Abstract
Receptor activity modifying proteins (RAMPs) associate with G-protein-coupled receptors (GPCRs) at the plasma membrane and together bind a variety of peptide ligands, serving as a communication interface between the extracellular and intracellular environments. The collection of RAMP-interacting GPCRs continues to expand and now consists of GPCRs from families A, B, and C, suggesting that RAMP activity is extremely prevalent. RAMP association with GPCRs can regulate GPCR function by altering ligand binding, receptor trafficking and desensitization, and downstream signaling pathways. Here, we elaborate on these RAMP-dependent mechanisms of GPCR regulation, which provide opportunities for pharmacological intervention.
Keywords: GPCR, CLR, ligand, trafficking, desensitization, signaling, G-protein
Introduction
G-protein-coupled receptors (GPCRs) constitute approximately 2% of the human genome and activate prominent cellular signaling pathways (Fredriksson et al., 2003). Due to their physiological importance and their cell surface localization, GPCRs are the most pharmacologically tractable proteins known. Approximately 40% of drugs currently marketed for the clinic target GPCRs (Rask-Andersen et al., 2011). However, only a small fraction of these receptors has been selected for drug development thus far, while the complex biology of the rest remain under intense study (Sexton et al., 2009).
Recently, there has been much excitement about the many modalities through which GPCR signals can be regulated, including: 1) biased agonism – differential signaling through a single receptor based on the bound ligand (Nagi and Pineyro, 2015); 2) allosteric modulation – changes in ligand affinity and efficacy by a substance bound to the receptor at a site distinct from the orthosteric ligand binding site (Christopoulos, 2014); 3) pleiotropic signaling through different G-proteins and other non-G-proteins, such as arrestin; and 4) heterodimerization. The ability of GPCRs to homodimerize and to heterodimerize with different GPCRs and other proteins complicates GPCR biology. How GPCR oligomers affect signaling and function is of particular interest, as these protein-protein interactions may enable the development of drugs with very specific targets and effects (George et al., 2002).
Receptor activity modifying proteins (RAMPs) are single-pass transmembrane proteins that heterodimerize with seven-transmembrane GPCRs belonging to each of the three main classes of GPCRs: the class A/Rhodopsin-like family, the largest of the three; the class B/secretin family, containing the majority of identified RAMP-interacting GPCRs; and the class C family (Table 1). The association of RAMPs with GCPRs affects ligand specificity, receptor trafficking, receptor desensitization, and signaling capabilities, profoundly expanding the repertoire of targets available for modifying clinical disease. Since their discovery over 15 years ago, much has been learned about the effects of RAMPs on the function of some of their GPCR partners. However, a dearth of knowledge still exists regarding additional GPCR-RAMP pairs and, more importantly, the biological significance of both known and unknown GPCR-RAMP partners. Here, we discuss what is currently known about how RAMPs modulate GPCR activity and address what important questions remain.
Table 1.
RAMP-associating GPCRs.
| Receptor Family | GPCR | Associating RAMPs |
|---|---|---|
| A | GPER | RAMP3 |
| B | CLR | RAMP1 RAMP2 RAMP3 |
| B | CTR | RAMP1 RAMP2 RAMP3 |
| B | CRF | RAMP2 |
| B | Glucagon Receptor | RAMP2 |
| B | PTH1R | RAMP2 |
| B | PTH2R | RAMP3 |
| B | Secretin Receptor | RAMP3 |
| B | VPAC1R | RAMP1 RAMP2 RAMP3 |
| B | VPAC2R | RAMP1 RAMP2 RAMP3 |
| C | CaSR | RAMP1 RAMP3 |
RAMP discovery
RAMPs were first identified during an effort to understand signaling of the neuropeptide calcitonin (CT) gene-related peptide (CGRP). CGRP belongs to the CT peptide family, which consists of five peptide hormones: CGRP, CT, amylin (AMY), adrenomedullin (AM), and adrenomedullin 2/intermedin (AM2/IMD). These peptides have significant structural homology, and their often overlapping biological activities include gastric emptying (AMY, AM2/IMD), vasodilation (CGRP, AM, AM2/IMD), angiogenesis (CGRP, AM), and pain sensation (CGRP, CT) (Hong et al., 2012, Muff et al., 2004, Poyner et al., 2002). Despite sharing many physiological functions as well as affinity for GPCRs, different members of this peptide family mediate unique biological events, suggesting that they also must target specific and distinct receptors (Poyner et al., 2002).
Following the cloning of the GPCR CT receptor-like receptor (CLR), conflicting data emerged regarding whether the orphan receptor was able to bind CGRP. CLR exhibits 58% amino acid sequence similarity to the human CT receptor (CTR) and was therefore predicted to bind one of the CT family peptides (Fluhmann et al., 1995). Previous data suggested that CGRP acted through a Gαs-coupled GPCR to elicit cyclic AMP (cAMP) production (Seifert et al., 1985). However, transfection of human CLR or its rat homologue into COS-7 cells and subsequent CGRP treatment did not lead to cAMP accumulation (Fluhmann et al., 1995, Njuki et al., 1993). In contrast, when CLR was transfected into HEK293 cells, CGRP treatment elicited a 60-fold increase in cAMP production (Aiyar et al., 1996, Han et al., 1997). Taken together, these data led to the speculation that HEK293 cells expressed an additional, endogenous factor that was required to activate the CGRP receptor, while COS-7 cells did not express this factor.
Using an expression cloning approach, McLatchie and colleagues demonstrated that the CGRP receptor consists of a GPCR heterodimer: CLR and the 148 amino acid protein RAMP1 (McLatchie et al., 1998). Co-transfection of CLR and RAMP1 followed by CGRP treatment significantly increased cAMP production compared to transfection of either protein alone (McLatchie et al., 1998). Other studies demonstrated that cells expressing endogenous RAMPs can generate a cAMP response when transfected with CLR, whereas cell lines lacking endogenous RAMP1 cannot (Aiyar et al., 1996, Fluhmann et al., 1995, Njuki et al., 1993). Clearly, RAMP1 allows CLR to respond to CGRP. Subsequently, two additional RAMPs were identified and found to modify ligand specificity of CLR. While the CLR-RAMP1 complex binds CGRP, the CLR-RAMP2 and -3 complexes bind AM (McLatchie et al., 1998). These data established a novel mechanism for altering GPCR ligand specificity and illustrated the complexity of GPCR signaling, yielding a paradigm shift in the understanding of GPCR function (Hay et al., 2006, Parameswaran and Spielman, 2006).
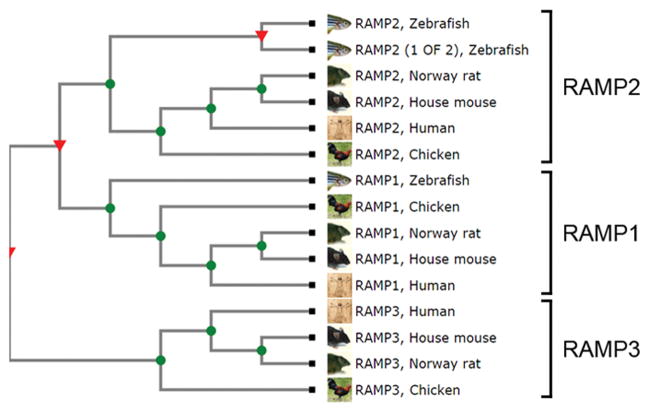
The RAMP family phylogenetic tree, assembled by TreeFam (TF333286), a database of the European Bioinformatics Institute, unveils 37% identity across 139 sequences from 53 species (Figure 1). RAMP-expressing species are extremely diverse and include many model organisms, such as zebrafish, mice, rats, guinea pigs, and non-human primates. Other eutherians or placental mammals (cats, dogs, ferrets, pigs, horses, and humans) also express RAMPs, as do marsupials (platypuses and Tasmanian devils); ruminants (cattle); birds (chicken, turkeys); fish; reptiles (frogs, turtles) and endangered species (giant pandas, elephants, and dolphins). In the vast majority of these species, RAMPs 1–3 are encoded by single genes. However, the genomes of several bony fishes encode two Ramp1 and Ramp2 genes, while RAMP3 is encoded by a single gene across all species and comprises a distinct outgroup to Ramp1 and Ramp2. Therefore, RAMP1 and RAMP2 are more closely related to each other than either protein is to RAMP3. To our knowledge, conservation of Ramp sequences across species has not been studied in depth, but further investigation into this topic could provide a lens through which we can interpret data and possibly use to explain functional differences between the three RAMPs.
Figure 1.
RAMP family phylogenetic tree. The RAMP family TreeFam (TF33386), assembled by a database of the European Bioinformatics Institute, contains 139 sequences from 53 species. Shown here is a condensed tree with a focus on model organisms and their relation to humans. Details on bioinformatic construction of this tree are provided in the literature (Guindon et al., 2010, Ruan et al., 2008). A color version of the figure is available online.
Each RAMP has a large extracellular N-terminal domain, a single transmembrane domain, and a cytoplasmic C-terminus (McLatchie et al., 1998). Studies using RAMP chimeras that exchanged RAMP domains concluded that the N-terminus determines ligand specificity, while the C-terminus modifies downstream signaling for CLR- and CTR-based receptors (Fraser et al., 1999, Udawela et al., 2006). However, despite sharing a common structure, RAMP proteins have only 30% sequence homology. RAMPs 1 and 3 are 148 amino acids long, while RAMP2 is considerably larger at 175 amino acids (McLatchie et al., 1998). Ultimately, variations between the three RAMPs enable them to differentially affect GPCR ligand specificity; GPCR trafficking and internalization; and signaling capabilities.
RAMP functionality
Ligand binding
As discussed above, the ability of RAMPs to modify CLR downstream signaling was essential to their discovery. Therefore, RAMP interaction with CLR has been particularly well-characterized. RAMP1 is necessary for CGRP binding, and RAMP2 or -3 facilitates AM binding (Figure 2A) (McLatchie et al., 1998). Crystal structures of the RAMP1 and -2 extracellular domains both individually or together with the CLR extracellular domain revealed several intermolecular interactions specific to CLR-RAMP2 that could explain the ability of AM to bind this receptor complex. Subsequent photo-crosslinking and mutagenesis demonstrated specific residues in the N-terminus of RAMP2 that are required for AM binding. These are not conserved in RAMP1, indicating that the N-terminus serves to define the specificity for ligand binding (Kusano et al., 2008, Kusano et al., 2012).
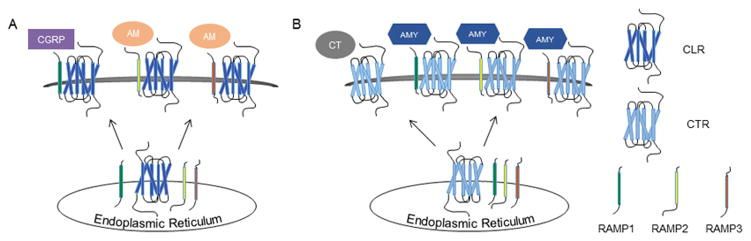
Figure 2.
Effects of RAMP association on CT receptor-like receptor (CLR) and CT receptor (CTR). (A) For cell surface expression, CLR requires the presence of one of the RAMP family members. CLR associates with RAMPs in the ER, facilitating plasma membrane expression. Association with RAMP1 yields a high affinity CT gene-related peptide (CGRP) receptor, whereas CLR association with RAMP2 or -3 results in a potent adrenomedullin (AM) receptor. (B) Conversely, CTR is trafficked to the plasma membrane independent of RAMPs. In the absence of a RAMP, CTR binds CT. In association with RAMP1, -2, -3, the CTR-RAMP complex binds amylin (AMY). A color version of the figure is available online.
Recently, Booe and colleagues solved the crystal structures of ligand-bound CLR-RAMP1 and CLR-RAMP2 and determined that RAMPs dictate ligand specificity via two mechanisms: RAMPs alter the GPCR ligand binding pocket first by allosterically augmenting the binding site and second by offering distinct contact sites for ligand interaction (Booe et al., 2015). Remarkably, single amino acids anchor the peptide C-termini and enable strong binding of specific peptides (Trp 84 in RAMP1 for CGRP binding; Glu 101 in RAMP2 for AM binding) (Booe et al., 2015). However, exchange of the C-terminal peptide residue between RAMP1 and -2 did not alter peptide binding, suggesting that additional mechanisms contribute to selectivity. Indeed, the crystal structures revealed that RAMP association with CLR allows for subtle changes in GPCR conformation that allosterically alter the CLR binding pocket. Whether the described RAMP residues are critical for determining ligand specificity for receptors other than CLR remains to be determined. Ultimately, these data demonstrate that subtle RAMP-induced changes in GPCR structure and ligand interactions can lead to dramatically different pharmacological profiles, providing support for future peptide-specific drug design (Booe et al., 2015).
Conversely, GPCR-RAMP associations may prevent ligand binding. For example, glucagon-like peptide-1 (GLP-1) acts as a partial agonist when bound to the glucagon receptor (Weston et al., 2015). However, association of the glucagon receptor with RAMP2 blocks this partial agonism by preventing GLP-1 binding, though the specific mechanism underlying this effect is unknown (Weston et al., 2015).
Receptor trafficking
In addition to dictating ligand specificity, RAMPs also act as chaperones for CLR, as co-expression of a RAMP is required for trafficking of CLR to the cell surface. Studies have shown that RAMPs have an endoplasmic reticulum (ER) retention signal within the C-terminus (Steiner et al., 2002). RAMP association with CLR overrides this retention signal, allowing translocation of the CLR-RAMP complex from the ER to the plasma membrane. RAMPs are therefore critical for plasma membrane localization of some GPCRs.
RAMPs also perform a similar trafficking function for a family C receptor, calcium sensing receptor (CaSR). When transfected alone into COS-7 cells, CaSR is retained in the ER (Bouschet et al., 2005). Co-expression of RAMP1 and RAMP3 with CaSR resulted in plasma membrane expression of CaSR, indicating that the CaSR-RAMP interaction promotes forward trafficking of CaSR to the cell surface (Bouschet et al., 2005). Although this finding elucidated a mechanism for CaSR regulation, these data in and of themselves were not groundbreaking. However, this was the first non-family B receptor identified to associate with RAMPs, opening the door to the possibility that RAMP effects are more pervasive than originally thought. A recent study has expanded this even further by identifying an association of RAMP3 with a class A receptor, GPR30, now known as G-protein-coupled estrogen receptor 1 (GPER) (Lenhart et al., 2013).
In contrast to CLR and CaSR, CTR does not require a RAMP for cell surface localization (Figure 2B) (Poyner et al., 2002). However, CTR-RAMP interaction in the ER allows for formation of a functional AMY receptor at the plasma membrane. Like the receptors for the CT peptide family, the AMY receptors proved challenging to isolate (Hay et al., 2006). While AMY appeared to associate with CTR, CTR transfection into cell lines did not consistently yield convincing AMY ligand binding (Hay et al., 2006, Perry et al., 1997). Following the discovery of RAMPs, several groups suspected that these inconsistencies were due to cell-endogenous expression of RAMPs and subsequently demonstrated that CTR association with all three RAMPs forms AMY receptors (Armour et al., 1999, Christopoulos et al., 1999, Muff et al., 1999). Interactions of CTR with RAMPs 1–3 have been classified as AMY1–3, which display varying affinities for AMY (Poyner et al., 2002).
Receptor desensitization
Not only can RAMPs facilitate GPCR plasma membrane localization, but they can affect GPCR internalization once at the cell surface, providing an additional context for pharmacological intervention. For example, early studies demonstrated that CLR-RAMP2 internalization was mediated by β-arrestin and dynamin (Hay et al., 2006, Hilairet et al., 2001, Kuwasako et al., 2000). Later, bioluminescence resonance energy transfer (BRET) studies indicated that CLR agonist-mediated β-arrestin recruitment was dependent on the presence of RAMP1 (Heroux et al., 2007a). Whether this was true for other RAMPs was not investigated.
More recent studies have suggested unique protein trafficking roles for RAMP3. The C-terminal tail of RAMP3 contains a type 1 PSD-95/Discs-large/ZO-1 (PDZ) domain that is absent in RAMP1 and -2. The PDZ domain allows for additional protein-protein interactions that alter receptor trafficking following receptor internalization. In contrast to the degradative pathway seen with RAMP1 and -2, Bomberger and colleagues showed that interaction of N-ethylmaleimide-sensitive factor (NSF) with the PDZ domain of RAMP3 causes CLR to be recycled back to the cell surface, allowing for rapid receptor resensitization (Bomberger et al., 2005a). Additionally, interaction of the PDZ domain with the Na+/H+ exchange regulatory factor (NHERF) tethers the CLR-RAMP3 complex to the actin cytoskeleton and inhibits internalization of CLR (Bomberger et al., 2005b). In contrast, NHERF had no effect on CLR-RAMP1 or CLR-RAMP2 complexes that lack this PDZ domain.
Cellular signaling
Because the discovery of RAMPs caused a major shift in the understanding of GPCR regulation, there was significant interest in other RAMP-interacting receptors. Given the structural similarities of family B GPCRs, such as CLR and CTR, and their affinity for peptide ligands, it stood to reason that other members of family B could interact with RAMPs. Using epitope-tagged constructs and immunofluorescence confocal microscopy, the Sexton laboratory discovered four novel GPCR-RAMP interactions (Christopoulos et al., 2003). Co-transfection of the glucagon receptor, parathyroid hormone 1 receptor (PTH1R), parathyroid hormone 2 receptor (PTH2R), or vasoactive intestinal peptide 1 receptor (VPAC1R) with RAMPs resulted in cell surface expression of RAMPs, whereas RAMP-only transfection did not. Interestingly, not all of these receptors interacted with all of the members of the RAMP family. The glucagon receptor and PTH1R associated only with RAMP2; PTH2R associated only with RAMP3; and VPAC1R associated with all three RAMPs (Christopoulos et al., 2003).
Further analysis of VPAC1R revealed that the interaction of a single GPCR with different RAMPs can elicit different downstream signals. VPAC1R couples to multiple G-proteins and is therefore able to stimulate cAMP accumulation and phosphoinositide hydrolysis (Christopoulos et al., 2003). While treatment of cells co-transfected with VPAC1R and RAMP1 or -3 did not alter cAMP accumulation, co-transfection of VPAC1R and RAMP2 enhanced phosphoinositide hydrolysis, suggesting that RAMP2 could modify G-protein coupling (Christopoulos et al., 2003). Whether this change in signaling is due to direct modification in G-protein coupling or due to changes in GPCR-RAMP localization remains to be determined (Sexton et al., 2012).
Studies of AMY1–3 offer evidence that RAMPs can indeed alter G-protein coupling. Morfis et al. observed that AMY treatment of cells co-transfected with CTR and RAMP1 or -3 (AMY1 and AMY3, respectively) elicited a 20- to 30-fold increase in the potency of cAMP production compared to transfection of CTR alone (Morfis et al., 2008). Conversely, only a 2- to 5-fold increase in the potency of intracellular Ca2+ production was observed, suggesting that AMY1 and AMY3 preferentially couple to Gαs versus Gαq relative to RAMP-independent CTR. Interestingly, while G-protein overexpression did not alter AMY binding in cells expressing CTR or CTR-RAMP1, overexpression of Gαs significantly increased AMY binding in CTR-RAMP2-expressing cells, and overexpression of Gαs and Gαq increased binding in CTR-RAMP3-expressing cells (Morfis et al., 2008). These data suggest that RAMPs are able to promote G-protein coupling compared to RAMP-independent GPCRs.
RAMPs may also promote G-protein uncoupling. A new study demonstrates that glucagon-mediated activation of the glucagon receptor is bolstered by co-expression of RAMP2 (Weston et al., 2015). This effect is not due to greater affinity of glucagon for the glucagon receptor or trafficking of the GPCR to the plasma membrane (Weston et al., 2015). Rather, this RAMP2-dependent enhancement in glucagon receptor activation is due to uncoupling of the GPCR to Gαi, facilitating unchecked Gαs activation (Weston et al., 2015). However, glucagon receptor G-protein coupling is not only RAMP-dependent but also ligand-dependent; when the glucagon receptor-RAMP2 complex binds to a glucagon-related peptide, oxyntomodulin, Gαi coupling is unaffected (Weston et al., 2015).
Finally, RAMP association with a GPCR may alter coupling to G-proteins other than Gαs, Gαq, and Gαi. For example, RAMPs interact with vasoactive intestinal peptide 2 receptor (VPAC2R) and corticotrophin releasing factor receptor-1 (CRF1R) and alter G-protein coupling of both receptors (Wootten et al., 2013). Here, co-expression of RAMP1 and -2 with VPAC2R in HEK293T and CHO-K1 cells significantly increased basal coupling to Gi/o/t/z compared with VPAC2R alone (Wootten et al., 2013). Similarly, co-expression of RAMP2 with CRF1R significantly enhanced Gi/o/t/z and Gq/11 coupling, leading to enhanced Ca2+ elevation in response to agonists (Wootten et al., 2013). Taken together, these data demonstrate the potential of RAMPs to alter downstream signaling, suggesting that an evaluation of the signaling profile is important to understanding the physiology of GPCR-RAMP interactions.
RAMP physiology
While the repertoire of GPCR-RAMP pairs continues to expand, studies of RAMP physiology in animal models lag behind advances in RAMP biochemistry and pharmacology and have addressed only a subset of demonstrated GPCR-RAMP associations (Table 2) (Kadmiel et al., 2012). Knockout mouse models of all three RAMPs have been generated and phenotyped (Caron and Smithies, 2001, Dackor et al., 2007, Tsujikawa et al., 2007). Only RAMP2 deficiency is embryonic lethal at mid-gestation due to cardiovascular defects and a hypoproliferative lymphatic vasculature, implying that, despite their structural similarity and common association with several GPCRs, RAMPs 1 and 3 are unable to compensate for loss of RAMP2 (Caron and Smithies, 2001, Dackor et al., 2007, Tsujikawa et al., 2007). Therefore, studies of RAMP2 in mice are limited to conditional, tissue-specific deletions, Ramp2 heterozygotes, and Ramp2-overexpressing transgenic animals.
Table 2.
Genetically engineered mouse models available for study of RAMP function in vivo.
| Mouse Model | RAMP1 | RAMP2 | RAMP3 | |
|---|---|---|---|---|
| LOSS OF FUNCTION | Global | Ramp1−/− (Li et al., 2014, Tsujikawa et al., 2007) | (Ramp2+/−)* (Dackor et al., 2007) | Ramp3−/− (Dackor et al., 2007) |
| Tissue-specific | N/A | Endothelial (VE-Cad) (Koyama et al., 2013) | N/A | |
| Drug-inducible, tissue-specific | N/A |
Cardiomyocyte (αMHC) (Yoshizawa et al., 2013) Endothelial (VE-Cad) (Koyama et al., 2013) |
N/A | |
| GAIN OF FUNCTION | Global transgenic | hRAMP1 (Zhang et al., 2007) | N/A | N/A |
| Tissue-specific transgenic | Neuronal (Chrissobolis et al., 2010) | N/A | N/A | |
| GPCR INTERACTIONS ADDRESSED | CLR | CLR CTR CRF Glucagon Receptor PTHR1 |
CLR GPER |
Ramp2−/− mice are embryonic lethal (Caron and Smithies, 2001).
To date, animal studies of RAMP function have largely focused on the three CLR-RAMP pairs and their ligands, CGRP and AM, with particular emphasis on blood pressure regulation and cardioprotection. RAMP1 function has been expanded to include neuronal and pulmonary inflammation with therapeutic implications for migraine and asthma, both prevalent clinical problems (Li et al., 2014, Walker et al., 2015, Zhang et al., 2007). Additionally, Ramp2 heterozygotes display a constellation of endocrine abnormalities consistent with pituitary dysfunction and dysregulation of several GPCR-RAMP2 ligands, prompting the obligation to consider all RAMP-associated GPCRs and ligands when interpreting RAMP under- or overexpression phenotypes (Kadmiel et al., 2011). Similarly, we should be cognizant of GPCR-RAMP pairs outside of the class B GPCR family, as a study examining a novel interaction between RAMP3 and the class A receptor GPER in Ramp3−/− mice illustrates (Lenhart et al., 2013).
Conclusions
Together, these data highlight the unique effects that RAMPs have on GPCR function. Many GPCRs interact with several RAMPs. Therefore, it is important to define the individual trafficking pathway, ligand affinity, and signaling profile of each GPCR-RAMP complex. Many additional RAMP effects remain undefined, so while the breadth of RAMP effects already seems widespread, we likely have only begun to uncover the capacity of RAMPs to alter GPCR physiology. However, it is worth noting that the majority of studies to date have not demonstrated direct interactions between GPCRs and RAMPs and that published observations about RAMP effects on GPCRs require further investigation.
Much remains to be understood about RAMP biochemistry and pharmacology. For example, the stoichiometry of GPCR-RAMP partners has been studied but is still debated. Evidence exists for a 1:1, 2:2, and 2:1 ratio of CLR-RAMP (Booe et al., 2015, Heroux et al., 2007b, Kusano et al., 2012, Watkins et al., 2013). However, whether the stoichiometry differs by cellular environment or between GPCRs is not known. These questions may be further complicated by the identification of RAMP-associating GPCR heterodimers. So far, studies have only focused on the interaction of one GPCR with one RAMP, but it is possible that two different GPCRs could complex with a RAMP, or two different RAMPs could bind to a single GPCR.
Figure 3 summarizes what is currently known about RAMP effects on GPCR function. In vitro biochemical studies have not only enabled the identification of novel GPCR-RAMP pairs but have also elucidated much about how RAMPs affect GPCR biology. GPCRs are pharmacologically tractable themselves, but the unique GPCR-RAMP interface may prove to be even more advantageous for the design of highly selective drugs (Sexton et al., 2009). Ultimately, in order to harness their potential as drug targets for disease, we must first understand how RAMPs affect physiology and what roles they play in physiological dysfunction in vivo.
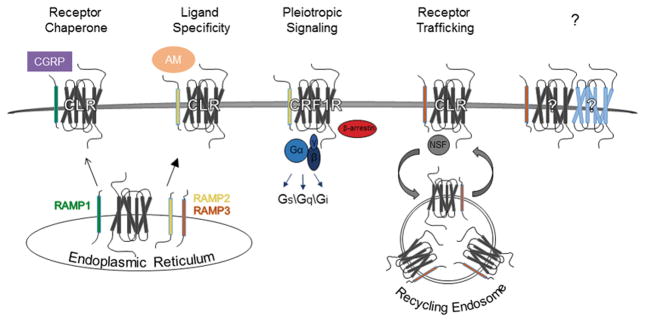
Figure 3.
Examples of effects of RAMP association on GPCR activity. GPCR-RAMP associations may promote GPCR plasma membrane localization; ligand specificity; functional selectivity; and trafficking. Receptor chaperone and ligand specificity: RAMPs can enable GPCRs to shuttle to the plasma membrane, and differential RAMP association may dictate preference for endogenous ligands. For example, RAMP association with CT receptor-like receptor (CLR) in the endoplasmic reticulum promotes localization of CLR at the plasma membrane, where CLR-RAMP1 can bind CT gene-related peptide (CGRP), and CLR-RAMP2 or –RAMP3 can bind adrenomedullin (AM). Functional selectivity: Depending on the GPCR-RAMP association, GPCRs may preferentially couple to certain G-proteins, such as Gs, Gq, and Gi, or other non-G-proteins, such as β-arrestin. For example, corticotrophin releasing factor receptor-1 (CRF1R)-RAMP2 preferentially couples to Gi/o/t/z and Gq/11. Receptor trafficking: GPCR-RAMP associations may affect whether the GPCR is degraded or recycled back to the plasma membrane following internalization. For example, RAMP3 contains a PSD-95/Discs-large/ZO-1 (PDZ) domain that interacts with N-ethylmaleimide-sensitive factor (NSF) to shunt CLR away from the degradative pathway it enters following internalization when associated with RAMP1 or RAMP2. ?: Other effects of GPCR-RAMP associations remain to be discovered. A color version of the figure is available online.
Acknowledgments
This work was supported by NIH grants DK099156 to K.M.C. and F30 HL118932 to K.R.K.
Footnotes
Declaration of Interest
The authors report no declarations of interest.
References
- Aiyar N, Rand K, Elshourbagy NA, Zeng Z, Adamou JE, Bergsma DJ, Li Y. A cDNA encoding the calcitonin gene-related peptide type 1 receptor. J Biol Chem. 1996;271:11325–9. doi: 10.1074/jbc.271.19.11325. [DOI] [PubMed] [Google Scholar]
- Armour SL, Foord S, Kenakin T, Chen WJ. Pharmacological characterization of receptor-activity-modifying proteins (RAMPs) and the human calcitonin receptor. J Pharmacol Toxicol Methods. 1999;42:217–24. doi: 10.1016/s1056-8719(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Bomberger JM, Parameswaran N, Hall CS, Aiyar N, Spielman WS. Novel function for receptor activity-modifying proteins (RAMPs) in post-endocytic receptor trafficking. J Biol Chem. 2005a;280:9297–307. doi: 10.1074/jbc.M413786200. [DOI] [PubMed] [Google Scholar]
- Bomberger JM, Spielman WS, Hall CS, Weinman EJ, Parameswaran N. Receptor activity-modifying protein (RAMP) isoform-specific regulation of adrenomedullin receptor trafficking by NHERF-1. J Biol Chem. 2005b;280:23926–35. doi: 10.1074/jbc.M501751200. [DOI] [PubMed] [Google Scholar]
- Booe JM, Walker CS, Barwell J, Kuteyi G, Simms J, Jamaluddin MA, Warner ML, Bill RM, Harris PW, Brimble MA, Poyner DR, Hay DL, Pioszak AA. Structural Basis for Receptor Activity-Modifying Protein-Dependent Selective Peptide Recognition by a G Protein-Coupled Receptor. Mol Cell. 2015;58:1040–52. doi: 10.1016/j.molcel.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouschet T, Martin S, Henley JM. Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J Cell Sci. 2005;118:4709–20. doi: 10.1242/jcs.02598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron KM, Smithies O. Extreme hydrops fetalis and cardiovascular abnormalities in mice lacking a functional Adrenomedullin gene. Proc Natl Acad Sci U S A. 2001;98:615–9. doi: 10.1073/pnas.021548898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrissobolis S, Zhang Z, Kinzenbaw DA, Lynch CM, Russo AF, Faraci FM. Receptor activity-modifying protein-1 augments cerebrovascular responses to calcitonin gene-related peptide and inhibits angiotensin II-induced vascular dysfunction. Stroke. 2010;41:2329–34. doi: 10.1161/STROKEAHA.110.589648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos A. Advances in G protein-coupled receptor allostery: from function to structure. Mol Pharmacol. 2014;86:463–78. doi: 10.1124/mol.114.094342. [DOI] [PubMed] [Google Scholar]
- Christopoulos A, Christopoulos G, Morfis M, Udawela M, Laburthe M, Couvineau A, Kuwasako K, Tilakaratne N, Sexton PM. Novel receptor partners and function of receptor activity-modifying proteins. J Biol Chem. 2003;278:3293–7. doi: 10.1074/jbc.C200629200. [DOI] [PubMed] [Google Scholar]
- Christopoulos G, Perry KJ, Morfis M, Tilakaratne N, Gao Y, Fraser NJ, Main MJ, Foord SM, Sexton PM. Multiple amylin receptors arise from receptor activity-modifying protein interaction with the calcitonin receptor gene product. Mol Pharmacol. 1999;56:235–42. doi: 10.1124/mol.56.1.235. [DOI] [PubMed] [Google Scholar]
- Dackor R, Fritz-Six K, Smithies O, Caron K. Receptor activity-modifying proteins 2 and 3 have distinct physiological functions from embryogenesis to old age. J Biol Chem. 2007;282:18094–9. doi: 10.1074/jbc.M703544200. [DOI] [PubMed] [Google Scholar]
- Fluhmann B, Muff R, Hunziker W, Fischer JA, Born W. A human orphan calcitonin receptor-like structure. Biochem Biophys Res Commun. 1995;206:341–7. doi: 10.1006/bbrc.1995.1047. [DOI] [PubMed] [Google Scholar]
- Fraser NJ, Wise A, Brown J, Mclatchie LM, Main MJ, Foord SM. The amino terminus of receptor activity modifying proteins is a critical determinant of glycosylation state and ligand binding of calcitonin receptor-like receptor. Mol Pharmacol. 1999;55:1054–9. doi: 10.1124/mol.55.6.1054. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- George SR, O’dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat Rev Drug Discov. 2002;1:808–20. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Han ZQ, Coppock HA, Smith DM, Van Noorden S, Makgoba MW, Nicholl CG, Legon S. The interaction of CGRP and adrenomedullin with a receptor expressed in the rat pulmonary vascular endothelium. J Mol Endocrinol. 1997;18:267–72. doi: 10.1677/jme.0.0180267. [DOI] [PubMed] [Google Scholar]
- Hay DL, Poyner DR, Sexton PM. GPCR modulation by RAMPs. Pharmacol Ther. 2006;109:173–97. doi: 10.1016/j.pharmthera.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Heroux M, Breton B, Hogue M, Bouvier M. Assembly and signaling of CRLR and RAMP1 complexes assessed by BRET. Biochemistry. 2007a;46:7022–33. doi: 10.1021/bi0622470. [DOI] [PubMed] [Google Scholar]
- Heroux M, Hogue M, Lemieux S, Bouvier M. Functional calcitonin gene-related peptide receptors are formed by the asymmetric assembly of a calcitonin receptor-like receptor homo-oligomer and a monomer of receptor activity-modifying protein-1. J Biol Chem. 2007b;282:31610–20. doi: 10.1074/jbc.M701790200. [DOI] [PubMed] [Google Scholar]
- Hilairet S, Belanger C, Bertrand J, Laperriere A, Foord SM, Bouvier M. Agonist-promoted internalization of a ternary complex between calcitonin receptor-like receptor, receptor activity-modifying protein 1 (RAMP1), and beta-arrestin. J Biol Chem. 2001;276:42182–90. doi: 10.1074/jbc.M107323200. [DOI] [PubMed] [Google Scholar]
- Hong Y, Hay DL, Quirion R, Poyner DR. The pharmacology of adrenomedullin 2/intermedin. Br J Pharmacol. 2012;166:110–20. doi: 10.1111/j.1476-5381.2011.01530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadmiel M, Fritz-Six K, Pacharne S, Richards GO, Li M, Skerry TM, Caron KM. Research resource: Haploinsufficiency of receptor activity-modifying protein-2 (RAMP2) causes reduced fertility, hyperprolactinemia, skeletal abnormalities, and endocrine dysfunction in mice. Mol Endocrinol. 2011;25:1244–53. doi: 10.1210/me.2010-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadmiel M, Fritz-Six KL, Caron KM. Understanding RAMPs through genetically engineered mouse models. Adv Exp Med Biol. 2012;744:49–60. doi: 10.1007/978-1-4614-2364-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Ochoa-Callejero L, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Iinuma N, Arai T, Yoshizawa T, Iesato Y, Lei Y, Uetake R, Okimura A, Yamauchi A, Tanaka M, Igarashi K, Toriyama Y, Kawate H, Adams RH, Kawakami H, Mochizuki N, Martinez A, Shindo T. Vascular endothelial adrenomedullin-RAMP2 system is essential for vascular integrity and organ homeostasis. Circulation. 2013;127:842–53. doi: 10.1161/CIRCULATIONAHA.112.000756. [DOI] [PubMed] [Google Scholar]
- Kusano S, Kukimoto-Niino M, Akasaka R, Toyama M, Terada T, Shirouzu M, Shindo T, Yokoyama S. Crystal structure of the human receptor activity-modifying protein 1 extracellular domain. Protein Sci. 2008;17:1907–14. doi: 10.1110/ps.036012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano S, Kukimoto-Niino M, Hino N, Ohsawa N, Okuda K, Sakamoto K, Shirouzu M, Shindo T, Yokoyama S. Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding. Protein Sci. 2012;21:199–210. doi: 10.1002/pro.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwasako K, Shimekake Y, Masuda M, Nakahara K, Yoshida T, Kitaura M, Kitamura K, Eto T, Sakata T. Visualization of the calcitonin receptor-like receptor and its receptor activity-modifying proteins during internalization and recycling. J Biol Chem. 2000;275:29602–9. doi: 10.1074/jbc.M004534200. [DOI] [PubMed] [Google Scholar]
- Lenhart PM, Broselid S, Barrick CJ, Leeb-Lundberg LM, Caron KM. G-protein-coupled receptor 30 interacts with receptor activity-modifying protein 3 and confers sex-dependent cardioprotection. J Mol Endocrinol. 2013;51:191–202. doi: 10.1530/JME-13-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wetzel-Strong SE, Hua X, Tilley SL, Oswald E, Krummel MF, Caron KM. Deficiency of RAMP1 attenuates antigen-induced airway hyperresponsiveness in mice. PLoS One. 2014;9:e102356. doi: 10.1371/journal.pone.0102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG, Foord SM. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–9. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- Morfis M, Tilakaratne N, Furness SG, Christopoulos G, Werry TD, Christopoulos A, Sexton PM. Receptor activity-modifying proteins differentially modulate the G protein-coupling efficiency of amylin receptors. Endocrinology. 2008;149:5423–31. doi: 10.1210/en.2007-1735. [DOI] [PubMed] [Google Scholar]
- Muff R, Born W, Lutz TA, Fischer JA. Biological importance of the peptides of the calcitonin family as revealed by disruption and transfer of corresponding genes. Peptides. 2004;25:2027–38. doi: 10.1016/j.peptides.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Muff R, Buhlmann N, Fischer JA, Born W. An amylin receptor is revealed following co-transfection of a calcitonin receptor with receptor activity modifying proteins-1 or -3. Endocrinology. 1999;140:2924–7. doi: 10.1210/endo.140.6.6930. [DOI] [PubMed] [Google Scholar]
- Nagi K, Pineyro G. Practical guide for calculating and representing biased signaling by GPCR ligands: A stepwise approach. Methods. 2015 doi: 10.1016/j.ymeth.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Njuki F, Nicholl CG, Howard A, Mak JC, Barnes PJ, Girgis SI, Legon S. A new calcitonin-receptor-like sequence in rat pulmonary blood vessels. Clin Sci (Lond) 1993;85:385–8. doi: 10.1042/cs0850385. [DOI] [PubMed] [Google Scholar]
- Parameswaran N, Spielman WS. RAMPs: The past, present and future. Trends Biochem Sci. 2006;31:631–8. doi: 10.1016/j.tibs.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Perry KJ, Quiza M, Myers DE, Morfis M, Christopoulos G, Sexton PM. Characterization of amylin and calcitonin receptor binding in the mouse alpha-thyroid-stimulating hormone thyrotroph cell line. Endocrinology. 1997;138:3486–96. doi: 10.1210/endo.138.8.5312. [DOI] [PubMed] [Google Scholar]
- Poyner DR, Sexton PM, Marshall I, Smith DM, Quirion R, Born W, Muff R, Fischer JA, Foord SM. International Union of Pharmacology. XXXII. The mammalian calcitonin gene-related peptides, adrenomedullin, amylin, and calcitonin receptors. Pharmacol Rev. 2002;54:233–46. doi: 10.1124/pr.54.2.233. [DOI] [PubMed] [Google Scholar]
- Rask-Andersen M, Almen MS, Schioth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10:579–90. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- Ruan J, Li H, Chen Z, Coghlan A, Coin LJ, Guo Y, Heriche JK, Hu Y, Kristiansen K, Li R, Liu T, Moses A, Qin J, Vang S, Vilella AJ, Ureta-Vidal A, Bolund L, Wang J, Durbin R. TreeFam: 2008 Update. Nucleic Acids Res. 2008;36:D735–40. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H, Sawchenko P, Chesnut J, Rivier J, Vale W, Pandol SJ. Receptor for calcitonin gene-related peptide: binding to exocrine pancreas mediates biological actions. Am J Physiol. 1985;249:G147–51. doi: 10.1152/ajpgi.1985.249.1.G147. [DOI] [PubMed] [Google Scholar]
- Sexton PM, Poyner DR, Simms J, Christopoulos A, Hay DL. Modulating receptor function through RAMPs: can they represent drug targets in themselves? Drug Discov Today. 2009;14:413–9. doi: 10.1016/j.drudis.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Sexton PM, Poyner DR, Simms J, Christopoulos A, Hay DL. RAMPs as drug targets. Adv Exp Med Biol. 2012;744:61–74. doi: 10.1007/978-1-4614-2364-5_6. [DOI] [PubMed] [Google Scholar]
- Steiner S, Muff R, Gujer R, Fischer JA, Born W. The transmembrane domain of receptor-activity-modifying protein 1 is essential for the functional expression of a calcitonin gene-related peptide receptor. Biochemistry. 2002;41:11398–404. doi: 10.1021/bi020279r. [DOI] [PubMed] [Google Scholar]
- Tsujikawa K, Yayama K, Hayashi T, Matsushita H, Yamaguchi T, Shigeno T, Ogitani Y, Hirayama M, Kato T, Fukada S, Takatori S, Kawasaki H, Okamoto H, Ikawa M, Okabe M, Yamamoto H. Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proc Natl Acad Sci U S A. 2007;104:16702–7. doi: 10.1073/pnas.0705974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udawela M, Christopoulos G, Tilakaratne N, Christopoulos A, Albiston A, Sexton PM. Distinct receptor activity-modifying protein domains differentially modulate interaction with calcitonin receptors. Mol Pharmacol. 2006;69:1984–9. doi: 10.1124/mol.105.021915. [DOI] [PubMed] [Google Scholar]
- Walker CS, Eftekhari S, Bower RL, Wilderman A, Insel PA, Edvinsson L, Waldvogel HJ, Jamaluddin MA, Russo AF, Hay DL. A second trigeminal CGRP receptor: function and expression of the AMY1 receptor. Ann Clin Transl Neurol. 2015;2:595–608. doi: 10.1002/acn3.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins HA, Au M, Bobby R, Archbold JK, Abdul-Manan N, Moore JM, Middleditch MJ, Williams GM, Brimble MA, Dingley AJ, Hay DL. Identification of key residues involved in adrenomedullin binding to the AM1 receptor. Br J Pharmacol. 2013;169:143–55. doi: 10.1111/bph.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C, Lu J, Li N, Barkan K, Richards GO, Roberts DJ, Skerry TM, Poyner D, Pardamwar M, Reynolds CA, Dowell SJ, Willars GB, Ladds G. Modulation of Glucagon Receptor Pharmacology by Receptor Activity-modifying Protein-2 (RAMP2) J Biol Chem. 2015;290:23009–22. doi: 10.1074/jbc.M114.624601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootten D, Lindmark H, Kadmiel M, Willcockson H, Caron KM, Barwell J, Drmota T, Poyner DR. Receptor activity modifying proteins (RAMPs) interact with the VPAC2 receptor and CRF1 receptors and modulate their function. Br J Pharmacol. 2013;168:822–34. doi: 10.1111/j.1476-5381.2012.02202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Kawate H, Iesato Y, Koyama T, Uetake R, Yang L, Yamauchi A, Tanaka M, Toriyama Y, Igarashi K, Nakada T, Kashihara T, Yamada M, Kawakami H, Nakanishi H, Taguchi R, Nakanishi T, Akazawa H, Shindo T. Novel regulation of cardiac metabolism and homeostasis by the adrenomedullin-receptor activity-modifying protein 2 system. Hypertension. 2013;61:341–51. doi: 10.1161/HYPERTENSIONAHA.111.00647. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Winborn CS, Marquez De Prado B, Russo AF. Sensitization of calcitonin gene-related peptide receptors by receptor activity-modifying protein-1 in the trigeminal ganglion. J Neurosci. 2007;27:2693–703. doi: 10.1523/JNEUROSCI.4542-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]