Abstract
In the RV144 vaccine trial, two antibody responses were found to correlate with HIV-1 acquisition. Because human leukocyte antigen (HLA) class II–restricted CD4+ T cells are involved in antibody production, we tested whether HLA class II genotypes affected HIV-1–specific antibody levels and HIV-1 acquisition in 760 individuals. Indeed, antibody responses correlated with acquisition only in the presence of single host HLA alleles. Envelope (Env)–specific immunoglobulin A (IgA) antibodies were associated with increased risk of acquisition specifically in individuals with DQB1*06. IgG antibody responses to Env amino acid positions 120 to 204 were higher and were associated with decreased risk of acquisition and increased vaccine efficacy only in the presence of DPB1*13. Screening IgG responses to overlapping peptides spanning Env 120–204 and viral sequence analysis of infected individuals defined differences in vaccine response that were associated with the presence of DPB1*13 and could be responsible for the protection observed. Overall, the underlying genetic findings indicate that HLA class II modulated the quantity, quality, and efficacy of antibody responses in the RV144 trial.
INTRODUCTION
The RV144 phase 3 vaccine trial conducted in Thailand resulted in an estimated 31.2% vaccine efficacy in the prevention of HIV-1 infection at 42 months after the initiation of vaccination (1, 2). A follow-up study identified two vaccine-induced immune responses that were associated with HIV-1 acquisition: high levels of immunoglobulin A (IgA) antibodies to HIV-1 envelope (Env) were associated with increased risk of infection, and high levels of IgG antibodies to Env amino acids 120 to 204 (reference sequence HXB2) were associated with decreased risk of infection (3). The IgA antibody response was a composite score of purified IgA binding to 14 Env gp120 and gp140 proteins from multiple subtypes, while the IgG response was binding to scaffolded Env comprising the variable 1 and 2 (V1 and V2) domains flanked by partial regions of the first and second conserved (C1 and C2) domains (4).
Several additional studies have investigated the mechanism of these two correlates of risk (5). IgA antibodies to the C1 region have been implicated in blocking antibody-dependent cellular cytotoxicity (ADCC) in RV144 vaccine recipients (6). Env (120–204)–specific IgG responses were mainly attributed to the V2 region; molecular sieve analysis identified amino acid residues in V2 under vaccine-induced immune pressure, and several monoclonal antibodies were isolated from vaccinees that bind to this region of Env (7, 8). Moreover, V2-specific IgG3 antibodies and associated nonneutralizing effector functions supported the role of V2 in the RV144 protective immune response (9, 10), and Env-specific CD4+ T cells directed against V2 were identified as the most common T cell response after vaccination (11).
Human leukocyte antigen (HLA) class II molecules (DR, DQ, and DP) found on the surface of antigen-presenting cells present foreign extracellular peptides to CD4+ T cells, which then induce B cells to produce antibodies. Several HLA class II genes encode these molecules, but polymorphisms in the DRB1, DQB1, and DPB1 genes are primarily responsible for enabling variable binding to different antigenic epitopes within the peptide-binding groove of the HLA class II molecule. These genes are highly polymorphic, and this variation can influence humoral immune responses. For example, several HLA alleles and haplotypes have been shown to be associated with humoral responses induced by vaccination: DRB1*03 has been implicated in nonresponse to vaccination with hepatitis B surface antigen (12, 13), the presence of DPB1*05 has been associated with increased magnitude of IgG responses to a malaria sporozoite vaccine (14), and individuals with DRB1*07-DQB1*03-DPB1*04 and DRB1*04-DQB1*03-DPB1*03 haplotypes had lower levels of measles and rubella virus–specific IgG antibody titers, respectively (15). Furthermore, lack of neutralizing antibody responses to the combined prime-boost HIV-1 vaccine used in RV144 has been attributed to certain HLA class II alleles in a phase 2 trial in Thailand that preceded RV144 (16). It is thus likely that the differences in vaccine-induced immune responses observed in the RV144 study could be due to the variation in HLA class II genes between individuals. The present study sought to identify whether specific DRB1, DQB1, or DPB1 alleles enhanced or reduced the effect of Env-specific IgA and Env (120–204)–specific IgG antibodies on HIV-1 acquisition.
RESULTS
Interaction of Env-specific IgA and DQB1*06 increases risk of acquisition
Thirty-one HLA class II alleles observed in the cohort with an allele frequency greater than 5% were investigated. High Env-specific IgA levels correlated with increased risk of HIV-1 acquisition only in the presence of DQB1*06 (P = 0.002, q = 0.11) (Fig. 1 and table S1). The impact of DQB1*06 on the association of vaccine-induced Env-specific IgA levels with HIV-1 acquisition was apparent when antibody responses of vaccinated individuals were stratified by HLA-DQB1 type (Fig. 1A). Anti-Env IgA levels were higher in HIV-1–infected compared to uninfected individuals only in the presence of DQB1*06. Higher levels of Env-specific IgA correlated with increased risk of HIV-1 infection only in the presence of DQB1*06 [odds ratio (OR), 14.51 per 1-SD increase; P = 0.002]. No effect of anti-Env IgA level was observed in the absence of DQB1*06 (OR, 1.07 per 1-SD increase; P = 0.74). Vaccinated individuals were further stratified into subgroups according to their IgA responses (low or medium versus high) corresponding to the lower two-thirds and upper one-third of responses at week 26 (3) and the presence or absence of DQB1*06. Cumulative incidence of HIV-1 infection over time showed that individuals with high levels of anti-Env IgA and DQB1*06 had accelerated time to HIV-1 infection compared to any other group (Fig. 1B).
Fig. 1. High Env-specific IgA (IgA) levels are associated with increased risk of HIV-1 acquisition only in the presence of HLA-DQB1*06.
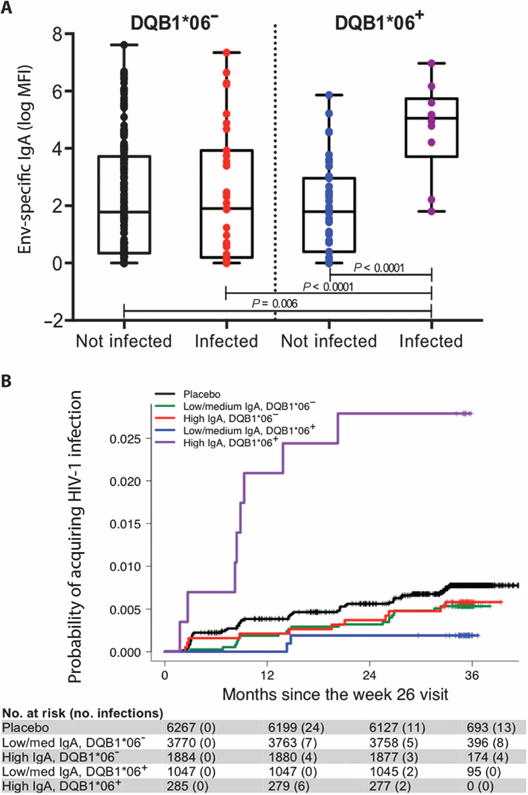
(A) Box plots of vaccinated individuals stratified according to HIV-1 infection status and the absence or presence of DQB1*06. Individual data points are indicated by colored circles within the box plots showing the 25th (bottom edge of the box), 50th (horizontal line in the box), and 75th percentiles (top edge of the box). Analysis of variance tested IgA distributions across subsets with differences further interrogated using Bonferroni-adjusted t tests. MFI, mean fluorescence intensity. (B) Estimated cumulative HIV-1 incidence curves for individuals stratified by IgA and DQB1*06 for the entire vaccinated RV144 cohort. Vaccinated individuals in the case-control study are stratified into subgroups according to their IgA responses (low/medium and high, corresponding to the lower two-thirds and upper one-third of responses) at week 26 and the absence or presence of DQB1*06. Individual curves represent the estimated cumulative incidence of HIV-1 infection over time since the measurement of IgA at week 26. Curves were estimated using the Kaplan-Meier method with inverse probability weighting accounting for the sampling design.
Env (120–204)–specific IgG responses are higher in individuals with DPB1*13
We also tested whether HLA class II alleles correlated with levels of HIV-1– specific antibody by linear regression. No class II allele was associated with Env-specific IgA levels, but the presence of DPB1*13 directly correlated with higher levels of anti–Env (120–204) IgG (P = 0.002, q = 0.09) (Fig. 2A and table S2). HLA-DPB1*13 was the only class II allele to show any interaction with vaccine-induced anti–Env (120–204) IgG levels and affect HIV-1 acquisition (P = 0.01, q = 0.26) (table S1). The protective effect of host DPB1*13 allele on HIV-1 acquisition conferred by vaccine-induced Env (120–204)–specific IgG responses was apparent when the vaccinated individuals’ antibody responses were plotted, stratified by HLA-DPB1 type (Fig. 2B). Higher levels of Env (120–204)–specific IgG correlated with decreased risk of HIV-1 infection only in the presence of DPB1*13 (OR, 0.29 per 1-SD increase; P = 0.006). Individuals with both higher levels of Env (120–204)–specific IgG and at least one DPB1*13 allele had the lowest HIV-1 incidence compared to other groups (Fig. 2C). Vaccine efficacy was estimated to be 71% for the group consisting of individuals with higher Env (120–204)–specific IgG levels and the DPB1*13 allele relative to the placebos (Fig. 2D).
Fig. 2. Env (120–204)–specific IgG is associated with the presence of HLA-DPB1*13 and is protective.
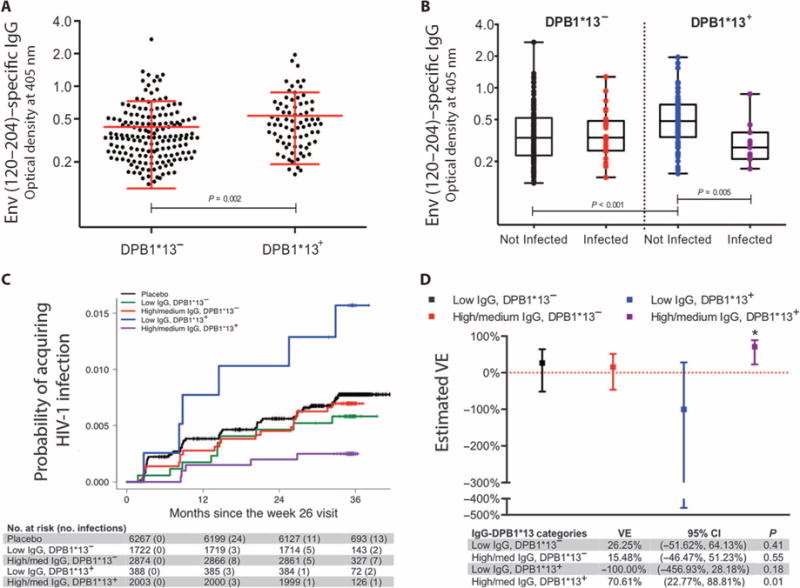
(A) Distribution of IgG stratified by absence or presence of DPB1*13. Red lines represent the mean and SD for the change in the measurement of IgG from absence to presence of DPB1*13. Means were compared using t tests. Individual data points are indicated by black circles. (B) Box plots of vaccinated individuals stratified according to HIV-1 infection status and the absence or presence of DPB1*13. Individual data points are indicated by colored circles within the box plots showing the 25th (bottom edge of the box), 50th (horizontal line in the box), and 75th percentiles (top edge of the box). Analysis of variance tested IgG distributions across subsets with differences further interrogated using Bonferroni-adjusted t tests. (C) Estimated cumulative HIV-1 incidence curves for individuals stratified by IgG and DPB1*13 for the entire vaccinated RV144 cohort. Vaccinated individuals in the case-control study are stratified into subgroups according to their IgG responses (low and high/medium corresponding to the lower one-third and upper two-thirds of responses) at week 26 and the absence or presence of DPB1*13. Individual curves represent the estimated cumulative incidence of HIV-1 infection over time. Curves were estimated using the Kaplan-Meier method with inverse probability weighting accounting for the sampling design. (D) Estimated vaccine efficacy (VE) for IgG and DPB1*13. Individuals were stratified into subgroups according to IgG response (low and high/medium, corresponding to the lower one-third and upper two-thirds of responses) and the absence or presence of DPB1*13 and were compared to the entire placebo-infected group. Points show the estimated VE values, and lines represent 95% confidence intervals (CI) that were estimated using logistic regression models that accounted for the sampling design.
Env (120–204)–specific IgG responses with DPB1*13 are not HIV subtype–specific
As a secondary analysis, we further investigated the gp120-specific IgG response associated with protection against HIV-1 acquisition in the presence of DPB1*13. The scaffolded Env antigen (positions 120 to 204) used to identify this antibody response as a correlate of risk in the Haynes et al. study (3) was derived from a subtype B–infected individual (4). We tested IgG responses for reactivity with several other Env (120–204) antigens from different HIV-1 group subtypes (A, B, C, or CRF01_AE) and a smaller region comprising only the V2 domain positions 160 to 183. For all the Env (120–204) and most of the V2 (160–183) antigens from varying subtypes, a higher level of IgG responses associated significantly with the presence of DPB1*13 (table S3). However, these antibody responses were not equally effective. Anti–Env (120–204) IgG responses to all subtypes were associated with decreased risk of HIV-1 acquisition in the presence of DPB1*13 (OR, 0.07 to 0.31; P = 0.006 to 0.02). In contrast, no anti-V2 IgG responses were associated with decreased risk of HIV-1 acquisition (P > 0.7).
IgG responses with DPB1*13 to the N terminus of Env (120–204) are protective
To further define the region responsible for conferring protection, Env-specific plasma IgG responses were assessed by overlapping peptides (15-mers overlapping by 12 amino acids) spanning the Env (120–204) region. Responses to 181 peptides from multiple HIV-1 subtypes were tested in all vaccinated individuals. For five of these epitopes, an IgG response was observed to be significantly associated with the presence of DPB1*13: 116–130 (P = 0.04), 119–133 (P = 0.01), 163–177 (P = 0.02), 166–180 (P = 0.04), and 169–183 (P = 0.01) (table S4). Of these epitopes, only peptide 119–133 demonstrated a significantly greater frequency of response in uninfected compared to infected individuals with DPB1*13 allele (Fig. 3A). The magnitude of IgG responses was also interrogated as a continuous variable. In a comparison between infected and uninfected DPB1*13 positive individuals, a greater magnitude of IgG response to peptide 119–133 was associated with protection from HIV-1 acquisition (Fig. 3B). Peptide 119–133 represents the N terminus of the Env (120–204) sequence, with 11 and 3 residues located in the C1 and V1 regions, respectively. Sequence alignments from different Env (120–204) antigens confirm that region 120–133 is fairly conserved between different viral subtypes (fig. S1).
Fig. 3. Frequency and magnitude of IgG responses to Env epitope (119–133) are associated with HIV-1 acquisition among DPB1*13 vaccinated individuals.
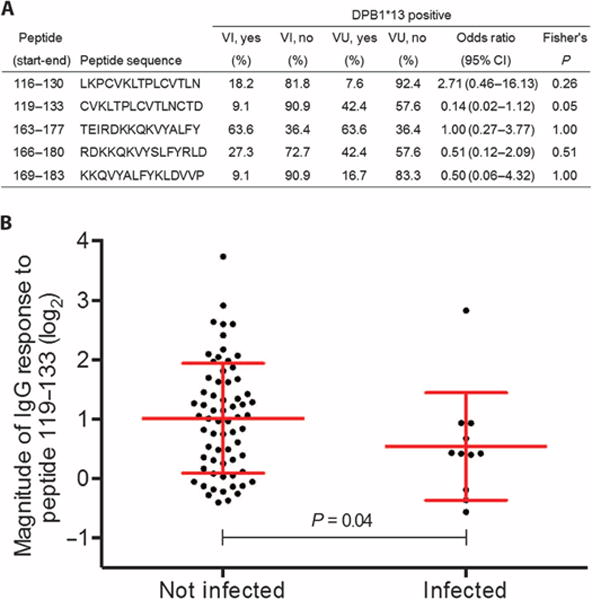
(A) Frequency of DPB1*13 restricted peptide responses comparing vaccinated infected (VI) and vaccinated uninfected (VU) individuals that have at least one DPB1*13 allele. Fisher’s exact test compared the frequency of antibody response between VU DPB1*13 and VI DPB1*13 individuals. (B) Red lines represent the mean and SD for the change in the magnitude in responses from absence to presence of DPB1*13. Means were compared using t tests.
Viral sequences differ in DPB1*13 positive vaccine recipients
That the host HLA type interacts with vaccine response to affect acquisition implies a functionally different immune response to vaccination in the presence versus absence of DPB1*13. This is supported by a sieve analysis of breakthrough Env-gp70 sequences from RV144-infected participants. Amino acid distributions in the vaccine and placebo groups were compared separately for subjects with or without the DPB1*13 allele. The following 15 sites were tested for their known function as contact residues and antibody epitopes (7): HXB2 amino acids 120, 124, 160, 165, 166, 168–173, 178, 179, 181, and 197 (table S5). Of these, position 173 differed significantly between vaccinees with or without DPB1*13. Histidine at position 173 was conserved in all eight DPB1*13 positive vaccinees but was found only in 15 of the 36 DPB1*13 negative vaccinees (P = 0.004) (Table 1). In contrast, histidine at 173 was present at a lower frequency in placebo recipients both with and without DPB1*13. Among subjects carrying DPB1*13, site 173 also distinguished vaccine and placebo recipients (P = 0.02), but there was no difference between vaccine and placebo recipients among subjects without DPB1*13.
Table 1.
Frequencies of amino acids at site 173 of Env-gp70 differ in breakthrough infections in the presence of the DPB1*13 allele.
| Vaccine (n = 44)
|
Fisher’s P | Placebo (n = 66)
|
Fisher’s P | |||
|---|---|---|---|---|---|---|
| DPB1*13+ | DPB1*13− | DPB1*13+ | DPB1*13− | |||
| H173 | 8 | 15 | 0.004 | 9 | 19 | 0.57 |
| H173X* | 0 | 21 | — | 9 | 29 | — |
|
| ||||||
|
DPB1*13+ (n = 26)
|
DPB1*13− (n = 84)
|
|||||
| H173 | H173X | H173 | H173X | |||
|
| ||||||
| Vaccine | 8 | 0 | 0.02 | 15 | 19 | 1.00 |
| Placebo | 9 | 9 | — | 21 | 29 | — |
X designates amino acid residues other than histidine (H) at position 173.
Interactions between HLA class II and vaccine-induced responses independently affect acquisition
Four immune variables that were part of the primary analysis in the immune correlate study, including IgG avidity, ADCC, neutralizing antibodies, and CD4+ T cell responses to Env, did not show associations with HIV-1 acquisition previously (3). We show that the two significant HLA interactions with Env-specific IgA (DQB1*06) and Env (120–204)–specific IgG (DPB1*13) remained significant and had independent effects on HIV-1 acquisition in a multivariable analysis, including all the other immune variables (Table 2). Our findings thus indicate an effect of the humoral immune response on HIV-1 acquisition in the presence of specific HLA class II alleles.
Table 2. Interactions of HLA class II alleles and vaccine-induced responses have an independent effect on HIV-1 acquisition.
ORs for HIV-1 acquisition in a multivariable analysis of all primary immune response variables from the immune correlate study (3) and the two significant HLA interactions observed in this study.
| Variable | Multivariable model*
|
|
|---|---|---|
| OR (95% CI) | P value | |
| Avidity of IgG antibodies for Env | 0.94 (0.56–1.57) | 0.80 |
| ADCC | 0.97 (0.62–1.51) | 0.89 |
| Neutralizing antibodies | 1.08 (0.63–1.85) | 0.78 |
| Env-specific CD4+ T cells | 1.07 (0.77–1.50) | 0.68 |
| Env-specific IgA-DQB1*06 interaction† | 8.24 (2.07–32.75) | 0.003 |
| Env (120–204)–specific IgG-DPB1*13 interaction† | 0.27 (0.09–0.80) | 0.02 |
For each primary immune variable (3), the OR is reported per 1-SD increase; sex, baseline behavioral risk score, and one significant principal component axis were included as covariates in the model. Env-specific IgA, DQB1*06, Env (120–204)–specific IgG, and DPB1*13 were also included as main effects.
Reported ORs and 95% CIs reflect the results from logistic regression analysis including a two-way interaction term for Env-specific IgA or Env (120–204)–specific IgG and the absence or presence of the given allele.
DISCUSSION
The RV144 vaccine produced two Env-specific antibody responses that correlated with HIV-1 acquisition (3). Because HLA class II molecules are important in the initiation of antibody responses, we hypothesized that variation in HLA class II genes might influence response to the vaccine. DRB1, DQB1, and DPB1 loci were typed in the RV144 vaccine recipients, and the alleles found were tested for an effect on magnitude of vaccine-induced responses and for interactions with the two vaccine-elicited immune responses that significantly affected HIV-1 acquisition. Both Env-specific antibody responses previously identified were found to correlate with HIV-1 acquisition only in the presence of specific HLA class II alleles.
Haynes et al. (3) observed that high levels of IgA antibody binding to Env correlated with reduced vaccine efficacy that was not distinguishable from the placebos in RV144. Here, we report that it is only in the presence of a single allele, DQB1*06, that high levels of Env-specific IgA antibodies are associated with HIV-1 infection. This finding remained even after normalization for total plasma IgA levels (OR, 4.06; P < 0.001). The effect of high anti-Env IgA levels in the presence of DQB1*06 on HIV-1 acquisition was stronger than that of high levels of anti-Env IgA antibodies alone. Env-specific IgA antibody levels did not differ between individuals with or without DQB1*06, suggesting that the presence of DQB1*06 influences the quality, not quantity, of anti-Env IgA antibody response to confer susceptibility to infection. The most parsimonious explanation for this finding is that differences in IgA isotypes or epitope specificity could be influenced by CD4+ T cell responses and may contribute to these qualitative changes. This is consistent with previous reports of class II alleles associating with both CD4+ T cell and specific antibody epitopes targeted by an immune response (16–18). Most of the 14 recombinant Env variables comprising the composite anti-Env IgA variable (3) interacted significantly with DQB1*06, showing that this finding was not restricted to the common Thai CRF01_AE viral subtype (table S6). The simplest interpretation of our data is that, in the presence of DQB1*06, anti-Env IgA antibodies are associated with increased risk of HIV-1 acquisition. This finding is of concern given the higher allele frequency of DQB1*06 in South Africa (32.4%) (19) than in Thailand (10.3%) (20), where an RV144-like regimen will be tested in a phase 2b HIV-1 efficacy trial (5).
Haynes et al. (3) also found that high IgG binding to the Env (120–204) B-subtype antigen correlated with decreased risk of HIV-1 acquisition in RV144 vaccinees compared with placebo recipients. We found that this association occurred only in the presence of a single host allele, DPB1*13, and identified a significant direct correlation between the presence of DPB1*13 and Env (120–204) IgG antibody levels. This association was replicated with Env antigens across multiple HIV-1 subtypes. IgG antibody responses to all these varying Env (120–204) sequences were also associated with decreased risk of HIV-1 acquisition. In addition to an increased quantity of IgG response to Env (120–204) in the presence of DPB1*13, we also determined differences in the specific Env antigens targeted. Analysis of IgG responses in 242 vaccinated individuals to overlapping peptides spanning the Env 120–204 region identified five linear epitopes to which a response was associated with the presence of DPB1*13. Of these, both the frequency and magnitude of IgG response to one epitope, Env positions 119 to 133, were associated with decreased risk of HIV-1 acquisition among the DPB1*13-vaccinated individuals. This antibody epitope overlaps with the C terminus of the C1 region and contains three residues from the V1 region. It contains the PLCV motif with side-chain and main-chain CD4 contact residues. Therefore, IgG binding to this epitope is likely to block Env interaction with CD4 (fig. S2) (21). This is further supported by the well-characterized monoclonal antibody (mAb) 17b that binds this epitope. Antibody 17b binds lysine at position 121, blocks chemokine co-receptor CCR5 binding (22, 23), and modifies the gp120 V1V2 loop structure to partially block the CD4 binding site (fig. S2) (22–24). Thus, only in the presence of DPB1*13 is high IgG response to Env (120–204) associated with reduced risk of HIV-1 acquisition. This could be due to the greater magnitude of IgG response observed in DPB1*13 positive vaccine recipients. It may also result from response to particular regions of the Env antigen in the presence of DPB1*13. Here, we defined a candidate in a linear epitope screen, but others such as conformational epitopes may also exist.
Further evidence that immune responses induced by vaccination in individuals carrying DPB1*13 are different from those without DPB1*13 was apparent in significant HIV-1 sequence differences between vaccine and placebo recipients when specifically subjects with DPB1*13 were considered (or when comparing vaccinees with or without DPB1*13). Only in the presence of both vaccine and DPB1*13 was a high frequency of H173 observed, suggesting that the vaccine responses in DPB1*13 positive individuals can select specific HIV-1 variants. That both the DQB1*06 and DPB1*13 alleles implicated in this study can mediate functional differences in immune response is also shown by their well-established associations with antibody response and autoimmune diseases (25, 26). Both DQB1*06 and DPB1*13 are associated with selective IgA deficiency (27–29). DQB1*06 is associated with several autoimmune diseases such as type 1 diabetes and narcolepsy (25, 30).
Growing awareness that host diversity may have contributed to the differences observed in vaccine-induced immune responses and efficacy in the RV144 study has generated increased interest in host genetic studies (20, 31–35). Data presented herein show clear differences in vaccine-induced immune responses among individuals having DQB1*06 and DPB1*13 alleles. These findings are significant because both of these alleles were identified as common with allele frequencies greater than 10% in 450 uninfected healthy controls from RV144 (20). Furthermore, the associations with HIV-1–specific antibodies are most likely vaccine-induced because there was no observed effect in the absence of vaccine (table S7) and none of the class II alleles had a direct association with HIV-1 acquisition (table S8).
The main limitation of this study is the relatively small sample size, with no possibility of replicating the genetic associations. This is due to the absence of other efficacious HIV-1 vaccine trials. Moreover, the secondary analyses were exploratory in nature, and the mechanisms postulated require confirmation in independent studies. Vaccine trials using the RV144 vaccine–based regimen, such as the one planned in southern Africa and Thailand, could address these limitations (5).
The RV144 vaccine trial has advanced understanding of HIV-1 vaccine–induced protective immune responses. The prime-boost vaccine showed an efficacy of 31.2% at 42 months after the beginning of the primary vaccination series (1). Vaccine efficacy was 58% among individuals with high levels of Env (120–204)–specific IgG antibodies and absent among individuals with high levels of anti-Env IgA antibodies (3). We further show that vaccine efficacy among individuals with high levels of Env (120–204)–specific IgG increased to 71% in conjunction with a specific class II allele. Thus, interactions of certain HLA class II genes with antibody responses to the RV144 vaccine regimen affect HIV-1 acquisition in a vaccine efficacy trial. The primary implication of these findings is that differences in vaccine-induced responses elicited by individuals with HLA-DPB1*13 should be further examined to determine the mechanism of protection of the vaccine. Understanding how variation in the host relates to vaccine-induced responses can help in the interpretation of vaccine efficacy studies and could prospectively improve vaccine design.
MATERIALS AND METHODS
Study design
We examined HLA class II genotypes in the 41 vaccinated HIV-1–infected cases and 205 matched vaccinated uninfected control subjects defined in the RV144 correlates of risk study. The study design, including selection of patients and immune response variables for this trial, has been previously described (1, 3). Briefly, the RV144 study was a community-based, randomized, multicenter, double-blind, placebo-controlled efficacy trial of the prime-boost combination of a vaccine containing ALVAC-HIV and AIDSVAX B/E (ClinicalTrials.gov number NCT00223080). For the correlates of risk study, vaccinated cases free of HIV-1 infection at week 24 of the vaccination period were selected for evaluation of immune responses at peak immunogenicity (week 26). Patients were stratified by sex, number of vaccinations received, and per-protocol status. For each HIV-1–infected case within a stratum, five vaccinated uninfected controls were randomly selected for comparison of immune responses. We also studied 517 placebo control samples that were administered placebo in the RV144 study. Placebo uninfected controls (n = 450) were selected for comparison to placebo infected (n = 67) cases using the same selection criteria described above for the vaccinated case-control cohort. The RV144 study was approved by the Human Subjects Research Review Board of the U.S. Army Medical Research and Material Command, the Thai Ministry of Public Health Ethical Committee, and the Institutional Review Boards of the Mahidol University and the Royal Thai Army. All individuals gave informed consent for participation in this study.
HLA genotyping
Genomic DNA was extracted and purified from human peripheral blood mononuclear cells using the QIAamp DNA Blood Mini Kit (Qiagen). DRB1 and DQB1 genotyping was performed using sequence-based typing (SBT) according to the International Histocompatibility Workshop Group protocol and as described previously (20, 36). Because SBT generated ambiguous genotypes for the DPB1 locus, next-generation sequencing was used for HLA typing (20, 36). Four-digit HLA typing was generated for DRB1, DQB1, and DPB1 loci for all samples. For the purposes of this paper, HLA typing data for all alleles with a population frequency (2N) greater than 5% were assessed. There were 18 alleles that had a frequency greater than 5% at the four-digit designation. To enable testing of more HLA alleles, we also included 13 alleles that met this criterion when combined to a two-digit designation. Four alleles at four-digit designations were identical at two-digit designations and thus were not included separately for analysis (DRB1*03:01/DRB1*03, DRB1*09:01/DRB1*09, DPB1*05:01/DPB1*05, and DPB1*13:01/DPB1*13). One individual was assigned as having a blank genotype for the DPB1 locus because DNA was unavailable for resolving ambiguity. A dominant genetic effect was assumed for HLA class II alleles.
Population stratification
A panel of 96 single nucleotide polymorphisms (SNPs) was selected to identify population stratification as described previously (35, 37). PLINK was used for data cleaning and quality control. No SNPs were excluded on the basis of missing data (call rate <0.985) or deviation from Hardy-Weinberg equilibrium. Two HIV-1–uninfected individuals with a high degree of missing data (call rate <0.95) were excluded from analysis. Two individuals, one HIV-1–infected and one uninfected, were identified as twins. To avoid losing power in the infected group, only the uninfected twin was excluded from analysis. Because genetic admixture has previously been observed in the RV144 cohort (35), population stratification was assessed through principal components analysis (PCA) using EIGENSTRAT (38). One significant axis of variation was identified through PCA analysis.
Statistical analysis
Primary objectives
For our primary analysis, to test whether the effect of Env-specific IgA or Env (120–204)–specific IgG on HIV-1 acquisition differed depending on the presence of an individual HLA class II allele, a two-way interaction term was included in univariate logistic regression models that accounted for the sampling design (39, 40). Sex, baseline self-reported behavioral risk category, and the one significant EIGENSTRAT axis identified through PCA were all included as covariates in the regression models. Taking into consideration the sample size, a minimal false discovery rate (q value) was used to correct for multiplicity testing of all individual alleles in two-way interaction analysis of both Env-specific IgA and Env (120–204)–specific IgG (41). A minimal false discovery rate is a conservative approach to multiplicity testing because it is presumed that all of the null hypotheses are true. A Holm-Bonferroni–adjusted P value (42) of less than 0.05 and a q value of less than 0.20 were considered statistically significant. To interpret a statistically significant two-way interaction identified through two-phase logistic regression models, the effect of the immune correlate of interest on HIV-1 acquisition was presented as a function of the given HLA class II allele (presence or absence) using linear regression models. Significant univariate interactions were also evaluated in a multivariable model including all original immune variables such as IgG avidity, ADCC, neutralizing antibodies, and CD4+ T cell responses to Env to identify independent associations (3). As an additional primary analysis, direct associations of HLA on Env-specific IgA or Env (120–204)–specific IgG were also compared using univariate linear regression models. A two-sided P value of less than 0.05 and a q value of less than 0.20 were used for considering statistical significance as described above.
Descriptive methods have been described previously (3). Briefly, immune correlate variables were modeled quantitatively and categorically based on the thirds of response (low, medium, and high subgroups). Quantitative variables were mean-centered and scaled to have an SD of 1. In the present descriptive analyses, the low and medium subgroups were collapsed together for Env-specific IgA-related analyses, and the high and medium subgroups were collapsed together for Env (120–204)–specific IgG-related analyses to increase statistical power in categorical subgroups stratified by presence or absence of a given allele. Box plots describe the distribution of each immune correlate for infection status (infected versus not infected) stratified by allele status. Analysis of variance was used to assess immune correlate distribution across subsets with differences further interrogated using Bonferroni-adjusted t tests. Cumulative HIV-1 incidence curves following peak immunogenicity (week 26) were plotted for different response subgroups in the presence or absence of an allele, as well as for the entire placebo group who were HIV-1 negative at the week 24 visit (n = 6267 subjects) for reference. Curves were estimated via the Kaplan-Meier method with inverse probability weighting accounting for the sampling design. Vaccine efficacy for response strata in the presence or absence of an allele in vaccine recipient subgroups versus the entire HIV-1–negative placebo group at the week 24 visit was estimated using logistic regression models that accounted for the sampling design.
Secondary analyses
As secondary analyses, alleles were also checked for any direct association on HIV-1 acquisition using the same logistic regression models as in the primary analysis. In further secondary analyses, binding antibody multiplex assays for IgG antibody response to seven Env (120–204) or Env-V2 (160–183) peptides were performed as described previously (43). All variables were tested for (i) the presence of antibody response associated with the allele of interest using the same univariate linear regression models as Env (120–204)–specific IgG, and (ii) the effect of this antibody response on HIV-1 acquisition using the same logistic regression models in two-way interaction analysis as the Env (120–204)–specific IgG variable.
Linear B cell epitopes from all vaccinated individuals in the study were identified using overlapping peptides of Env sequences from six HIV-1 group M subtypes as described previously (3). Env-specific plasma IgG was assessed with 181 overlapping peptides (15-mers overlapping by 12 amino acids) spanning the Env (120–204) region. Epitopes were identified using Fisher’s exact test by comparing the frequency of responses to each peptide in the presence and absence of an allele of interest. Significant epitopes were then tested using Fisher’s exact test for an effect on HIV-1 acquisition by comparing the frequency of antibody response between vaccinated infected and uninfected individuals with DPB1*13. Antibody responses to epitopes were also examined as a continuous variable, and the distribution of responses between infected and uninfected vaccinees with DPB1*13 allele were compared using the two-sample Kolmogorov-Smirnov test.
HIV-1 breakthrough infections in the RV144 trial were previously sequenced (7). An alignment of Env-gp70 sequences from subjects infected with HIV-1 CRF01_AE (n = 110) was created, and potential antibody contact sites that showed sufficient but tolerable variation to detect a signal were identified. The amino acid distribution between vaccine and placebo recipients with or without the DPB1*13 allele was compared at selected sites using Fisher’s exact tests.
All P values for secondary analysis were uncorrected for the number of tests performed because they were exploratory in nature. Statistical analysis was completed using the R Statistical Computing Environment with the R package tpsDesign and SAS. Descriptive box plots, scatter plots, cumulative incidence curves, and vaccine efficacy curves were created using R or GraphPad Prism 6.
Supplementary Material
Fig. S1. Multiple alignment of different Env (120–204) and partial V2 sequences (160–183).
Fig. S2. Cocrystal structure of HIV-1 gp120 (red) binding to CD4 (green) and the Fab of mAb 17b (blue) (23).
Table S1. Odds ratios for two-way interaction analysis of HLA class II alleles with Env-specific IgA and Env (120–204)–specific IgG.
Table S2. Association between Env-specific IgA or Env (120–204)–specific IgG and HLA class II alleles present in more than 5% of the study population.
Table S3. High IgG binding to multiple Env (120–204) antigens correlates with presence of DPB1*13 and is associated with decreased HIV-1 acquisition across multiple subtypes.
Table S4. IgG responses to five overlapping peptides spanning Env (120–204) were significantly associated with presence of DPB1*13.
Table S5. Comparison of frequency of amino acid (AA) sites in Env-gp70 in RV144 breakthrough infections stratified by DPB1*13.
Table S6. Odds ratios for IgA binding to different recombinant Env proteins on HIV-1 acquisition after stratification by absence or presence of HLA-DQB1*06.
Table S7. Odds ratios for HIV-1 acquisition in univariate analyses of all HLA class II alleles present in the placebo controls.
Table S8. Odds ratios for HIV-1 acquisition in univariate analyses of all HLA class II alleles present in the vaccinated volunteers.
Acknowledgments
We thank C. Andrews, M. Milazzo, and K. Baldwin (all from the U.S. Military HIV Research Program) for operational support, assistance with sample selection, and genotyping. The views expressed are those of the authors and should not be construed as official or representing the positions of the U.S. Departments of the Army or Defense.
Funding: This work was supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine Inc. and the U.S. Department of Defense, Bill & Melinda Gates Foundation (Collaboration for AIDS Vaccine Discovery, grant OPP1032144), and the NIH/NIAID (UM1 AI068618 and Duke Center for AIDS Research AI064518). This research was funded, in part, by the U.S. National Institute of Allergy and Infectious Disease.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/7/296/296ra112/DC1
Author contributions: The project was conceptualized and supervised by R.T. The study was designed by H.A.P. and R.T. Data were analyzed and interpreted by G.D.T., P.B.G., R.A., M.R., J.H.K., N.L.M., and R.T. Statistical analysis was performed by H.A.P., Y.F., and P.B.G. Data on immune correlates were provided by A.D.C., S.Z.-P., D.C.M., M.J.M., R.T.B., R.A.K., and G.F. HLA genotyping/sequencing was performed by P.K.E., D.E.G., G.H.K., W.N., and R.T. Binding assays were performed by N.L.Y., S.J.K., X.S., and G.D.T. The vaccine trial was designed and conducted by R.J.O., S.N., S.R.-N., J.K., P.P., N.L.M., and J.H.K. The manuscript was written by H.A.P., R.A., and R.T. and edited by S.K., G.D.T., P.B.G., M.L.R., J.H.K., P.K.E., Y.F., G.F., and N.L.M.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. MOPH-TAVEG Investigators, Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Robb ML, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Kunasol P, Khamboonruang C, Thongcharoen P, Morgan P, Benenson M, Paris RM, Chiu J, Adams E, Francis D, Gurunathan S, Tartaglia J, Gilbert P, Stablein D, Michael NL, Kim JH. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: A post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect Dis. 2012;12:531–537. doi: 10.1016/S1473-3099(12)70088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao H-X, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinter A, Honnen WJ, Kayman SC, Trochev O, Wu Z. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine. 1998;16:1803–1811. doi: 10.1016/s0264-410x(98)00182-0. [DOI] [PubMed] [Google Scholar]
- 5.Kim JH, Excler J-L, Michael NL. Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection. Annu Rev Med. 2015;66:423–437. doi: 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- 6.Tomaras GD, Ferrari G, Shen X, Alam SM, Liao H-X, Pollara J, Bonsignori M, Moody MA, Fong Y, Chen X, Poling B, Nicholson CO, Zhang R, Lu X, Parks R, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Gilbert PB, Kim JH, Michael NL, Montefiori DC, Haynes BF. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc Natl Acad Sci USA. 2013;110:9019–9024. doi: 10.1073/pnas.1301456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O’Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O’Connell RJ, deSouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao H-X, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang K-K, Chen X, Tsao C-Y, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang Z-Y, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yates NL, Liao H-X, Fong Y, deCamp A, Vandergrift NA, Williams WT, Alam SM, Ferrari G, Yang Z-Y, Seaton KE, Berman PW, Alpert MD, Evans DT, O’Connell RJ, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Pinter A, Zolla-Pazner S, Gilbert PB, Nabel GJ, Michael NL, Kim JH, Montefiori DC, Haynes BF, Tomaras GD. Vaccine-induced Env V1–V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med. 2014;6:228–239. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, Dugast A-S, Schoen MK, Rolland M, Suscovich TJ, Mahan AE, Liao L, Streeck H, Andrews C, Rerks-Ngarm S, Nitayaphan S, de Souza MS, Kaewkungwal J, Pitisuttithum P, Francis D, Michael NL, Kim JH, Bailey-Kellogg C, Ackerman ME, Alter G. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci Transl Med. 2014;6:228–238. doi: 10.1126/scitranslmed.3007736. [DOI] [PubMed] [Google Scholar]
- 11.de Souza MS, Ratto-Kim S, Chuenarom W, Schuetz A, Chantakulkij S, Nuntapinit B, Valencia-Micolta A, Thelian D, Nitayaphan S, Pitisuttithum P, Paris RM, Kaewkungwal J, Michael NL, Rerks-Ngarm S, Mathieson B, Marovich M, Currier JR, Kim JH, Ministry of Public Health–Thai AIDS Vaccine Evaluation Group Collaborators The Thai phase III trial (RV144) vaccine regimen induces T cell responses that preferentially target epitopes within the V2 region of HIV-1 envelope. J Immunol. 2012;188:5166–5176. doi: 10.4049/jimmunol.1102756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alper CA, Kruskall MS, Marcus-Bagley D, Craven DE, Katz AJ, Brink SJ, Dienstag JL, Awdeh Z, Yunis EJ. Genetic prediction of nonresponse to hepatitis B vaccine. N Engl J Med. 1989;321:708–712. doi: 10.1056/NEJM198909143211103. [DOI] [PubMed] [Google Scholar]
- 13.Caillat-Zucman S, Gimenez J-J, Wambergue F, Albouze G, Lebkiri B, Naret C, Moynot A, Jungers P, Bach JF. Distinct HLA class II alleles determine antibody response to vaccination with hepatitis B surface antigen. Kidney Int. 1998;53:1626–1630. doi: 10.1046/j.1523-1755.1998.00909.x. [DOI] [PubMed] [Google Scholar]
- 14.Stephens HA, Brown AE, Chandanayingyong D, Webster HK, Sirikong M, Longta P, Vangseratthana R, Gordon DM, Lekmak S, Rungruang E. The presence of the HLA class II allele DPB1*0501 in ethnic Thais correlates with an enhanced vaccine-induced antibody response to a malaria sporozoite antigen. Eur J Immunol. 1995;25:3142–3147. doi: 10.1002/eji.1830251123. [DOI] [PubMed] [Google Scholar]
- 15.Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. J Infect Dis. 2006;193:655–663. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- 16.Paris R, Bejrachandra S, Thongcharoen P, Nitayaphan S, Pitisuttithum P, Sambor A, Gurunathan S, Francis D, Ratto-Kim S, Karnasuta C, de Souza MS, Polonis VR, Brown AE, Kim JH, Stephens HA, Thai AIDS Vaccine Evaluation Group HLA class II restriction of HIV-1 clade-specific neutralizing antibody responses in ethnic Thai recipients of the RV144 prime-boost vaccine combination of ALVAC-HIV and AIDSVAX® B/E. Vaccine. 2012;30:832–836. doi: 10.1016/j.vaccine.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Ranasinghe S, Cutler S, Davis I, Lu R, Soghoian DZ, Qi Y, Sidney J, Kranias G, Flanders MD, Lindqvist M, Kuhl B, Alter G, Deeks SG, Walker BD, Gao X, Sette A, Carrington M, Streeck H. Association of HLA-DRB1–restricted CD4+ T cell responses with HIV immune control. Nat Med. 2013;19:930–933. doi: 10.1038/nm.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spencer SJ, Burns A, Gaskin G, Pusey CD, Rees AJ. HLA class II specificities in vasculitis with antibodies to neutrophil cytoplasmic antigens. Kidney Int. 1992;41:1059–1063. doi: 10.1038/ki.1992.161. [DOI] [PubMed] [Google Scholar]
- 19.Julg B, Moodley ES, Qi Y, Ramduth D, Reddy S, Mncube Z, Gao X, Goulder PJ, Detels R, Ndung’u T, Walker BD, Carrington M. Possession of HLA class II DRB1*1303 associates with reduced viral loads in chronic HIV-1 clade C and B infection. J Infect Dis. 2011;203:803–809. doi: 10.1093/infdis/jiq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldwin KM, Ehrenberg PK, Geretz A, Prentice HA, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, O’Connell RJ, Kim JH, Thomas R. HLA class II diversity in HIV-1 uninfected individuals from the placebo arm of the RV144 Thai vaccine efficacy trial. Tissue Antigens. 2015;85:117–126. doi: 10.1111/tan.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizzuto CD, Wyatt R, Hernández-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 23.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Godillot AP, Wyatt R, Sodroski J, Chaiken I. Antibody 17b binding at the coreceptor site weakens the kinetics of the interaction of envelope glycoprotein gp120 with CD4. Biochemistry. 2001;40:1662–1670. doi: 10.1021/bi001397m. [DOI] [PubMed] [Google Scholar]
- 25.Klein J, Sato A. The HLA system. Second of two parts. N Engl J Med. 2000;343:782–786. doi: 10.1056/NEJM200009143431106. [DOI] [PubMed] [Google Scholar]
- 26.Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, Vyse TJ, Rioux JD. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLOS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machulla HK, Schönermarck U, Schaaf A, Müller LP, Kloß C, Krüger J, Kunze G, Schönermarck G, Langner J. HLA-A, B, Cw and DRB1, DRB3/4/5, DQB1, DPB1 frequencies in German immunoglobulin A-deficient individuals. Scand J Immunol. 2000;52:207–211. doi: 10.1046/j.1365-3083.2000.00765.x. [DOI] [PubMed] [Google Scholar]
- 28.Kralovicova J, Hammarström L, Plebani A, Webster AD, Vorechovsky I. Fine-scale mapping at IGAD1 and genome-wide genetic linkage analysis implicate HLA-DQ/DR as a major susceptibility locus in selective IgA deficiency and common variable immunodeficiency. J Immunol. 2003;170:2765–2775. doi: 10.4049/jimmunol.170.5.2765. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen GH, Ornolfsson AE, Johannesson A, Gudmundsson S, Janzi M, Wang N, Hammarström L, Ludviksson BR. Association of immunoglobulin A deficiency and elevated thyrotropin-receptor autoantibodies in two Nordic countries. Hum Immunol. 2010;72:166–172. doi: 10.1016/j.humimm.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Todd JA, Bell JI, McDevitt HO. HLA-DQb gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 31.O’Connell RJ, Excler J-L. HIV vaccine efficacy and immune correlates of risk. Curr HIV Res. 2013;11:450–463. doi: 10.2174/1570162x113116660052. [DOI] [PubMed] [Google Scholar]
- 32.Tomaras GD, Haynes BF. Advancing toward HIV-1 vaccine efficacy through the intersections of immune correlates. Vaccines. 2014;2:15–35. doi: 10.3390/vaccines2010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gartland AJ, Li S, McNevin J, Tomaras GD, Gottardo R, Janes H, Fong Y, Morris D, Geraghty DE, Kijak GH, Edlefsen PT, Frahm N, Larsen BB, Tovanabutra S, Sanders-Buell E, deCamp AC, Magaret CA, Ahmed H, Goodridge JP, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O’Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O’Connell RJ, deSouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, Sidney J, Sette A, Zolla-Pazner S, Montefiori D, McElrath MJ, Mullins JI, Kim JH, Gilbert PB, Hertz T. Analysis of HLA A*02 association with vaccine efficacy in the RV144 HIV-1 vaccine trial. J Virol. 2014;88:8242–8255. doi: 10.1128/JVI.01164-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li SS, Gilbert PB, Tomaras GD, Kijak G, Ferrari G, Thomas R, Pyo C-W, Zolla-Pazner S, Montefiori D, Liao H-X, Nabel G, Pinter A, Evans DT, Gottardo R, Dai JY, Janes H, Morris D, Fong Y, Edlefsen PT, Li F, Frahm N, Alpert MD, Prentice H, Rerks-Ngarm S, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Robb ML, O’Connell RJ, Haynes BF, Michael NL, Kim JH, McElrath MJ, Geraghty DE. FCGR2C polymorphisms associate with HIV-1 vaccine protection in RV144 trial. J Clin Invest. 2014;124:3879–3890. doi: 10.1172/JCI75539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prentice HA, Ehrenberg PK, Baldwin KM, Geretz A, Andrews C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, O’Connell RJ, Robb ML, Kim JH, Michael NL, Thomas R. HLA class I, KIR, and genome-wide SNP diversity in the RV144 Thai phase 3 HIV vaccine clinical trial. Immunogenetics. 2014;66:299–310. doi: 10.1007/s00251-014-0765-6. [DOI] [PubMed] [Google Scholar]
- 36.Ehrenberg PK, Geretz A, Baldwin KM, Apps R, Polonis VR, Robb ML, Kim JH, Michael NL, Thomas R. High-throughput multiplex HLA genotyping by next-generation sequencing using multi-locus individual tagging. BMC Genomics. 2014;15:864. doi: 10.1186/1471-2164-15-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 39.Borgan O, Langholz B, Samuelsen SO, Goldstein L, Pogoda J. Exposure stratified case-cohort designs. Lifetime Data Anal. 2000;6:39–58. doi: 10.1023/a:1009661900674. [DOI] [PubMed] [Google Scholar]
- 40.Breslow NE, Holubkov R. Weighted likelihood, pseudo-likelihood and maximum likelihood methods for logistic regression analysis of two-stage data. Stat Med. 1997;16:103–116. doi: 10.1002/(sici)1097-0258(19970115)16:1<103::aid-sim474>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 42.Holm S. A simple sequentially rejective multiple test procedure. Scand J Statist. 1979;6:65–70. [Google Scholar]
- 43.Zolla-Pazner S, deCamp A, Gilbert PB, Williams C, Yates NL, Williams WT, Howington R, Fong Y, Morris DE, Soderberg KA, Irene C, Reichman C, Pinter A, Parks R, Pitisuttithum P, Kaewkungwal J, Rerks-Ngarm S, Nitayaphan S, Andrews C, O’Connell RJ, Yang Z-Y, Nabel GJ, Kim JH, Michael NL, Montefiori DC, Liao H-X, Haynes BF, Tomaras GD. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLOS One. 2014;9:e87572. doi: 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Multiple alignment of different Env (120–204) and partial V2 sequences (160–183).
Fig. S2. Cocrystal structure of HIV-1 gp120 (red) binding to CD4 (green) and the Fab of mAb 17b (blue) (23).
Table S1. Odds ratios for two-way interaction analysis of HLA class II alleles with Env-specific IgA and Env (120–204)–specific IgG.
Table S2. Association between Env-specific IgA or Env (120–204)–specific IgG and HLA class II alleles present in more than 5% of the study population.
Table S3. High IgG binding to multiple Env (120–204) antigens correlates with presence of DPB1*13 and is associated with decreased HIV-1 acquisition across multiple subtypes.
Table S4. IgG responses to five overlapping peptides spanning Env (120–204) were significantly associated with presence of DPB1*13.
Table S5. Comparison of frequency of amino acid (AA) sites in Env-gp70 in RV144 breakthrough infections stratified by DPB1*13.
Table S6. Odds ratios for IgA binding to different recombinant Env proteins on HIV-1 acquisition after stratification by absence or presence of HLA-DQB1*06.
Table S7. Odds ratios for HIV-1 acquisition in univariate analyses of all HLA class II alleles present in the placebo controls.
Table S8. Odds ratios for HIV-1 acquisition in univariate analyses of all HLA class II alleles present in the vaccinated volunteers.


