Abstract
Aims
This study was carried to reveal the genetic mechanisms of trimethoprim/sulfamethoxazole (SXT) resistance.
Methods
Among 300 clinical Stenotrophomonas maltophilia isolates from China, resistance determinants such as sul and dfrA genes, integrons and transposase were examined using PCR, DNA sequencing and thermal asymmetric interlaced PCR (TAIL-PCR). Data were analyzed using SPSS 20.0.
Results
Of the 300 isolates, 116 (38.7%) were resistant to SXT. An alarming trend of increased resistance to SXT were found over the 10-year period. The positive rates of sul and class 1 integrase (intI1) increased gradually with the development of SXT resistance over the 10-year period. Multiple logistic regression analyses indicated that the genes of qacEΔ1-sul1 (81% vs 46.2%, p = 0.000), sul2 (50.9% vs 9.8%, p = 0.000), intI1 (83.6% vs 65.8%, p = 0.000), dfrA12 (25% vs 3.3%, p = 0.000), dfrA17 (15.5% vs 3.8%, p = 0.000) and dfrA27 (4.3% vs 1.6%, p = 0.01) were more prevalent in SXT-resistant isolates than SXT-susceptible isolates except dfrA1(p = 0.83) and dfrA5(p = 0.18). Sequencing data revealed 12 types of resistance gene cassettes (aar-3-dfrA27, dfrA12–aadA2, dfrA17–aadA5, cmlA1, aacA4, aadA5, arr-3-aacA4, aadA1, aadB–aadA4, aacA4–catB8–aadA1, aadB–aac(6′)-II–blaCARB-8 and aac(6′)-II–blaCARB-8) located in the class 1 integron in 163 isolates (87% SXT-resistant vs 33.7% SXT-susceptible isolates, p = 0.000). A novel finding was the aar-3-dfrA27 (KC748137) gene cassette. The gene of sul2 linked to transposase in 50 SXT- resistant and 7 SXT- susceptible isolates was detected by TAIL-PCR.
Conclusions
The findings demonstrated a higher prevalence of sul, dfrA, intI1 and resistance gene cassettes in class 1 integron in SXT-resistant clinical S. maltophilia isolates in China. The sul1 and dfrA genes located in integrons and the sul2 linked to transposase may imply wide and rapid dissemination of resistance gene in bacteria.
Introduction
S. maltophilia, a non-fermentative gram-negative bacterium, is generally regarded as an important opportunistic pathogen, especially in immunocompromised patients with underlying disease, and can cause a number of clinical syndromes, such as bacteraemia, sepsis, pneumonia, meningitis, endocarditis, septic arthritis, urinary infections, and endophthalmitis [1–3].
S. maltophilia has been recognized as one of the leading multidrug resistant organisms in hospital settings due to its resistance to a broad array of antimicrobial agents afforded by the existence of intrinsic and acquired resistance mechanisms [4]. Trimethoprim/sulfamethoxazole (SXT) is traditionally recommended as the first choice against S. maltophilia infections; however, increasing resistance to SXT has complicated the treatment. Resistance determinants such as sul and dfrA genes, class 1 integrons and mobile genetic elements have been reported to contribute to SXT resistance [5–6], but these determinants were sometimes also detected in SXT- susceptible S. maltophilia isolates. To our knowledge, there is no detailed study describing the association between SXT resistance and resistance genes in a large collection by multivariate statistics.
In this study, the in vitro susceptibility of SXT against 300 clinical S. maltophilia isolates were examined to reveal the evolution of SXT resistance, SXT resistance determinants such as the sul and dfrA genes and resistance gene cassettes in integrons were determined, and their contribution to SXT resistance were analysed by multivariate statistics.
Materials and Methods
Bacterial isolates
A total of 300 clinical S. maltophilia isolates were collected from different patients after 72 h of hospitalization in 25 hospitals in Anhui, China in 2005–2014. The 300 isolates were comprised of 236 (78.67%) from sputum specimens, 26 from secretion (8.67%), 14 from urine (4.67%), 11 from blood (3.67%), 10 from drainage (3.33%), and 3 from cerebrospinal fluid (1.0%). All S. maltophilia isolates were identified using the MicroscanWalkaway-40 System (Dade Behring, Deerfield, IL, USA) and were re-identified via 16S rRNA sequencing with specific primers in Table 1. Quality control strain Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 35218, and Escherichia coli ATCC 25922 were stored at the Anhui Center for Surveillance of Bacterial Resistance.
Table 1. Oligonucleotide primers used in this study.
| primers | Sequences(5'-3') | Target | Reference |
|---|---|---|---|
| intI1-R | CAGTGGACATAAGCCTGTTC | class1 integrase | 6 |
| intI1-F | CCCGAGGCATAGACTGTA | ||
| In1-R | AAGCAGACTTGACCTGA | class 1 integron | 8 |
| In1-F | GGCATCCAAGCAGCAAG | ||
| hep74 | CGGGATCCCGGACGGCATGCAC | class 2 integron | 9 |
| hep51 | GATGCCATCGCAAGTACGAG | ||
| int3-F | AGTGGGTGGCGAATGAGTG | class 3 integranse | 10 |
| int3-F | TGTTCTTGTATCGGCAGGTG | ||
| sul1-R | ATTCAGAATGCCGAACACCG | sul1 | 6 |
| sul1-F | TAGCGAGGGCTTTACTAAGC | ||
| sul2-R | GAAGCGCAGCCGCAATTCAT | sul2 | 6 |
| sul2-F | CCTGTTTCGTCCGACACAGA | ||
| dfrA1-R | TTGTGAAACTATCACTAATGGTAG | dfrA1 | 6 |
| dfrA1-F | CTTGTTAACCCTTTTGCCAGA | ||
| dfrA5-R | TCCACACATACCCTGGTCCG | dfrA5 | 6 |
| dfrA5-F | ATCGTCGATATATGGAGCGTA | ||
| dfrA12-R | ATGAACTCGGAATCAGTACGC | dfrA12 | 6 |
| dfrA12-F | TTAGCCGTTTCGACGCGCAT | ||
| dfrA13-R | GAAACTATCACTAATGGCAGC | dfrA13 | 6 |
| dfrA13-F | CTCATCTGCTGGCTATCTCA | ||
| dfrA17-R | TTGAAAATATTATTGATTTCTGCAGTG | dfrA17 | 6 |
| dfrA17-F | GTTAGCCTTTTTTCCAAATCTGGTATG | ||
| dfrA27-R | TAAAGCAATAACTTACAATC | dfrA27 | This study |
| dfrA27-F | AAGAGTCTGATCGCCCATGCCG | ||
| sul2-SP1 | AAAGAACGCCGCAATGTGATCC | 3’flanking nucleotide of sul2 | This study |
| sul2-SP2 | GGATAGAACGCAGCGTCTGGAAAA | ||
| sul2-SP3 | CATCTGCCTTGAGCGCGTCC | ||
| sul2-SP4 | GTGGGAATGAGTTGGAAGAA | 5’flanking nucleotide of sul2 | This study |
| sul2-SP5 | GTTGCCGTTGTGGGTGCTGT | ||
| sul2-SP6 | TGCGCTATCGTGGGGGAATG | ||
| 16s RNA-R | ACGGCAGCACAGAAGAGC | 16s RNA | This study |
| 16s RNA-F | AGTTCGCATCGTTTAGGG |
SP, special primer; F, forward; R, reverse.
Antimicrobial susceptibility testing
Minimal inhibitory concentration (MIC) of trimethoprim/sulfamethoxazole against each isolate was determined by the agar dilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [7]. Quality control strains were included in each batch of antimicrobial testing to ensure the accuracy of the results. Isolates with MIC to trimethoprim/sulfamethoxazole < 2/38 mg/L were defined as susceptible isolates and resistant isolates with MIC to trimethoprim/sulfamethoxazole > 4/76 mg/L suggested by the CLSI [7].
Polymerase chain reaction (PCR)
The presence of class 1, 2 and 3 integrons as well as qacEΔ1-sul1, sul2 and dfrA1, dfrA5, dfrA12, dfrA17 and dfrA27 genes in each strain were assessed using the primers shown in Table 1[6, 8–10] using previously described conditions [6] using PCR (Biometra, Germany). Gene cassettes embedded within the class 1 integrons were also determined using the primers specific to the 5'conserved segment (CS) and the 3' CS listed in Table 1.
Thermal asymmetric interlaced PCR (TAIL-PCR)
For TAIL-PCR, three nested sequence-specific primers were utilized in consecutive reactions together with arbitrary degenerate (AD) primers to enhance the amplification efficiency of specific products [11]. Three specific primers (Table 1) and four AD primers (AD1, AD2, AD3 and AD4) from the Genome Walking Kit (TaKaRa Bio, Dalian, China) were used to amplify the 3' flanking sequences and 5' flanking sequences of sul2. The flanking sequences of sul2 were amplified by PCR according to the protocol of the Genome Walking Kit (TaKaRa Bio). PCR products were cloned into the pMD19-T vector and sequenced.
DNA sequencing and analysis
The nucleotide sequences of gene cassettes embedded in class 1 integrons were sequenced on both strands by the dideoxy chain termination method using an ABI 3130 DNA Sequencer (Applied Biosystem, Foster City, California, USA). Sequence analysis was performed with DNA Sequencing Analysis Software v5.1 (Applied Biosystems). Sequence alignment was conducted using the Nucleotide BLAST program (http://www.ncbi.nlm.nih.gov/nucleotide/).
Nucleotide sequence accession number
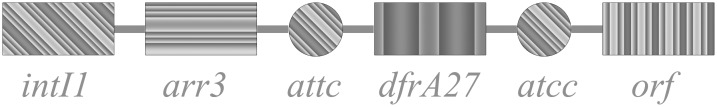
The sequences of the novel gene cassettes (aar-3-dfrA27, Fig 1) embedded in class 1 integron were deposited in GenBank and assigned the accession number: KC748137.
Fig 1. Schematic diagram of resistance gene cassettes located in the class 1 integron from Stenotrophomonas maltophilia isolates.
Statistical analyses
Multiple logistic regression analyses and chi-squared tests focused on the association of SXT resistance phenotype and resistance determinants using SPSS software, 20.0 (SPSS Inc., Chicago, IL, USA). A two-sided p-value < 0.05 was considered to be statistically significant.
Results
Antimicrobial susceptibility
An alarming trend of decreased susceptibility to SXT was found over the 10-year period in clinical S. maltophilia isolates (Tables 2 or 3). MIC50 (MIC for 50% of the isolates) was also increasing during the 10-year period. According to the comparison of resistance rates between the periods of 2005 to 2009 and 2010 to 2014, the percentage of isolates resistant to SXT were significantly changed: from 29.7% in the period 2005 to 2009 to 47.1% in the period 2010 to 2014 (p = 0.02).
Table 2. Distribution of resistance determinants to trimethoprim/sulfamethoxazole resistance in clinical Stenotrophomonas maltophilia isolates.
| year (n) | genes | SXT | MIC(mg/L) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| qacEΔ1-sul1(%) | sul2(%) | dfra1(%) | dfra5(%) | dfra12(%) | dfra17(%) | dfra27(%) | int1(%) | S(%) | R(%) | range | MIC50 | MIC90 | |
| 2005(26) | 11(42.3) | 2(7.7) | 1(3.8) | 11(42.3) | 21(80.8) | 5(19.2) | 0.25/4.75-32/608 | 0.25/4.75 | 8/152 | ||||
| 2006(26) | 12(46.2) | 2(7.7) | 1(3.8) | 2(7.7) | 13(50) | 18(69.2) | 8(30.8) | 0.25/4.75-32/608 | 0.25/4.75 | 16/304 | |||
| 2007(26) | 18(69.2) | 3(11.5) | 1(3.8) | 2(7.7) | 2(7.7) | 17(65.4) | 17(65.4) | 9(34.6) | 0.25/4.75-32/608 | 0.25/4.75 | 16/304 | ||
| 2008(24) | 13(54.2) | 6(25) | 3(12.5) | 3(12.5) | 15(62.5) | 16(66.7) | 8(33.3) | 0.25/4.75-32/608 | 0.25/4.75 | 32/608 | |||
| 2009(43) | 23(53.5) | 5(11.6) | 5(11.6) | 26(60.5) | 30(69.8) | 13(30.2) | 0.25/4.75-32/608 | 0.5/9.5 | 32/608 | ||||
| 2010(43) | 23(53.5) | 15(34.9) | 2(4.7) | 3(7.0) | 2(4.7) | 35(81.4) | 24(55.8) | 19(44.2) | 0.25/4.75-32/608 | 0.5/9.5 | 32/608 | ||
| 2011(20) | 12(60) | 10(50) | 1(5) | 1(5) | 4(20) | 2(10) | 3(15) | 17(85) | 12(60) | 8(40) | 0.25/4.75-32/608 | 1/19 | 32/608 |
| 2012(32) | 17(53.1) | 16(50) | 1(3.1) | 1(3.1) | 4(12.5) | 5(15.6) | 4(12.5) | 30(93.8) | 15(46.9) | 17(53.1) | 0.25/4.75-32/608 | 4/76 | 32/608 |
| 2013(28) | 25(89.3) | 13(46.4) | 1(3.6) | 6(21.4) | 5(17.9) | 1(3.6) | 24(85.7) | 14(50) | 14(50) | 0.25/4.75-32/608 | 4/76 | 32/608 | |
| 2014(32) | 25(78.1) | 7(21.9) | 5(15.6) | 3(9.4) | 30(93.7) | 17(53.1) | 15(46.9) | 0.25/4.75-32/608 | 4/76 | 32/608 | |||
| Total (300) | 179(59.7) | 77(25.7) | 4(1.3) | 4(1.3) | 35(11.7) | 25(8.3) | 8(2.7) | 218(72.7) | 184(61.4) | 116(38.7) | 0.25/4.75-32/608 | 0.5/9.5 | 32/608 |
SXT, trimethoprim-sulfamethoxazole; S, susceptible; R, resistant; MIC, Minimal inhibitory concentration; MIC50, the concentration needed to inhibit 50% of the strains; MIC90, the concentration needed to inhibit 90% of the strains
Table 3. Resistance determinants contributing to trimethoprim/sulfamethoxazole resistance in clinical Stenotrophomonas maltophilia isolates.
| Genes | SXT-resistant isolates (%) | SXT-susceptible isolates (%) | Multivariate Analysis | ||
|---|---|---|---|---|---|
| n = 116 | n = 184 | OR | 95%CI | p | |
| qacEΔ1-sul1 | 94(81.0) | 85(46.2) | 22 | (2.08–4.24) | 0.000 |
| sul2 | 59(50.9) | 18(9.8) | 49 | (2.87–5.12) | 0.000 |
| dfra1 | 2(1.7) | 2(1.1) | 1 | (-2.05–2.53) | 0.830 |
| dfra5 | 3(2.6) | 1(0.5) | 11 | (-1.17–6.09) | 0.180 |
| dfra12 | 29(25) | 6(3.3) | 20 | (1.84–4.20) | 0.000 |
| dfra17 | 18(15.5) | 7(3.8) | 10 | (1.10–3.53) | 0.000 |
| dfra27 | 5(4.3) | 3(1.6) | 11 | (0.49–4.33) | 0.010 |
| int1 | 97(83.6) | 121(65.8) | 4 | (0.70–2.46) | 0.000 |
SXT, trimethoprim-sulfamethoxazole.
Prevalence of resistance genes
In the present study, the genes for qacEΔ1-sul1, sul2 and class 1 integrase (intI1) were detected in 179 (59.7%), 77 (25.7%) and 218 (72.7) of the 300 S. maltophilia isolates, and the positive rates of the three genes increased gradually with the development of SXT resistance (Table 2). No class 2 or 3 integrons were found. The dfrA genes were detected in 76 isolates (dfrA1, 4 isolate; dfrA5, 4 isolate; dfrA12, 35 isolates; dfrA17, 25 isolates; dfrA27, 8 isolates) (Table 2). In addition, 85(46.2%), 18(9.8%), 121(65.8%), 2(1.1%), 1(0.5%), 6(3.3%), 7(3.8%), and 3(1.6) SXT-susceptible isolates were detected to contain the qacEΔ1-sul1, sul2, intI1, dfrA1, dfrA5, dfrA12, dfrA17, and dfrA27 genes, respectively. Considering all the resistance determinants contributing to SXT resistance between the two groups of SXT-resistant isolates and SXT-susceptible isolates, the results of multiple logistic regression analysis indicated that qacEΔ1-sul1(OR: 22, 95% CI: 2.08–4.24, p = 0.000), sul2(OR: 49, 95% CI: 2.87–5.12, p = 0.000), intI1(OR: 4, 95% CI: 0.70–2.46, p = 0.000), dfrA12(OR: 20, 95% CI: 1.84–4.20, p = 0.000), dfrA17 (OR: 10, 95% CI: 1.10–3.53, p = 0.000)and dfrA27 (OR: 11, 95% CI: 0.49–4.33, p = 0.01) genes were significantly more prevalent in SXT-resistant isolates than SXT-susceptible isolates except dfrA1(p = 0.83) and dfrA5(p = 0.18) (Table 3).
Among the 77 sul2-positive isolates, according to the results of TAIL-PCR and sequence analysis, the sul2 gene was detected to be associated with a transposase gene in 50 SXT- resistant isolates and 7 SXT- susceptible isolates.
Detection of resistance gene cassettes in class 1 integron
Of the 218 class 1 integrase-positive isolates, 74.8% (163 isolates) were shown to contain resistance gene cassettes, which were found in 87% (101 isolates) of the SXT-resistant isolates and 33.7% (62 isolates) of the SXT-susceptible isolates. The positive rate of resistance gene cassettes in SXT-resistant isolates was significantly higher than that in SXT-susceptible isolates by chi-squared tests (p = 0.000). The gene cassettes in the 163 S. maltophilia isolates included those resistant to aminoglycosides, trimethoprim, β-lactams, rifampicin and chloramphenicol: aar-3-dfrA27 (8 isolates), dfrA17–aadA5 (25 isolates), dfrA12–aadA2 (35 isolates), aacA4–catB8–aadA1(8 isolates), aadB–aac(6′)-II–blaCARB-8 (8 isolates), arr-3-aacA4 (11 isolates), aadB–aadA4 (9 isolates), aacA4 (15 isolates), aadA5(12 isolates), aadA1(10 isolates), aac(6′)-II–blaCARB-8 (7 isolates) and cmlA1(15 isolates), The complete sequence of gene was deposited in the Genebank (S1 Data).
Discussion
Due to its natural resistance to multiple classes of antimicrobial agents in S. maltophilia isolates, SXT is recommended as the first-line therapy for S. maltophilia infection. However, the treatment of S. maltophilia infections is being threatened due to the recent development of resistance to SXT. Reports from different countries have demonstrated 4%-84% resistance to SXT in S. maltophilia isolates [12–16]. In the present study, our data demonstrated a growing frequency of isolates resistant to SXT over the 10-year period. SXT-resistant isolates made up 19% of the isolates in 2005, increased to 30% in the period 2006 to 2009, and then increased to 40% from 2010 to 2014. There are statistically significant differences in SXT-resistant rates between the 2005 to 2009 period, and the 2010 to 2014 period (p = 0.02).
SXT resistance in S. maltophilia has been reported to be associated with the qacEΔ1-sul1 and sul2 genes encoding the dihydrofolate synthetase enzyme which confer resistance to sulfamethoxazole [6, 16–17]. We have found that qacEΔ1-sul1 in combination with sul2 confer high-level resistance to SXT in previous research [6]. However, as the study progressed, some SXT-susceptible isolates were detected to contain both qacEΔ1-sul1 and sul2 genes. In this research, among the 300 isolates collected during the 10-year period, the qacEΔ1-sul1 genes were detected in 81% (n = 94) isolates that were SXT-resistant. However, 46.2% of the SXT-susceptible isolates were found to contain qacEΔ1-sul1 genes, which may imply that resistance to SXT is caused by multiple drug resistance genes confirmed by multiple logistic regression analysis. Because resistance to SXT is coselected with resistance to quaternary ammonium compound (QAC), the presence of the QAC resistance gene qacEΔ1 has been associated with SXT resistance [18–19]. Therefore, the use of disinfectants containing QACs might increase the risk of SXT resistance in hospital settings and natural environments.
Class 1 integrase genes were detected in 83.6% (97 isolates) of SXT-resistant isolates and 65.8% (121 isolates) of SXT-susceptible isolates; however, more resistance genes were embedded in SXT-resistant isolates. Typically, a class 1 integron structure contains IntI1 on the 5’-conserved segment (5’CS) and qacEΔ1-sul1 on the 3’-conserved segment. In this study, thirteen isolates were detected to contain qacEΔ1-sul1 genes without intI1, and the qacEΔ1-sul1 gene was found absent in 52 intI1-positive isolates, which is in agreement with previous findings on the unusual structures of the 3’- conserved region of class 1 integrons [20].
The sul2 genes were found in 50.9% (59 isolates) of SXT-resistant isolates and 9.8% (18 isolates) of SXT-susceptible isolates, and sul2 genes in 50 SXT- resistant isolates and 7 SXT- susceptible isolates were detected to be associated with transposase gene. The presence of sul2 gene has increased SXT resistance, when it is associated with transposons, the gene could be further disseminated among bacteria through horizontal gene transfer in S. maltophilia isolates.
49.1% (57 isolates) of SXT-resistant isolates and 10.3% (19 isolates) of SXT-susceptible isolates were positive for dfrA genes encoding the dihydrofolate reductase enzyme, which confer resistance to trimethoprim. Of the dfrA gene subtypes, dfrA12 and dfrA17 were more prevalent than other genes. Besides, dfrA12, dfrA17, and dfrA27 were found to be embedded in class 1 integron, which may imply the wide and rapid dissemination of these genes in S. maltophilia isolates in this region.
Among the 300 isolates, multiple logistic regression analysis was used in determining the contribution of resistance determinants to SXT resistance against S. maltophilia isolates among the qacEΔ1-sul, dfra and intI1 genes. The resistance determinants including qacEΔ1-sul1, sul2, intI1, dfrA12, dfrA17, and dfrA27, but not dfrA1 and dfrA5, were found to be significantly more frequent in the SXT-resistant isolates than in the SXT-susceptible isolates (p < 0.05). This might be attributed to fewer dfrA1- positive and dfrA5—positive strains. The difference in the prevalence of the two genes between SXT- resistant isolates and SXT-susceptible isolates did not reveal any statistical significance.
Furthermore, among 218 intI1-positive isolates, a total of 12 kinds of resistance gene cassettes were detected in 163 (74.8%) of S. maltophilia isolates. The positive rate of resistance gene cassettes was significantly higher in SXT-resistant isolates (87%, 101 isolates) than that in SXT-susceptible isolates (33.7%, 62 isolates) (p = 0.000). The most common gene cassettes located in the class 1 integrons of S. maltophilia isolates in this region were aar-3-dfrA27, dfrA12–aadA2, dfrA17–aadA5, cmlA1, aacA4, aadA5, arr-3-aacA4, aadA1, aadB–aadA4, aacA4–catB8–aadA1, aadB–aac(6′)-II–blaCARB-8, and aac(6′)-II–blaCARB-8. Among the resistance gene cassettes in this research, a novel finding was the discovery of aar-3-dfrA27 genes which confer resistance to rifampin and trimethoprim. As far as we know, this is the first report of the dfrA27 gene being embedded in class 1 integron in S. maltophilia isolates, following the discovery of the dfrA17 and dfrA12 gene cassettes in clinical S. maltophilia isolates [6]. In addition, the prevalence of intI1 gene and resistance gene cassettes contained in class 1 integrons have increased year by year since the development of SXT resistance, to which clinicians may need pay more attention.
In conclusion, the findings of the current study revealed an increasing presence of qacEΔ-sul1, sul2, dfra12, dfra17 and intI1 in clinical S. maltophilia isolates in China during a 10-year period. The present findings also indicated a higher prevalence of these resistance determinants in SXT-resistant isolates than in SXT-susceptible isolates. Moreover, the qacEΔ1-sul1, dfrA12, dfrA17, and dfrA27 genes located in class 1 integron, and the sul2 gene linked to transposase gene may imply the wide and rapid dissemination of resistance genes in bacteria in this region. In our opinion, this is a serious medical problem which should be of concern.
Supporting Information
(DOCX)
Acknowledgments
The authors thank the 25 participating hospitals for help in collecting the isolates and Yan-Yan Liu, Xue-Jiao Ma and Nian Sun for technical assistance in antimicrobial susceptibility testing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Natural Science Foundation of China (No. 81101313, Li-Fen Hu, https://isis.nsfc.gov.cn), and by Natural Science Foundation of China (No. 81172737, Jia-Bin Li, https://isis.nsfc.gov.cn). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Denton M, Kerr KG (1998) Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 11, 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang JS, Flynn HW Jr, Miller D, Smiddy WE (2013) Stenotrophomonas maltophilia endophthalmitis following cataract surgery: clinical and microbiological results. Clin Ophthalmol. 7, 771–777. 10.2147/OPTH.S39608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Looney WJ, Narita M, Mühlemann K (2009) Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis. 9, 312–323. 10.1016/S1473-3099(09)70083-0 [DOI] [PubMed] [Google Scholar]
- 4.Brooke JS. New strategies against Stenotrophomonas maltophilia: a seriousworldwide intrinsically drug-resistant opportunistic pathogen (2014) Expert Rev Anti Infect Ther. 12, 1–4. 10.1586/14787210.2014.864553 [DOI] [PubMed] [Google Scholar]
- 5.Barbolla R, Catalano M, Orman BE, Famiglietti A, Vay C, Smayevsky J, et al. (2004) Class 1 integrons increase trimethoprim-sulphamethaxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 48, 666–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu LF, Chang X, Ye Y, Wang ZX, Shao YB, Shi W, et al. (2011) Stenotrophomonas maltophilia resistance to trimethoprim/ sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int J Antimicrob Agents 37, 230–234. 10.1016/j.ijantimicag.2010.10.025 [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (2013) Performance standards for antimicrobial susceptibility testing. Twenty-third informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA, M100-S23. 33, 68–69. [Google Scholar]
- 8.Levesque C, Pyche L, Larose C, Roy PH (1995) PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 39, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White PA, McIver CJ, Rawlinson WD (2001) Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother 45, 2658–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Registe B, et al. (2001) Incidence of Class 1 and 2 Integrases in Clinical and Commensal Bacteria from Livestock, Companion Animals, and Exotics. Antimicrob Agents Chemother 45, 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trinh Q, Zhu P, Shi H, Xu W, Hao J, Luo Y, et al. (2014) A-T linker adapter polymerase chain reaction for determining flanking sequences by rescuing inverse PCR or thermal asymmetric interlaced PCR products. Anal Biochem 466, 24–26. 10.1016/j.ab.2014.07.022 [DOI] [PubMed] [Google Scholar]
- 12.Farrell DJ, Sader HS, Jones RN (2010) Antimicrobial susceptibilities of a worldwide collection of Stenotrophomonas maltophilia isolates tested against tigecycline and agents commonly used for S. maltophilia infections. Antimicrob Agents Chemother 54, 2735–2737. 10.1128/AAC.01774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Wang JT, Shiau YR, Wang HY, Lauderdale TL, Lauderdale TL, et al. (2012) A multicenter surveillance of antimicrobial resistance on Stenotrophomonas maltophilia inTaiwan. J Microbiol Immunol Infect 45, 120–126. 10.1016/j.jmii.2011.09.028 [DOI] [PubMed] [Google Scholar]
- 14.Cho SY, Lee DG, Choi SM, Park C, Chun HS, Park YJ, et al. (2015) Stenotrophomonas maltophilia bloodstream infection in patients with hematologic malignancies: a retrospective study and in vitro activities of antimicrobial combinations. BMC Infect Dis 19, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Church D, Lloyd T, Peirano G, Pitout J (2013) Antimicrobial susceptibility and combination testing of invasive Stenotrophomonas maltophilia isolates. Scand J Infect Dis 45, 265–270. 10.3109/00365548.2012.732240 [DOI] [PubMed] [Google Scholar]
- 16.Kaur P, Gautam V, Tewari R (2015) Distribution of class 1 Integrons, sul1 and sul2 genes among clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in North India. Microb Drug Resist 21, 380–385. 10.1089/mdr.2014.0176 [DOI] [PubMed] [Google Scholar]
- 17.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR (2007) Global emergence of trimethoprim/ sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis. 13, 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang LL, Lin HH, Chang CY, Lu PL (2007) Increased incidence of class 1 integrons in trimethoprim/sulfamethoxazole resistant clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother 59, 1038–1039. [DOI] [PubMed] [Google Scholar]
- 19.Gaze WH, Abdouslam N, Hawkey PM, Wellington EMH (2005) Incidence of class 1 integrons in quaternary ammonium compound-polluted environment. Antimicrob Agents Chemother 49, 1802–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MF, Peng CF, Hsu HJ, Toh HS (2011) Use of inverse PCR for analysis of class 1 integrons carrying an unusual 3′conserved segment structure. Antimicrob Agents Chemother 55, 943–945. 10.1128/AAC.00988-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.