Abstract
Background:
The present study focuses on biodistribution profile and pharmacokinetic parameters of EGFR-targeted chitosan nanoparticles (TG CS nanoparticles) for siRNA/cisplatin combination therapy of lung cancer.
Material & methods:
Mad2 siRNA was encapsulated in EGFR targeted and nontargeted (NTG) CS nanoparticles by electrostatic interaction. The biodistribution of the nanoparticles was assessed qualitatively and quantitatively in cisplatin (DDP) sensitive and resistant lung cancer xenograft model.
Results:
TG nanoparticles showed a consistent and preferential tumor targeting ability with rapid clearance from the plasma to infiltrate and sustain within the tumor up to 96 h. They exhibit a sixfold higher tumor targeting efficiency compared with the NTG nanoparticles.
Conclusion:
TG nanoparticles present as an attractive drug delivery platform for RNAi therapeutics against NSCLC.
Keywords: : biodistribution, chitosan nanoparticles, EGFR targeting, Mad2 siRNA delivery, non-small-cell lung cancer models, pharmacokinetic analysis
Lung cancer is one of the deadliest types of malignancy worldwide, accounting for more than one quarter of all cancer related deaths with 85% of the cases being non-small-cell lung cancer (NSCLC) [1]. Most cases of lung cancer are diagnosed in their advanced stages, compromising long-term survival (1-year survival rate of ∼10%) [2]. Platinum-based therapy is the recommended first-line treatment for advanced NSCLC. However, systemic toxicity of platinates and development of acquired drug resistance have become an increasing problem in clinic, leading to high rate of mortality in NSCLC [3]. As such, there is an urgent need for development of novel treatment approaches against NSCLC, especially in improving clinical outcomes in refractory disease. Some current therapies, such as the use of taxanes and vinca alkaloids; take advantage of those neoplastic cells with high proliferation rate to inhibit their division through targeting microtubules [4]. The disadvantage on this strategy is the nonspecific toxicity, such as nerve damage, associated with these drugs [5,6]. A promising alternative is the use of molecular targeted strategies to disrupt mitosis without interfering with microtubule dynamics. As the targeted proteins only intervene in actively dividing cells, these strategies would not affect nondividing cells thereby decreasing the treatment-related adverse effects [7].
The mad2 gene is an essential component of the mitotic checkpoint, a surveillance mechanism that inhibits the metaphase-to-anaphase transition whenever chromosomes are not properly attached to the mitotic spindle [8,9]. Overexpression of mad2 has been observed in several types of cancer including NSCLC, oral cancer, cervical carcinogenesis and urothelial bladder cancer [10–13]. Mad2 depletion disrupts mitotic checkpoint function leading to premature mitotic exit, increased chromosome fragmentation and extensive cell death, and also sensitizes cancer cells to anticancer drugs. [14–16]. We recently used RNA interference (RNAi) approach to highlight the potential of mad2 gene knockdown as antitumor therapeutic strategy [14]. A major challenge in applying this technology is to effectively deliver the small interfering RNAs (siRNAs) to the tumor in vivo. Enzymatic degradation, removal from circulation by renal excretion or mononuclear phagocyte system (MPS), poor cellular uptake and endosomal release are examples of physiological barriers that siRNAs need to overcome [17,18]. Several viral and nonviral delivery vectors, such as adenovirus, polyplexes, liposomes, and micelles, amongst others, have been developed to overcome these obstacles and improve RNAi therapeutic efficacy in vivo [19–25].
Chitosan (CS) has gained increasing interest as a safer and more cost-effective vehicle for delivery of gene materials. The deacetylated derivative of chitin, is one of the most abundant carbohydrate polymers with several essential features that make it a useful natural material for medical and pharmaceutical applications [26]. As a random copolymer of poly(D-glucosamine) and poly(acetyl-D-glucosamine), CS is characterized by the degree of deacetylation and primary amine groups which, at low pH, are protonated and afford water solubility and cationic properties [27]. The presence of positive charges on CS backbone is related to its improved mucoadhesive properties and hemostatic activity [28]. The polymer interacts with membrane negative charges leading to a translocation of tight junction proteins from the membrane to the cytoskeleton, resulting in tight junction disruption and enhanced permeability [29–31]. Many reports have shown that CS can be enzymatically degraded in vivo due to the cleavable glycosidic bonds, and has minimal toxicity upon systemic administration [26]. CS has been widely used as a nonviral gene carrier since it can form complexes with nucleic acids by electrostatic interactions [32]. This way, nucleic acids are protected from nucleases and there is an increased efficiency of gene delivery to the cells where these complexes can be released from endosomes to enter the nucleus [14,32].
Chitosan may be delivered to the tumor tissues by taking advantage of ‘leaky’ and heterogeneous vascularization, which allows the migration of particles with diameter around 200 nm into the surrounding tumor region. Modifications such as anchoring poly(ethylene glycol) (PEG) to the nanoparticle surface can be utilized to prolong in vivo circulation and improve tumor infiltration. This process refers to the enhanced permeability and retention (EPR) effect that allows for passive delivery of long circulating nanoparticle systems to the solid tumor [33,34]. Other surface alterations, such as the incorporation of targeting moieties (e.g., antibodies, proteins, polysaccharides, and small molecules) provide enhanced efficacy and selectivity to tumor tissue and cells, thereby enhancing specificity and reducing off-target effects. The active targeting approach takes advantage of the overexpression of certain ligands and receptors on the tumor cell surfaces that permit a molecular recognition and a more efficient uptake of functionalized nanoparticles [35].
Overexpression and activation of EGF receptors (EGFRs) have been strongly indicated in tumorigenesis, tumor growth, progression, invasiveness and metastasis [36,37]. Due to their high overexpression, they have been extensively used as cancer cell selective targeting receptor. We have previously used a synthetic 12 amino acids peptide that has demonstrated efficient targeting of EGFR receptor and its targeting capability has been successfully validated in vitro as well as in vivo in various EGFR-overexpressing tumor cells such as A549 [38–43]. Most importantly, this peptide is capable of inducing nanoparticles cellular uptake without activating EGFR signaling pathway and therefore was included as targeting ligand in our formulation approach [44].
We recently demonstrated efficient in vitro delivery and mad2 gene silencing by siRNA encapsulated in nontargeted (NTG) and EGFR-targeted (TG) CS-based self-assembling nanoparticles system, in cisplatin sensitive A549-WT and resistant A549-DDP NSCLC cells [14].
Here, we have investigated the biodistribution profile of these Mad2 siRNA (siMad2)-loaded TG and NTG nanoparticles, in mice bearing subcutaneous, cisplatin sensitive or resistant, human lung adenocarcinoma xenograft tumors. The differences in the biodistribution and pharmacokinetics parameters between TG and NTG nanoparticles has been studied qualitatively and quantitatively.
Materials & methods
Materials
Chitosan (CS) with a viscosity-average molecular weight of 55 kDa and a degree of deacetylation of 75–85% was purchased from Sigma-Aldrich Inc. (MO, USA). Fluorescent dye DyLight 680 NHS-Ester and Pico-Green fluorescence reagent were obtained from Life Technologies (CA, USA). Succinimidyl-([N-maleimidopropionamido]-ethyleneglycol) ester (MAL-PEG2000-NHS, MW 2,000 Da) was purchased from JenKem (TX, USA). EGFR specific peptide was synthesized at Tufts University's Peptide Synthesis Core Facility (MA, USA). SiMad2 and its corresponding scrambled siRNA were obtained from Santa Cruz Biotechnology Inc. (TX, USA). All primers were ordered from Eurofins Scientific (Luxembourg City, Luxembourg). AgPath-ID One step RT-PCR kit was purchased from Thermo Scientific (IL, USA) to perform RT-PCR. All other reagents were obtained at high purity (>99%) from Sigma-Aldrich Inc. or Thermo Scientific.
Cisplatin sensitive & resistant cell lines & tumor models
Cisplatin sensitive (parenteral) NSCLC cell line (A549-WT) was obtained from American Type Culture Collections (ATCC, VA, USA). Cisplatin resistant NSCLC cell line (A549-DDP) was obtained from our collaborator, Massachusetts General Hospital (MA, USA). Both cell lines were cultured at 37°C in 5% CO2 environment in DMEM/F12 medium from Life Technologies supplemented with 10% fetal bovine serum and penicillin/streptomycin (100 U/ml; Thermo Fisher Scientific, MA, USA). A549-DDP were cultured in the presence of 2 μg/ml cisplatin to maintain the drug resistance phenotype. Three days before use, A549-DDP media was changed to a medium without cisplatin.
Six weeks old female nu/nu (athymic) mice, strain CrTac:NCr-Foxn1nu from Taconic Biosciences, Inc. (NY, USA), weighing approximately 20 g, were group-housed upon arrival in the Division of Laboratory Animal. The animals were allowed to acclimate for at least 72 h prior to any experimentation, raised under specific pathogen-free conditions, kept in individually ventilated cage racks and supplied with sterile rodent pellets and water ad libitum. Mice were housed under a 12 h light/dark cycle.
For A549-WT and A549-DDP tumor model development, mice were injected subcutaneously with 3 × 106 cells in a mixture of 50 µl DMEM/F12 medium and 50 µl Matrigel on the right flank, under mild anesthesia. Tumor volume was measured with caliper every 3 days and calculated by the modified ellipsoid formula:
 |
Each study commenced when the tumors reached an average size of 200 mm3 and the animals were randomly assigning to a specific group. The animals were monitored daily for food/water intake, body weight and any physical signs of discomfort. In all the experiments performed in this study, sample size was determined by power analysis using the software G*Power.
Synthesis of chitosan derivatives
For NTG and TG nanoparticles encapsulating Mad2 siRNA, CS derivatives were synthesized as previously described [14]. Briefly, to a 2 mg/ml CS solution in 2% acetic acid, a 10% molar equivalent of Mal-PEG2000-NHS was added and left to react overnight at room temperature. Dialysis (molecular weight cutoff - 10 kDa) in water was performed in order to purify the conjugate. Cysteine was reacted in excess under N2 environment, to inactivate the maleimide group to obtain the NTG CS derivative. The synthesis of TG CS derivative was achieved by using a 17-amino acid peptide consisting of 12-amino acid EGFR recognition peptide, four glycine residues spacer and a terminal cysteine for conjugation via the thiol group (i.e., YHWYGYTPQWVI-GGGG-C). The peptide was added in a twofold molar excess to the CS derivative with a reactive maleimide group and left to react overnight at 4°C in nitrogen atmosphere to allow the cysteine group from the peptide to react with the maleimide group. NTG and TG derivatives were purified by dialysis (molecular weight cutoff: 10 kDa) against water, freeze-dried and stored at -20°C until their use. Degree of PEG substitution on the CS backbone was estimated by NMR analysis in D2O using 400 MHz 1H NMR spectroscopy (Varian Inc., CA, USA).
Chitosan labeling with NIR dye for in vivo imaging
Near-infrared (NIR) labeled CS was obtained by mixing NTG CS derivative with the amine-reactive dye DyLight 680 NHS-Ester (DL680) in an aqueous solutions at an equivalent amount to achieve 5% substitution of the CS amine groups. DL680 is a near-infrared imaging dye with 684/707 nm as excitation/emission maxima.
After reacting overnight in the dark, the NIR derivative was dialyzed overnight using 10 kDa MW cut-off membrane (Spectrapore, Spectrum Labs, CA, USA) and freeze-dried. The degree of modification of the CS backbone with the dye was estimated against a standard curve obtained from the absorbance of the known concentrations of the dye measured at 680 nm.
siMad2-loaded chitosan nanoparticles
Both NTG and TG CS nanoparticles were obtained by self-assembly in aqueous solution maintaining a N:P ratio (ratio of CS bearing cationic amines to siRNA bearing anionic phosphates) of 50. Independent of the desired type of nanoparticles (NTG, TG or NIR-labeled), CS derivative was dissolved in water (1 mg/ml). SiMad2 was slowly added to this solution under magnetic stirring and incubated for 30 min at room temperature. This procedure allowed CS and siMad2 complexation and nanoparticles self-assembly prior to use. If we intended to produce TG CS NPS, the initial mixture would be a 50% (w/w) mixture of CS-PEG and CS-EGFR (1 mg/ml). In case of NIR-labeled nanoparticles, CS-PEG would be substituted by NIR-labeled CS-PEG. For every study, nanoparticles were freshly prepared and dissolved in PBS in order to obtain a desired concentration and also achieve an osmolarity of 300 mOsm/kg and a pH of 7.2.
Hydrodynamic diameter (particle size), surface charge and polydispersity index (PDI) of freshly prepared siMad2 loaded CS (CS-Mad2) nanoparticles were measured using a ZetaSizer Nano ZS (Malvern Instruments, Worcestershire, UK). Each sample was obtained from four independent batches in different days. They were diluted in PBS, analyzed in triplicate at 25°C and the different parameters reported as mean ± SD. The encapsulation efficiency (ratio of encapsulated siRNA over total siRNA added) was determined using Quant-iT Pico-Green kit (Life Technologies).
Whole body & ex vivo NIR imaging
A549-WT and A549-DDP tumor bearing mice were intravenously injected into the lateral tail veins with a dose of 3 mg/kg of siMad2 encapsulated in NIR labeled TG and NTG CS nanoparticles (n = 4/group). Intravenous injection was the route of nanoparticles administration in all of the experiments in order to avoid an absorption phase which would compromise an accurate estimation of distribution and elimination parameters [45]. Mice were imaged every 24 h until 96 h after the injection, to monitor the distribution of the nanoparticles using Xenogen IVIS® Imaging System (Xenogen Corporation, CA, USA; Ex: 685 nm, Em: 720 nm). Along with these formulations, free NIR dye in PBS at identical concentration was also injected into tumor bearing mice. For ex vivo imaging, the same procedure was adapted but animals were sacrificed at different time points. Tumor and major organs (liver, spleen, kidneys, heart and lungs) were collected for imaging purposes.
Quantitative analysis of siMad2 in blood & tissues
siMad2 was encapsulated in NTG and TG CS nanoparticles as described above and injected once at time zero into A549-WT and A549-DDP tumor bearing mice at 3 mg/kg (n = 5/group). At different time points (12, 24, 48, 72, 96 and 120 h), animals were sacrificed and blood samples, the major organs (liver, spleen, lung, heart, kidney), and tumors were collected. Blood samples from the facial vein were also collected 6 h after injection. Plasma was isolated from the blood samples by centrifugation at 1200 g for 15 min at 4°C, followed by siMad2 quantification.
Tumor and organs were homogenized in RNALater solution in order to preserve RNA integrity and the tissue lysates were diluted at 1:1000 dilution. Mad2 siRNA was then quantified in the samples based on an antiprimer quenching PCR method [46]. The diluted samples were used for subsequent annealing step followed by qRT-PCR. The primers used were: Reverse: 5′ GGA AGC CGA TGG CAG T; Forward:/56-FAM/- 5′ ACT CCC TCC CTC GAT TTT CAA TAT CAA AC; and antiprimer: 5′ AAA TCG AGG GAG GGA GT/3BHQ_1/.
The procedure was performed according to the previously optimized protocol published by our group [47]. Briefly, 6 µl of diluted tissue sample was mixed with 18 μl of 100 nM reverse primer, and put through a cycle of denaturation by incubating at 95°C for 5 min and another cycle of annealing for 2 min at each temperature of 80, 70, 60 and 45°C. From this mixture, 3.5 µl were used and mixed with 8.5 μl from a AgPath-ID One step RT-PCR master mix constituted by the following components: RT-PCR buffer (6.25 ml), forward primer (10 mmol/l, 0.12 ml), reverse primer (10 mmol/l, 0.12 ml), antiprimer (100 mmol/l, 0.12 ml), 25 U RT-PCR enzyme (0.5 ml) and water (1.5 ml). The PCR conditions were as follows: 50°C (10 min), 9°C (10 min), 40 cycles, 95°C (15 s), 45°C (60 s). Using lysate from untreated mouse tissue and spiked with known siRNA concentrations, we were able to do a standard curve and quantify. The quantitated siRNA in each tissue was then normalized toward the percentage of input dose per volume of plasma or per mass of tissue.
Quantitative pharmacokinetic analysis
SiMad2 plasma pharmacokinetic parameters and organs exposure were determined by noncompartmental analysis using Phoenix® WinNonLin® version 1.3 software (Certara, MO, USA). Plasma half-life (HL) was calculated by log-linearly fitting the three last time points. The maximum observed concentration (Cmax) and the last observed concentration (Clast) were calculated as the mean of observed concentrations at the time when the mean concentration was the highest, and at the last time point respectively. Exposure, quantified as area under concentrations versus time curve from time zero to the last collection time (i.e., AUClast), was calculated using the linear trapezoidal method. AUC extrapolated to infinity (AUC∞) was the sum of AUClast and Clast/(ln2/HL). The associated standard error of the mean was estimated using equations provided by Nedelman and Jia, and later corrected by Holder [48,49]. The mean residence time was calculated as the ratio of AUMC∞ (Area under the first moment (time * concentration) versus time curve) and AUC∞. Clearance (CL) was calculated as the ratio of dose (100%ID) and AUC. The volume of distribution at the steady-state (Vss), in other words, at the time when equilibrium is reached for exchanges between distribution compartments, was calculated using the formula: Vss = CL * mean residence time. Noncompartmental analysis for sparse data being a naive pooled method, no variability could be estimated for pharmacokinetic parameters. Tumor targeting efficiency was evaluated by comparing tumor exposure to plasma exposure, and to the sum of other organs’ exposure. For each time-point and each condition, five animals were used.
Data analysis
With the exception of the quantitative pharmacokinetic analysis, statistical analyses were performed using GraphPad Prism software (CA, USA). Paired comparisons were performed by Student's t-test. A p value of 0.05 was considered to be statistically significant. Data presented are means ± standard deviation.
Results & discussion
In our previous in vitro studies, we designed and prepared EGFR targeted chitosan nanoparticles as a delivery system for Mad2 siRNA in lung adenocarcinoma cells [14]. This nanoparticles were modified with PEG in order to provide a hydrophilic shielding and attain passive targeting to tumor site. NTG and TG nanoparticles loaded with siMad2 were able to efficiently silence mad2 gene, leading to increased cytotoxicity by induction of apoptosis [14]. In the current study, our goal was to evaluate the biodistribution pattern of these two types of CS nanoparticles in a subcutaneous xenograft model of human drug sensitive A549-WT and platinum resistant A549-DDP tumors. A 3 mg/kg dose of siMad2 was used for all the experiments to ascertain accurate and consistent quantitative detection of the siRNA from various tissues samples.
Whole body & ex vivo qualitative biodistribution studies
Qualitative nanoparticles biodistribution studies were evaluated using an NIR dye, DL680, covalently bound to the CS skeleton. Due to the dye's strong signal, CS was modified in a very low percentage (4%) which did not significantly impact the nanoparticles characteristics (Table 1). Both TG and NTG particles were prepared and used to encapsulate siMad2 using the same method, independently of the presence or absence of conjugated dye. NIR labeled CS/siRNA nanoparticles were intravenously injected via tail vein in A549-WT and A549-DDP tumor bearing mice at a single dose of 3 mg/kg of siMad2. Mice were imaged at different time points and the NIR signal was measured to capture the whole-body distribution pattern. Using the same conditions, a different set of animals were sacrificed and their major organs collected and imaged. Although studies were performed in A549-WT and A549-DDP tumor bearing mice, the results were identical in both models and even though we will refer to the results from the A549-WT model, the conclusions apply to both tumor models.
Table 1. . Near-infrared dye-loaded chitosan nanoparticles characterization.
| Size (nm) ± SD | PDI ± SD | Zeta potential (mV) ± SD | siMad2 encapsulation efficiency (%) | |
|---|---|---|---|---|
| NTG |
106.8 ± 2.1 |
0.551 ± 0.10 |
+35.6 ± 3.5 |
105.1 ± 4 |
| NTG-DL680 |
113.1 ± 5.3 |
0.472 ± 0.53 |
+32.8 ± 2.4 |
101.6 ± 2.3 |
| TG |
227.3 ± 1.8 |
0.362 ± 0.02 |
+28.3 ± 2.0 |
99.5 ± 2.4 |
| TG-DL680 | 230.1 ± 4.1 | 0.341 ± 0.21 | +29.9 ± 2.9 | 97.5 ± 3.3 |
NTG: Nontargeted; PDI: Polydispersity index; SD: Standard deviation; TG: Targeted; WT: Wild-type.
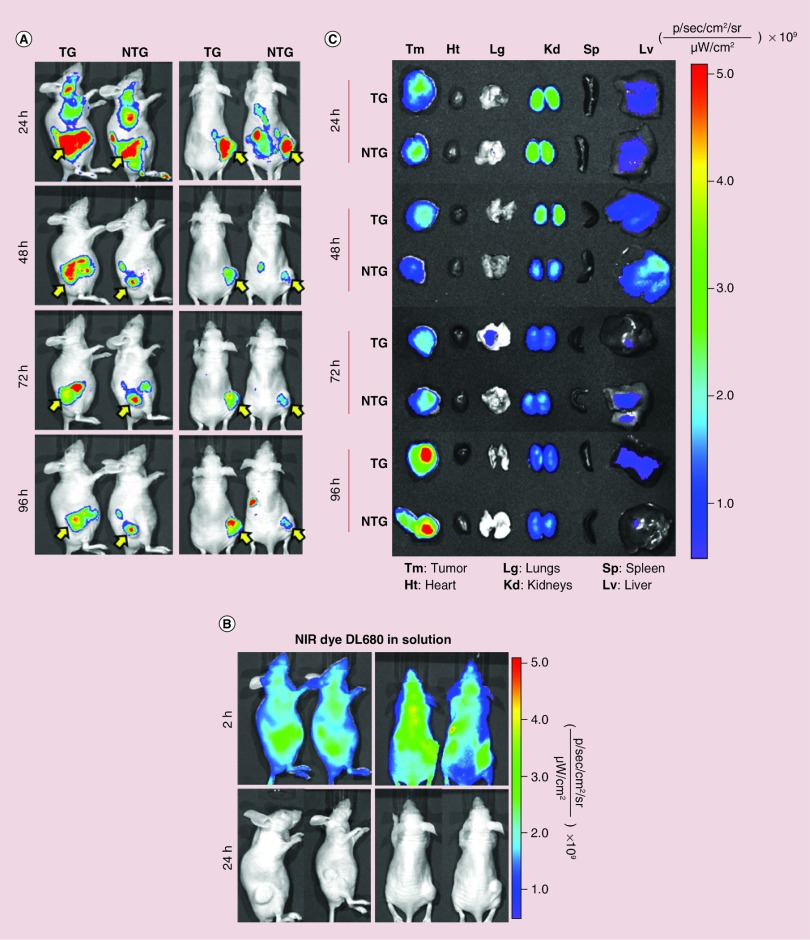
Within the first 12 h post-injection, a very strong NIR signal was observed throughout the whole body of animals injected with both systems, and only after 24 h the signal started to accumulate in certain areas (Figure 1). The whole body images (Figure 1A) show a strong signal at 24 h for both TG and NTG nanoparticles in the liver and kidneys, which are the significant sites of CS nanoparticles accumulation and metabolism, mainly when using CS of low molecular weight [50]. Tumors show nanoparticle accumulation, and the signal appeared to be stronger for TG nanoparticles compared with the NTG nanoparticles, particularly in posterior view images. Due to the limited depth penetration of the optical source through the tissue in the whole body imaging, mice were sacrificed and the organs were excised for ex vivo imaging. Previous annotations were confirmed by ex vivo fluorescent imaging where liver showed a strong signal until 48 h, while kidneys demonstrate higher accumulation throughout but mainly within the first 48 h (Figure 1C). These results are supported by the fact that CS has been described to have relatively long circulation time [50,51]. Contrarily, heart, lungs and spleen did not show any nanoparticles accumulation with the only exception being lungs that show low signal with TG nanoparticles at 72 h which could be due to some contamination during tissue collection, but a very low signal is detected. The excised tumors showed a strong signal after 24 h post-injection for both nanoparticles, although TG nanoparticles seemed to show a stronger signal at 48 h. For the following time points, both systems seemed to have similar signal intensity and they reach their maximum at 96 h. Although both NTG and TG nanoparticles have shown a high level of signal in the tumor site, the imaging technique is not sensitive enough to discern an improvement in tumor accumulation due to the presence of EGFR targeting peptide. In spite of this, both nanoparticle systems presented better delivery efficiency compared with free dye, which was not detected within the tumor. The body distribution of free NIR dye was monitored in order to ensure that the signal seen for NTG and TG nanoparticles was due to intact nanoparticles and not due to free dye released from nanoparticles. At 24 h, there was no detectable fluorescence signal in any of the mice injected with free dye and therefore mice had to be imaged at 1 and 2 h post-injection (Figure 1B). The rapid in vivo signal decrease of free NIR imaging dye could be attributed to fluorescence quenching in physiological environments and also indicative of a rapid hepatic clearance from the systemic circulation [52–54]. Nanoparticles allow a long circulation of the dye, providing it with an effective shielding and preserving its fluorescence [55]. An increased circulation associated with small sizes is beneficial since the nanoparticles will be able to extravasate across the fenestrated endothelium of the cancer vasculature and accumulate in tumor tissue through the EPR effect [34].
Figure 1. . Whole body and ex vivo optical imaging of near-IR labeled chitosan/siMad2 nanoparticles in mice A549-WT tumor bearing mice for up to 96 h.
Prelabeled chitosan with a near-infrared Cy 5.5 dye was used to encapsulate siMad2 using a N:P ratio of 50:1. A549 tumor bearing mice were injected once in a concentration of 3 mg/kg of siMad2 encapsulated in nontargeted (NTG) or targeted (TG) CS nanoparticles. (A-B) Mice were imaged at different time points up to 96 h, on their posterior and lateral view using IVIS live imaging system. In these images we have two representative animals although a total of four animals per tumor model were used. A solution of free dye was also administered at equivalent concentration but it was only detected until 2 h after injection. Yellow arrow indicates tumor localization. (C) Ex vivo NIR images of major tissues excised from A549 tumor bearing mice at different time points post-injection.
NTG: Nontargeted; TG: Targeted; WT: Wild-type.
Plasma pharmacokinetic analysis
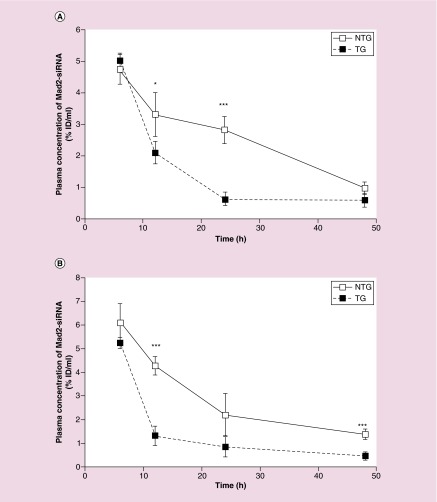
SiMad2 encapsulated in NTG or TG CS nanoparticles were administered by intravenous injection at a single dose of 3 mg/kg in mice bearing A549-WT and A549-DDP tumors. Later Mad2 siRNA was quantified in different organs, tumor and plasma, using the antiprimer quenching based RT-PCR method [46]. Data were processed and expressed as percentage of the injected dose per ml of plasma or mg of tissue (% ID/ml or % ID/mg). For all conditions, siMad2 was detected in plasma up to 48 h after administration. This long circulation could be related to the presence of positive charges on CS structure. These charges can interact with negative charges on red blood cell membranes and allow the nanoparticles to circulate in the bloodstream for long periods of time [56,57]. SiMad2 concentrations were below the limit of quantification at 72 h post-injection. Regarding siRNA concentration in plasma in A549-WT tumor model (Figure 2A), both NTG and TG nanoparticles produced similar concentrations after 6 h, but then concentrations decreased faster for TG nanoparticles than for NTG nanoparticles, with concentrations at 12 and 24 h being significantly lower for TG nanoparticles. A similar trend was obtained in A549-DDP tumor model (Figure 2B) where siMad2 being at a similar concentration at 6 h for both type of particles and significantly lower for TG nanoparticles at 12 and 48 h.
Figure 2. . Plasma concentrations of siMad2 (%ID/ml) ± SD versus time (h) in A549-WT (A) and A549-DDP (B) tumor models, after IV injection of NTG and TG nanoparticles at 3 mg/kg equivalent siRNA.
n = 5 mice.
*p < 0.05; **p < 0.01; ***p < 0.001 (t-test TG vs NTG).
DDP: Cisplatin; NTG: Nontargeted; SD: Standard deviation; TG: Targeted; WT: Wild-type.
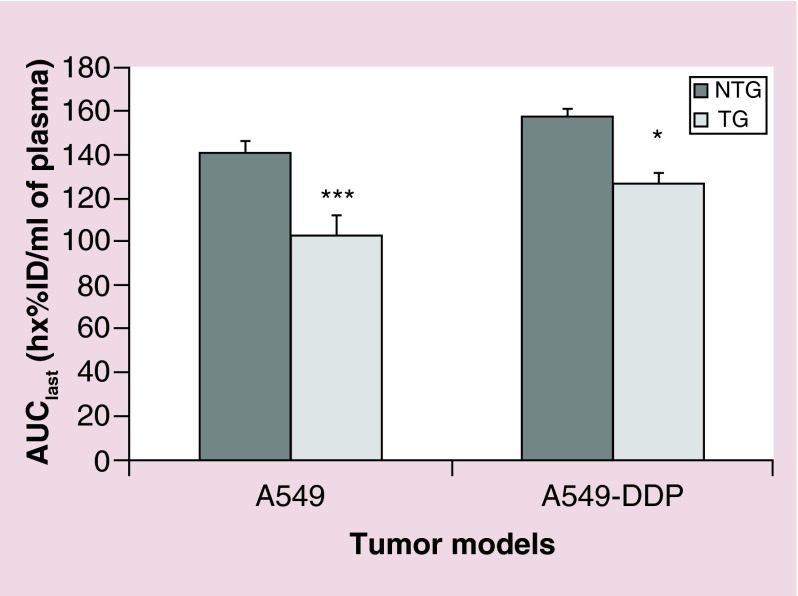
AUClast is an important pharmacokinetic parameter that expresses the concentration of the siRNA in the interstitium as a function of time [58]. This parameter reflects the tissue degree of exposure to siRNA and also its clearance from the body. AUClast was significantly higher in NTG nanoparticles than in TG nanoparticles, independent of the tumor models (Figure 3). This observation was consistent with the observed kinetics and exposures. The clearance of TG nanoparticles was higher than the clearance of NTG nanoparticles for both models studied, leading to a shorter mean residence time in the body (Table 2). Accordingly, Clast value was lower for TG nanoparticles than for NTG nanoparticles, while Cmax values were similar. The volume of distribution appeared reduced by the targeting peptide in the A549-DDP model only. Surprisingly, half-life calculated from the last three time points showed no clear difference between nanoparticles. This might be ascribed to the small number of time points, which would not enable the characterization of a biphasic kinetics and a relevant elimination half-life. Overall, the addition of EGFR targeting peptide induced a significantly higher exposure to tumor tissue, which is in line with most published results [41,59,60].
Figure 3. . Plasma exposure to siMad2 over the duration of the study.
AUClast +SE (in h*%ID/ml of plasma) for A549-WT and A549-DDP tumor models, after IV injection of NTG and TG nanoparticles at 3 mg/kg equivalent siRNA.
n = 5 mice.
*p < 0.05; **p < 0.01; ***p < 0.001 (t-test TG vs NTG).
AUClast: Area under the concentration versus time curve until the last time point; DDP: Cisplatin; NTG: Nontargeted; SE: Standard error; TG: Targeted; WT: Wild-type.
Table 2. . Plasma PK parameters calculated by noncompartmental analysis using Phoenix WinNonLin software.
| A549-NTG | A549-TG | A549 DDP-NTG | A549 DDP-TG | |
|---|---|---|---|---|
| HL (h) |
19.5 |
22.6 |
23.4 |
24.1 |
| Cmax (SE) (%ID/ml) |
4.73 (0.23) |
5.02 (0.10) |
6.10 (0.40) |
5.25 (0.12) |
| Clast (%ID/ml) |
0.970 |
0.588 |
1.388 |
0.459 |
| AUC∞ (h x%ID/ml) |
168 |
122 |
205 |
143 |
| MRT (h) |
26.1 |
20.9 |
30.7 |
16.0 |
| CL (ml/h) |
0.595 |
0.817 |
0.489 |
0.700 |
| Vss (ml) | 15.5 | 17.1 | 15.0 | 11.2 |
AUC∞: Area under the time-concentration curve from time zero to infinity; CL: Total body clearance; Clast: Concentration at the last time-point; Cmax: Maximum observed concentration; DDP: Cisplatin; HL: Half-life of the log-linear terminal part of the curve; MRT: Mean residence time; NTG: Nontargeted; SE: Standard error of the mean; TG: Targeted; Vss: Volume of distribution at the steady-state.
Tumor & other tissue pharmacokinetic analysis
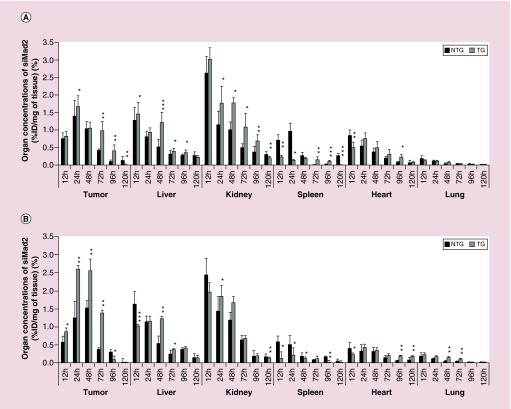
Whole body images were informative regarding the nanoparticles biodistribution and ex vivo imaging allowed to identify the most exposed organs, but results needed to be confirmed by quantitative measurements. In A549-WT model treated with NTG nanoparticles, the concentrations at 12 h were the highest in kidneys, followed by liver, tumor, heart, spleen and lungs (Figure 4A). At 24 h, the order was slightly different, with tumor displaying the highest concentration, followed by kidneys, spleen, liver, heart and lungs. The addition of the targeting peptide increased siMad2 concentration in tumor (significantly at 24, 72 and 96 h), but also in liver and kidneys. A significant decrease was seen in spleen and heart at early time points with TG nanoparticles, but not maintained later. No significant changes were observed in lungs. Interestingly, the quantitative approach allowed to observe differences, for which imaging was not sensitive enough. The quantitative approach also showed that tumor concentrations were peaking at 24 h and then decreasing, while ex vivo images suggested an increase of concentrations until 96 h. It is important to take into account that in one study we are quantifying siMad2 while in the imaging studies, we have labeled CS nanoparticles. Also, siMad2 is rapidly used by the cellular silencing mechanism once in the cytoplasm while the fate of CS molecule is unsatisfactorily understood [17]. In the A549-DDP tumor model (Figure 4B), the trends were very similar to the A549-WT model. However, a few differences could be observed. First, with NTG nanoparticles, concentration in spleen was not lower than in tumor at 12 h, and was higher than in heart. At 24 h, concentration in kidney was still higher than in tumor, and spleen was also less exposed than, in A549-WT model. When comparing TG to NTG particles, the most striking differences were a much higher increase of tumor concentrations, while liver and kidney concentrations were lower at 12 h by the addition of the targeted peptide. These results enhance the influence of active targeting on increasing target-site accumulation.
Figure 4. . Biodistribution in tumor and major organs.
Organ concentrations of siMad2 (%ID/mg of tissue) ± SD at various times after injection (from 12 to 120 h) in A549-WT (A) and A549-DDP (B) tumor models, after IV injection of NTG and TG nanoparticles at 3 mg/kg equivalent siRNA. n = 5 mice.
*p < 0.05; **p < 0.01; ***p < 0.001 (t-test TG vs NTG).
DDP: Cisplatin; ID: Injected dose; NTG: Nontargeted; TG: Targeted.
For some of the organs, it was not possible to properly estimate a log-linear terminal phase half-life and a mean residence time, due to an unclear decrease slope. For example, due to concentrations measurements variability, the spleen concentration at 120 h in the A549-WT model after NTG nanoparticles administration was higher than earlier concentrations in the same conditions. In order to compare the residence times for the different conditions, we chose to use an indirect parameter for residence time, which was available for all conditions, in other words, the Clast/Cmax ratio (Table 3). In plasma, the ratio was lower for TG particles, suggesting a faster elimination and a shorter residence time, which is in line with the PK parameters (Table 2). In tumor, the ratio was particularly low compared with other tissues, suggesting a rapid clearance of siMad2 from the tumor. This might be due to consumption of the siRNA by its target, leading to a ‘target-mediated drug disposition’. The differences were rarely significant between TG and NTG particles, except for kidney and spleen in A549 model where the ratio was significantly lower for TG particles, and for plasma and heart in A549-DDP model where the ratio was significantly lower and higher respectively. The clearest difference between nanoparticle types, confirmed in both models, was a decrease of the Clast/Cmax ratio in spleen for TG vs NTG nanoparticles, suggesting that the targeting peptide might increase the clearance of the siRNA from the spleen.
Table 3. . Ratio of the last time-point concentration (Clast) versus the maximum concentration reached over the duration of the study (Cmax), expressed in% (SD).
|
Clast/Cmax (%) (SD) |
||||
|---|---|---|---|---|
| A549-WT NTG | A549-WT TG | A549-DDP NTG | A549-DDP TG | |
| Plasma |
21.1 (6.4) |
11.9 (4.7) |
22.7 (0.6) |
8.71 (3.11)*** |
| Tumor |
8.62 (6.73) |
0.619 (0.195) |
0.452 (0.269) |
0.544 (0.346) |
| Liver |
20.8 (5.4) |
15.1 (0.7) |
9.07 (3.05) |
11.8 (1.1) |
| Kidney |
11.8 (2.8) |
6.92 (1.15)* |
7.19 (1.32) |
7.11 (0.98) |
| Spleen |
33.3 (13.5) |
1.50 (0.12)** |
11.0 (12.0) |
1.84 (0.66) |
| Heart |
6.89 (6.06) |
16.5 (5.5) |
19.8 (6.9) |
50.3 (16.0)* |
| Lung | 13.1 (6.7) | 15.6 (6.7) | 11.4 (5.8) | 6.77 (1.73) |
n = 5 mice.
*p < 0.05; **p < 0.01; ***p < 0.001 (t-test TG vs NTG).
Clast: Last time-point concentration; Cmax: Maximum concentration reached over the duration of the study; DDP: Cisplatin; NTG: Nontargeted; SD: Standard deviation; TG: Targeted; WT: Wild-type.
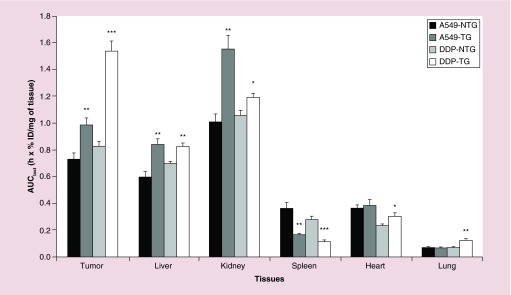
Regarding the global exposure of the various tissues to siRNA over the study course (estimated as AUClast), it appeared in both tumor models that kidneys were the most exposed to siRNA, followed by tumor, liver, spleen, heart, and lungs, when NTG nanoparticles were used (Figure 5). Introduction of the targeting peptide significantly increased exposure of tumor, liver, and kidneys for both models, of heart and lungs in A549-DDP model only, and significantly decreased spleen exposure in both models. In A549-DDP model, the enhancement of tumor exposure was far higher than in A549-WT model and this influence was felt not only in a higher tumor accumulation but also in a lower accumulation in other tissues. The difference between the tumor models can be ascribed to the slightly higher vascularization that had been detected in the A549-DDP model. Indeed, a higher permeability of the tumor allows nanoparticles to travel more deeply into the tumor tissues after extravasation [61]. Another reason that could contribute to such difference and probably more significantly, is the EGFR expression levels in these two models. We have used flow cytometry to measure EGFR) cell surface expression levels on A549-WT and A549-DDP and results showed that the cisplatin resistant cell line, A549-DDP has a significantly higher expression. This fact is of great influence on the amount of nanoparticles being taken up by tumor cells.
Figure 5. . Tissues exposure to siMad2 over the duration of the study.
AUClast +SE (in h x%ID/mg of tissue) for A549-WT and A549-DDP tumor models, after IV injection of NTG and TG nanoparticles at 3 mg/kg equivalent siRNA. n = 5 mice.
*p < 0.05; **p < 0.01; ***p < 0.001 (t-test TG vs NTG).
AUClast: Area under the concentration versus time curve until the last time point; DDP: Cisplatin; NTG: Nontargeted; TG: Targeted.
Evaluation of the tumor targeting efficiency
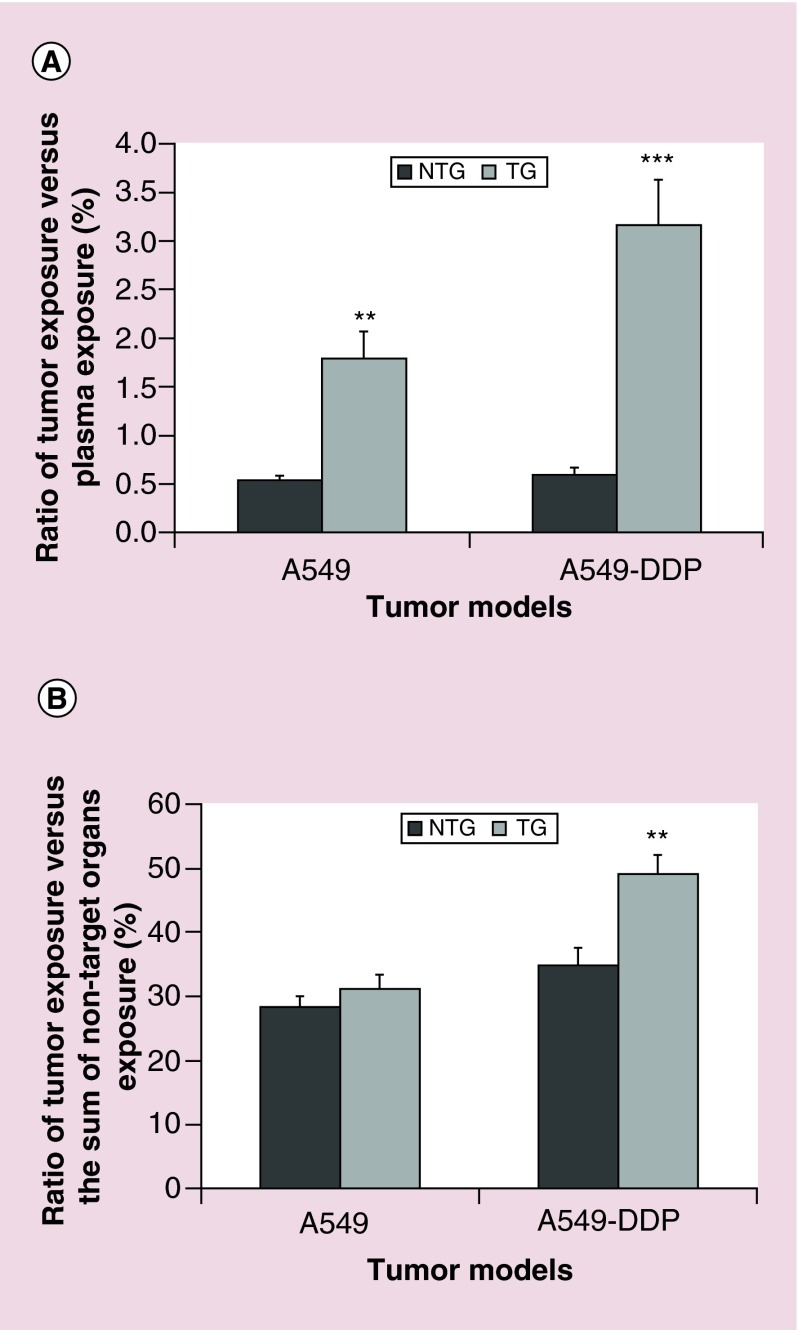
Since not only tumor exposure but also other main organs exposure was increased by the presence of targeting peptide, especially in A549-WT; we calculated a new parameter, the targeting efficiency (TE). With this parameter we wanted to check if the presence of the peptide in the TG nanoparticles was able to increase tumor exposure, in other words, the ability of the particle to deliver the siRNA preferably to the tumor than to other tissues. TE was calculated as the ratio of tumor exposure versus plasma exposure or nontarget organs exposure. In all conditions, plasma was far more exposed than tumor, the ratio reaching a maximum of 3.16% (Figure 6A). However, TE was significantly increased by the targeting strategy in both models, a 3.4-fold increase in A549-WT and 5.5 in A549-DDP model. It is apparent that the presence of the peptide in the TG nanoparticles has assisted the process of concentrating them into the cancer tissue in comparison to the NTG nanoparticles. We should also keep in mind that both nanoparticles are PEGylated, which by itself promotes tumor accumulation based on the EPR effect [34]. Many studies have associated PEG with greater tumor uptake and longer circulation time [62,63]. Besides, PEG contributes to increased particle hydrophilicity, stability, avoidance of plasma protein identification and escape from opsonization and clearance [64].
Figure 6. . Targeting efficiency.
Ratio (expressed in%) + SE of tumor exposure versus plasma exposure (A) or versus the sum of nontarget organs exposure (liver, kidney, spleen, heart, lung) for A549-WT and A549-DDP tumor models, after IV injection of NTG and TG nanoparticles at 3 mg/kg equivalent siRNA.
n = 5 mice.
*p < 0.05; **p < 0.01; ***p < 0.001 (t-test TG vs NTG).
DDP: Cisplatin; NTG: Nontargeted; SE: standard error; TG: Targeted.
When comparing tumor exposure to the sum of other organs exposure (Figure 6B), we observed that TE was high, with tumor exposure ranging between 28.3 and 49.0% of the global organ exposure. TE was increased by the targeting strategy in both models, increasing from 28.3 to 31.0% in A549 model, and from 34.8 to 49.0% in A549-DDP model. However, the difference was statistically significant only in the A549-DDP model. These results confirm the influence of EGFR-peptide targeted nanoparticles to target the cancer cells, which is in agreement with previous studies from our group where the same peptide was successfully used [40,65,66].
Based only on the imaging data, the targeting effect could not be clearly detected from the whole body and ex vivo images. These comes from the fact that near-IR images are not sensitive enough to allow for visualization of the difference that was later detected from the quantitative data. Based on the siMad2 quantification study, we could conclude that the presence of the targeting peptide leads to a significant decrease in plasma exposure of targeted nanoparticles and a higher targeting efficiency, in other words, a higher tumor exposure to Mad2 siRNA associated to no or a moderate increase of off-target organs exposure. The advantage of the targeting strategy was particularly clear for the A549-DDP model, with a higher increase of tumor exposure and targeting efficiency, probably due in some part to its higher EGFR cell surface expression levels.
Conclusion
The imaging and distribution studies together with the pharmacokinetic parameters showed that the tested CS formulations were efficient in delivering siMad2 to the tumor tissue in both models. In both tumor models, the addition of the targeting peptide resulted in a higher clearance, reflecting a faster elimination from the organism, leading to decreased plasma concentrations. Since tumor exposure was simultaneously increased, the targeting strategy proved efficient in both models to favor the entry of the siRNA inside the tumor. However, the entry of the siRNA was also increased in other tissues. Calculating the ratio of tumor exposure versus nontarget organs exposure, we proved that the tumor targeting efficiency was enhanced by the EGFR peptide, giving a specific advantage for delivery to the tumor when compared with other tissues, particularly in A549-DDP model. With an increased tumor targeting efficiency demonstrated in vivo and an efficient induction of apoptosis shown in vitro, our targeted delivery strategy proved to be a promising approach for NSCLC therapy in vivo [14].
Executive summary.
We have developed an EGF receptor (EGFR) targeted chitosan (CS) nanoparticle system for efficient delivery of siRNA targeted against mitotic checkpoint gene mad2.
Pharmacokinetics and biodistribution was assessed in A549-WT and cisplatin resistant A549-DDP tumor-bearing mice after intravenous delivery of 3 mg/kg dose of siMad2 encapsulated in nontargeted (NTG) or targeted (TG) CS nanoparticles.
Qualitative biodistribution profile based on near-IR labeled CS nanoparticles showed a long-lasting exposure of the tumor (up to 96 h) to the nanoparticles, independent of the tumor model or targeting property.
A quantitative pharmacokinetic study was performed based on siMad2 quantification using an antiprimer quenching based PCR method.
Noncompartmental analysis for both tumor models showed that the presence of targeting peptide leads to a significant decrease in plasma exposure of targeted nanoparticles.
The targeting strategy led to a higher targeting efficiency, in other words, a higher tumor exposure to Mad2 siRNA associated to no or a moderate increase of off-target organs exposure. The advantage of the targeting strategy was particularly clear for the A549-DDP model, with a higher increase of tumor exposure and targeting efficiency.
We conclude that our targeted siRNA delivery strategy is a promising approach to efficiently treat NSCLC tumors in vivo.
Supplementary Material
Footnotes
Financial & competing interests disclosure
This study was supported by a grant from the National Cancer Institute's (NCI) Alliance for Nanotechnology in Cancer Platform Partnership (CNPP) grant CA-151452 and an NCI R21 grant CA-179652. This work was also partially supported by CESPU under the project 02-GCQF-CICS- 2011N. AV Nascimento greatly appreciates Fundação para a Ciência e Tecnologia (FCT), Portugal for the PhD Fellowship Grant #: SFRH/BD/69271/2010. This work was financed by the European Regional Development Fund (ERDF) through the Programa Operacional Factores de Competitividade – COMPETE, by Portuguese funds through FCT in the framework of the project PEst-C/SAU/LA0002/2013, and co-financed by the North Portugal Regional Operational Programme (ON.2 – O Novo Norte) in the framework of project SAESCTN-PIIC&DT/2011, under the National Strategic Reference Framework (NSRF). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J. Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Islam KM, Jiang X, Anggondowati T, Lin G, Ganti AK. Comorbidity and survival in lung cancer patients. Cancer Epidemiol. Biomarkers Prev. 2015;24(7):1079–1085. doi: 10.1158/1055-9965.EPI-15-0036. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Solomides C, Parekh H, Simpkins F, Simpkins H. Cisplatin resistance in human cervical, ovarian and lung cancer cells. Cancer Chemother. Pharmacol. 2015;75(6):1217–1227. doi: 10.1007/s00280-015-2739-2. [DOI] [PubMed] [Google Scholar]
- 4.Ganguly A, Cabral F. New insights into mechanisms of resistance to microtubule inhibitors. Biochim. Biophys. Acta. 2011;1816(2):164–171. doi: 10.1016/j.bbcan.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivera E, Cianfrocca M. Overview of neuropathy associated with taxanes for the treatment of metastatic breast cancer. Cancer Chemother. Pharmacol. 2015;75(4):659–670. doi: 10.1007/s00280-014-2607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swain SM, Arezzo JC. Neuropathy associated with microtubule inhibitors: diagnosis, incidence, and management. Clin. Adv. Hematol. Oncol. 2008;6(6):455–467. [PubMed] [Google Scholar]
- 7.Huang M, Shen A, Ding J, Geng M. Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol. Sci. 2014;35(1):41–50. doi: 10.1016/j.tips.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Silva P, Barbosa J, Nascimento AV, Faria J, Reis R, Bousbaa H. Monitoring the fidelity of mitotic chromosome segregation by the spindle assembly checkpoint. Cell Prolif. 2011;44(5):391–400. doi: 10.1111/j.1365-2184.2011.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbosa J, Nascimento AV, Faria J, Silva P, Bousbaa H. The spindle assembly checkpoint: perspectives in tumorigenesis and cancer therapy. Front. Biol. 2011;6(2):147–155. [Google Scholar]
- 10.Kato T, Daigo Y, Aragaki M, et al. Overexpression of MAD2 predicts clinical outcome in primary lung cancer patients. Lung Cancer. 2011;74(1):124–131. doi: 10.1016/j.lungcan.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Teixeira JH, Silva P, Faria J, et al. Clinicopathologic significance of BubR1 and Mad2 overexpression in oral cancer. Oral Dis. 2015;21(6):713–720. doi: 10.1111/odi.12335. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y, Choi JW, Lee JH, Kim YS. MAD2 and CDC20 are upregulated in high-grade squamous intraepithelial lesions and squamous cell carcinomas of the uterine cervix. Int. J. Gynecol. Pathol. 2014;33(5):517–523. doi: 10.1097/PGP.0000000000000082. [DOI] [PubMed] [Google Scholar]
- 13.Choi JW, Kim Y, Lee JH, Kim YS. High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch. 2013;463(5):681–687. doi: 10.1007/s00428-013-1473-6. [DOI] [PubMed] [Google Scholar]
- 14.Nascimento AV, Singh A, Bousbaa H, Ferreira D, Sarmento B, Amiji MM. Mad2 checkpoint gene silencing using epidermal growth factor receptor-targeted chitosan nanoparticles in non-small cell lung cancer model. Mol. Pharm. 2014;11(10):3515–3527. doi: 10.1021/mp5002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgess A, Rasouli M, Rogers S. Stressing mitosis to death. Front. Oncol. 2014;4:140. doi: 10.3389/fonc.2014.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel L, Diaz-Rodriguez E, Narayan G, Hernando E, Murty VV, Benezra R. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc. Natl Acad. Sci. USA. 2004;101(13):4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borna H, Imani S, Iman M, Azimzadeh Jamalkandi S. Therapeutic face of RNAi: in vivo challenges. Expert Opin. Biol. Ther. 2015;15(2):269–285. doi: 10.1517/14712598.2015.983070. [DOI] [PubMed] [Google Scholar]
- 18.Haussecker D. Current issues of RNAi therapeutics delivery and development. J. Control. Release. 2014;195:49–54. doi: 10.1016/j.jconrel.2014.07.056. [DOI] [PubMed] [Google Scholar]
- 19.Chaturvedi K, Ganguly K, Kulkarni AR, et al. Cyclodextrin-based siRNA delivery nanocarriers: a state-of-the-art review. Expert Opin. Drug Deliv. 2011;8(11):1455–1468. doi: 10.1517/17425247.2011.610790. [DOI] [PubMed] [Google Scholar]
- 20.Eloy JO, Petrilli R, Lopez RF, Lee RJ. ‘Stimuli-responsive nanoparticles for siRNA delivery’. Curr. Pharm. Des. 2015;21(29):4131–4144. doi: 10.2174/1381612821666150901095349. [DOI] [PubMed] [Google Scholar]
- 21.Hattori Y, Nakamura A, Arai S, Kawano K, Maitani Y, Yonemochi E. siRNA delivery to lung-metastasized tumor by systemic injection with cationic liposomes. J. Liposome Res. 2015;25(4):1–8. doi: 10.3109/08982104.2014.992024. [DOI] [PubMed] [Google Scholar]
- 22.Kim MJ, Park JS, Lee SJ, et al. Notch1 targeting siRNA delivery nanoparticles for rheumatoid arthritis therapy. J. Control. Release. 2015;216:140–148. doi: 10.1016/j.jconrel.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 23.Ngamcherdtrakul W, Morry J, Gu S, et al. Cationic polymer modified mesoporous silica nanoparticles for targeted SiRNA delivery to HER2+ breast cancer. Adv. Funct. Mater. 2015;25(18):2646–2659. doi: 10.1002/adfm.201404629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yhee JY, Song S, Lee SJ, et al. Cancer-targeted MDR-1 siRNA delivery using self-cross-linked glycol chitosan nanoparticles to overcome drug resistance. J. Control. Release. 2015;198:1–9. doi: 10.1016/j.jconrel.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Li H, Wu R, et al. Antitumor effects of oncolytic adenovirus-carrying siRNA targeting potential oncogene EphA3. PLoS ONE. 2015;10(5):e0126726. doi: 10.1371/journal.pone.0126726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrade F, Antunes F, Nascimento AV, et al. Chitosan formulations as carriers for therapeutic proteins. Curr. Drug Disc. Technol. 2011;8(3):157–172. doi: 10.2174/157016311796799035. [DOI] [PubMed] [Google Scholar]
- 27.Sarmento B, Das Neves J. Chitosan-Based Systems For Biopharmaceuticals: Delivery, Targeting And Polymer Therapeutics. John Wiley & Sons; 2012. [Google Scholar]
- 28.Croisier F, Jérôme C. Chitosan-based biomaterials for tissue engineering. Eur. Polym. J. 2013;49(4):780–792. [Google Scholar]
- 29.Smith J, Wood E, Dornish M. Effect of chitosan on epithelial cell tight junctions. Pharm. Res. 2004;21(1):43–49. doi: 10.1023/b:pham.0000012150.60180.e3. [DOI] [PubMed] [Google Scholar]
- 30.Rai M. College Rutgers University. CABI; 2011. Natural antimicrobials in food safety and quality. [Google Scholar]
- 31.Kim S-K. Chitin And Chitosan Derivatives: Advances In Drug Discovery And Developments. CRC Press; USA, FL: 2013. [Google Scholar]
- 32.Lai WF, Lin MC. Nucleic acid delivery with chitosan and its derivatives. J. Control. Release. 2009;134(3):158–168. doi: 10.1016/j.jconrel.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Nehoff H, Parayath NN, Domanovitch L, Taurin S, Greish K. Nanomedicine for drug targeting: strategies beyond the enhanced permeability and retention effect. Int. J. Nanomed. 2014;9:2539–2555. doi: 10.2147/IJN.S47129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabhakar U, Maeda H, Jain RK, et al. Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Res. 2013;73(8):2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong Y, Meng F, Deng C, Zhong Z. Ligand-directed active tumor-targeting polymeric nanoparticles for cancer chemotherapy. Biomacromolecules. 2014;15(6):1955–1969. doi: 10.1021/bm5003009. [DOI] [PubMed] [Google Scholar]
- 36.Gaber R, Watermann I, Kugler C, et al. Correlation of EGFR expression, gene copy number and clinicopathological status in NSCLC. Diagn. Pathol. 2014;9:165. doi: 10.1186/s13000-014-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirsch FR, Scagliotti GV, Langer CJ, Varella-Garcia M, Franklin WA. Epidermal growth factor family of receptors in preneoplasia and lung cancer: perspectives for targeted therapies. Lung Cancer. 2003;41(Suppl. 1):S29–S42. doi: 10.1016/s0169-5002(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 38.Ganta S, Singh A, Patel NR, et al. Development of EGFR-targeted nanoemulsion for imaging and novel platinum therapy of ovarian cancer. Pharm. Res. 2014;31(9):2490–2502. doi: 10.1007/s11095-014-1345-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milane L, Duan Z, Amiji M. Therapeutic efficacy and safety of paclitaxel/lonidamine loaded EGFR-targeted nanoparticles for the treatment of multi-drug resistant cancer. PLoS ONE. 2011;6(9):e24075. doi: 10.1371/journal.pone.0024075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh A, Xu J, Mattheolabakis G, Amiji M. EGFR-targeted gelatin nanoparticles for systemic administration of gemcitabine in an orthotopic pancreatic cancer model. Nanomedicine. 2015 doi: 10.1016/j.nano.2015.11.010. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, Gattacceca F, Amiji M. Biodistribution and pharmacokinetics of EGFR-targeted thiolated gelatin nanoparticles following systemic administration in pancreatic tumor-bearing mice. Mol. Pharm. 2013;10(5):2031–2044. doi: 10.1021/mp400054e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusnak DW, Alligood KJ, Mullin RJ, et al. Assessment of epidermal growth factor receptor (EGFR, ErbB1) and HER2 (ErbB2) protein expression levels and response to lapatinib (Tykerb, GW572016) in an expanded panel of human normal and tumour cell lines. Cell Prolif. 2007;40(4):580–594. doi: 10.1111/j.1365-2184.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schafer A, Pahnke A, Schaffert D, et al. Disconnecting the yin and yang relation of epidermal growth factor receptor (EGFR)-mediated delivery: a fully synthetic, EGFR-targeted gene transfer system avoiding receptor activation. Hum. Gene Ther. 2011;22(12):1463–1473. doi: 10.1089/hum.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Zhao R, Wu X, et al. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J. 2005;19(14):1978–1985. doi: 10.1096/fj.05-4058com. [DOI] [PubMed] [Google Scholar]
- 45.Turner PV, Brabb T, Pekow C, Vasbinder MA. Administration of substances to laboratory animals: routes of administration and factors to consider. J. Am. Assoc. Lab. Anim. Sci. 2011;50(5):600. [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Makrigiorgos GM. Anti-primer quenching-based real-time PCR for simplex or multiplex DNA quantification and single-nucleotide polymorphism genotyping. Nat. Protoc. 2007;2(1):50–58. doi: 10.1038/nprot.2007.11. [DOI] [PubMed] [Google Scholar]
- 47.Ganesh S, Iyer AK, Gattacceca F, Morrissey DV, Amiji MM. In vivo biodistribution of siRNA and cisplatin administered using CD44-targeted hyaluronic acid nanoparticles. J. Control. Release. 2013;172(3):699–706. doi: 10.1016/j.jconrel.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holder DJ. Comments on Nedelman and Jia's extension of Satterthwaite's approximation applied to pharmacokinetics. J. Biopharm. Stat. 2001;11(1–2):75–79. doi: 10.1081/bip-100104199. [DOI] [PubMed] [Google Scholar]
- 49.Nedelman JR, Jia X. An extension of Satterthwaite's approximation applied to pharmacokinetics. J. Biopharm. Stat. 1998;8(2):317–328. doi: 10.1080/10543409808835241. [DOI] [PubMed] [Google Scholar]
- 50.Kean T, Thanou M. Biodegradation, biodistribution and toxicity of chitosan. Adv. Drug Del. Rev. 2010;62(1):3–11. doi: 10.1016/j.addr.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 2008;5(4):505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao J, Dragulescu-Andrasi A, Yao H. Fluorescence imaging in vivo: recent advances. Curr. Opin. Biotechnol. 2007;18(1):17–25. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Bloch S, Akers W, Achilefu S. Near-infrared molecular probes for in vivo imaging. Curr. Protoc. Cytom. 2012;12(Unit 12):27. doi: 10.1002/0471142956.cy1227s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi X, Wang F, Qin W, Yang X, Yuan J. Near-infrared fluorescent probes in cancer imaging and therapy: an emerging field. Int. J. Nanomedicine. 2014;9:1347–1365. doi: 10.2147/IJN.S60206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo S, Zhang E, Su Y, Cheng T, Shi C. A review of NIR dyes in cancer targeting and imaging. Biomaterials. 2011;32(29):7127–7138. doi: 10.1016/j.biomaterials.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 56.Qi L, Xu Z. In vivo antitumor activity of chitosan nanoparticles. Bioorg. Med. Chem. Lett. 2006;16(16):4243–4245. doi: 10.1016/j.bmcl.2006.05.078. [DOI] [PubMed] [Google Scholar]
- 57.Jayakumar R, Muzzarelli R, Prabaharan M. Chitosan for Biomaterials I. Springer-Verlag; Berlin, Heidelberg, Germany: 2011. [Google Scholar]
- 58.Wang J, Sui M, Fan W. Nanoparticles for tumor targeted therapies and their pharmacokinetics. Curr. Drug Metab. 2010;11(2):129–141. doi: 10.2174/138920010791110827. [DOI] [PubMed] [Google Scholar]
- 59.Leucuta SE. Subcellular drug targeting, pharmacokinetics and bioavailability. J. Drug Target. 2014;22(2):95–115. doi: 10.3109/1061186X.2013.848453. [DOI] [PubMed] [Google Scholar]
- 60.Yamashita F, Hashida M. Pharmacokinetic considerations for targeted drug delivery. Adv. Drug Deliv. Rev. 2013;65(1):139–147. doi: 10.1016/j.addr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 61.Cabral H, Matsumoto Y, Mizuno K, et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol. 2011;6(12):815–823. doi: 10.1038/nnano.2011.166. [DOI] [PubMed] [Google Scholar]
- 62.Choi KY, Min KH, Yoon HY, et al. PEGylation of hyaluronic acid nanoparticles improves tumor targetability in vivo . Biomaterials. 2011;32(7):1880–1889. doi: 10.1016/j.biomaterials.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Tobin LA, Xie Y, Tsokos M, et al. Pegylated siRNA-loaded calcium phosphate nanoparticle-driven amplification of cancer cell internalization in vivo . Biomaterials. 2013;34(12):2980–2990. doi: 10.1016/j.biomaterials.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen S, Yang K, Tuguntaev RG, et al. Targeting tumor microenvironment with PEG-based amphiphilic nanoparticles to overcome chemoresistance. Nanomed. Nanotechnol. Biol. Med. 2015 doi: 10.1016/j.nano.2015.10.020. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganta S, Singh A, Kulkarni P, et al. EGFR targeted theranostic nanoemulsion for image-guided ovarian cancer therapy. Pharm. Res. 2015;32(8):2753–2763. doi: 10.1007/s11095-015-1660-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Talekar M, Trivedi M, Shah P, et al. Combination wt-p53 and MicroRNA-125b transfection in a genetically-engineered lung cancer model using dual CD44/EGFR targeting nanoparticles. Mol. Ther. 2015 doi: 10.1038/mt.2015.225. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.