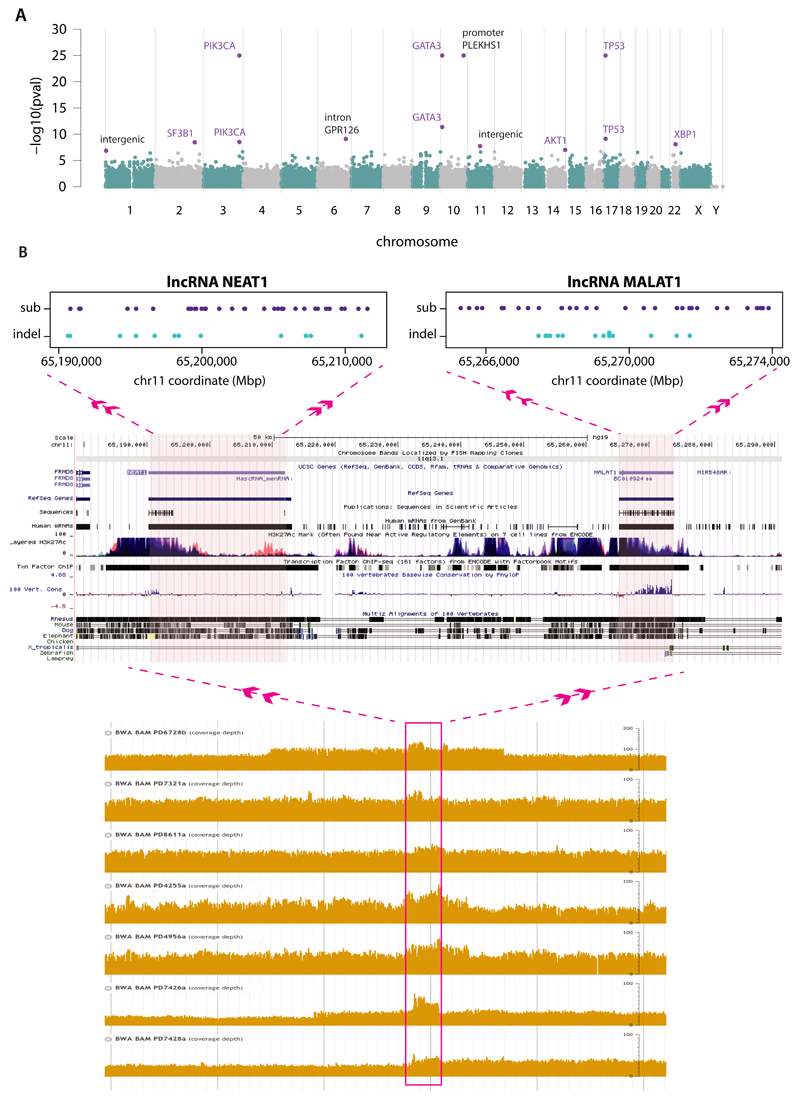
To investigate non-coding somatic driver substitutions and indels, we searched for non-coding genomic regions with more mutations than expected by chance (Fig.2A, Supplementary Table 16, Extended Data Figure 3). The promoter of PLEKHS1 (pleckstrin homology domain containing, family S member 1) exhibited recurrent mutations at two genomic positions30 (Fig.2A), the underlined bases in the sequence CAGCAAGC TGAACA GCTTGCTG (as previously reported30). The two mutated bases are flanked on either side by 9bp of palindromic sequence forming inverted repeats31. Most cancers with these mutations showed many base substitutions of mutational signatures 2 and 13 that have been attributed to activity of APOBEC DNA-editing proteins that target the TCN sequence motif. One of the mutated bases is a cytosine in a TCA sequence context (shown above as the reverse complement, TGA) at which predominantly C>T substitutions were found. The other is a cytosine in ACA context which showed both C>T and C>G mutations.
Figure 2. Non-coding analyses of breast cancer genomes.
(A) Distributions of substitution (purple dots) and indel (blue dots) mutations within the footprint of five regulatory regions identified as being more significantly mutated than expected is provided on the left. The proportion of base substitution mutation signatures associated with corresponding samples carrying mutations in each of these non-coding regions, is displayed on the right.
(B) Mutability of TGAACA/TGTTCA motifs within inverted repeats of varying flanking palindromic sequence length compared to motifs not within an inverted repeat.
(C) Variation in mutability between loci of TGAACA/TGTTCA inverted repeats with 9bp palindromes.
The TGAACA core sequence was mutated at the same two positions at multiple locations elsewhere in the genome (Supplementary Table 16C) where the TGAACA core was also flanked by palindromes (inverted repeat), albeit of different sequences and lengths (Supplementary Table 16C). These mutations were also usually found in cancers with many signature 2 and 13 mutations (Fig.2A). TGAACA core sequences with longer flanking palindromes generally exhibited a higher mutation rate, and TGAACA sequences flanked by 9bp palindromes exhibited a ~265-fold higher mutation rate than sequences without them (Fig.2B, Supplementary Table 16D). However, additional factors must influence the mutation rate because it varied markedly between TGAACA core sequences with different palindromes of the same length (Fig.2C). Some TGAACA-inverted repeat sites were in regulatory regions but others were intronic or intergenic without functional annotation (examples in Supplementary Table 16C) or exonic. The propensity for mutation recurrence at specific positions in a distinctive sequence motif in cancers with numerous mutations of particular signatures renders it plausible that these are hypermutable hotspots32–34, perhaps through formation of DNA hairpin structures35, which are single stranded at their tips enabling attack by APOBEC enzymes, rather than driver mutations.
Two recurrently mutated sites were also observed in the promoter of TBC1D12 (TBC1 domain family, member 12) (q-value 4.5e-2) (Fig.2A). The mutations were characteristic of signatures 2 and 13 and enriched in cancers with many signature 2 and 13 mutations (Fig.2A). The mutations were within the TBC1D12 Kozak consensus sequence (CCCCAGATGGTGGG)) shifting it away from the consensus36. The association with particular mutational signatures suggests that these may also be in a region of hypermutability rather than drivers.
The WDR74 (WD repeat domain 74) promoter showed base substitutions and indels (q-value 4.6e-3) forming a cluster of overlapping mutations (Fig.2A)20. Coding sequence driver mutations in WDR74 have not been reported. No differences were observed in WDR74 transcript levels between cancers with WDR74 promoter mutations compared to those without. Nevertheless, the pattern of this non-coding mutation cluster, with overlapping and different mutation types, is more compatible with the possibility of the mutations being drivers.
Two long non-coding RNAs, MALAT1 (q-value 8.7e-11, as previously reported12) and NEAT1 (q-value 2.1e-2) were enriched with mutations. Transcript levels were not significantly different between mutated and non-mutated samples. Whether these mutations are drivers, or result from local hypermutability, is unclear.
Mutational signatures
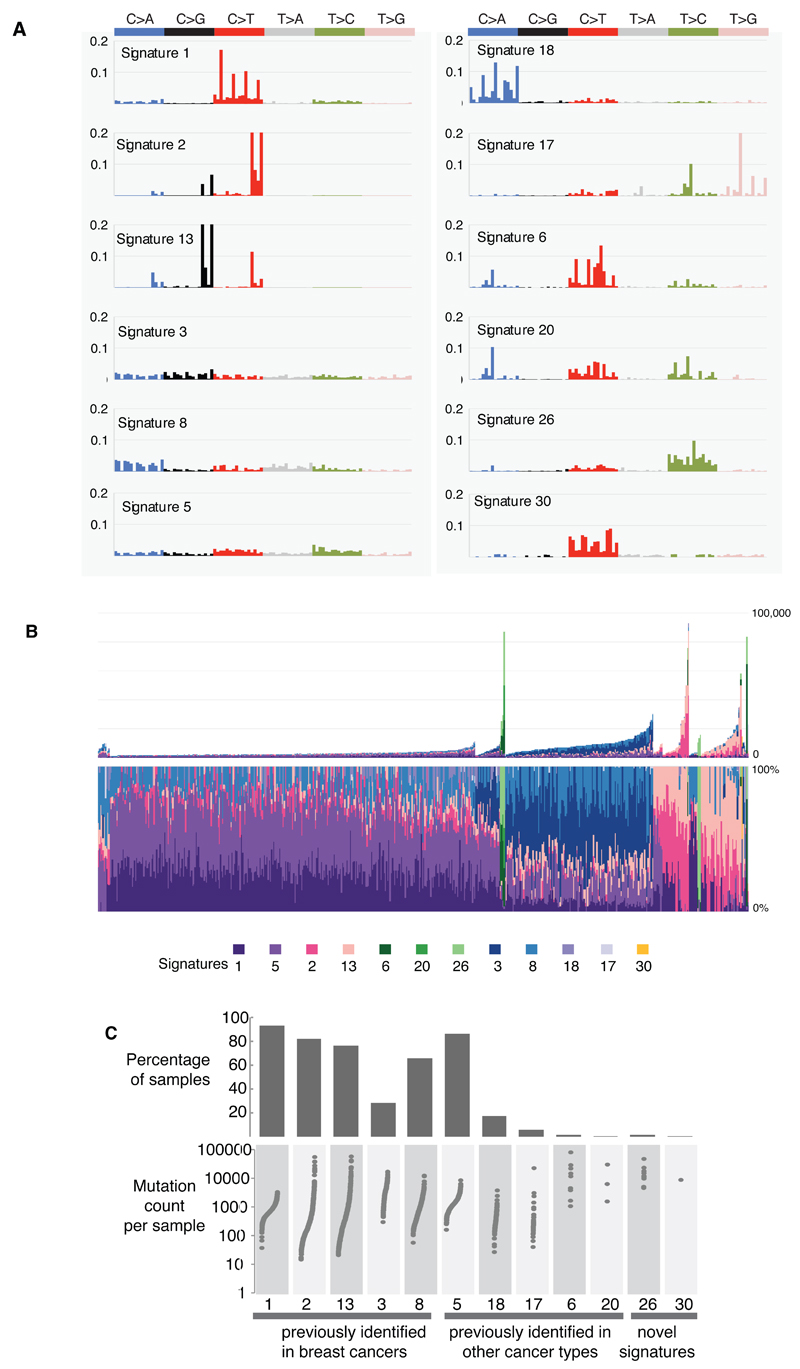
Mutational processes generating somatic mutations imprint particular patterns of mutations on cancer genomes, termed signatures2,24,37. Applying a mathematical approach25 to extract mutational signatures previously revealed five base substitution signatures in breast cancer; signatures 1, 2, 3, 8 and 132,24. Using this method in the 560 cases revealed 12 signatures, including those previously observed and a further seven, of which five have formerly been detected in other cancer types (signatures 5, 6, 17, 18 and 20) and two are new (signatures 26 and 30) (Fig.3A-B, Fig.4A, Supplementary Table 21A-C, Supplementary Methods 15 for further details). Two indel signatures were also found2,24.
Figure 3. Extraction and contributions of base substitution signatures in 560 breast cancers.
(A) Twelve mutation signatures extracted using Non-Negative Matrix Factorization. Each signature is ordered by mutation class (C>A/G>T, C>G/G>C, C>T/G>A, T>A/A>T, T>C/A>G, T>G/A>C), taking immediate flanking sequence into account. For each class, mutations are ordered by 5’ base (A,C,G,T) first before 3’ base (A,C,G,T).
(B) The spectrum of base substitution signatures within 560 breast cancers. Mutation signatures are ordered (and coloured) according to broad biological groups: Signatures 1 and 5 are correlated with age of diagnosis, Signatures 2 and 13 are putatively APOBEC-related, Signatures 6, 20 and 26 are associated with MMR deficiency, Signatures 3 and 8 are associated with HR deficiency, Signatures 18, 17 and 30 have unknown etiology. For ease of reading, this arrangement is adopted for the rest of the manuscript. Samples are ordered according to hierarchical clustering performed on mutation signatures. Top panel shows absolute numbers of mutations of each signature in each sample. Lower panel shows proportion of each signature in each sample.
(C) Distribution of mutation counts for each signature in relevant breast cancer samples. Percentage of samples carrying each signature provided above each signature.
Figure 4. Additional characteristics of base substitution signatures and novel rearrangement signatures in 560 breast cancers.
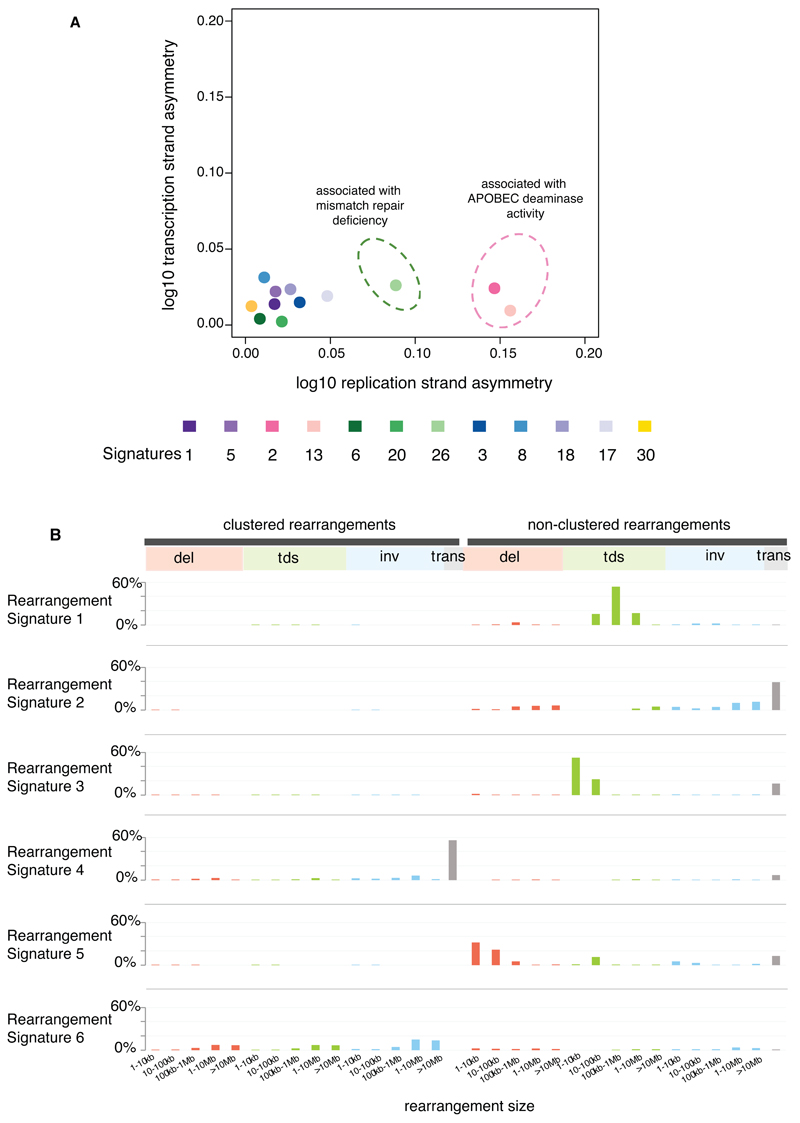
(A) Contrasting transcriptional strand asymmetry and replication strand asymmetry between twelve base substitution signatures.
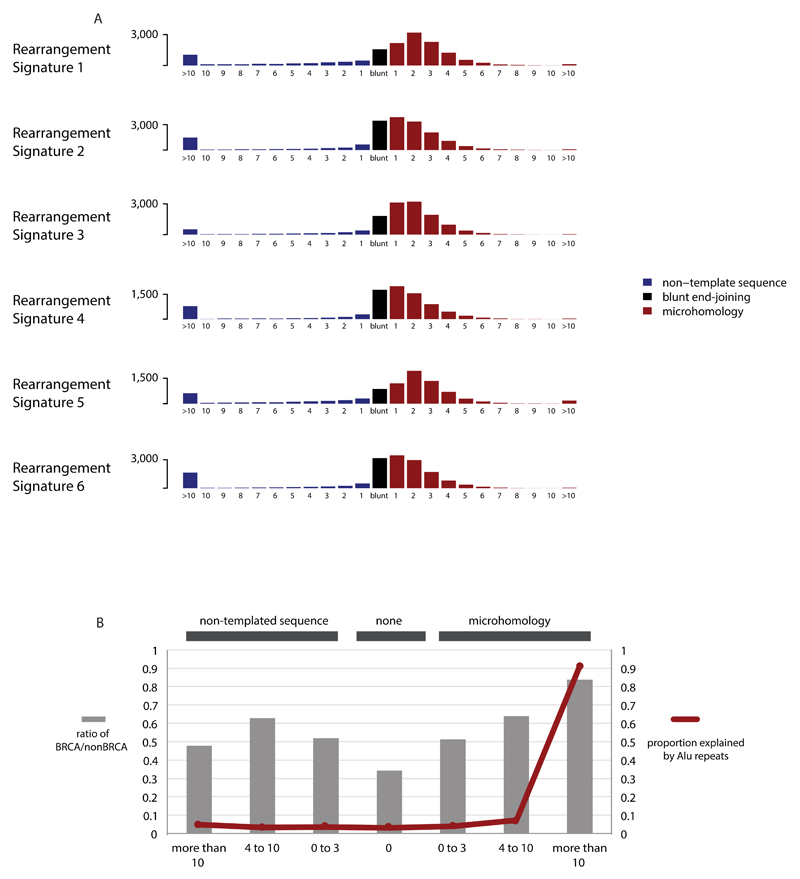
(B) Six rearrangement signatures extracted using Non-Negative Matrix Factorization. Probability of rearrangement element on y-axis. Rearrangement size on x-axis. Del= deletion, tds = tandem duplication, inv = inversion, trans = translocation.
Signatures of rearrangement mutational processes have not previously been formally investigated. To enable this we adopted a rearrangement classification incorporating 32 subclasses. In many cancer genomes, large numbers of rearrangements are regionally clustered, for example in zones of gene amplification. Therefore, we first classified rearrangements into those inside and outside clusters, further subclassified them into deletions, inversions and tandem duplications, and then according to the size of the rearranged segment. The final category in both groups was interchromosomal translocations.
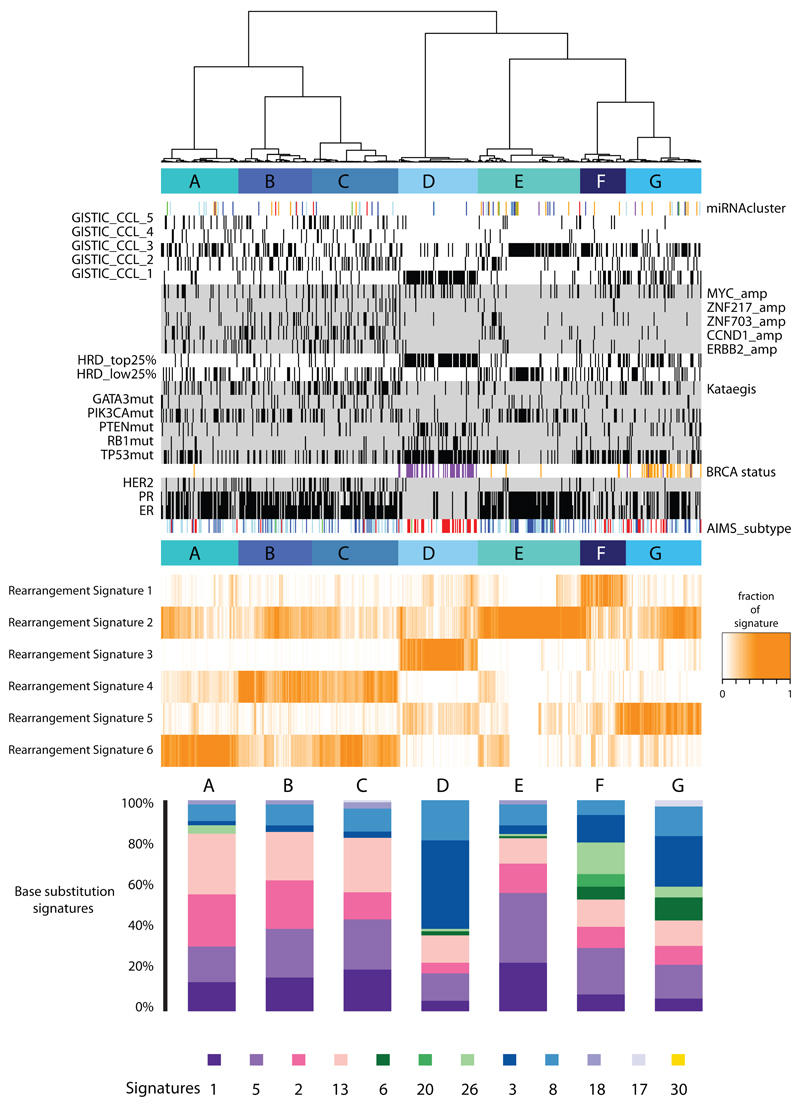
Application of the mathematical framework used for base substitution signatures2,24,25 extracted six rearrangement signatures (Fig.4B, Supplementary Table 21). Unsupervised hierarchical clustering on the basis of the proportion of rearrangements attributed to each signature in each breast cancer yielded seven major subgroups exhibiting distinct associations with other genomic, histological or gene expression features (Fig.5, Extended Data Figure 4-6).
Figure 5. Integrative analysis of rearrangement signatures.
Heatmap of rearrangement signatures (RS) following unsupervised hierarchical clustering based on proportions of RS in each cancer. 7 cluster groups (A-G) noted and relationships with expression (AIMS) subtype (basal=red, luminal B=light blue, luminal A=dark blue), immunohistopathology status (ER, PR, HER2 status – black=positive), abrogation of BRCA1 (purple) and BRCA2 (orange) (whether germline, somatic or through promoter hypermethylation), presence of 3 or more foci of kataegis (black=positive), HRD index (top 25% or lowest 25% - black=positive), GISTIC cluster group (black=positive) and driver mutations in cancer genes. miRNA cluster groups : 0=red, 1=purple, 2=blue, 3=light blue, 4=green, 5=orange. Contribution of base substitution signatures in these 7 cluster groups is provided in the lowermost panel.
Rearrangement Signature 1 (9% of all rearrangements) and Rearrangement Signature 3 (18% rearrangements) were characterised predominantly by tandem duplications (Fig.4B). Tandem duplications associated with Rearrangement Signature 1 were mostly >100kb (Fig.4B), and those with Rearrangement Signature 3 <10kb (Fig.4B, Extended Data Figure 7). More than 95% of Rearrangement Signature 3 tandem duplications were concentrated in 15% of cancers (Cluster D, Fig.5), many with several hundred rearrangements of this type. Almost all cancers (91%) with BRCA1 mutations or promoter hypermethylation were in this group, which was enriched for basal-like, triple negative cancers and copy number classification of a high Homologous Recombination Deficiency (HRD) index38–40. Thus, inactivation of BRCA1, but not BRCA2, may be responsible for the Rearrangement Signature 3 small tandem duplication mutator phenotype.
More than 35% of Rearrangement Signature 1 tandem duplications were found in just 8.5% of the breast cancers and some cases had hundreds of these (Cluster F, Fig.5). The cause of this large tandem duplication mutator phenotype (Fig.4B) is unknown. Cancers exhibiting it are frequently TP53-mutated, relatively late diagnosis, triple-negative breast cancers, showing enrichment for base substitution signature 3 and a high Homologous Recombination Deficiency (HRD) index (Fig.5) but do not have BRCA1/2 mutations or BRCA1 promoter hypermethylation.
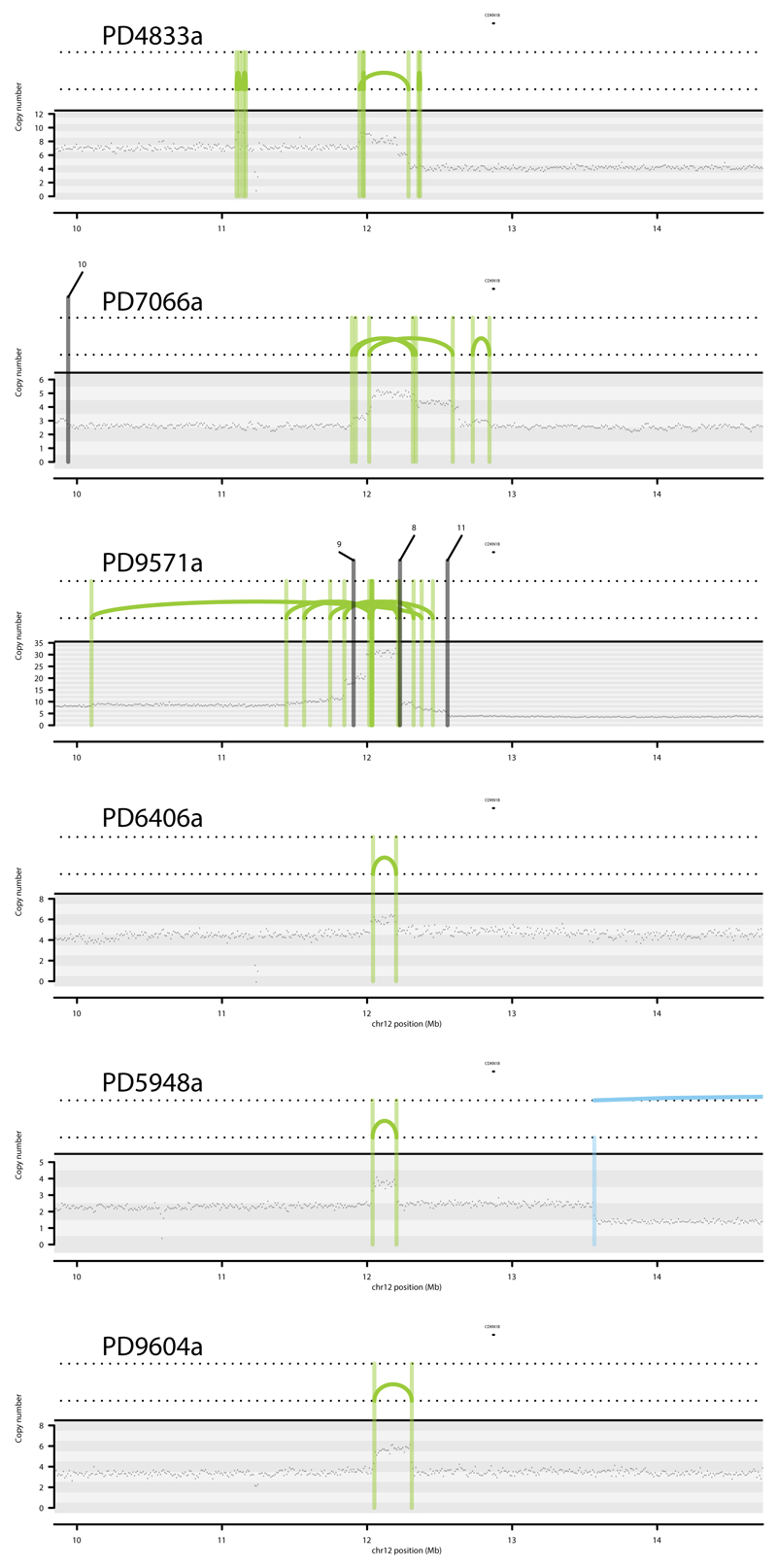
Rearrangement Signature 1 and 3 tandem duplications (Extended Data Figure 7) were generally evenly distributed over the genome. However, there were nine locations at which recurrence of tandem duplications was found across the breast cancers and which often showed multiple, nested tandem duplications in individual cases (Extended Data Figure 8). These may be mutational hotspots specific for these tandem duplication mutational processes although we cannot exclude the possibility that they represent driver events.
Rearrangement Signature 5 (accounting for 14% rearrangements) was characterised by deletions <100kb. It was strongly associated with the presence of BRCA1 mutations or promoter hypermethylation (Cluster D, Fig.5), BRCA2 mutations (Cluster G, Fig.5) and with Rearrangement Signature 1 large tandem duplications (Cluster F, Fig.5).
Rearrangement Signature 2 (accounting for 22% rearrangements) was characterised by non-clustered deletions (>100kb), inversions and interchromosomal translocations, was present in most cancers but was particularly enriched in ER positive cancers with quiet copy number profiles (Cluster E, GISTIC Cluster 3, Fig.5). Rearrangement Signature 4 (accounting for 18% of rearrangements) was characterised by clustered interchromosomal translocations while Rearrangement Signature 6 (19% of rearrangements) by clustered inversions and deletions (Clusters A, B, C, Fig.5).
Short segments (1-5bp) of overlapping microhomology characteristic of alternative methods of end joining repair were found at most rearrangements2,14. Rearrangement Signatures 2, 4 and 6 were characterised by a peak at 1bp of microhomology while Rearrangement Signatures 1, 3 and 5, associated with homologous recombination DNA repair deficiency, exhibited a peak at 2bp (Extended Data Figure 9). Thus, different end-joining mechanisms may operate with different rearrangement processes. A proportion of breast cancers showed Rearrangement Signature 5 deletions with longer (>10bp) microhomologies involving sequences from short-interspersed nuclear elements (SINEs), most commonly AluS (63%) and AluY (15%) family repeats (Extended Data Figure 9). Long segments (more than 10bp) of non-templated sequence were particularly enriched amongst clustered rearrangements.
Localised hypermutation: kataegis
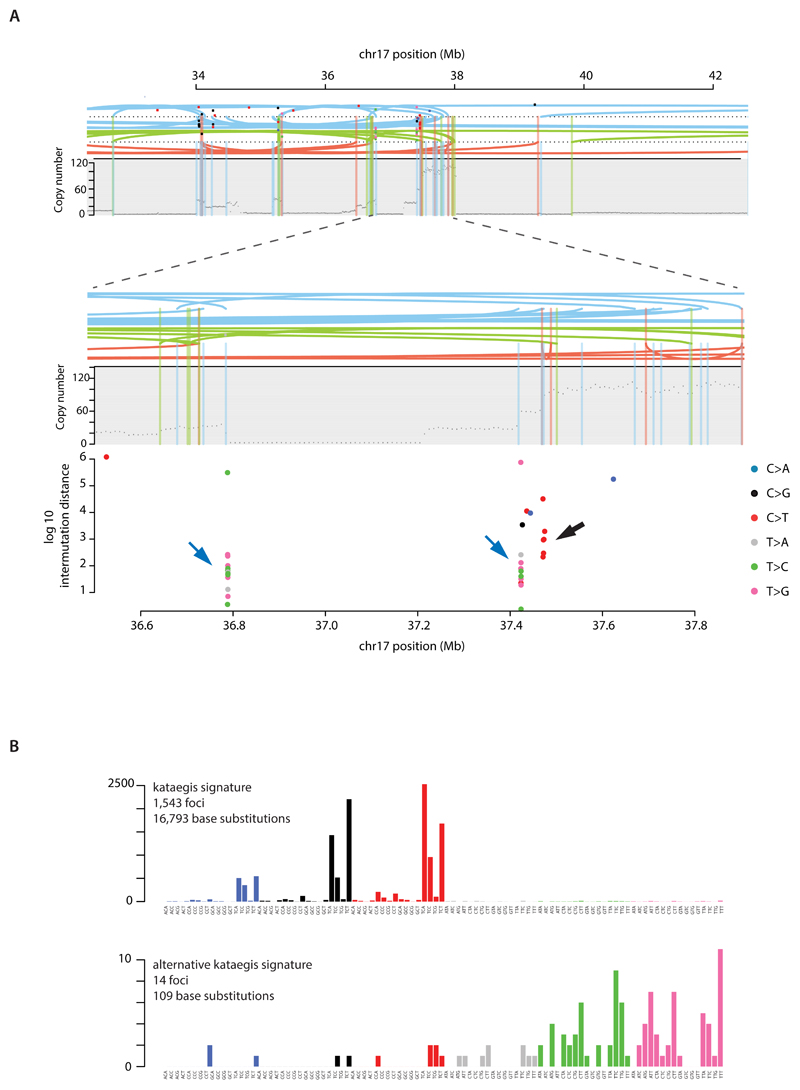
Focal base substitution hypermutation, termed kataegis, is generally characterised by substitutions with characteristic features of signatures 2 and 132,24. Kataegis was observed in 49% breast cancers, with 4% exhibiting 10 or more foci (Supplementary Table 21C). Kataegis colocalises with clustered rearrangements characteristic of rearrangement signatures 4 and 6 (Fig.4B). Cancers with tandem duplications or deletions of rearrangement signatures 1, 3 and 5 did not usually demonstrate kataegis. However, there must be additional determinants of kataegis since only 2% of rearrangements are associated with it. A rare (14/1,557 foci, 0.9%), alternative form of kataegis colocalising with rearrangements but with a base substitution pattern characterised by T>G and T>C mutations predominantly at NTT and NTA sequences was also observed (Extended Data Figure 10). This pattern of base substitutions most closely matches Signature 9 (Extended Data Figure 10) (http://cancer.sanger.ac.uk/cosmic/signatures), previously observed in B lymphocyte neoplasms and attributed to polymerase eta activity 41.
Mutational signatures exhibit distinct DNA replication strand biases
The distributions of mutations attributable to each of the 20 mutation signatures (12 base substitution, two indel and six rearrangement) were explored42 with respect to DNA replication strand. We found an asymmetric distribution of mutations between leading and lagging replication strands for many, but not all signatures42 (Fig.4A). Notably, Signatures 2 and 13, due to APOBEC deamination, showed marked lagging replication strand bias (Fig.4A) suggesting that lagging strand replication provides single-stranded DNA for APOBEC deamination. Of the three signatures associated with mismatch repair deficiency (Signatures 6, 20, 26), only Signature 26 exhibited replicative strand bias, highlighting how different signatures arising from defects of the same pathway can exhibit distinct relationships with replication.
Mutational signatures associated with BRCA1 and BRCA2 mutations
Of the 560 breast cancers, 90 had germline (60) or somatic (14) inactivating mutations in BRCA1 (35) or BRCA2 (39) or showed methylation of the BRCA1 promoter (16). Loss of the wild-type chromosome 17 or 13 was observed in 80/90 cases. The latter exhibited many base substitution mutations of signature 3, accompanied by deletions of >3bp with microhomology at rearrangement breakpoints, and signature 8 together with CC>AA double nucleotide substitutions. Cases in which the wild type chromosome 17 or 13 was retained did not show these signatures. Thus signature 3 and, to a lesser extent, signature 8 are associated with absence of BRCA1 and BRCA2 functions.
Cancers with inactivating BRCA1 or BRCA2 mutations usually carry many genomic rearrangements. Cancers with BRCA1, but not BRCA2, mutations exhibit large numbers of Rearrangement Signature 3 small tandem duplications. Cancers with BRCA1 or BRCA2 mutations show substantial numbers of Rearrangement Signature 5 deletions. No other Rearrangement Signatures were associated with BRCA1 or BRCA2 null cases (Clusters D and G, Fig.5). Some breast cancers without identifiable BRCA1/2 mutations or BRCA1 promoter methylation showed these features and segregated with BRCA1/2 null cancers in hierarchical clustering analysis (Fig.5). In such cases, the BRCA1/2 mutations may have been missed or other mutated or promoter methylated genes may be exerting similar effects (Please see http://cancer.sanger.ac.uk/cosmic/sample/genomes for examples of whole genome profiles of typical BRCA1 null (e.g. PD6413a, PD7215a) and BRCA2 null tumours (e.g. PD4952a, PD4955a)).
A further subset of cancers (Cluster F, Fig.5) show similarities in mutational pattern to BRCA1/2 null cancers, with many Rearrangement Signature 5 deletions and enrichment for base substitution signatures 3 and 8. However, these do not segregate together with BRCA1/2 null cases in hierarchical clustering analysis, have Rearrangement Signature 1 large tandem duplications and do not show BRCA1/2 mutations. Somatic and germline mutations in genes associated with the DNA double-strand break repair pathway including ATM, ATR, PALB2, RAD51C, RAD50, TP53, CHEK2 and BRIP1, were sought in these cancers. We did not observe any clear-cut relationships between mutations in these genes and these mutational patterns.
Cancers with BRCA1/2 mutations are particularly responsive to cisplatin and PARP inhibitors43–45. Combinations of base substitution, indel and rearrangement mutational signatures may be better biomarkers of defective homologous recombination based DNA double strand break repair and responsiveness to these drugs46 than BRCA1/2 mutations or promoter methylation alone and thus may constitute the basis of future diagnostics.
Conclusions
A comprehensive perspective on the somatic genetics of breast cancer is drawing closer (please see website for individual patient genome profiles: http://cancer.sanger.ac.uk/cosmic/sample/genomes, Methods Section 10 for orientation). At least 12 base substitution mutational signatures and six rearrangement signatures contribute to the somatic mutations found. 93 mutated cancer genes (31 dominant, 60 recessive, 2 uncertain) are implicated in genesis of the disease. However, dominantly-acting activated fusion genes and non-coding driver mutations appear rare. Additional infrequently mutated cancer genes probably exist. However, the genes harbouring the substantial majority of driver mutations are now known.
Nevertheless, important questions remain to be addressed. Recurrent mutational events including whole chromosome copy number changes and unexplained regions with recurrent rearrangements could harbour additional cancer genes. Identifying non-coding drivers is challenging and requires further investigation. Although almost all breast cancers have at least one identifiable driver mutation, the number with only a single identified driver is perhaps surprising. The roles of viruses or other microbes have not been exhaustively examined. Thus, further exploration and analysis of whole genome sequences from breast cancer patients will be required to complete our understanding of the somatic mutational basis of the disease.
Methods
1. Sample selection
DNA was extracted from 560 breast cancers and normal tissue (peripheral blood lymphocytes, adjacent normal breast tissue or skin) and total RNA extracted from 268 of the same individuals. Samples were subjected to pathology review and only samples assessed as being composed of > 70% tumor cells, were accepted for inclusion in the study (Supplementary Table 1).
2. Massively-parallel sequencing and alignment
Short insert 500bp genomic libraries and 350bp poly-A selected transcriptomic libraries were constructed, flowcells prepared and sequencing clusters generated according to Illumina library protocols47. 108 base/100 base (genomic), or 75 base (transcriptomic) paired-end sequencing were performed on Illumina GAIIx, Hiseq 2000 or Hiseq 2500 genome analyzers in accordance with the Illumina Genome Analyzer operating manual. The average sequence coverage was 40.4 fold for tumour samples and 30.2 fold for normal samples (Supplementary Table 2).
Short insert paired-end reads were aligned to the reference human genome (GRCh37) using Burrows-Wheeler Aligner, BWA (v0.5.9)48. RNA-seq data was aligned to the human reference genome (GRCh37) using TopHat (v1.3.3) (http://ccb.jhu.edu/software/tophat/index.shtml).
3. Processing of genomic data
CaVEMan (Cancer Variants Through Expectation Maximization: http://cancerit.github.io/CaVEMan/) was used for calling somatic substitutions.
Indels in the tumor and normal genomes were called using a modified Pindel version 2.0. (http://cancerit.github.io/cgpPindel/) on the NCBI37 genome build 49.
Structural variants were discovered using a bespoke algorithm, BRASS (BReakpoint AnalySiS) (https://github.com/cancerit/BRASS) through discordantly mapping paired-end reads. Next, discordantly mapping read pairs that were likely to span breakpoints, as well as a selection of nearby properly-paired reads, were grouped for each region of interest. Using the Velvet de novo assembler50, reads were locally assembled within each of these regions to produce a contiguous consensus sequence of each region. Rearrangements, represented by reads from the rearranged derivative as well as the corresponding non-rearranged allele were instantly recognisable from a particular pattern of five vertices in the de Bruijn graph (a mathematical method used in de novo assembly of (short) read sequences) of component of Velvet. Exact coordinates and features of junction sequence (e.g. microhomology or non-templated sequence) were derived from this, following aligning to the reference genome, as though they were split reads.
Supplementary Table 3 for summary of somatic variants. Annotation was according to ENSEMBL version 58.
Single nucleotide polymorphism (SNP) array hybridization using the Affymetrix SNP6.0 platform was performed according to Affymetrix protocols. Allele-specific copy number analysis of tumors was performed using ASCAT (v2.1.1), to generate integral allele-specific copy number profiles for the tumor cells51 (Supplementary Table 4 and 5). ASCAT was also applied to NGS data directly with highly comparable results.
12.5% of the breast cancers were sampled for validation of substitutions, indels and/or rearrangements in order to make an assessment of the positive predictive value of mutation-calling (Supplementary Table 6).
Further details of these processing steps as well as processing of transcriptomic and miRNA data (Supplementary Table 7 and 8) can be found in Supplementary Methods.
4. Identification of novel breast cancer genes
To identify recurrently mutated driver genes, a dN/dS method that considers the mutation spectrum, the sequence of each gene, the impact of coding substitutions (synonymous, missense, nonsense, splice site) and the variation of the mutation rate across genes52,53 was used for substitutions (Supplementary Table 9). Owing to the lack of a neutral reference for the indel rate in coding sequences, a different approach was required (Supplementary Table 10, Supplementary Methods for details). To detect genes under significant selective pressure by either point mutations or indels, for each gene the P-values from the dN/dS analysis of substitutions and from the recurrence analysis of indels were combined using Fisher’s method. Multiple testing correction (Benjamini-Hochberg FDR) was performed separately for the 600+ putative driver genes and for all other genes, stratifying the FDR correction to increase sensitivity (as described in Sun et al. 200654). To achieve a low false discovery rate a conservative q-value cutoff of <0.01 was used for significance (Supplementary Table 11).
This analysis was applied to the new whole genome sequences of 560 breast cancers as well as a further 772 breast cancers that have been sequenced previously by other institutions.
Please see Supplementary Methods for detailed explanations of these methods.
5. Recurrence in the non-coding regions
5.1. Partitioning the genome into functional regulatory elements/gene features
To identify non-coding regions with significant recurrence, we used a method similar to the one described for searching for novel indel drivers (Supplementary Methods for detailed description).
The genome was partitioned according to different sets of regulatory elements/gene features, with a separate analysis performed for each set of elements, including exons (n=20,245 genes), core promoters (n=20,245 genes, where a core promoter is the interval [−250,+250] bp from any transcription start site (TSS) of a coding transcript of the gene, excluding any overlap with coding regions), 5’ UTR (n=9,576 genes), 3’ UTR (n=19,502 genes), intronic regions flanking exons (n=20,212 genes, represents any intronic sequence within 75bp from an exon, excluding any base overlapping with any of the above elements. This attempts to capture recurrence in essential splice site or proximal splicing-regulatory elements), any other sequence within genes (n=18,591 genes, for every protein-coding gene, this contains any region within the start and end of transcripts not included in any of the above categories), ncRNAs (n=10,684, full length lincRNAs, miRNAs or rRNAs), enhancers (n=194,054) 55, ultra-conserved regions (n=187,057, a collection of regions under negative selection based on 1,000 genomes data 20.
Every element set listed above was analysed separately to allow for different mutation rates across element types and to stratify the FDR correction 54. Within each set of elements, we used a negative binomial regression approach to learn the underlying variation of the mutation rate across elements. The offset reflects the expected number of mutations in each element assuming uniform mutation rates across them (i.e. Esubs,element = Σ j∈{1,2,…,192}Extende (t*rj*Sj), and, Eindels,element = μindel * Sindel,element ). As covariate here we used the local density of mutations in neighbouring non-coding regions, corrected for sequence composition and trinucleotide mutation rates, that is, the t parameter of the dN/dS equations described in section 7.1 of Supplementary Methods. Normalised local rates were pre-calculated for 100kb non-overlapping bins of the genome and used in all analyses. Other covariates (expression, replication time or HiC) were not used here as they were not found to substantially improve the model once the local mutation rate was used as a covariate. A separate regression analysis was performed for substitutions and indels, to account for the different level of uncertainty in the distribution of substitution and indel rates across elements.
modelsubs = glm.nb(formula = nsubs ~ offset(log(Esubs)) + μlocal,subs )
modelindels = glm.nb(formula = nindels ~ offset(log(Eindels)) + μlocal,indels )
The observed counts for each element (nsubs,element and nindels,element) are compared to the background distributions using a negative binomial test, with the estimated overdispersion parameters (θsubs and θindels) estimated by the negative binomial regression, yielding P-values for substitution and indel recurrence for each element. These P-values were combined using Fisher’s method and corrected for multiple testing using FDR (Supplementary Table 16A).
5.2. Partitioning the genome into discrete bins
We performed a genome-wide screening of recurrence in 1kb non-overlapping bins. We employed the method described in earlier section, using as covariate the local mutation rate calculated from 5Mb up and downstream from the bin of interest and excluding any low-coverage region from the estimate (Supplementary Table 16B, Extended Data Figure 3A for example). Significant hits were subjected to manual curation to remove false positives caused by sequencing or mapping artefacts.
6. Mutational signatures analysis
Mutational signatures analysis was performed following a three-step process: (i) hierarchical de novo extraction based on somatic substitutions and their immediate sequence context, (ii) updating the set of consensus signatures using the mutational signatures extracted from breast cancer genomes, and (iii) evaluating the contributions of each of the updated consensus signatures in each of the breast cancer samples. These three steps are discussed in more details in the next sections.
6.1. Hierarchical de novo extraction of mutational signatures
The mutational catalogues of the 560 breast cancer whole genome sequences were analysed for mutational signatures using a hierarchical version of the Wellcome Trust Sanger Institute mutational signatures framework 25. Briefly, we converted all mutation data into a matrix, M, that is made up of 96 features comprising mutations counts for each mutation type (C>A, C>G, C>T, T>A, T>C, and T>G; all substitutions are referred to by the pyrimidine of the mutated Watson–Crick base pair) using each possible 5’ (C, A, G, and T) and 3’ (C, A, G, and T) context for all samples. After conversion, the previously developed algorithm was applied in a hierarchical manner to the matrix M that contains K mutation types and G samples. The algorithm deciphers the minimal set of mutational signatures that optimally explains the proportion of each mutation type and then estimates the contribution of each signature across the samples. More specifically, the algorithm makes use of a well-known blind source separation technique, termed nonnegative matrix factorization (NMF). NMF identifies the matrix of mutational signature, P, and the matrix of the exposures of these signatures, E, by minimizing a Frobenius norm while maintaining non-negativity:
The method for deciphering mutational signatures, including evaluation with simulated data and list of limitations, can be found in ref 25. The framework was applied in a hierarchical manner to increase its ability to find mutational signatures present in few samples as well as mutational signatures exhibiting a low mutational burden. More specifically, after application to the original matrix M containing 560 samples, we evaluated the accuracy of explaining the mutational patterns of each of the 560 breast cancers with the extracted mutational signatures. All samples that were well explained by the extracted mutational signatures were removed and the framework was applied to the remaining sub-matrix of M. This procedure was repeated until the extraction process did not reveal any new mutational signatures. Overall, the approach extracted 12 unique mutational signatures operative across the 560 breast cancers (Figure 3, Supplementary Table 21).
6.2. Updating the set of consensus mutational signatures
The 12 hierarchically extracted breast cancer signatures were compared to the census of consensus mutational signatures 25. 11 of the 12 signatures closely resembled previously identified mutational patterns. The patterns of these 11 signatures, weighted by the numbers of mutations contributed by each signature in the breast cancer data, were used to update the set of consensus mutational signatures as previously done in ref 25. 1 of the 12 extracted signatures is novel and at present, unique for breast cancer. This novel signature is consensus signature 30 (http://cancer.sanger.ac.uk/cosmic/signatures).
6.3. Evaluating the contributions of consensus mutational signatures in 560 breast cancers
The complete compendium of consensus mutational signatures that was found in breast cancer includes: signatures 1, 2, 3, 5, 6, 8, 13, 17, 18, 20, 26, and 30. We evaluated the presence of all these signatures in the 560 breast cancer genomes by re-introducing them into each sample. More specifically, the updated set of consensus mutational signatures was used to minimize the constrained linear function for each sample:
Here, represents a vector with 96 components (corresponding to a consensus mutational signature with its six somatic substitutions and their immediate sequencing context) and Exposurei is a nonnegative scalar reflecting the number of mutations contributed by this signature. N is equal to 12 and it reflects the number of all possible signatures that can be found in a single breast cancer sample. Mutational signatures that did not contribute large numbers (or proportions) of mutations or that did not significantly improve the correlation between the original mutational pattern of the sample and the one generated by the mutational signatures were excluded from the sample. This procedure reduced over-fitting the data and allowed only the essential mutational signatures to be present in each sample (Supplementary Table 21B).
7. Kataegis
Kataegis or foci of localized hypermutation has been previously defined 25 as 6 or more consecutive mutations with an average intermutation distance of less than or equal to 1,000 bp. Kataegis were sought in 560 whole-genome sequenced breast cancers from high-quality base substitution data using the method described previously 25. This method likely misses some foci of kataegis sacrificing sensitivity of detection for a higher positive predictive value of kataegic foci (Supplementary Table 21C).
8. Rearrangement signatures
8.1. Clustered vs non-clustered rearrangements
We sought to separate rearrangements that occurred as focal catastrophic events or focal driver amplicons from genome-wide rearrangement mutagenesis using a piecewise constant fitting (PCF) method. For each sample, both breakpoints of each rearrangement were considered individually and all breakpoints were ordered by chromosomal position. The inter-rearrangement distance, defined as the number of base pairs from one rearrangement breakpoint to the one immediately preceding it in the reference genome, was calculated. Putative regions of clustered rearrangements were identified as having an average inter-rearrangement distance that was at least 10 times greater than the whole genome average for the individual sample. PCF parameters used were γ = 25 and kmin = 10. The respective partner breakpoint of all breakpoints involved in a clustered region are likely to have arisen at the same mechanistic instant and so were considered as being involved in the cluster even if located at a distant chromosomal site. Extended Data Table 4A summarises the rearrangements within clusters (“clustered”) and not within clusters (“non-clustered”).
8.2. Classification – types and size
In both classes of rearrangements, clustered and non-clustered, rearrangements were subclassified into deletions, inversions and tandem duplications, and then further subclassified according to size of the rearranged segment (1-10kb, 10kb-100kb, 100kb-1Mb, 1Mb-10Mb, more than 10Mb). The final category in both groups was interchromosomal translocations.
8.3. Rearrangement signatures by NNMF
The classification produces a matrix of 32 distinct categories of structural variants across 544 breast cancer genomes. This matrix was decomposed using the previously developed approach for deciphering mutational signatures by searching for the optimal number of mutational signatures that best explains the data without over-fitting the data 25 (Supplementary Table 21D-E).
8.4. Consensus clustering of rearrangement signatures
To identify subgroups of samples sharing similar combinations of six identified rearrangement signatures derived from whole genome sequencing analysis we performed consensus clustering using the ConsensusClusterPlus R package 56. Input data for each sample (n=544, a subset of the full sample cohort) was the proportion of rearrangements assigned to each of the six signatures. Thus, each sample has 6 data values, with a total sum of 1. Proportions for each signature were mean-centred across samples prior to clustering. The following settings were used in the consensus clustering:
-
-
Number of repetitions: 1000
-
-
pItem = 0.9 (resampling frequency samples)
-
-
pFeature = 0.9 (resampling frequency)
-
-
Pearson distance metric
-
-
Ward linkage method
9. Distribution of mutational signatures relative to genomic architecture
Following extraction of mutational signatures and quantification of the exposures (or contributions) of each signature to each sample, a probability for each mutation belonging to each mutation signature (for a given class of mutation e.g. substitutions) was assigned42.
The distribution of mutations as signatures were assessed across multiple genomic features including replication time, strands, transcriptional strands and nucleosome occupancy. Please see Morganella et al for technical details, per signature results.
10. Individual patient whole genome profiles
Breast cancer whole genome profiles were adapted from the R Circos package57. Features depicted in circos plots from outermost rings heading inwards: Karyotypic ideogram outermost. Base substitutions next, plotted as rainfall plots (log10 intermutation distance on radial axis, dot colours: blue=C>A, black=C>G, red=C>T, grey=T>A, green=T>C, pink=T>G). Ring with short green lines = insertions, ring with short red lines = deletions. Major copy number allele (green = gain) ring, minor copy number allele ring (pink=loss), Central lines represent rearrangements (green= tandem duplications, pink=deletions, blue=inversions and gray=interchromosomal events. Top right hand panel displays the number of mutations contributing to each mutation signature extracted using NNMF in individual cancers. Middle right hand panel represents indels. Bottom right corner shows histogram of rearrangements present in this cancer. Bottom left corner shows all curated driver mutations, top and middle left panels show clinical and pathology data respectively.
Extended Data
Extended Data Figure 1. Landscape of driver mutations.
(A) Summary of subtypes of cohort of 560 breast cancers
(B) Driver mutations by mutation type
(C) Distribution of rearrangements throughout the genome. Black line represents background rearrangement density (calculation based on rearrangement breakpoints in intergenic regions only). Red lines represent frequency of rearrangement within breast cancer genes.
Extended Data Figure 2. Rearrangements in oncogenes.
(A) Variation in rearrangement and copy number events affecting ESR1. Clear amplification in topmost panel, transection of ESR1 in middle panel and focused tandem duplication events in lower panel.
(B) Predicted outcomes of some rearrangements affecting ETV6. Red crosses indicate exons deleted as a result of rearrangements within the ETV6 genes, black dotted lines indicate rearrangement break points resulting in fusions between ETV6 and ERC, WNK1, ATP2B1 or LRP6
Extended Data Figure 3. Recurrent non-coding events in breast cancers.
(A) Manhattan plot demonstrating sites with most significant p-values as identified by binning analysis. Purple highlighted sites were also detected by the method seeking recurrence when partitioned by genomic features.
(B) Locus at chr11:65Mb which was identified by independent analyses as being more mutated than expected by chance. In the lowermost panel, a rearrangement hotspot analysis identified this region as a tandem duplication hotspot, with nested tandem duplications noted at this site. Partitioning the genome into different regulatory elements, an analysis of substitutions and indels identified lncRNAs MALAT1 and NEAT1 (topmost panels) with significant p-values.
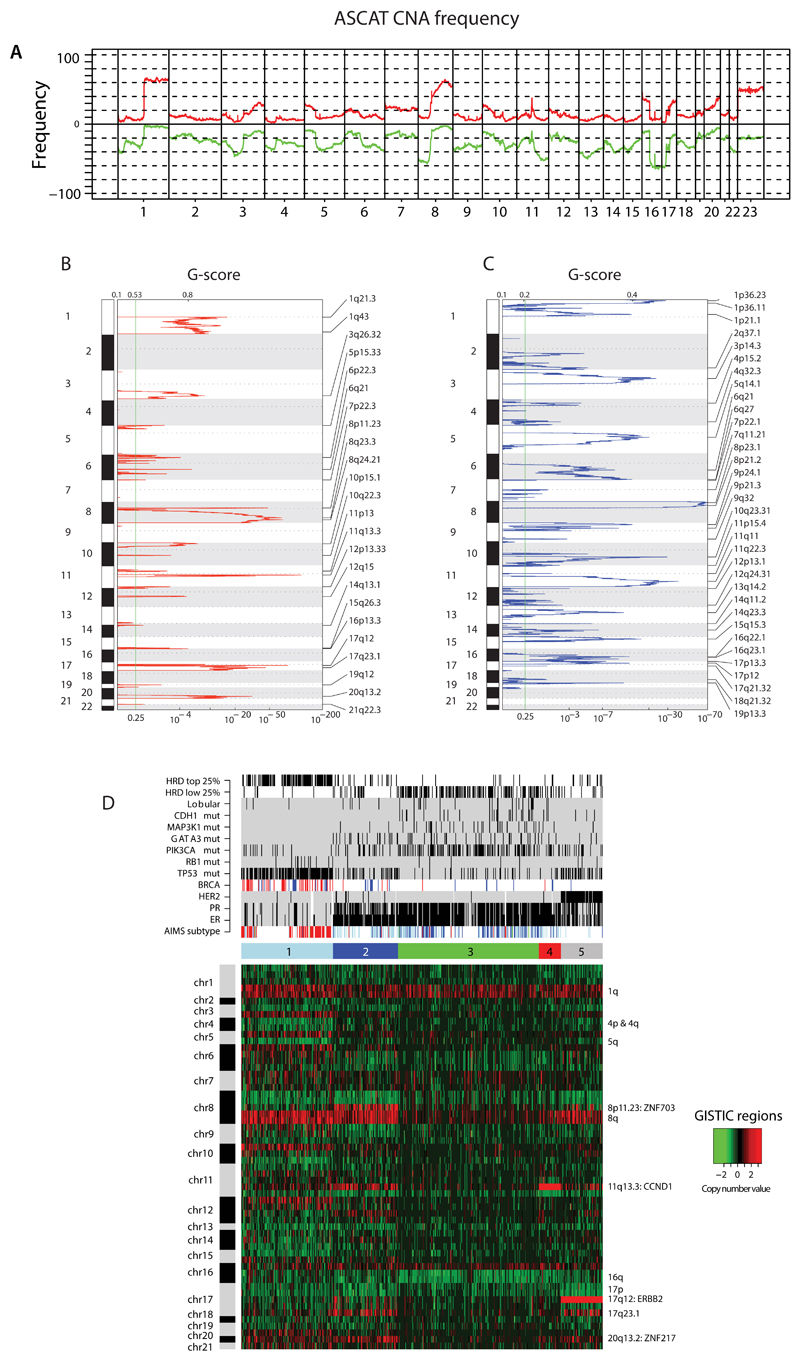
Extended Data Figure 4. Copy number analyses.
(A) Frequency of copy number aberrations across the cohort. Chromosome position along x-axis, frequency of copy number gains (red) and losses (green) y-axis.
(B) Identification of focal recurrent copy number gains by the GISTIC method (Supplementary Methods)
(C) Identification of focal recurrent copy number losses by the GISTIC method
(D) Heatmap of GISTIC regions following unsupervised hierarchical clustering. 5 cluster groups are noted and relationships with expression subtype (basal=red, luminal B=light blue, luminal A=dark blue), immunohistopathology status (ER, PR, HER2 status – black=positive), abrogation of BRCA1 (red) and BRCA2 (blue) (whether germline, somatic or through promoter hypermethylation), driver mutations (black=positive), HRD index (top 25% or lowest 25% - black=positive).
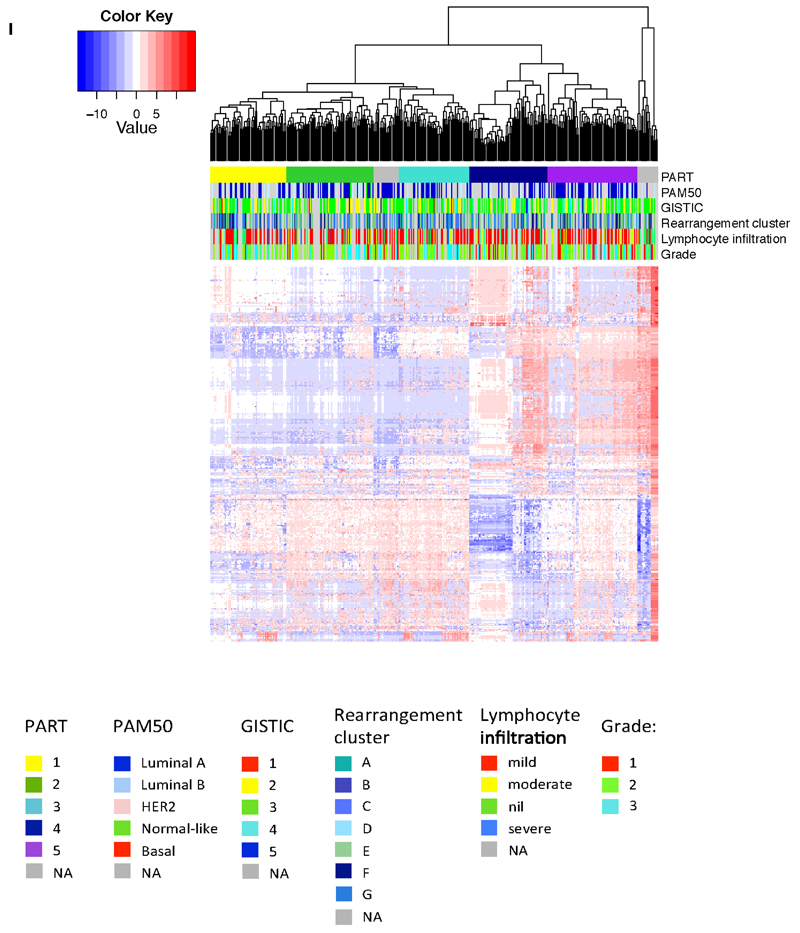
Extended Data Figure 5. miRNA analyses.
Hierarchical clustering of the most variant miRNAs using complete linkage and Euclidean distance. miRNA clusters were assigned using the Partitioning Algorithm using Recursive Thresholding (PART) method. Five main patient clusters were revealed. The horizontal annotation bars show (from top to bottom): PART cluster group, PAM50 mRNA expression subtype, GISTIC cluster, rearrangement cluster, lymphocyte infiltration score and histological grade. The heatmap shows clustered and centered miRNA expression data (log2 transformed). Details on colour coding of the annotation bars are presented below the heatmap.
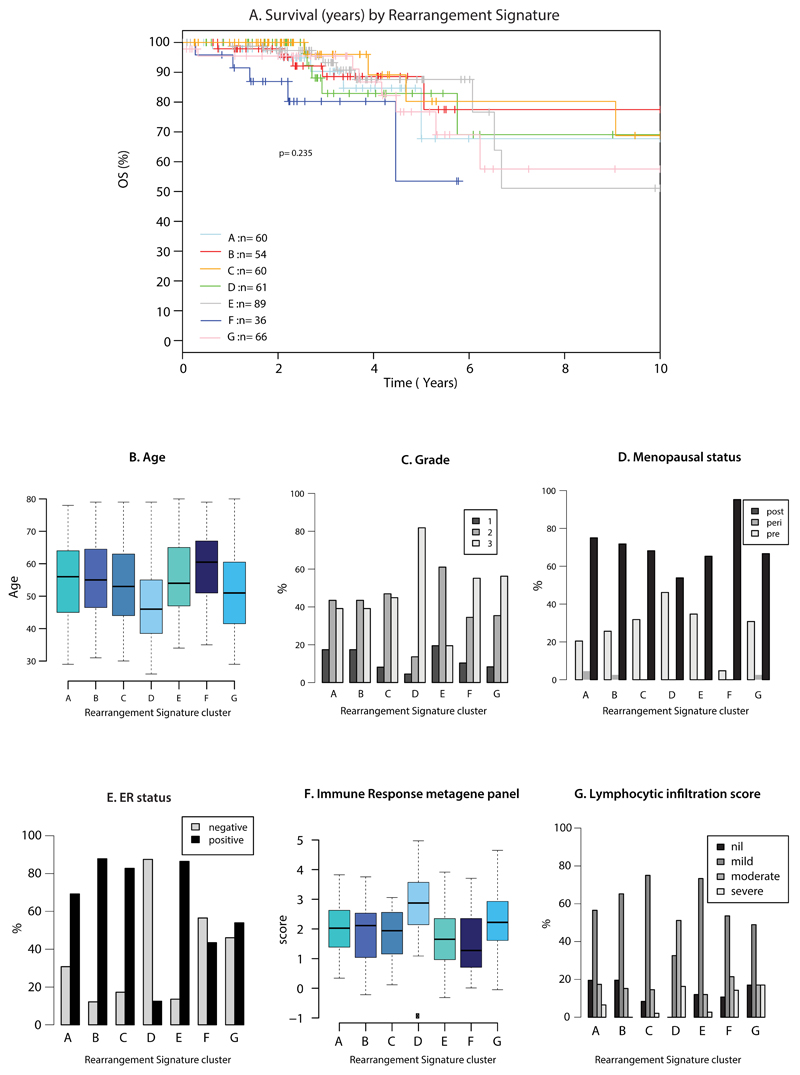
Extended Data Figure 6. Rearrangement cluster groups and associated features.
(A) Overall survival by rearrangement cluster group
(B) Age of diagnosis
(C) Tumor grade
(D) Menopausal status
(E) ER status
(F) Immune response metagene panel
(G) Lymphocytic infiltration score
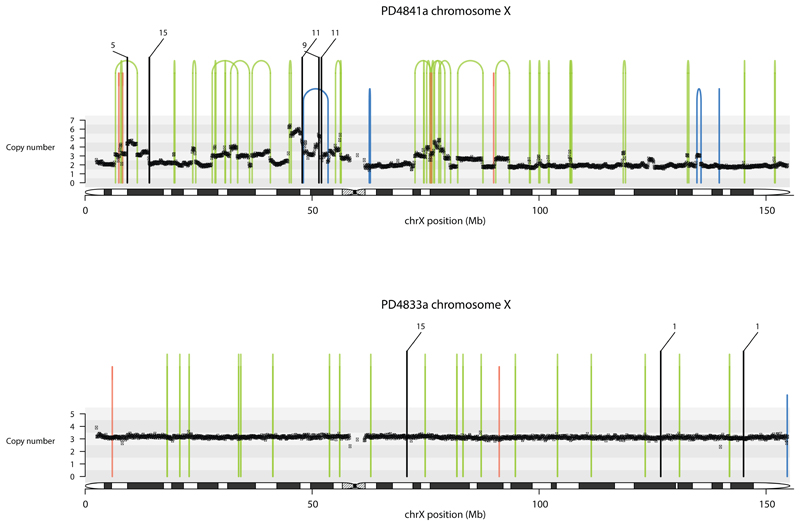
Extended Data Figure 7. Contrasting tandem duplication phenotypes.
Contrasting tandem duplication phenotypes of two breast cancers using chromosome X. Copy number (y-axis) depicted as black dots. Lines represent rearrangements breakpoints (green=tandem duplications, pink=deletions, blue=inversions, black=translocations with partner breakpoint provided). Top panel, PD4841a, is overwhelmed by large tandem duplications (>100kb, RS1) while PD4833a has many short tandem duplications (< 10kb, RS3) appearing as “single” lines in its plot.
Extended Data Figure 8. Hotspots of tandem duplications.
A tandem duplication hotspot occurring in 6 different patients
Extended Data Figure 9. Rearrangement breakpoint junctions.
(A) Breakpoint features of rearrangements in 560 breast cancers by Rearrangement Signature.
(B) Breakpoint features in BRCA and non-BRCA cancers
Extended Data Figure 10. Signatures of focal hypermutation.
(A) Kataegis and alternative kataegis occurring at the same locus (ERBB2 amplicon in PD13164a). Copy number (y-axis) depicted as black dots. Lines represent rearrangements breakpoints (green=tandem duplications, pink=deletions, blue=inversions). Topmost panel showing a ~10Mb region including the ERBB2 locus. Second panel from top zooms in 10-fold to a ~1Mb window highlighting co-occurrence of rearrangement breakpoints, with copy number changes and three different kataegis loci. Third panel from top demonstrates kataegis loci in more detail. Log10 intermutation distance on y axis. Black arrow highlighting kataegis. Blue arrows highlighting alternative kataegis.
(B) Sequence context of kataegis and alternative kataegis identified in this dataset.
Supplementary Material
Acknowledgements
This work has been funded through the ICGC Breast Cancer Working group by the Breast Cancer Somatic Genetics Study (BASIS), a European research project funded by the European Community’s Seventh Framework Programme (FP7/2010-2014) under the grant agreement number 242006; the Triple Negative project funded by the Wellcome Trust (grant reference 077012/Z/05/Z) and the HER2+ project funded by Institut National du Cancer (INCa) in France (Grants N° 226-2009, 02-2011, 41-2012, 144-2008, 06-2012). The ICGC Asian Breast Cancer Project was funded through a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A111218-SC01).
Personally funded by grants above: FGR-G, SM, KR, SM were funded by BASIS.
Recruitment was performed under the auspices of the ICGC breast cancer projects run by the UK, France and Korea.
For contributions towards instruments, specimens and collections: Tayside Tissue Bank (funded by CRUK, University of Dundee, Chief Scientist Office & Breast Cancer Campaign), Asan Bio-Resource Center of the Korea Biobank Network, Seoul, South Korea, OSBREAC consortium, The Icelandic Centre for Research (RANNIS), The Swedish Cancer Society and the Swedish Research Council, and Fondation Jean Dausset-Centre d’Etudes du polymorphisme humain. Icelandic Cancer Registry, The Brisbane Breast Bank (The University of Queensland, The Royal Brisbane & Women’s Hospital and QIMR Berghofer), Breast Cancer Tissue and Data Bank at KCL and NIHR Biomedical Research Centre at Guy’s and St Thomas’s Hospitals. Breakthrough Breast Cancer and Cancer Research UK Experimental Cancer Medicine Centre at KCL.
For pathology review: The Mouse Genome Project and Department of Pathology, Cambridge University Hospitals NHS Foundation Trust for microscopes. Andrea Richardson, Anna Ehinger, Anne Vincent-Salomon, Carolien Van Deurzen, Colin Purdie, Denis Larsimont, Dilip Giri, Dorthe Grabau, Elena Provenzano, Gaetan MacGrogan, Gert Van den Eynden, Isabelle Treilleux, Jane E Brock, Jocelyne Jacquemier, Jorge Reis-Filho, Laurent Arnould, Louise Jones, Marc van de Vijver, Øystein Garred, Roberto Salgado, Sarah Pinder, Sunil R Lakhani, Torill Sauer, Violetta Barbashina.
Illumina UK Ltd for input on optimisation of sequencing throughout this project.
Wellcome Trust Sanger Institute Sequencing Core Facility, Core IT Facility and Cancer Genome Project Core IT team and Cancer Genome Project Core Laboratory team for general support.
Personal funding: SN-Z is a Wellcome Beit Fellow and personally funded by a Wellcome Trust Intermediate Fellowship (WT100183MA). LBA is supported through a J. Robert Oppenheimer Fellowship at Los Alamos National Laboratory. ALR is partially supported by the Dana-Farber/Harvard Cancer Center SPORE in Breast Cancer (NIH/NCI 5 P50 CA168504-02). DG was supported by the EU-FP7-SUPPRESSTEM project. AS was supported by Cancer Genomics Netherlands (CGC.nl) through a grant from the Netherlands Organisation of Scientific research (NWO). MS was supported by the EU-FP7-DDR response project. CS and CD are supported by a grant from the Breast Cancer Research Foundation. EB was funded by EMBL. CS is funded by FNRS (Fonds National de la Recherche Scientifique). SJJ is supported by Leading Foreign Research Institute Recruitment Program through the National Research Foundation of Republic Korea (NRF 2011-0030105). GK is supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (NRF 2015R1A2A1A10052578). John Foekens received funding from an ERC Advanced grant (No 322737).
For general contribution and administrative support: Fondation Synergie Lyon Cancer in France. Jon G Jonasson, Department of Pathology, University Hospital & Faculty of Medicine, University of Iceland. Kaltin Ferguson, Tissue Bank Manager, Brisbane Breast Bank and The Breast Unit, The Royal Brisbane and Women’s Hospital, Brisbane, Australia. The Oslo Breast Cancer Consortium of Norway (OSBREAC). Angelo Paradiso, IRCCS Istituto Tumori “Giovanni Paolo II”, Bari Italy. Antita Vines for administratively supporting to identifying the samples, organizing the bank, and sending out the samples. Margrete Schlooz-Vries, Jolien Tol, Hanneke van Laarhoven, Fred Sweep, Peter Bult in Nijmegen for contributions in Nijmegen. This research used resources provided by the Los Alamos National Laboratory Institutional Computing Program, which is supported by the U.S. Department of Energy National Nuclear Security Administration under Contract No. DE-AC52-06NA25396. Research performed at Los Alamos National Laboratory was carried out under the auspices of the National Nuclear Security Administration of the United States Department of Energy. Nancy Miller (in memoriam) for her contribution in setting up the clinical database.
Finally, we would like to acknowledge all members of the ICGC Breast Cancer Working Group and ICGC Asian Breast Cancer Project.
Footnotes
Accession Numbers
Raw data have been submitted to the European-Genome Phenome Archive under the overarching accession number EGAS00001001178 (please see Supplementary Notes for breakdown by data type).
Author Contributions
S.N-Z, M.R.S designed the study, analysed data and wrote the manuscript.
H.R.M., J.S, M.Ramakrishna, D.G, X.Z. performed curation of data and contributed towards genomic and copy number analyses.
M.S., A.B.B., M.R.A., O.C.L., A.L., M.Ringner, contributed towards curation and analysis of non-genomic data (transcriptomic, miRNA, methylation).
I.M., L.B.A., D.C.W., P.V.L., S.Morganella, Y.S.J., contributed towards specialist analyses.
G.T., G.K., A.L.R., A-L.B-D., J.W.M.M., M.J.v.d.V., H.G.S., E.B., A.Borg., A.V., P.A.F., P.J.C., designed the study, drove the consortium and provided samples.
S.Martin was project coordinator.
S.McL., S.O.M., K.R., contributed operationally.
S-M.A., S.B., J.E.B., A.Brooks., C.D., L.D., A.F., J.A.F., G.K.J.H., S.J.J., H.-Y.K., T.A.K., S.K., H.J.L., J.-Y.L., I.P., X.P., C.P., F.G.R.-G., G.R., A.M.S., P.T.S., O.A.S., S.T., I.T., G.G.V.d.E., P.V., A.V.-S., L.Y., C.C., L.v.V., A.T., S.K., B.K.T.T., J.J., N.t.U., C.S., P.N.S., S.V.L., S.R.L., J.E.E., A.M.T contributed pathology assessment and/or samples.
A.Butler., S.D., M.G., D.R.J., Y.L., A.M., V.M., K.R., R.S., L.S., J.T. contributed IT processing and management expertise.
All authors discussed the results and commented on the manuscript.
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicks J, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome research. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergamaschi A, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. The Journal of pathology. 2008;214:357–367. doi: 10.1002/path.2278. [DOI] [PubMed] [Google Scholar]
- 6.Ching HC, Naidu R, Seong MK, Har YC, Taib NA. Integrated analysis of copy number and loss of heterozygosity in primary breast carcinomas using high-density SNP array. International journal of oncology. 2011;39:621–633. doi: 10.3892/ijo.2011.1081. [DOI] [PubMed] [Google Scholar]
- 7.Fang M, et al. Genomic differences between estrogen receptor (ER)-positive and ER-negative human breast carcinoma identified by single nucleotide polymorphism array comparative genome hybridization analysis. Cancer. 2011;117:2024–2034. doi: 10.1002/cncr.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu YM, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer discovery. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giacomini CP, et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS genetics. 2013;9:e1003464. doi: 10.1371/journal.pgen.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson DR, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nature medicine. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson J, et al. Activation of human telomerase reverse transcriptase through gene fusion in clear cell sarcoma of the kidney. Cancer letters. 2015;357:498–501. doi: 10.1016/j.canlet.2014.11.057. [DOI] [PubMed] [Google Scholar]
- 20.Khurana E, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Molecular cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinagre J, et al. Frequency of TERT promoter mutations in human cancers. Nature communications. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 24.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell reports. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natrajan R, et al. Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast. The Journal of pathology. 2014;232:553–565. doi: 10.1002/path.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalyana-Sundaram S, et al. Gene fusions associated with recurrent amplicons represent a class of passenger aberrations in breast cancer. Neoplasia. 2012;14:702–708. doi: 10.1593/neo.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tubio JM. Somatic structural variation and cancer. Briefings in functional genomics. 2015 doi: 10.1093/bfgp/elv016. [DOI] [PubMed] [Google Scholar]
- 30.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nature genetics. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ussery DW, Binnewies TT, Gouveia-Oliveira R, Jarmer H, Hallin PF. Genome update: DNA repeats in bacterial genomes. Microbiol-Sgm. 2004;150:3519–3521. doi: 10.1099/Mic.0.27628-0. [DOI] [PubMed] [Google Scholar]
- 32.Lu S, et al. Short Inverted Repeats Are Hotspots for Genetic Instability: Relevance to Cancer Genomes. Cell reports. 2015 doi: 10.1016/j.celrep.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wojcik EA, et al. Direct and inverted repeats elicit genetic instability by both exploiting and eluding DNA double-strand break repair systems in mycobacteria. PloS one. 2012;7:e51064. doi: 10.1371/journal.pone.0051064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson CE, Zorbas H, Price GB, Zannis-Hadjopoulos M. Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication. Journal of cellular biochemistry. 1996;63:1–22. doi: 10.1002/(SICI)1097-4644(199610)63:1<1::AID-JCB1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mammalian genome : official journal of the International Mammalian Genome Society. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 37.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nature reviews Genetics. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birkbak NJ, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer discovery. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abkevich V, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. British journal of cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popova T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer research. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 41.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morganella SALB. The topography of mutational processes in breast cancer. 2015 doi: 10.1038/ncomms11383. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. The New England journal of medicine. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 44.Forster MD, et al. Treatment with olaparib in a patient with PTEN-deficient endometrioid endometrial cancer. Nature reviews Clinical oncology. 2011;8:302–306. doi: 10.1038/nrclinonc.2011.42. [DOI] [PubMed] [Google Scholar]
- 45.Turner N, Tutt A, Ashworth A. Targeting the DNA repair defect of BRCA tumours. Current opinion in pharmacology. 2005;5:388–393. doi: 10.1016/j.coph.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nature methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome research. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics. 2006;173:2187–2198. doi: 10.1534/genetics.105.044677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun L, Craiu RV, Paterson AD, Bull SB. Stratified false discovery control for large-scale hypothesis testing with application to genome-wide association studies. Genetic epidemiology. 2006;30:519–530. doi: 10.1002/gepi.20164. [DOI] [PubMed] [Google Scholar]
- 55.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. btq170 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC bioinformatics. 2013;14:244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.