ABSTRACT
This study was aimed to detect the correlation among EGFR/KRAS status and PD-1/PD-L1 expression in non-small-cell lung cancer (NSCLC) patients. PD-1 and PD-L1 expressions were detected by immunohistochemistry in 100 surgically resected lung adenocarcinoma tissues and were statistically correlated with clinicopathological characteristics including EGFR and KRAS statuses. Besides, the overall survival (OS) times were analyzed. There was a statistical significances between PD-1 expression in tumor and KRAS status (P = 0.043), with a higher mutation rate in with lower PD-1 expression patients. There was a statistical significance between PD-L1 expression in tumor and EGFR status (P = 0.012), with a higher mutation rate in patients with lower PD-L1 expression. The OS of patients with EGFR mutation was significantly longer than those without EGFR mutation. The OS of patients with lower PD-L1 in tumor was significantly longer than those with higher PD-L1 expression. We found negative associations between PD-L1 expression in tumor and mutated EGFR status, as well as between PD-1 expression in tumor and mutated KRAS status.
KEYWORDS: EGFR, KRAS, non-small-cell lung cancer, PD-1, PD-L1
Introduction
Globally, lung cancer is the most common malignancy and also the leading cause of cancer-related deaths.1 Particularly in China, both of the incidence and mortality rates of lung cancer occupy the first place.2 Of all the lung cancer cases, non-small-cell lung cancer (NSCLC) accounts for 80% – 85%.1,3
Although much progresses have been achieved in the optimization of NSCLC treatment, platinum-based chemotherapy can only provide a response rate of 20% – 35% and a median overall survival of 8 – 12 months.4,5 Even with the application of mutated genes targeted drugs, the the 5-year OS rate remains only 15% of all stages.1 Thus, it is urgent to develop effective treatment strategies for NSCLC.
NSCLC is a disease characterized by driver mutation-defined molecular subsets. Two most important driver genes are EGFR and KRAS.6 Approximately 10% of pts with NSCLC and 35% in East Asia have tumor associated EGFR mutations.7-9 About 50–65% of patients with EGFR mutations respond to EGFR-TKIs. And KRAS mutation is usually mutually exclusive with EGFR mutation though the coexistence of EGFR and KRAS mutations has been rarely observed.10,11 In murine melanoma models, activation of EGFR and KRAS pathways might be involved in immune response suppression either through activating regulatory T cells 12 or reducing the levels of the T cell chemoattractant.13 A connection between driver gene and T cell regulator can be established.
Recently, a crucial breakthrough in cancer immunotherapy is the discovery of so called “immune checkpoints” including PD-1 and PD-L1.14 In NSCLC, PD-1 overexpression on CD8+T cells suggests a reduced production of cytokines and T cell proliferation.15 Aberrant expression of PD-L1 has been found ranging from 19% to 100% of NSCLC patients 16-19 and associated with poor prognosis.17,18 Blockade of PD-1/PD-L1 interaction allows the tumor-specific T cells to exact their effector reactivity on tumor cells of NSCLC and have shown their efficacy and feasibility in NSCLC immunotherapy.20-24 However, an objective response rate of only 10%–20% of NSCLC patients has been observed.24,25
It is unclear whether specific genomic subsets of NSCLC utilize the PD-1/PD-L1 pathway to achieve immune evasion. The correlations among EGFR/KRAS and PD-1/PD-L1 have been reported in several articles but the results vary 26-30 (Table 1). But high mutational rates seem to contribute to enhanced immunogenicity,27 indicating the sensitivity to immune checkpoint blockage.28
Table 1.
Correlation between PD-1/PD-L1 expression and EGFR/KRAS status.
| Driver gene |
||||
|---|---|---|---|---|
| Immune checkpoint | EGFR | KRAS | Author | Reference |
| PD-1 | / | + | D'Incecco et al. | 26 |
| PD-L1 | + | / | ||
| PD-L1 | / | / | Cooper et al. | 30 |
| PD-L1 | + | / | Lin et al. | 41 |
| PD-L1 | + | / | Tang et al. | 42 |
| PD-L1 | + | / | Azuma et al. | 43 |
| PD-L1 | / | / | Zhang et al. | 44 |
| PD-1 | + | / | Akbay et al. | 45 |
| PD-L1 | + | / | ||
| PD-L1 | / | / | Yang et al. | 46 |
+: PD-1/PD-L1 expression is correlated with the presence of EGFR/KRAS mutation; /: PD-1/PD-L1 expression is not correlated with the presence of EGFR/KRAS mutation
Based on these premises we propose a hypothesis that there could be a possibility of correlation between EGFR/KRAS status and PD-1/PD-L1 expression, which is still unconfirmed. In the current study, we detected the EGFR/KRAS status and PD-1/PD-L1 expression in NSCLC patients and analyzed the correlation among them and the clinicopathological features and survival outcome.
Results
Patient characteristics
A total of 100 patients with primary lung adenocarcinoma were enrolled and samples from each patient were obtained. Among them, 51 patients were male and 26 were smokers. There were 60 (60 %) EGFR mutated and 10 (10%) KRAS mutated cases. Among cases with EGFR mutation, there were 19 cases with exon 21 mutation, 5 with exon 21/20 mutation, 3 with exon 21/19 mutation, 2 with exon 21/20/19 mutation, 2 with exon 21/18 mutation, 27 with exon 19 mutation, and 2 with exon 20/18 mutation. Among cases with KRAS mutation, there were 4 cases with codon 1 mutation, 1 with codon 4 mutation, 3 with codon 5 mutation, 1 with codon 6 mutation, and 2 with codon 7 mutation. The positive rates of PD-1 in tumor, PD-L1 in tumor, PD-L1 in TILs were 53%, 40% and 23%, respectively. See Table 2.
Table 2.
Clinicalpathological characteristic in the whole population.
| Characteristic | Low PD-1 in tumor | High PD-1 in tumor | P | Low PD-L1 in tumor | High PD-L1 in tumor | P | Low PD-L1 in TILs | High PD-L1 in TILs | P |
|---|---|---|---|---|---|---|---|---|---|
| Age | 0.732 | 0.682 | 0.431 | ||||||
| < 60 | 22 | 23 | 28 | 17 | 33 | 12 | |||
| ≥ 60 | 25 | 30 | 32 | 23 | 44 | 11 | |||
| Sex | 0.990 | 0.060 | 0.729 | ||||||
| Male | 24 | 27 | 26 | 25 | 40 | 11 | |||
| Female | 23 | 26 | 34 | 15 | 37 | 12 | |||
| Smoking | 0.416 | 0.852 | 0.283 | ||||||
| No | 33 | 41 | 44 | 30 | 55 | 19 | |||
| Yes | 14 | 12 | 16 | 10 | 22 | 4 | |||
| EGFR mutation | 0.252 | 0.012* | 0.174 | ||||||
| No | 16 | 24 | 18 | 22 | 28 | 12 | |||
| Yes | 31 | 29 | 42 | 18 | 49 | 11 | |||
| KRAS mutation | 0.043* | 0.515 | 0.446 | ||||||
| No | 39 | 51 | 55 | 35 | 68 | 22 | |||
| Yes | 8 | 2 | 5 | 5 | 9 | 1 | |||
| PD-1 expression | – | 0.744 | 0.181 | ||||||
| Low | – | – | 29 | 18 | 39 | 8 | |||
| High | – | – | 31 | 22 | 38 | 15 | |||
| PD-L1 in tumor | 0.744 | – | 0.065 | ||||||
| Low | 29 | 31 | – | – | 50 | 10 | |||
| High | 18 | 22 | – | – | 27 | 13 | |||
| PD-L1 in TILs | 0.181 | 0.065 | – | ||||||
| Low | 39 | 38 | 50 | 27 | – | – | |||
| High | 8 | 15 | 10 | 13 | – | – | |||
| T | 0.681 | 0.169 | 0.332 | ||||||
| T1-2 | 45 | 49 | 58 | 36 | 71 | 23 | |||
| T3-4 | 2 | 4 | 2 | 4 | 6 | 0 | |||
| N | 0.715 | 0.252 | 0.928 | ||||||
| N0 | 23 | 24 | 31 | 16 | 36 | 11 | |||
| N+ | 24 | 29 | 29 | 24 | 41 | 12 | |||
| AJCC stage | 0.437 | 0.943 | 0.785 | ||||||
| I | 20 | 22 | 26 | 16 | 31 | 11 | |||
| II | 15 | 12 | 16 | 11 | 21 | 6 | |||
| III | 12 | 19 | 28 | 13 | 25 | 6 |
Abbreviations: EGFR = epidermal growth factor receptor; KRAS = Kirsten rat sarcoma viral oncogene homolog; TIL = tumor-infiltrating lymphocyte
P < 0.05.
Correlation between PD-1/PD-L1 expression and clinicopathologic features
PD-1 expression was successfully evaluated in all the 100 samples. The expression of PD-L1 and PD-L2 mainly located in the cell membrane and cytoplasm of tumor cells. Scattered expression of PD-L1 (weak to moderate) and PD-L2 (weak) was also observed in macrophages (Fig. 1).
Figure 1.
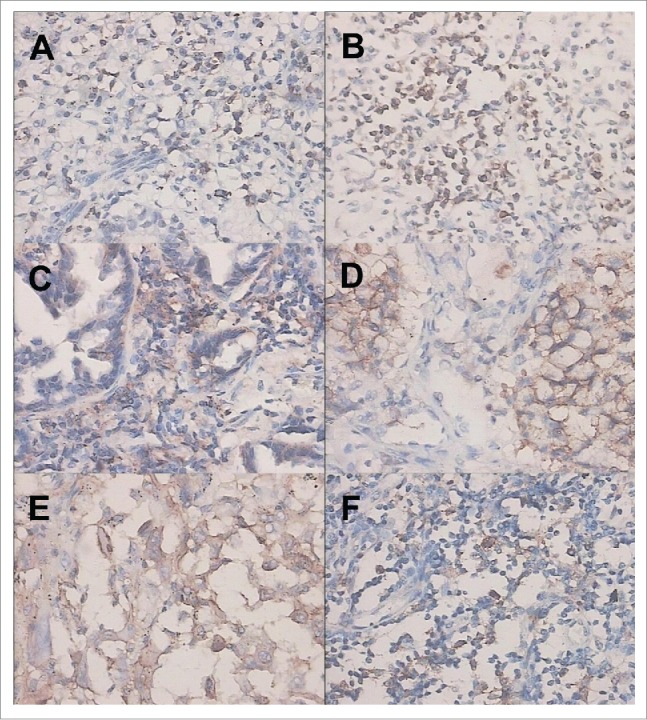
Immunohistochemical staining of PD-1 and PD-L1. (A) Weak positive expression of PD-1 in tumor-infiltrating lymphocytes (TILs). (B) Strong positive expression of PD-1 in TILs. (C) Weak positive expression of PD-L1 in tumor. (D) Strong positive expression of PD-L1 in tumor. (E) Weak positive expression of PD-L1 in TILs. (F). Strong positive expression of PD-L1 in TILs.
Median PD-1 expression was higher in male, or in never/former smokers, or in patients with wild type EGFR status, or in patients with wild type KRAS status. There was a statistical significance between PD-1 expression and KRAS status (P = 0.043), patients with lower PD-1 expression having a higher KRAS mutation rate. Median PD-L1 expression on tumor was higher in male, or in never/former smokers, or in patients harboring EGFR mutation, or in patients with wild type KRAS status. Median PD-L1 expression on TILs was higher in male, or in smokers, or in patients wild type EGFR status, or in patients with wild type KRAS status. There was a statistical significances between PD-L1 expression in tumor and EGFR status (P = 0.012), patients with lower PD-L1 expression had a higher EGFR mutation rate (Table 2).
Survival
EGFR status (P = 0.030), PD-L1 expression in tumor (P = 0.026), T stage (P = 0.021) and N stage (P = 0.005) were correlated with survival outcome, with a risk ratio of 0.043 (mutated/wild), 2.205 (lower/higher PD-L1 expression), 3.237 (T3+4/ T1+2) and 1.865 (N3+4/ N1+2), respectively. See Table 3.
Table 3.
Cox multivariate analysis for survival.
| Log-rank analysis |
Cox multivariate analysis |
||||
|---|---|---|---|---|---|
| χ2 | ρ | RR | 95% CI | ρ | |
| Sex | 2.217 | 0.136 | |||
| Age | 0.569 | 0.451 | |||
| Smoking | 0.26 | 0.610 | |||
| EGFR | 4.560 | 0.033 | 0.043 | 0.201–0.923 | 0.030 |
| KRAS | 0.640 | 0.424 | |||
| PD-1 expression | 0.429 | 0.512 | |||
| PD-L1 expression in TILs | 0.061 | 0.805 | |||
| PD-L1 expression in tumor | 4.512 | 0.032 | 2.205 | 1.099–4.423 | 0.026 |
| T | 12.041 | < 0.001 | 3.237 | 1.190–8.804 | 0.021 |
| N | 5.574 | 0.018 | 1.865 | 1.209–2.875 | 0.005 |
RR: relative risk.
The OS of patients with EGFR mutation was significantly longer than that of those without EGFR mutation. The OS of patients with lower PD-L1 in tumor was significantly longer than that of those with higher PD-L1. The OS of patients with tumor size of T1/2 was significantly longer than that of those with tumor size of T3/4. The OS of patients without lymph node metastasis was significantly longer than that of those with lymph node metastasis. See Fig. 2.
Figure 2.
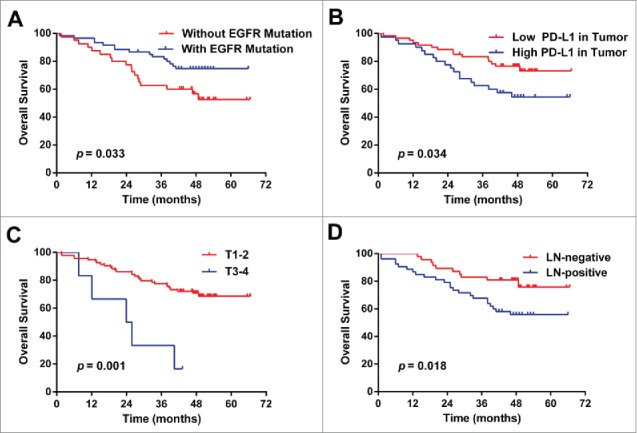
Overall survival curves. (A) The OS of patients without EGFR mutation is significantly longer than that with EGFR mutation. (B) The OS of patients with low PD-L1 in tumor is significantly longer than that with high PD-L1. (C) The OS of patients with tumor size of T1/2 is significantly longer than that with tumor size of T3/4. (D) The OS of patients without lymph node metastasis is significantly longer than that with lymph node metastasis. Fig 1.
Patients with mutated EGFR status, lower PD-L1 expression in tumor, T1/2, N0 or stage I/II had a significant increase in 3-year survival rate than those with wild type EGFR status (P = 0.033), higher PD-L1 expression in tumor (P = 0.034), T3/4 (P = 0.001), N1-3 (P = 0.018) or stage III/IV (P = 0.027), respectively. See Table 4.
Table 4.
Survival rates.
| Characteristic | 3-year survival rate (%) | P |
|---|---|---|
| Age | 0.451 | |
| < 60 | 75.6 | |
| ≥ 60 | 74.5 | |
| Sex | 0.136 | |
| Male | 66.7 | |
| Female | 83.6 | |
| Smoking | 0.610 | |
| without | 75.7 | |
| with | 73.1 | |
| EGFR | 0.033* | |
| Without mutation | 62.5 | |
| With mutation | 83.3 | |
| KRAS | 0.424 | |
| Without mutation | 74.4 | |
| With mutation | 80.0 | |
| PD-1 expression | 0.512 | |
| Low | 72.2 | |
| High | 77.4 | |
| PD-L1 in TILs | 0.805 | |
| Low | 75.2 | |
| High | 73.9 | |
| PD-L1 in tumor | 0.034* | |
| Low | 83.3 | |
| High | 62.5 | |
| T | 0.001* | |
| T1-2 | 77.6 | |
| T3-4 | 33.3 | |
| N | 0.018* | |
| N0 | 83 | |
| N+ | 67.8 | |
| AJCC stage | 0.027* | |
| I-II | 79.6 | |
| III | 64.5 |
P < 0.05.
Discussion
In the present study, we found that expression of PD-1 and PD-L1 vary according to the patient characteristics. Patients without EGFR mutation or without KRAS mutation had higher level of PD-1 expression comparing with those harboring EGFR mutation or KRAS mutation. Patients harboring EGFR mutation or without KRAS mutation had higher level of PD-L1 expression on tumor comparing with those without EGFR mutation or harboring KRAS mutation. Patients without EGFR mutation or without KRAS mutation had higher level of PD-L1 expression on TILs comparing with those harboring EGFR mutation or KRAS mutation. Moreover, the OS of patients with EGFR mutation was significantly longer than that without EGFR mutation. The OS of patients with lower PD-L1 in tumor was significantly longer than that with higher PD-L1. Importantly, we found negative associations between PD-L1 expression in tumor and mutated EGFR status, as well as between PD-1 expression in tumor and mutated KRAS status.
The frequency of EGFR mutation has been studied most extensively in East Asian populations, where it varies from 36.4 to 66.3 % in lung adenocarcinoma.31,32 For KRAS, the mutation frequency ranges from 2.3 to 9.4 % in East Asian.31,33
EGFR is overexpressed by 40 to 80% of NSCLC, and the expression levels are correlated with the EGFR tyrosine kinase domain mutations.34 KRAS mutation is present in approximately 30% of lung adenocarcinomas and uncommon in squamous carcinomas (< 5%).35 In our study, the mutation rates of the 2 target genes were in line with these reported data.
Cox multivariate analysis reveals that 4 factors associate with survival outcome of NSCLC patients. Patients with mutated EGFR status have lower relative risk of NSCLC-related deaths, possibly because of these patients have the opportunity of receiving TKIs. Contrary to other findings, we did not find smoking status associated with high PD-1/PD-L1 possibly due to the relatively small sample size because only a small portion of patients in our study were smokers.26 And due to the immune inhibitory regulation on T cells, patients with higher PD-L1 expression have higher relative risk. As for T and N stage, it is a consensus that later stages predict poorer outcome. Moreover, these observations are confirmed by the comparison of 3-year survival rates. Notably, more than half of the enrolled patients were still alive after 3 y so the median survival time can not be reached. The follow-up is still going on and data are being collected.
There have been a few other studies detecting EGFR/KRAS status and PD-1/PD-L1 expression in the same NSCLC samples and the results from different groups vary. The variety of the results of other researches and ours are possibly because of 1) the limit of sample size; 2) the heterogeneity the subjects, which means certain results can only be obtained in specific populations (as shown in Table 1); 3) the non-major roles of PD-1/PD-L1 and EGFR/KRAS on each other's pathways, which means there are multiple other mechanisms of immunosuppression in the complicated network of human immunity; and 4) the biologic difference between the mouse models and human patients.
However, our Kras story is not strong enough since it was based on only 8 samples. So the data on Kras is suggestive but not conclusive. Additional prospective studies with larger samples are warranted.
Conclusively, we found that the negative wild type EGFR or KRAS status cannot be satisfactory biomarkers for assessing the effects of blockage of PD-1/PD-L1 pathway based on the results of existing studies. Though a few mentioned studies come to different opinions from ours, the association between mutated driver genes and immune checkpoints is a definite interest of future investigations.
Material and methods
Patients
This retrospective study was conducted in a cohort of 100 primary NSCLC patients who had received lung tumor resection between April 2009 and December 2011 and been followed in The Third Affiliated Hospital of Soochow University. Eligible cases were required to have sufficient tissue for immunohistochemical staining and mutational analyses. Patients who received neoajuvant chemotherapy or had a history of malignant tumors before the enrollment were excluded. The study had been approved by The Ethics Committee of The Third Affiliated Hospital of Soochow University, and all patients provided written informed consent before the enrollment.
Clinicopathological variables collected for analyses included sex, age at diagnosis, smoking history, tumor histology, tumor differentiation, pathologic TNM stage according to the seventh edition of the lung cancer staging system 36 after operation, adenocarcinoma subtypes confirmed by 2 pathologists according to the new International Association for the Study of Lung Cancer/American Thoracic Society/ European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma.33 Data of survival outcome were observed in the follow-up.
Immunohistochemical staining of PD-1 and PD-L1
Formalin-fixed, paraffin embedded 5-micron sections of 100 primary NSCLC samples were used throughout this study. Reagents for immunohistochemical analyses were purchased from Abcam®, Inc., Cambridge, MA, USA (CAT No. ab137132 for anti-PD-1 antibody and ab174838 for anti-PD-L1 antibody). For PD-1 and PD-L1 immunostaining with mouse polyclonal antibodies, tissue sections were deparaffinized in xylene and rehydrated in an ethanol series. The sections were then treated for 30 min with 0.3% hydrogen peroxide to block endogenous peroxidase activity, then subsequently washed with phosphate-buffered saline (PBS) and unmasked in citrate antigen unmasking solution in an autoclave for 20 min at 120°C for antigen recovery. The sections were incubated with goat serum for 15 min at room temperature (RT) and then were incubated with the primary antibodies [polyclonal antibody to PD-1 (1/4000); polyclonal antibody to PD-L1 (1/4000)] for 1 h at RT. The bound primary antibodies were detected by adding anti-goat secondary antibodies (1/2000) and avidin/biotin/horseradish peroxidase complex for 30 min at RT. The sections were visualized using solid diaminobenzine diluted in PBS, counterstained with Mayer's hematoxylin, and finally mounted. After that, they were incubated with HRP-labeled anti-mouse immunoglobulin G as the secondary antibody. Substrate chromogen was added and the specimens were counterstained with hematoxylin.
Two independent well-experienced pathologists (LQ and TY) assessed PD-1 and PD-L1 positivity semiquantitatively without prior information on the clinicopathological features of the samples and follow-up data.
Percentages of PD-L1 and PD-1 positive tumor cells and staining intensity were evaluated for each sample. The staining intensity was scored as 0 (negative or trace), 1 (weak), 2 (moderate) and 3 (high) (Fig. 1). In absence of standardized scoring system, cases with staining intensity ≥ 2 in more than 5% of tumor cells were considered as positive as described in previous studies.37-40 A semiquantitative approach was used to generate a total score for each tissue core. The percentage of stained cells (0 – 100%) was multiplied by the staining score of dominant intensity pattern. Therefore, the total score ranged from 0 to 300.
Mutation analysis of EGFR and KRAS
All patients were analyzed for presence of EGFR and KRAS mutations. EGFR mutations and KRAS mutations were evaluated using polymerase chain reaction and direct sequencing. Ribonucleic acid was extracted as per standard protocol after frozen tissues were dissected into TRIzol® (Life Technologies, Carlsbad, CA, USA), and was reverse transcribed into cDNA (cDNA). EGFR (exons 18–22) and KRAS (exons 2–3) were amplified using cDNA. Amplified products were analyzed by direct dideoxynucleotide sequencing.
Statistical analysis
A sample size of at least 49 patient's was required for each group with the assumption of one-sided 10% α and 80% power. The sample size was determined according to the formula as follows: n1 = n2 = 2[(μα + μβ)/(δ/σ)]2+ (μα)2/4
Here, 2 side α = 0.05, β = 0.1, δ/σ = 0.8, μα=1.96, μβ= 1.282. Finally, n1 = n2 = 33.8. Statistical analyses were performed to compare differences between patients with and without PD-1 and PD-L1 expression. Comparison between the 2 groups with or without PD-1 or PD-L1 expression was performed by log rank test. A multivariable analysis was performed using a logistic regression model to explore the association of PD-1/PD-L1 expression with patient characteristics (sex, smoke, histology, EGFR and KRAS). Correlations between clinicopathological variables and EGFR/KRAS or PD-1/PD-L1 were compared by the rank sum tests. Differences between median score were performed by U-Mann–Witney test. OS was defined as the time from the date therapy started to the date of death from any cause or the date of the last follow-up., with 95% confidence intervals calculated using the Kaplan–Meier method, and comparisons between groups were performed by the log rank test. P values < 0.05 were considered statistical significant. All statistical analyses were performed using SPSS version 13.0 software (SPSS, Inc., USA).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63:11-30; PMID:23335087; http://dx.doi.org/ 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 2.Stewart BW, Wild CP. World Cancer Report 2014. Lyon: International Agency for Research on Cancer, 2014. [Google Scholar]
- 3.Xu W, Yang G, Xu Y, Zhang Q, Fu Q, Yu J, Yu M, Zhao W, Yang Z, Hu F, et al.. The possibility of traditional chinese medicine as maintenance therapy for advanced nonsmall cell lung cancer. Evid Based Complement Alternat Med 2014; 2014:278917; PMID:25165478; http://dx.doi.org/ 10.1155/2014/278917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi K, Hagiwara K. Epidermal growth factor receptor (EGFR) mutation and personalized therapy in advanced nonsmall cell lung cancer (NSCLC). Target Oncol 2013; 8:27-33; PMID:23361373; http://dx.doi.org/ 10.1007/s11523-013-0258-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azzoli CG, Temin S, Giaccone G. 2011 Focused Update of 2009 American Society of Clinical Oncology Clinical Practice Guideline Update on Chemotherapy for Stage IV Non-Small-Cell Lung Cancer. J Oncol Pract 2012; 8:63-6; PMID:22548014; http://dx.doi.org/ 10.1200/JOP.2011.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer 2001; 37 Suppl 4:S9-15; PMID:11597399; http://dx.doi.org/ 10.1016/S0959-8049(01)00231-3 [DOI] [PubMed] [Google Scholar]
- 7.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al.. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350:2129-39; PMID:15118073; http://dx.doi.org/ 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 8.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al.. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304:1497-500; PMID:15118125; http://dx.doi.org/ 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 9.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al.. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004; 101:13306-11; PMID:15329413; http://dx.doi.org/ 10.1073/pnas.0405220101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Jänne PA, Januario T, Johnson DH, et al.. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005; 23:5900-9; PMID:16043828; http://dx.doi.org/ 10.1200/JCO.2005.02.857 [DOI] [PubMed] [Google Scholar]
- 11.Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res 2009; 15:4554-60; PMID:19584155; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-0089 [DOI] [PubMed] [Google Scholar]
- 12.Zaiss DM, van Loosdregt J, Gorlani A, Bekker CP, Grone A, Sibilia M, van Bergen en Henegouwen PM, Roovers RC, Coffer PJ, Sijts AJ. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity 2013; 38:275-84; PMID:23333074; http://dx.doi.org/ 10.1016/j.immuni.2012.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pivarcsi A, Muller A, Hippe A, Rieker J, van Lierop A, Steinhoff M, Seeliger S, Kubitza R, Pippirs U, Meller S, et al.. Tumor immune escape by the loss of homeostatic chemokine expression. Proc Natl Acad Sci U S A 2007; 104:19055-60; PMID:18025475; http://dx.doi.org/ 10.1073/pnas.0705673104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Intlekofer AM, Thompson CB. At the bench: preclinical rationale for CTLA-4 and PD-1 blockade as cancer immunotherapy. J Leukoc Biol 2013; 94:25-39; PMID:23625198; http://dx.doi.org/ 10.1189/jlb.1212621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Huang S, Gong D, Qin Y, Shen Q. Programmed death-1 upregulation is correlated with dysfunction of tumor-infiltrating CD8+ T lymphocytes in human non-small cell lung cancer. Cell Mol Immunol 2010; 7:389-95; PMID:20514052; http://dx.doi.org/ 10.1038/cmi.2010.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirahara K, Ghoreschi K, Yang XP, Takahashi H, Laurence A, Vahedi G, Sciumè G, Hall AO, Dupont CD, Francisco LM, et al.. Interleukin-27 priming of T cells controls IL-17 production in trans via induction of the ligand PD-L1. Immunity 2012; 36:1017-30; PMID:22726954; http://dx.doi.org/ 10.1016/j.immuni.2012.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konishi J, Yamazaki K, Azuma M, Kinoshita I, Dosaka-Akita H, Nishimura M. B7-H1 expression on non-small cell lung cancer cells and its relationship with tumor-infiltrating lymphocytes and their PD-1 expression. Clin Cancer Res 2004; 10:5094-100; PMID:15297412 [DOI] [PubMed] [Google Scholar]
- 18.Mu CY, Huang JA, Chen Y, Chen C, Zhang XG. High expression of PD-L1 in lung cancer may contribute to poor prognosis and tumor cells immune escape through suppressing tumor infiltrating dendritic cells maturation. Med Oncol 2011; 28:682-8; PMID:20373055; http://dx.doi.org/ 10.1007/s12032-010-9515-2 [DOI] [PubMed] [Google Scholar]
- 19.Wolfle SJ, Strebovsky J, Bartz H, Sahr A, Arnold C, Kaiser C, Dalpke AH, Heeg K. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur J Immunol 2011; 41:413-24; PMID:21268011; http://dx.doi.org/ 10.1002/eji.201040979 [DOI] [PubMed] [Google Scholar]
- 20.Brahmer J. Nivolumab (anti-PD-1; BMS-936558; ONO-4538) in patients with non-small cell lung cancer (NSCLC): overall survival and longterm safety in a phase 1 trial. Presented at: IASLC 15th World Conference on Lung Cancer; October 2013; Sydney, Australia. MO18.03. [Google Scholar]
- 21.Sosman J, Sznol M, McDermott D, Carvsjal R, Lawrence D, Topalian SL, et al.. Clinical activity and safety of anti-programmed death-1 (PD-1) (BMS-936558/MDX-1106/ONO-4538) in patients (PTS) with advanced melanoma (MEL) in ESMO. 2012. [Google Scholar]
- 22.Rizvi N. A phase I study of nivolumab (anti-PD-1; BMS-936558, ONO-4538) plus platinum-based doublet chemotherapy (PT-doublet) in chemotherapy-naive non-small cell lung cancer (NSCLC) patients (pts). J Clin Oncol 2013; 31(suppl; abstr 8072); 3935-43; PMID:2404374524043745 [Google Scholar]
- 23.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, et al.. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 2013; 369:134-44; PMID:23724846; http://dx.doi.org/ 10.1056/NEJMoa1305133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al.. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012; 366:2455-65; PMID:22658128; http://dx.doi.org/ 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al.. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366:2443-54; PMID:22658127; http://dx.doi.org/ 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Incecco A, Andreozzi M, Ludovini V, Rossi E, Capodanno A, Landi L, Tibaldi C, Minuti G, Salvini J, Coppi E, et al.. PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br J Cancer 2015; 112:95-102; PMID:25349974; http://dx.doi.org/ 10.1038/bjc.2014.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen DS, Irving BA, Hodi FS. Molecular pathways: next-generation immunotherapy–inhibiting programmed death-ligand 1 and programmed death-1. Clin Cancer Res 2012; 18:6580-7; PMID:23087408; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-1362 [DOI] [PubMed] [Google Scholar]
- 28.Soria JC, Cruz C, Bahleda R, Delord JP, Horn L, Herbst RS, et al.. Clinical activity, safety and biomarkers of PD-L1 blockade in non-small cell lung cancer (NSCLC): additional analyses from a clinical study of the engineered antibody MPDL3280A (anti-PDL1). Eur J Cancer 2013; 49(suppl):abstract 3408 [Google Scholar]
- 29.Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, Zhang Y, He X, Zhou T, Qin T, et al.. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients with EGFR Mutation. J Thorac Oncol 2015; 10:910-23; PMID:25658629; http://dx.doi.org/ 10.1097/JTO.0000000000000500 [DOI] [PubMed] [Google Scholar]
- 30.Cooper WA, Tran T, Vilain RE, Madore J, Selinger CI, Kohonen-Corish M, Yip P, Yu B, O'Toole SA, McCaughan BC, et al.. PD-L1 expression is a favorable prognostic factor in early stage non-small cell carcinoma. Lung Cancer 2015; 89:181-8; PMID:26024796; http://dx.doi.org/ 10.1016/j.lungcan.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 31.Gao B, Sun Y, Zhang J, Ren Y, Fang R, Han X, Shen L, Liu XY, Pao W, Chen H, et al.. Spectrum of LKB1, EGFR, and KRAS mutations in chinese lung adenocarcinomas. J Thorac Oncol 2010; 5:1130-5; PMID:20559149; http://dx.doi.org/ 10.1097/JTO.0b013e3181e05016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae NC, Chae MH, Lee MH, Kim KM, Lee EB, Kim CH, Park TI, Han SB, Jheon S, Jung TH, et al.. EGFR, ERBB2, and KRAS mutations in Korean non-small cell lung cancer patients. Cancer Genet Cytogenet 2007; 173:107-13; PMID:17321325; http://dx.doi.org/ 10.1016/j.cancergencyto.2006.10.007 [DOI] [PubMed] [Google Scholar]
- 33.Lee SY, Kim MJ, Jin G, Yoo SS, Park JY, Choi JE, Jeon HS, Cho S, Lee EB, Cha SI, et al.. Somatic mutations in epidermal growth factor receptor signaling pathway genes in non-small cell lung cancers. J Thorac Oncol 2010; 5:1734-40; PMID:20881644; http://dx.doi.org/ 10.1097/JTO.0b013e3181f0beca [DOI] [PubMed] [Google Scholar]
- 34.Suzuki M, Shigematsu H, Hiroshima K, Iizasa T, Nakatani Y, Minna JD, Gazdar AF, Fujisawa T. Epidermal growth factor receptor expression status in lung cancer correlates with its mutation. Hum Pathol 2005; 36:1127-34; PMID:16226114; http://dx.doi.org/ 10.1016/j.humpath.2005.08.007 [DOI] [PubMed] [Google Scholar]
- 35.Slebos RJ, Kibbelaar RE, Dalesio O, Kooistra A, Stam J, Meijer CJ, Wagenaar SS, Vanderschueren RG, van Zandwijk N, Mooi WJ. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med 1990; 323:561-5; PMID:2199829; http://dx.doi.org/ 10.1056/NEJM199008303230902 [DOI] [PubMed] [Google Scholar]
- 36.Arribalzaga EB. New tumor, node, metastasis staging system for lung cancer. J Thorac Oncol 2009; 4:1301; author reply-2; PMID:20197739; PMID:24714771; http://dx.doi.org/ 10.1097/JTO.0b013e3181b5a867 [DOI] [PubMed] [Google Scholar]
- 37.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014; 20:5064-74.; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antonia S, Grosso J, Horak C, Harbison C, Kurland J, Inzunza D, et al.. Association of tumor PD-L1 expression and immune biomarkers with clinical activity in patients with non-small cell lung cancer treated with nivolumab. J Thorac Oncol 2013; 8(suppl):abstract P2.11-035 [Google Scholar]
- 39.Grosso J, Horak CE, Inzunza D, Cardona DM, Simon JS, Gupta AK, et al.. Association of tumor PD-L1 expression and immune biomarkers with clinical activity in patients (pts) with advanced solid tumors treated with nivolumab (anti-PD-1; BMS-936558; ONO-4538). J Clin Oncol 2013; 31(suppl):abstract 3016 [Google Scholar]
- 40.Soria JC, Cruz C, Bahleda R, Delord JP, Horn L, Herbst RS, et al.. Clinical activity, safety and biomarkers of PD-L1 blockade in non-small cell lung cancer (NSCLC): additional analyses from a clinical study of the engineered antibody MPDL3280A (anti-PDL1). Eur J Cancer 2013; 49(suppl):abstract 3408 [Google Scholar]
- 41.Lin K, Cheng J, Yang T, Li Y, Zhu B. EGFR-TKI down-regulates PD-L1 in EGFR mutant NSCLC through inhibiting NF-kappaB. Biochem Biophys Res Commun 2015; 463:95-101; PMID:25998384; http://dx.doi.org/ 10.1016/j.bbrc.2015.05.030 [DOI] [PubMed] [Google Scholar]
- 42.Tang Y, Fang W, Zhang Y, Hong S, Kang S, Yan Y, Chen N, Zhan J, He X, Qin T, et al.. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs. Oncotarget 2015; 6:14209-19; PMID:25895031; http://dx.doi.org/ 10.18632/oncotarget.3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azuma K, Ota K, Kawahara A, Hattori S, Iwama E, Harada T, Matsumoto K, Takayama K, Takamori S, Kage M, et al.. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol 2014; 25:1935-40; PMID:25009014; http://dx.doi.org/ 10.1093/annonc/mdu242 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Wang L, Li Y, Pan Y, Wang R, Hu H, Li H, Luo X, Ye T, Sun Y, et al.. Protein expression of programmed death 1 ligand 1 and ligand 2 independently predict poor prognosis in surgically resected lung adenocarcinoma. Onco Targets Ther 2014; 7:567-73; PMID:24748806; http://dx.doi.org/ 10.2147/OTT.S59959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, et al.. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 2013; 3:1355-63; PMID:24078774; http://dx.doi.org/ 10.1158/2159-8290.CD-13-0310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang CY, Lin MW, Chang YL, Wu CT, Yang PC. Programmed cell death-ligand 1 expression in surgically resected stage I pulmonary adenocarcinoma and its correlation with driver mutations and clinical outcomes. Eur J Cancer 2014; 50:1361-9; PMID:24548766; http://dx.doi.org/ 10.1016/j.ejca.2014.01.018 [DOI] [PubMed] [Google Scholar]


