ABSTRACT
Invasive zygomycosis in immunocompromised patients results in a high mortality rate, and early identification is crucial to optimize therapy and to reduce morbidity. However, diagnosing specific species of zygomycetes fungi possess challenge in the clinical laboratories. A need for a rapid and sensitive diagnostic tool for early recognition of a zygomycetes fungus in clinical samples to the species level will lead to prompt and accurate therapy and the PathoChip provides one such platform. We utilized a pathogen array technology referred to as PathoChip, comprised of oligonucleotide probes that can detect all the sequenced viruses as well as known pathogenic bacteria, fungi and parasites and family-specific conserved probes, thus providing a means for detecting previously uncharacterized members of a family. We rapidly identified a zygomycetous fungus, Rhizomucor pusillus, an otherwise challenge for the clinical laboratories, predominantly in a patient with acute myelogenous leukemia. This report highlights the value of PathoChip as a diagnostic tool to identify micro-organisms to the species level, especially for those difficult to identify in most clinical laboratories. It will also help clinicians to obtain a critical snapshot of the infection profile of a patient to plan treatment strategies.
KEYWORDS: Acute myelogenous leukemia, diagnostic, opportunistic fungal infection, PathoChip, pan-pathogen array, Rhizomucor, zygomycete
Introduction
Zygomycosis, an infection caused by fungi in the class zygomycetes occurs in immunocompromised states, as a result of either therapy or underlying disease.1,2 Invasive infection due to these molds results in a high mortality rate, and early identification is crucial to optimizing therapy. However, it has been a challenge to diagnose specific species of zygomycetes fungi in the clinical laboratories using phenotypic methods.3 Difficulty in the recognition of microscopic differences among species and the lack of tester strains to perform mating studies, make the detection of species specific zygomycetes fungi impossible in the clinical laboratories and so most rely on identification of the genus. In zygomycosis, identification of species is very important from the therapeutic perspective. Studies have shown in vitro susceptibility to anti-fungal agents to vary within the zygomycetes family and thus, identification of the species provides clues for optimization of treatment.4 Thus, molecular assays were developed to identify specific species of zygomycetes. These include sequencing of internal transcribed spacer (ITS) region,5-8 28S rRNA 9 and non-sequencing based tests like polymerase chain reaction (PCR) with restriction fragment length polymorphism (RFLP),10,11 real- time PCR using melting curve analysis,12 multiplex PCR using specific DNA probes in a microarray based assay,13 and PCR assay using the Luminex xMAP hybridization method.14 While, the sequencing based assays have become more popular to identify fungal species, most of the species identification using sequencing was done from culture isolates, though direct sequencing from tissue has shown some success.5,8,9,15,16 Although these molecular methods could detect species of a particular zygomycetes genus, they are limited by their detection range of pathogenic fungi, which is an important limiting factor in the context of diagnosis of invasive fungal infections. Further, the sensitivity of detection of fungal species by PCR or sequencing is very low in formalin fixed tissues compared to fresh tissues,16 thus making diagnosis from fixed tissue section unreliable. More recently, to specifically identify species of fungi/zygomycetes, a combined approach of both morphologic and molecular testing methods have been explored when a histological diagnosis is not always possible.15 Neither of the 2 methods is rapid and sensitive and both are restricted by limitations to identify only a small range of species.
A rapid and sensitive diagnostic tool for early recognition of a zygomycetes fungus in clinical samples (using fresh or fixed) to the species level will lead to prompt accurate therapy. This reduces morbidity and ensures the best chance of survival. PathoChip provides one such platform17 using a microarray-based approach containing probe sets for parallel DNA and RNA detection of viruses, bacteria, fungi, parasites and other human pathogenic microorganisms. The current version of the PathoChip has 60,000 probes per array, representing all known viruses, 250 helminths, 130 protozoa, 360 fungi and 320 bacteria.17 The array contains 2 types of probes: unique probes for each specific virus and microorganism, and conserved probes which target genomic regions that are conserved between members of a family of viruses, thereby providing a means for detection of previously uncharacterized members of the family. The PathoChip screening technology includes an amplification step that allows detection of microorganisms and viruses present in low genomic copy number in samples and also fragmented genomes extracted from formalin-fixed paraffin embedded archival tissues. We have used the PathoChip to detect microorganisms associated with different cancer samples that are obtained as paraffin fixed tissues.17 Thus, PathoChip allows multiple clinical samples to be rapidly and sensitively screened for the presence of microbial agents. There are 37,704 specific probes for pathogenic microorganisms, including 896 specific probes (from 28s rRNA, ITS, 18S rRNA genes) for the fungal species available in the GenBank.17
Using the PathoChip, we detected a zygomycetous fungal species, Rhizomucor pusillus from FFPE samples obtained from a patient with acute myelogenous leukemia. The results of the PathoChip screen were further validated by PCR, sequencing and staining.
The patient, a 47 year old male who presented to the hospital of University of Pennsylvania with relapsed acute myelogenous leukemia (AML) (containing inv(16)(p13.1q22) and FLT3 D835 mutation). He received induction chemotherapy and was subsequently neutropenic (ANC = 0). He developed sudden onset of bilateral lower extremity weakness which rapidly progressed to T4 paraplegia and acute urinary retention. Both serum galactomanann and β-D-glucan were negative prior to surgery. Imaging studies (MRI) showed an epidural mass lesion at T4-T5 with prevertebral and paravertebral extension. He was taken to the operating room for resection of the epidural mass and an emergent T4-5 laminectomy for cord compression. Tissue from this procedure did not show infectious etiologies in their differential and thus microbiologic studies were not performed on this sample. However, staining of the tissue revealed angioinvasive fungal infection (Fig. 4). Therefore, to rapidly identify the invasive fungus, formalin fixed paraffin embedded (FFPE) tissue section was provided for PathoChip analysis.
Results
PathoChip screen identified Rhizomucor pusillus as the predominant agent in the patient sample
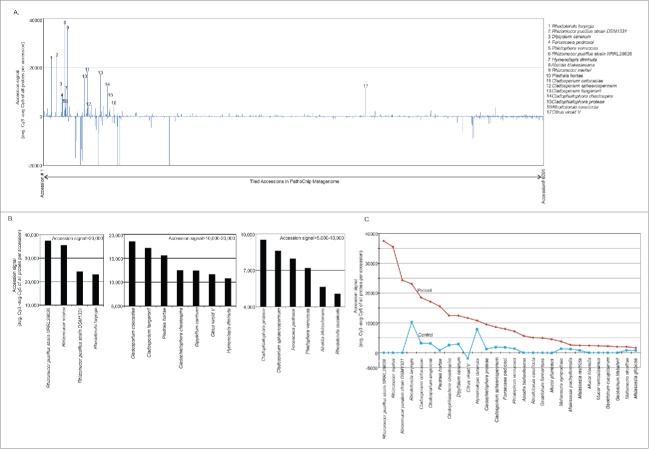
Accession analysis for all the signals from the hybridization of the PathoChip were first completed for the patient sample to see which organisms showed high accession signals in the patient sample screened. Accession signals determine the average hybridization signal of all the probes per accession. Out of the 6001 organisms only 17 accessions or organisms showed accession signals >5000 (Fig. 1A and B). This suggests that the majority of the associated probes in the PathoChip were detected for those particular organisms. Probes of Rhizomucor (zygomycetes) and Rhodotorula laryngis (Basidiomycetes) were detected with the highest accession signals of >20,000 (Fig. 1B, left panel). Probes of ascomycetous fungi (Cladosporium, Piedraia, Cladophialophora), parasites (Diplidium and Hymenolepis) and viroid were detected with accession signals ranging from >10,000 to 20,000 (Fig. 1B, middle panel). Other ascomycetous strains of Cladosporium, Cladophialophora, Fonsecaea and Phialophora along with the zygomycete Absidia blakesleeana and the Basidiomycete Rhodotorula cassicolla were detected with accession signals raging from >5000-10,000 (Fig. 1B, right panel). Organisms detected in the patient with accession signal less than 5000 included skin fungus Malassezia, yeast Geotrichum and species of Mucor (Fig. 1C). Accession signal analysis of the control sample was also carried out, and accession signals of those organisms that were detected in the patient were compared to that of the control (Fig. 1C and Table 1). We detected genomic signatures of species of Rhizomucor with the highest accession signal (Fig. 1 and Table 1) and importantly, the accession analysis of the control sample did not detect any species of Rhizomucor (Fig. 1C and Table 1). Except for Hymenolepis diminuta, others seem to have much higher accession signal in the patient sample compared to control (Fig. 1C and Table 1). Of those probes with lower (<5000) accession signal detected in the patient sample, the probes of Geotrichum and Mucor were found only to be associated with the patient and not with control, while probes of the skin fungus Malassezia were also detected in the control samples (Fig. 1C and Table 1).
Figure 1.
Accession analysis of samples screened by PathoChip. Figure A shows the accession signal (average Cy3 signal for probes-average Cy5 signal for probes/accession) of 6001 accesions of PathoChip metagenome, represented as bar graph. The organisms showing high accession signal (>5000) are numbered. Figure B shows the top hit accessions detected in the patient sample in decreasing order of accesion signals. Figure C represents the comparative accesion signals in cancer versus the control.
Table 1.
Candidate organisms detected in the patient sample with higher accession signal than in control.
| Accession signal |
|||
|---|---|---|---|
| (Mean g- mean r)/ accession |
|||
| Type | Organism | Cancer | Control |
| Fungus | Rhizomucor pusillus strain NRRL28626 | 37471 | −1 |
| Rhizomucor miehei | 35494 | 3 | |
| Rhizomucor pusillus strain DSM1331 | 24334 | 6 | |
| Rhodotorula laryngis | 23084 | 10256 | |
| Cladosporium colocasiae | 18532 | 3213 | |
| Cladosporium langeronii | 17132 | 3134 | |
| Piedraia hortae | 15602 | 817 | |
| Cladophialophora chaetospira | 12480 | 2570 | |
| Parasite | Dipylidium caninum | 12445 | 2912 |
| Viroid | Citrus viroid V | 11644 | −1840 |
| Parasite | Hymenolepis diminuta | 10797 | 7876 |
| Fungus | Cladophialophora proteae | 9512 | 1243 |
| Cladosporium sphaerospermum | 8598 | 1862 | |
| Fonsecaea pedrosoi | 7970 | 1819 | |
| Phialophora verrucosa | 7178 | 1360 | |
| Absidia blakesleeana | 5629 | 4 | |
| Rhodotorula cassiicola | 5065 | 2 | |
| Malassezia sympodialis | 3789 | 1340 | |
| Malassezia pachydermatis | 2625 | 1236 | |
| Prosthodendrium longiforme | 3574 | 856 | |
| Malassezia slooffiae | 1995 | 825 | |
| Malassezia globosa | 1624 | 797 | |
| Malassezia restricta | 2437 | 790 | |
| Geotrichum fermentans | 4949 | 11 | |
| Geotrichum cucujoidarum | 2226 | 1 | |
| Geotrichum klebahnii | 2018 | -2 | |
MAT analysis of the patient sample identified individual probes of the same organisms that were detected by accession analysis. Hybridization signals (Cy3-Cy5) of the probes for those organisms in the patient sample were then compared to that in the control sample, represented as a heat map in Fig. 2.
Figure 2.
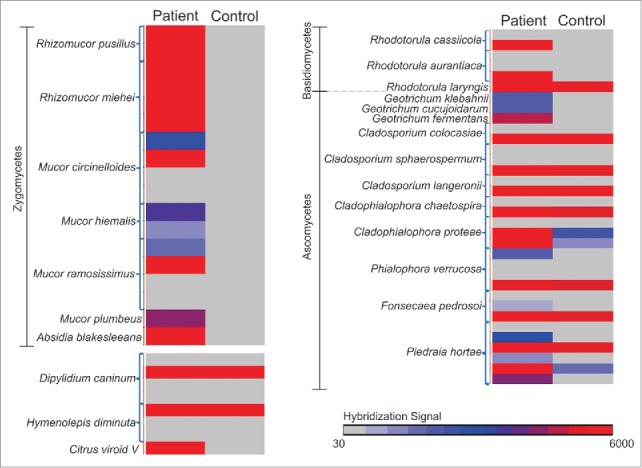
Hybridization signals (Cy3-Cy5) of the probes of the organisms detected in the patient sample screen compared to the control sample screen, represented in a heat map.
Individual probes of Rhizomucor (both Rhizomucor pusillus and Rhizomucor miehei) detected by MAT analysis, showed very high hybridization signal (Cy3-Cy5 >3000) in the patient screen and not in the controls (Fig. 2). All or few probes of related fungal organisms (other zygomycetes like Absidia and Mucor) were also detected by the PathoChip screen in the patient sample and not in the control (Fig. 1, Fig. 2), thus further suggesting an association of zygomycete infection in the patient. Fungal probes of Rhodotorula, Cladosporium, Cladophialophora, Phialophora, Fonsecaea and Piedraia and parasitic probes of Dipylidium and Hymenolepis, that were detected in the patient sample were also detected in the control samples (Fig. 2), and thus were not uniquely associated with the patient, although the hybridization signal for the probes of those organisms were higher in the patient sample compared to control. The leukemic condition of the patient might have provided an amiable microenvironment for those organisms that would not generally be present in normal individuals.
Validation of the PathoChip screen results by PCR and sequencing
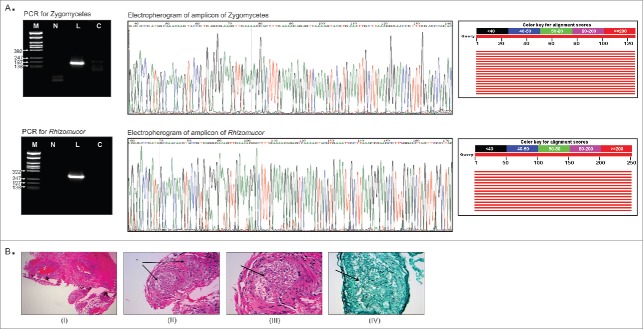
Primers were used to detect any zygomycetous fungi and also to specifically detect Rhizomucor by PCR (material and methods section). The primers that detect any zygomyceteous fungi yielded the expected 181bp amplicon in the patient sample, not in the control. Likewise, the specific PCR primers for Rhizomucor detected the fungus (305 bp amplicon) in the patient sample and not in the control. This was confirmed by sequencing the amplified PCR products (Fig. 3A). The sequenced amplicon was subjected to the BLAST tool of NCBI and it was determined that both amplicons showed 100% sequence homology (red lines representing high alignment scores) to the 28S rRNA gene Rhizomucor pusillus and Rhizomucor miehei (Fig. 3A).
Figure 3.
Validation of PathoChip screen results by PCR, sequencing and staining of infected tissue section. Fig. 3A shows the the PCR amplified products yielded using primers to detect any Zygomycetes fungi and primers to detect Rhizomucor, run on 2% agarose gel (left panel). Ethidium bromide stained gel picture of the amplicons are shown (left panel). M: φX174/Rsa1 DNA ladder, L: leukemic patient sample, C: control sample. Part of the electropherogram of the ampilcons sequenced is shown (middle panel). The NCBI BLAST tool showed alignement scores for each amplicon sequenced (right panel). The graphic summary shows alignments (as colored lines) of database matches to the amplicons sequenced (query). The red color lines represented the highest alignment scores, thus suggesting the query sequence to be significantly similar to the database hit Rhizomucor pusillus and Rhizomucor miehei 28S rRNA sequences. Fig. 3B shows the staining of tissue section from the patient for fungus. A Hematoxylin and Eosin (I-III) as well as Grocott Methanamine Silver (IV) stain were performed on the tissue sections from the paravertebral mass of the patient. Figure (I) shows the low power view of the soft tissue, (II) shows the 20X view, (III) shows the 40X view and (IV) shows the 40X view using silver stain (Grocott stain). Fungus in blood vessels is shown with arrows.
Staining results
On both the H&E and Grocott stain, fungal hyphae are visible. These hyphae show variable thickness and no discrete septation within the hyphae which was interpreted as most consistent with a zygomycoses infection (Fig. 3B).
Discussions
Rhizomucor involves 2 ubiquitous species: R. pusillus and R. miehei, that can lead to Zygomycosis in humans.18 It is a rare disease, often found in the patients with a weakened immune system and can often lead to a fatal outcome.18 It occurs most often in patients with hematological malignancies,19-26 and diabetes mellitus.27 Zygomycosis presents a major clinical challenge, both during diagnosis and patient treatment.28 Invasive fungal infections are often diagnosed by histopathology without identification of the causative fungi, which show significant different antifungal susceptibilities.4 Frequently these fungi fail to grow in clinical assays or culture systems, and since some fungi require several weeks to grow, detection of the fungal agent at an advanced stage of disease becomes difficult.29,30 Mortality rates approach 100%, especially in the absence of immune reconstitution.31 Given the expanding list of available antifungal drugs, it is hoped that early diagnosis will lead to prompt initiation of directed antifungal therapy and enhance the chance of survival in patients with zygomycetes. We report the rapid detection of R. pusillus and R. miehei in a patient with acute myelogenous leukemia, using an array based high throughput pathogen detection system. The PathoChip, which was used to screen the patient sample, has been earlier proven to be sensitive, specific and useful in detecting cancer associated pathogens.17 We were able to rapidly and efficiently detect the fungal organisms from FFPE tissue sample of a patient with acute T4 paraplegia due to cord compression from an epidural mass due to a zygomycete.
All specific probes of Rhizomucor fungi showed very high hybridization signal. However, we also detected some probes of the related zygomycetes, Mucor and Absidia blakesleeana with moderate to high hybridization signal, associated with the patient sample thus demonstrating a zygomycete signature association. Additionally, probes for the species of Geotrichum were detected with moderate hybridization signal, suggesting other opportunistic fungal infection in the patient at much lower levels. It is not surprising that multiple infections are seen in an immunocompromised patient with a dominant agent driving the pathology. However, using this strategy a clinician can obtain a critical snapshot of the infection profile of a patient and so inform their intervention strategy.
Interestingly, probes for a plant viroid were detected in the patient sample (not in the control), which could be because of dietary raw fruits and vegetables consumption that expose us to large numbers of plant viruses and viriods, some of which may have persisted in the patient.
The importance of zygomycosis as an emerging infectious disease in compromised patients highlights the need for better methods to diagnose and treat this condition. The ability of the PathoChip to rapidly identify this fungus and other uncommon pathogenic fungi, viruses and other micro-organisms could be helpful for future diagnostics and intervention paradigms.
Methods
PathoChip design
The design of the 60,000 probe sets for all the available microorganisms used on the PathoChip Array has been previously described.17 The designed probe sets were manufactured as SurePrint glass slide microarrays (Agilent Technologies Inc.). Probes were represented as 60-nt DNA oligomers with 60,000 probes on 8 replicate arrays per slide.17 There are multiple probes for each pathogenic viral, prokaryotic, and eukaryotic organism.
Sample preparation and microarray processing
FFPE tissue section (10 micron) from the paravertebral mass of the patient with AML, was provided. Five non-matched control samples comprising of tissue from healthy individuals were used as control. Briefly, DNA and RNA was extracted in parallel from 3 FFPE tissue sections of the patient and from the control group as described earlier.17 The qualities of the extracted DNA/RNA were assessed by measuring the A260/280 ratio, and the size distributions of the extracted nucleic acids were determined by agarose gel electrophoresis. The partially degraded RNA and DNA samples extracted from the FFPE tissue samples were subjected to RNA/DNA amplification (Whole Transcriptome Amplification/ WTA) as previously described using 50 ng each of RNA and DNA as input.17 The sample preparation and amplification protocols to detect DNA and RNA of microorganisms from FFPE tissues has been previously described.17 The 5 non-matched controls were pooled as 1 sample during the WTA step (10ng each of RNA/DNA). The amplification products yielded amplicons ranging from 200-400bp as expected for FFPE samples, and monitored for integrity by agarose gel electrophoresis. 15ng each of human reference RNA and DNA extracted from the BJAB human B cell line was also subjected to WTA. All the amplified products (from the patient sample, control and human reference) were purified using a PCR purification kit (Qiagen, Germantown, MD, USA). 1μg of the amplified product from the FFPE tissues of the patient and control was used for Cy3 labeling by the SureTag labeling kit (Agilent Technologies, Santa Clara, CA), and Cy5 labeling was performed on 1 μg of human reference cDNA/DNA amplification product as a control to identiy any cross-hybridization of probes to human DNA. The labeled DNA were purified and the extent of labeling was determined by A550 for Cy3 (green) and A650 for Cy5 (red).17 The labeled samples were hybridized to the PathoChip as described by Agilent Technologies, Santa Clara, CA. Hybridization cocktail consisting of a CGH blocking agent, hybridization buffer (as per manufacturer's instruction), was added to the mix of labeled test sample (Cy3) and the reference (Cy5), denatured and hybridized to the PathoChip arrays. One array was used for the patient sample screening and another for the control screen. Hybridization was carried out at 65°C with rotation in an Agilent hybridization oven for 24 hours. Post-hybridization, the slides were washed using wash buffer and scanned using an Agilent SureScan G4900DA array scanner.
Microarray data analyses and statistical analysis
Data analysis was done using the Partek Genomics Suite (Partek Inc., St. Louis, MO, USA) as previously described.17 Post normalization of the data (as described earlier) analysis was initially performed to determine specific positive hybridization signals (Cy3-Cy5) for all the probes of an organism.17 Thus, the hybridization signal for each accession/organism (accession signal) was determined by subtracting the average Cy5 signal for all the probes per accession from average Cy3 signal of the same (accession signal = average Cy3 of all probes/accession minus average Cy5 of all probes/accession).17 Accession signal for each organism in the PathoChip was calculated both for the patient sample and the control and then compared. Model-based analysis of tiling arrays (MAT), which utilized a sliding window analysis of probe signals for each sample screened, was also performed. MAT analysis as previously described was helpful in detecting positive hybridization signal (Cy3-Cy5) even if a few and not all the probes of an organism are positive.17
PCR and sequence validation of the PathoChip results
PCR primers from the 28S rDNA sequences to detect Zygomycetes32 (zygo-F1: TTCAAAGAGTCAGGTTGTTTGG and zygo-R1: CAGTCTGGCTCCAAACGGTTC), and also to detect the specific genus, Rhizomucor 33 (Rm1-FP: TCTATTGCGATGCATGCTCCAAG and Rm2-RP: GGTCTCTTTAGACTCCAAAGCAC) were used to further validate the findings of the PathoChip screen. The PCR amplification reaction mixtures for each reaction contained 300 ng of WTA product and 10 pmol each of forward and reverse primers, 300μM of dNTPs and 2.5U of LongAmpTaq DNA polymerase (New England Biolabs). DNA was denatured at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 51°C for zygo primers and 52°C for Rm primers for 30 s, and 65°C for 30 s and a final extension of 65°C for 10 min. The amplified product was fractionated by electrophoresis on a 2% agarose gel with an appropriate DNA ladder to determine the amplicon size and the amplicons were gel extracted and subjected to Sanger sequencing with their respective forward primers. Post sequencing, the sequence chromatograms were visualised using the BioEdit program 34 and the sequences were put in the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) tool 35 (http://blast.ncbi.nlm.nih.gov) to identify the sequence match to the available sequences in GenBank.
Staining
A Hematoxylin and Eosin as well as Grocott Methanamine Silver stain were performed on the tissue sections from the paravertebral mass using standard methodologies.36 Polysaccharides in the fungal wall are oxidized by the grocott reaction and the oxidized sugars are then free to react with the metallic silver to form a dark black silver precipitate on the fungal cell wall (Fig. 3B).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Avon Foundation Grant no. [Avon-02-2012-053] (to Erle S. Robertson), and from the Abramson Cancer Center Director's fund.
References
- 1. Ameen M, Arenas R, Martinez-Luna E, Reyes M, Zacarias R. The emergence of mucormycosis as an important opportunistic fungal infection: five cases presenting to a tertiary referral center for mycology. Int J Dermatol 2007; 46:380-4; PMID:17442077; http://dx.doi.org/ 10.1111/j.1365-4632.2007.03057.x [DOI] [PubMed] [Google Scholar]
- 2. Chayakulkeeree M, Ghannoum MA, Perfect JR. Zygomycosis: the re-emerging fungal infection. Eur J Clin Microbiol Infect Dis 2006; 25:215–29; PMID:16568297; http://dx.doi.org/ 10.1007/s10096-006-0107-1 [DOI] [PubMed] [Google Scholar]
- 3. Iwen PC, Thapa I, Bastola D. Review of Methods for the Identification of Zygomycetes With an Emphasis on Advances in Molecular Diagnostics. Lab Medicine 2011; 42:260-6; http://dx.doi.org/ 10.1309/LMJ8Z0QPJ8BFVMZF [DOI] [Google Scholar]
- 4. Almyroudis NG, Sutton DA, Fothergill AW, Rinaldi MG, Kusne S. In vitro susceptibilities of 217 clinical isolates of zygomycetes to conventional and new antifungal agents. Antimicrob Agents Chemother 2007; 51:2587-90; PMID:17452481; http://dx.doi.org/ 10.1128/AAC.00452-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iwen PC, Hinrichs SH, Rupp ME. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med Mycol 2002; 40:87-109; PMID:11860017; http://dx.doi.org/ 10.1080/mmy.40.1.87.109 [DOI] [PubMed] [Google Scholar]
- 6. Iwen PC, Freifeld AG, Sigler L, Tarantolo SR. Molecular identification of Rhizomucor pusillus as a cause of sinus-orbital zygomycosis in a patient with acute myelogenous leukemia. J Clin Microbiol 2005; 43:5819-21; PMID:16272531; http://dx.doi.org/ 10.1128/JCM.43.11.5819-5821.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alvarez E, Sutton DA, Cano J, Fothergill AW, Stchigel A, Rinaldi MG, Guarro J. Spectrum of zygomycete species identified in clinically significant specimens in the United States. J Clin Microbiol 2009; 47:1650-6; PMID:19386856; http://dx.doi.org/ 10.1128/JCM.00036-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwarz P, Bretagne S, Gantier JC, Garcia-Hermoso D, Lortholary O, Dromer F, Dannaoui E. Molecular identification of zygomycetes from culture and experimentally infected tissues. J Clin Microbiol 2006; 44:340-9; PMID:16455881; http://dx.doi.org/ 10.1128/JCM.44.2.340-349.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vollmer T, Stormer M, Kleesiek K, Dreier J. Evaluation of novel broad-range real-time PCR assay for rapid detection of human pathogenic fungi in various clinical specimens. J Clin Microbiol 2008; 46:1919-26; PMID:18385440; http://dx.doi.org/ 10.1128/JCM.02178-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nyilasi I, Papp T, Csernetics A, Krizsan K, Nagy E, Vagvolgyi C. High-affinity iron permease (FTR1) gene sequence-based molecular identification of clinically important Zygomycetes. Clin Microbiol Infect 2008; 14:393-7; PMID:18190575; http://dx.doi.org/ 10.1111/j.1469-0691.2007.01932.x [DOI] [PubMed] [Google Scholar]
- 11. Machouart M, Larche J, Burton K, Collomb J, Maurer P, Cintrat A, Biava MF, Greciano S, Kuijpers AF, Contet-Audonneau N, et al. . Genetic identification of the main opportunistic Mucorales by PCR-restriction fragment length polymorphism. J Clin Microbiol 2006; 44:805-10; PMID:16517858; http://dx.doi.org/ 10.1128/JCM.44.3.805-810.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hata DJ, Buckwalter SP, Pritt BS, Roberts GD, Wengenack NL. Real-time PCR method for detection of zygomycetes. J Clin Microbiol 2008; 46:2353-8; PMID:18480229; http://dx.doi.org/ 10.1128/JCM.02331-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spiess B, Seifarth W, Hummel M, Frank O, Fabarius A, Zheng C, Mörz H, Hehlmann R, Buchheidt D. DNA microarray-based detection and identification of fungal pathogens in clinical samples from neutropenic patients. J Clin Microbiol 2007; 45:3743-53; PMID:17715373; http://dx.doi.org/ 10.1128/JCM.00942-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Landlinger C, Preuner S, Willinger B, Haberpursch B, Racil Z, Mayer J, Lion T. Species-specific identification of a wide range of clinically relevant fungal pathogens by use of Luminex xMAP technology. J Clin Microbiol 2009; 47:1063-73; PMID:19244466; http://dx.doi.org/ 10.1128/JCM.01558-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balajee SA, Sigler L, Brandt ME. DNA and the classical way: identification of medically important molds in the 21st century. Med Mycol 2007; 45:475-90; PMID:17710617; http://dx.doi.org/ 10.1080/13693780701449425 [DOI] [PubMed] [Google Scholar]
- 16. Dannaoui E, Schwarz P, Slany M, Loeffler J, Jorde AT, Cuenca-Estrella M, Hauser PM, Shrief R, Huerre M, Freiberger T, et al. . Molecular detection and identification of zygomycetes species from paraffin-embedded tissues in a murine model of disseminated zygomycosis: a collaborative European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Fungal Infection Study Group (EFISG) evaluation. J Clin Microbiol 2010; 48:2043-6; PMID:20375233; http://dx.doi.org/ 10.1128/JCM.02319-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baldwin DA, Feldman M, Alwine JC, Robertson ES. Metagenomic assay for identification of microbial pathogens in tumor tissues. MBio 2014; 5:e01714-14; PMID:25227467; http://dx.doi.org/ 10.1128/mBio.01714-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lukacs G, Papp T, Nyilasi I, Nagy E, Vagvolgyi C. Differentiation of Rhizomucor species on the basis of their different sensitivities to lovastatin. J Clin Microbiol 2004; 42:5400-2; PMID:15528755; http://dx.doi.org/ 10.1128/JCM.42.11.5400-5402.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bethge WA, Schmalzing M, Stuhler G, Schumacher U, Krober SM, Horger M, Einsele H, Kanz L, Hebart H. Mucormycoses in patients with hematologic malignancies: an emerging fungal infection. Haematologica 2005; 90 Suppl:ECR22; PMID:16266913 [PubMed] [Google Scholar]
- 20. Barnert J, Behr W, Reich H. An amphotericin B-resistant case of rhinocerebral mucor mycosis. Infection 1985; 13:134-6; PMID:3928496; http://dx.doi.org/ 10.1007/BF01642873 [DOI] [PubMed] [Google Scholar]
- 21. del Palacio,Hernanz A, Fereres J, Larregla Garraus S, Rodriguez-Noriega A, Sanz Sanz F. Nosocomial infection by Rhizomucor pusillus in a clinical haematology unit. J Hosp Infect 1983; 4:45-9; PMID:6190884; http://dx.doi.org/ 10.1016/0195-6701(83)90064-6 [DOI] [PubMed] [Google Scholar]
- 22. Kramer BS, Hernandez AD, Reddick RL, Levine AS. Cutaneous infarction. Manifestation of disseminated mucormycosis. Arch Dermatol 1977; 113:1075-6; PMID:268159; http://dx.doi.org/ 10.1001/archderm.1977.01640080077012 [DOI] [PubMed] [Google Scholar]
- 23. Meyer RD, Kaplan MH, Ong M, Armstrong D. Cutaneous lesions in disseminated mucormycosis. Jama 1973; 225:737-8; PMID:4515739; http://dx.doi.org/ 10.1001/jama.1973.03220340043014 [DOI] [PubMed] [Google Scholar]
- 24. Ryan ME, Ochs D, Ochs J. Primary cutaneous mucormycosis: superficial and gangrenous infections. Pediatr Infect Dis 1982; 1:110-4; PMID:6960330; http://dx.doi.org/ 10.1097/00006454-198203000-00009 [DOI] [PubMed] [Google Scholar]
- 25. Severo LC, Job F, Mattos TC. Systemic zygomycosis: nosocomial infection by Rhizomucor pusillus. Mycopathologia 1991; 113:79-80; PMID:2034262; http://dx.doi.org/ 10.1007/BF00442413 [DOI] [PubMed] [Google Scholar]
- 26. St-Germain G, Robert A, Ishak M, Tremblay C, Claveau S. Infection due to Rhizomucor pusillus: report of four cases in patients with leukemia and review. Clin Infect Dis 1993; 16:640-5; PMID:8507755; http://dx.doi.org/ 10.1093/clind/16.5.640 [DOI] [PubMed] [Google Scholar]
- 27. Wickline CL, Cornitius TG, Butler T. Cellulitis caused by Rhizomucor pusillus in a diabetic patient receiving continuous insulin infusion pump therapy. South Med J 1989; 82:1432-4; PMID:2814631; http://dx.doi.org/ 10.1097/00007611-198911000-00024 [DOI] [PubMed] [Google Scholar]
- 28. Kivivuori SM, Karikoski R, Koukila-Kahkola P, Anttila VJ, Saarinen-Pihkala UM. Zygomycosis presenting a major clinical challenge: case report on Rhizomucor pusillus infection in a stem-cell-transplant recipient. Mycopathologia 2011; 172:241-5; PMID:21475989; http://dx.doi.org/ 10.1007/s11046-011-9424-8 [DOI] [PubMed] [Google Scholar]
- 29. Chen SC, Halliday CL, Meyer W. A review of nucleic acid-based diagnostic tests for systemic mycoses with an emphasis on polymerase chain reaction-based assays. Med Mycol 2002; 40:333-57; PMID:12230214; http://dx.doi.org/ 10.1080/mmy.40.4.333.357 [DOI] [PubMed] [Google Scholar]
- 30. Kontoyiannis DP, Wessel VC, Bodey GP, Rolston KV. Zygomycosis in the 1990s in a tertiary-care cancer center. Clin Infect Dis 2000; 30:851-6. Epub 2000 Jun 13 ; PMID:10852735; http://dx.doi.org/ 10.1086/313803 [DOI] [PubMed] [Google Scholar]
- 31. Gonzalez CE, Rinaldi MG, Sugar AM. Zygomycosis. Infect Dis Clin North Am 2002; 16:895-914, vi; PMID:12512186; http://dx.doi.org/ 10.1016/S0891-5520(02)00037-5 [DOI] [PubMed] [Google Scholar]
- 32. Francesconi A, Kasai M, Harrington SM, Beveridge MG, Petraitiene R, Petraitis V, Schaufele RL, Walsh TJ. Automated and manual methods of DNA extraction for Aspergillus fumigatus and Rhizopus oryzae analyzed by quantitative real-time PCR. J Clin Microbiol 2008; 46:1978-84; PMID:18353931; http://dx.doi.org/ 10.1128/JCM.02246-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Voigt K, Cigelnik E, O'Donnell K. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal-DNA sequence data. J Clin Microbiol 1999; 37:3957-64; PMID:10565914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Series 1999; 41:95-8; PMID:17267571726757 [Google Scholar]
- 35. Madden T. The BLAST sequence analysistTool. In: McEntyre J, Ostell J, editors The NCBI Handbook [Internet] Bethesda (MD: ): National Center for Biotechnology Information (US); 2002- Chapter 16 Available from: http://wwwncbinlmnihgov/books/NBK21097/2002 Oct 9 [Updated 2003 Aug 13]. [Google Scholar]
- 36. Grocott RG. A stain for fungi in tissue sections and smears using Gomori's methenamine-silver nitrate technic. Am J Clin Pathol 1955; 25:975-9; PMID:14398663 [DOI] [PubMed] [Google Scholar]