Abstract
Chronic Hepatitis B Virus (HBV) infections can progresses to liver cirrhosis and hepatocellular carcinoma (HCC). The HBV covalently-closed circular DNA cccDNA is a key to HBV persistence, and its degradation can be induced by the cellular deaminase APOBEC3. This study aimed to measure the distribution of intrahepatic cccDNA levels and evaluate the association between levels of cccDNA and APOBEC3 in HCC patients. Among 49 HCC patients, 35 matched cancerous and contiguous noncancerous liver tissues had detectable cccDNA, and the median intrahepatic cccDNA in the cancerous tissues (CT) was significantly lower than in the contiguous noncancerous tissues (CNCT) (p = 0.0033). RCA (rolling circle amplification), followed by 3D-PCR identified positive amplification in 27 matched HCC patients. Sequence analysis indicated G to A mutations accumulated to higher levels in CT samples compared to CNCT samples, and the dinucleotide context showed preferred editing in the GpA context. Among 7 APOBEC3 genes, APOBEC3B was the only one up-regulated in cancerous tissues both at the transcriptional and protein levels (p < 0.05). This implies APOBEC3B may contribute to cccDNA editing and subsequent degradation in cancerous tissues.
Introduction
Hepatitis B virus (HBV) infection is a major health problem world-wide. Up to 350 million people have chronic infection and are at high risk of progressing to liver cirrhosis and hepatocellular carcinoma (HCC) [1,2].
HBV is a partially double-stranded DNA virus that replicates by reverse transcription which occurs within viral core particles in the cytoplasm [3,4]. Newly synthesized viral genomes can either be secreted as virions, or they can be transported into the nucleus where the relaxed circular DNA (RC DNA) is converted to covalently-closed circular DNA (cccDNA). The nuclear cccDNA accumulates to about 1 to 50 copies per hepatocyte in animal models as fairly stable mini-chromosomes [5,6]. Despite this low accumulation level, the cccDNA is key to HBV persistence because it is the template for the all HBV mRNAs, including the pregenomic RNA (pgRNA) that is converted to DNA during reverse transcription [7].
Previously, extensive studies have shown that HBV cccDNA persists throughout the various clinical phases of chronic hepatitis [8,9]. Even in patients with occult HBV infection who are negative for hepatitis B surface antigen (HBsAg) but have detectable HBV DNA in sera or liver tissues, cccDNA is still present [10,11], although at low levels. Clinical reports stressed the importance of measuring levels of cccDNA due to its key position in the viral replication cycle [8]. In addition, intrahepatic cccDNA levels vary between the immune-tolerant phase and immune-clearance phage of chronic HBV infection [12]. This implies that host factors such as hepatocyte turnover and inflammatory cytokines may affect cccDNA levels [7,13].
The apolipoprotein B mRNA-editing catalytic polypeptide 3 (APOBEC3) family is comprised of seven DNA cytidine deaminases (APOBEC3A, B, C, D, F, G and H) in humans. These proteins can bind to target DNA and convert cytosine to uracil [14–16]. As innate antiviral factors, APOBEC3 proteins can edit HBV genome and reduce HBV replication in vitro and in vivo [17–19]. HBV DNA carrying C to U modifications can subsequently be degraded, or alternatively, the large number of G to A mutations that accumulate in the plus strand DNA during reverse transcription could render it non-infectious. Recently, Kitamura et al demonstrated that Duck HBV cccDNA accumulated G to A hypermutations that were induced by APOBEC3G, and the lesions were repaired by the uracil-DNA glycosylase (UNG)- mediated base-excision repair (BER) pathway [20]. Another intriguing study reported IFN -α and lymphotoxin-β-receptor could up-regulate APOBEC3A and 3B, respectively, leading to cytidine deamination- dependent cccDNA degradation [21]. These two reports imply that APOBEC3 could target double-strand cccDNA and decrease its levels in the nucleus.
Considering important role of cccDNA in chronic hepatitis B infection (CHB), conducting clinical research to verify this association of cccDNA levels and APOBEC3 proteins is important. Therefore, we measured the levels of cccDNA in HCC tissues and then examined whether APOBEC3B may be a candidate host factor for cccDNA editing that could decrease levels cccDNA in HCC. These studies employed cccDNA quantification, analysis cccDNA mutation patterns, evaluation of APOBEC3B expression levels in paired CT and CNCT samples of HCC patients.
Materials and Methods
Patients
Matched cancerous and contiguous noncancerous liver tissues were obtained from 49 HCC patients who underwent surgical resection at the Chongqing Medical University First and Second Hospitals. All patients were ethnic Han Chinese. HBsAg-positive carriers (n = 45) and HBsAg-negative/ hepatitis B core antibody (HBcAb) -positive patients (n = 4) were enrolled. Diagnosis of HCC for these samples was confirmed by histopathology. Hepatitis C virus (HCV) or Human Immunodeficiency Virus (HIV) infected patients defined by anti-HCV or anti -HIV positivity were excluded as APOBEC3 expression could be affected by coinfection [22]. Serum HBsAg, anti-HBs, HBeAg, anti-HBe, anti-HBc were measured by ELISA (Kehua, Shanghai, China) according to the manufacturer’s instruction. Tissue samples were rapidly frozen in liquid nitrogen and stored at –80°C until use.
Plasmids employed
The expression vector for hemagglutinin (HA)-tagged APOBEC3B was constructed by Genecopoeia Company. The human APOBEC3B (NM_004900.4) was cloned with a C-terminal HA-tag in pReceiver vector.
pCH9/3091 is an HBV expression vector contains 1.1 copies of HBV (genotype D) genome. It was obtained from Dr. Lin Lan (The Third Military Medical University, Chongqing, China).The YMHA derivative of this plasmid was made by introducing YMDD to YMHA mutations into the HBV P gene at this key active site motif.
Quantitation of intrahepatic HBV cccDNA and HBV total DNA
Intrahepatic HBV total DNA was extracted from 20mg of liver tissues using QIAamp
DNA Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions, then the extracted DNAs were treated with DNA-safe ATP dependent enzyme (PSAD) (Epicentre, Madison, WI) to digest HBV linear double-strands (dsl) and single-strand (ss) DNA in preparation for cccDNA detection and amplification.
We then performed real-time PCR with a TagMan probe to quantify intrahepatic HBV cccDNA and total HBV DNA. For the HBV cccDNA, a pair of cccDNA-selective primers and a probe were selected that targets the gap region between the two direct repeat regions (DR1 and DR2) of the viral genome [8]. Intrahepatic HBV cccDNA and total DNA levels were normalized by the amount of β-Globin DNA in the samples. Cell numbers were calculated based on an estimation of 6.667 pg/hgDNA per cell [9]. The primers and probe for HBV total DNA and β-Globin are shown in S1 Table.
Mutation analysis of cccDNA by RCA and 3D-PCR
HBV cccDNA was preferentially amplified relative to the other HBV DNA forms by rolling circle amplification (RCA) [23]. Briefly, cccDNA from liver tissue was amplified by phi-29 DNA polymerase mixed with HBV specific primers as described, then the concatemerized RCA product was amplified with full-length HBV genomic PCR.
The core region was amplified by employing 3D-PCR for mutational analysis of the cccDNA: Concatemerized PCR products from RCA reaction were used as templates for 3D-PCR. The amplification conditions were: 5 min (82–86°C), then 20 sec at 82–86°C; 30 sec at 51.5°C; and 50 sec at 72°C for 35 cycles, then at 10 min 72°C. The primers for full-length genomic PCR and 3D-PCR are in S1 Table.
Quantitation of AP sites of cccDNA
To measure the AP sites in cccDNA-containing samples, 1μg of cccDNA-containing extracts was treated with apuricic/apyrimidinic endonuclease (APE1) (New England Biolabs, Ipswich, MA, USA) for 4h at 37°C, then the levels of cccDNA were measured before and after treatment by real time PCR with a cccDNA specific probe described above.
Quantitation of APOBEC3 transcript profiles
Total RNA was isolated from the liver samples using Trizol reagent (Life Technologies, Carlsbad, CA, USA). 1 ug RNA was reverse transcribed into cDNA using GoScriptTM Reverse Transcription System (Promega, WI, USA), then the mRNA levels of the 7 APOBEC3 genes were measured by RT-qPCR using SYBRPremix Ex Taq (Takara Bio Inc, Ostu, Japan) as described earlier [24]. The measurement of each target mRNA level was calculated relative to constitutive housekeeping gene GAPDH using the 2-ΔΔCT method.
Immunoblotting analysis
Total protein was isolated from liver samples using RIPA (Beyotime, Beijing, China) with protease inhibitors cocktail (Roche, Indianapolis, USA). Expression level of APOBEC3A and APOBEC3B were analyzed by immunoblotting using primary antibodies specific for APOBEC3A and APOBEC3B (sc-130688, sc-130955; Santa Cruz Biotechnology, Dallas, USA), followed by secondary peroxidase-conjugated antibodies (ZSGB-BIO, Beijing, China) and chemiluminescence (Millipore Corporation, Billerica, USA).
Isolation of viral DNA and Southern blot
For RCA control experiment, HBV cccDNA extraction from HepAD38 cells employed a modified Hirt procedure [25]. HBV DNA from core particles were isolated by mild detergent lysis of the cells and sedimentation through sucrose density gradient as described [26,27]. To detect HBV DNAs of RCA product, Southern blot was performed as reported [26] with minor modifications by using the DIG high prime DNA labeling and detection starter kit (Roche Diagnostics GmbH, Mannheim, German).
For APOBEC3B edit RNA experiments, Huh7.0 cells were maintained in Dulbecco’s modified Eagle medium with 10% fetal bovine serum. All transfections were performed using Mirus LT1 transfection reagent (Mirus Bio, Madison,WI, USA). Total RNA in the cell lysates was isolated using Trizol reagent (Life Technologies, Carlsbad, CA, USA). After digesting DNA in the samples with DNase I (Takara Bio Inc, Ostu, Japan), 1 μg of RNA was reverse transcribed into cDNA using GoScriptTM Reverse Transcription System (Promega, WI, USA), and analyzed by 3D-PCR.
Statistical Analysis
All statistical analyses were performed by R language (The R Project for Statistical Computing). Comparisons of categorical variables (ratio of male: female, HBeAg, HBcAb) between groups were performed by Fisher exact test, while the Wilcoxon test was used for continuous variables (age, ALT AFP, serum DNA load). Differences in levels of HBV cccDNA or APOBEC3 in matched samples were analyzed by paired-Wilcoxon test, and correlations were by Pearson’s correlation analysis. The number of mutations was calculated by a perl script. A P value of < 0.05 was considered as statistically significant.
Ethics
The study was approved by the Chinese Ethics Committee of Human Resources at the First and Second Hospitals of Chongqing Medical University. The participants provide their written informed consent to participate in this study. The Ethics Committee approved this consent procedure.
Results
Clinical Characteristics of the Patients
Demographic data for the 49 HCC patients are in Table 1. The average age was 51 years, and the average serum alanine aminotransferase (ALT) and HBV DNA levels were 87.65 IU/L and 9.37 x105 copies/ml, respectively. The mean serum HBV DNA levels in the HBsAg-positive group were significantly higher than in the HBsAg-negative group (9.65 x105 copy/ml versus 1.0 x 103, p = 0.015).
Table 1. Clinical information of patients.
| Characteristic | HBsAg-positive(n = 45) | HBsAg-negative(n = 4) | p value |
|---|---|---|---|
| Male:Female ratio | 39:6 | 2:2 | 0.1195b |
| HBeAg(+/-) | 13/32 | 1/3 | 0.629b |
| HBcAb(+/-) | 33/2 | 4/0 | 0.3376b |
| Mean age ± SD (years) | 49.64±12.90 | 69.5±9.57 | 0.0088a |
| ALT(IU/L) | 92.51±152.28 | 34.75±18.52 | 0.433a |
| AFP(ng/ml) | 1.94x 104±8.54x 104 | 42.1±67.86 | 0.212a |
| Mean serum HBV DNA levels(copies/ml) | 9.65x 105 | 1.0x 103 | 0.015a |
—: no detective data
a: Wilcox Test
b: Fisher exact test.
Intrahepatic levels of HBV DNA and cccDNA in patients
We detected intrahepatic cccDNA and total HBV DNA in the matched CT and CNCT samples using qPCR. All 49 HCC patients, including both HBsAg-positive and -negative individuals had detectable total intrahepatic HBV DNA, and the majority of the samples had intrahepatic total HBV DNA levels above 1 copy/cell, 88% (43/49) in the CT group and 94% (46/49) in the CNCT groups.
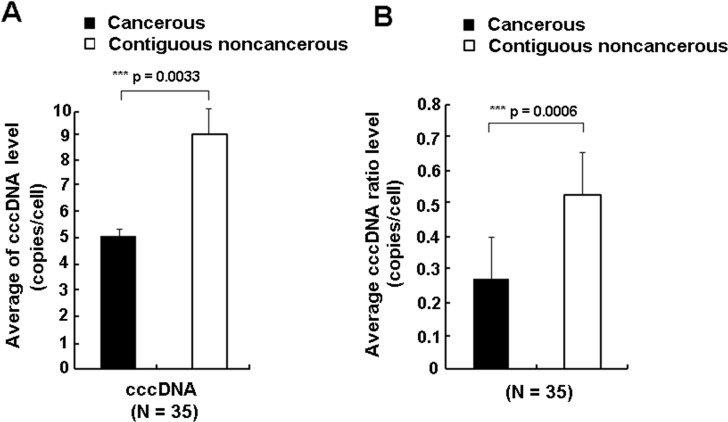
Intrahepatic levels of cccDNA were relatively low in the 49 HCC patients. For the 4 HBsAg- negative but anti-HBc positive patients, cccDNA was detected in 2 CT and 3 CNCT samples, while among the 45 HBsAg-positive patients, cccDNA was detected in 33 CT and 45 CNCT samples. About 41% (20/49) of the CT group with cccDNA had values lower than 0.1copies/cell, compared to 12% (6/49) of the CNCT group. The mean intrahepatic cccDNA in the CT group was about 1.8-fold lower than in the CNCT group (5.20 copies/cell versus 9.35 copies/cell, p = 0.0033, Fig 1A), and the mean ratio of cccDNA/total HBV DNA in CT group was also significantly lower than in the contiguous CNCT group (0.27 vs 0.53, p = 0.0006, Fig 1B).
Fig 1.
Comparison of detectable intrahepatic cccDNA levels (A) and ratio of cccDNA/total HBV DNA (B) between the CT group (solid box) and matched CNCT group (empty box) for 35 HCC patients by paired-Wilcoxon test. The levels of total DNA and cccDNA were normalized to copies/ cell.
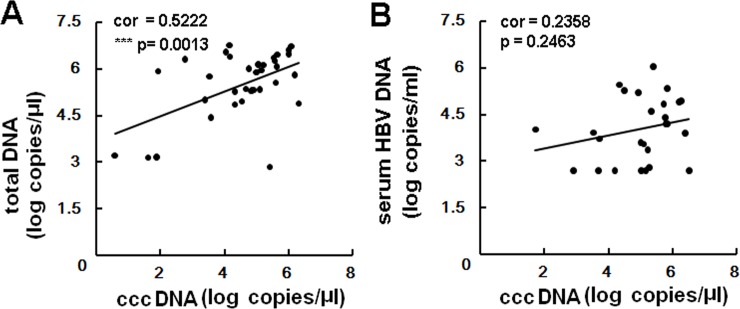
We also analyzed the correlation between intrahepatic cccDNA levels and total HBV levels or serum HBV DNA levels. The values of intrahepatic cccDNA, total HBV DNA, serum DNA for this correlation analysis complied with standard normal distribution employing R language Shapiro-Wilk Normality Test (S2 Table). Among patients who had detectable cccDNA, a positive correlation was observed between intrahepatic cccDNA levels and HBV DNA (r2 = 0.5221, p = 0.0013), but not for serum HBV DNA levels (r2 = 0.2358, p = 0.2463) (Fig 2).
Fig 2. Correlation between intrahepatic HBV cccDNA and HBV total DNA or serum HBV DNA.
A significant correlation was observed between total intrahepatic cccDNA levels and HBV DNA (panel A), but not for serum HBV DNA level (panel B) among 35 HCC patients with detectable cccDNA in paired samples.
Analysis of G to A mutations in HBV cccDNA in liver tissues
APOBEC3 proteins can inhibit HBV replication, especially APOBEC3A and APOBEC3B, by inducing HBV cccDNA degradation via a cytidine-deamination mechanism [21]. Therefore, we analyzed a possible correlation between APOBEC3 and HBV cccDNA levels in HCC patients by assessing the G to A mutation levels of cccDNA in the 49 HCC samples employed for cccDNA quantification.
RCA can be used not only for amplification of HBV cccDNA from liver biopsies [23], but also for preparation of randomly mutated plasmid libraries [28]. This let us to hypothesize RCA could be employed for cccDNA mutation analysis. The specificity of this method was confirmed with following validation assays. As shown in S1A Fig, only HBV cccDNA from HepAD38 or clinical liver tissues could be amplified by RCA, yielding a high molecular weight DNA product consisting of tandem repeats of HBV genome. As controls, HBV replication intermediates from cytoplasmic core particles or genomic liver DNA from HBV transgenic mice were both unable to be amplified by RCA due to lack of cccDNA. To recover sequence from cccDNA, the high molecular weight RCA products were amplified by HBV full-genomic PCR, yielding a clear band at 3.2kb (S1B Fig). Southern blot with HBV specific probe confirmed this band came from HBV (S1C Fig). Therefore, cccDNA sequence could be analyzed by combining RCA and PCR methods.
Next, to analyze G to A mutation levels in the paired 35 HCC samples with the detectable cccDNA, we employed RCA to amplify cccDNA specifically in these tissue samples first, then the RCA products were amplified by 3D-PCR as the 3D-PCR method can selectively amplify hypermutated DNA by lowering the denaturation temperature [29]. These 35 paired HCC samples were divided into 2 groups based on the ratio of cccDNA in the paired CT and CNCT samples (cccDNA value in CT / cccDNA value in CNCT) being either <1 or >1 (Table 2). Among these samples, 27 paired samples were amplified successfully in both tissue types, while 8 paired samples were not able to be amplified (Table 2, S2 Fig). For the 25 paired samples with ratio of cccDNA levels <1 (CT/CNCT), amplification of cccDNA from 16 paired CT group (such as #110, #178) employing a low denaturation temperature resulted in a relatively strong PCR signals compared with those from matched CNCT tissues (Fig 3A). For the 10 paired samples with ratio of cccDNA levels >1 (CT/CNCT), this 3D-PCR result was observed in 6 paired samples (Fig 3A, #161). Therefore, the positive results in 3D-PCR which were accordance with that ratio of cccDNA value between CT and CNCT are 62.85% (22/35, Table 2). In addition, there was no distinct difference in 3D-PCR patterns for 3 paired samples (Fig 3A, such as #155).
Table 2. 3D-PCR results of HCC patients.
| Ratio of cccDNA (CT/CNCT) | Results of 3D-PCR | |||
|---|---|---|---|---|
| CT amplified at lower temperatures than CNCT | CT amplified at higher temperatures than CNCT | No difference in amplification at lower temperatures between CT and CNCT | Not amplifiable | |
| Ratio<1 (n = 25) | 16 | 2 | 2 | 5 |
| Ratio>1 (n = 10) | 0 | 6 | 1 | 3 |
We define positive change for the following results: the denaturation temperature is lower in CT sample than that in CNCT sample with cccDNA ratio <1 (example like #110, 178 in Fig 3) or the denaturation temperature is higher in CT sample than that in the CNCT sample for the group with cccDNA ratio>1 (example like # 161 in Fig 3).
Fig 3. Mutational analysis of HBV cccDNA in HCC patients.
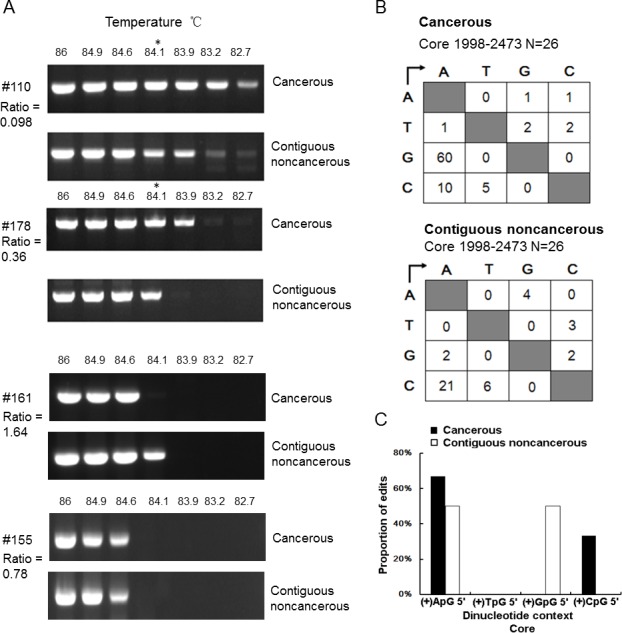
A. Agarose gel analysis of 3D-PCR of cccDNA from CT and CNCT of samples from patients #110, #178, #161and #155. cccDNA was specifically amplified by RCA, then the concatemerized product was analysis by 3D-PCR using a denaturation temperature gradient of 86°C-82°C. B. Mutation matrices of cccDNA from CT and CNCT groups. DNA fragments amplified at 94°C (as reference wild type sequence) or 84.1°C in each group were cloned into T vectors, 13 clones were randomly selected, and the core region was sequenced. C. Dinucleotide editing profile of G to A mutations of plus-polarity strand DNA. Asterisks indicate the 3D-PCR products cloned and sequenced.
Next, to confirm the predication that PCR products amplified from the cccDNA in the CT group had more AT-rich DNA due to deamination than the CNCT group, DNAs amplified at 84.1°C from matched CT and CNCT samples (#110 and #178) were cloned and sequenced. Sequence analysis of 13 individual clones revealed that more G to A mutations accumulated in CT group than in the CNCT group (Fig 3B). The frequency of G to A changes was 4.85 per 1000 sites in CT group, while it was only 0.16 in the CNCT group. These results demonstrated that G to A mutations accumulated 30-fold more frequently in the CT samples than in matched CNCT samples.
We noted the dinucleotide context for these G to A hypermutations in CT group showed preferred editing in the GpA motifs (66.7%, Fig 3C). This implies the editing may have been performed by one or more of the APOBEC3 proteins, consistent with previous studies that revealed that editing in GpA and GpG of dinucleotide context was a hallmark for APOBEC3 deaminases [18,22].
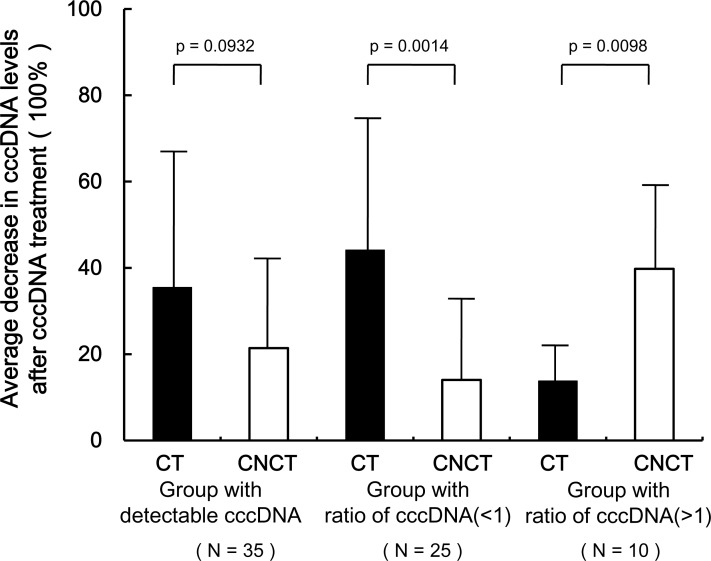
To further investigate whether the cccDNA contain AP sites whose levels may correlate with the loss of cccDNA in the CT samples, we quantified cccDNA levels before and after treatment by APE1 in 35 paired HCC samples because APE1 cleaves the phosphodiester backbone within an AP site. The mean decrease in cccDNA levels in the CT group following AP1 treatment was 1.7-fold greater than that in CNCT group (35.48% vs 21.39%, p = 0.0932, Fig 4), and was 3.1 fold higher in the group of samples where the CT/CNCT cccDNA ratio was < 1 (44.15% vs 14.04%, p = 0.0014).
Fig 4. Effect of APE1 treatment on detection of cccDNA levels in 35 paired CT and CNCT samples.
The 35 paired HCC samples with detectable cccDNA values (left column, N = 35) were divided into two group according to ratio of cccDNA in the paired CT and CNCT samples (CT/CNCT) < 1 (middle column, N = 25) or >1 (right column, N = 10). Extracted tissue DNAs containing the cccDNA were treated with APE1, and cccDNA levels were measured before and after treatment. The decrease in cccDNA levels in each sample was calculated as = (value before APE1 treatment—value after APE1 treatment) / value before APE1 treatment. CT: cancerous tissues, CNCT: Contiguous noncancerous tissues.
Expression levels of APOBEC3 deaminases in HCC liver tissues
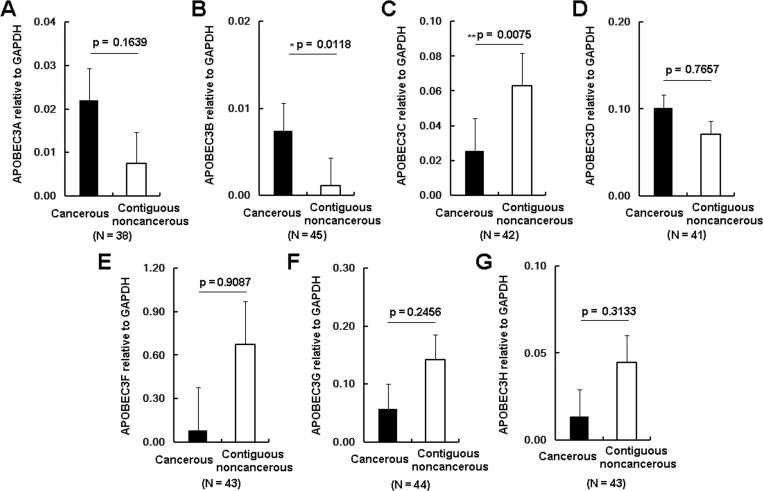
Next, the transcription profile of the APOBEC3 genes in these 49 HCC samples was measured by RT-qPCR with normalization to GAPDH. As shown in Fig 4, APOBEC3A, APOBEC3B and APOBEC3D were up-regulated in the CT group compared to the CNCT group, while APOBEC3C, APOBEC3E, APOBEC3F and APOBEC3G were down-regulated. Changes in the levels of APOBEC3B and APOBEC3C achieved statistical significance (APOBEC3B, p = 0.01, Fig 5B; APOBEC3C, p = 0.0075, Fig 5C). The proportion of samples in which APOBEC3B was up-regulated was 65% (29/45) in the CT tissues, while the proportion of up-regulated APOBEC3A was 37.5% (15/40). Considering APOBEC3A and APOBEC3B had been reported to be factors in limiting cccDNA levles, we conducted Western blot analysis of APOBEC3A and APOBEC3B from these liver tissues. Western blot from 21 paired HCC samples indicated the levels of APOBEC3B protein in CT samples were also widely up-regulated compared to the matched CNCT tissues (APOBEC3B, p = 0.0358, Fig 6). Again, the levels of APOBEC3A protein were up-regulated in the CT group, but this did not achieve statistical significance (APOBEC3A, p = 0.1942, Fig 6). Combined with the G to A mutations analysis, these data are consistent with up-regulation of APOBEC3B contributing to editing and degradation of the cccDNA in HCC patients.
Fig 5. The transcription profile of the 7 APOBEC3 genes in the CT group and matched CNCT group in HCC patients.
Levels of the target APOBEC3 mRNAs were measured by qRT-PCR and normalized to the expression levels of GAPDH. N represents the number of HCC patients with detectable expression of the 7 target APOBEC3 mRNAs. Asterisks indicate statistically significant differences: * 0.01< p < 0.05, ** 0.005< p < 0.01.
Fig 6. APOBEC3B is widely up-regulated in HCC.
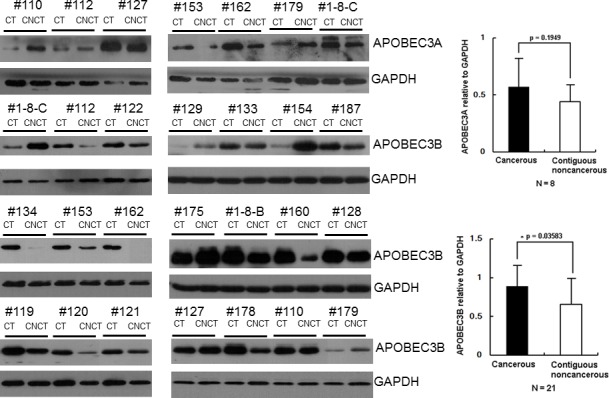
A. Protein levels of APOBEC3A and APOBEC3B in HCC samples were measured by Western blotting using anti-APOBEC3A and APOBEC3B antibodies; GAPDH was used as control for protein loading. CT: cancerous tissues, CNCT: Contiguous noncancerous tissues. Qualification of protein levels of APOBEC3A (panel B) and APOBEC3B (panel C) in HCC by normalization to the expression levels of GAPDH.
Discussion
HCC is one of the most frequent fatal cancer in the world, and it is estimated about 55% HCC are related to chronic infection with HBV [30,31]. Although the exact mechanism of HBV-induced HCC remains unclear, accumulated evidence has shown HBx protein is a multifunctional regulator in the pathogenesis of HCC [32]. In addition, HBV integration into host genome and alters expression of several genes that contribute to transforming hepatocytes and promote HCC development [33]. The essential role of the cccDNA in maintaining HBV infection indicated that it should be central in HCC development, albeit indirectly [34]. This indicates that investigating intrahepatic cccDNA levels of HCC patients is important.
Here, we found that about 71% (35/49) and 96% (47/49) of HCC patients had detectable cccDNA in CT and CNCT tissues, respectively. In the CT group, the levels of cccDNA ranged from 0.00114 copies/cell to 41.02 copies/cell, and this correlated with the amount of total intrahepatic HBV DNA (Fig 2). A similar result was observed by Wang et al [13]. When comparing the levels of cccDNA, the mean intrahepatic cccDNA in the CT group was significantly decreased by 1.8 fold than in the CNCT group (p = 0.0033, Fig 1), and cccDNA levels in the CT group tended to be distributed among the lower values. Previously, Wong et al had measured and compared the cccDNA levels in CT and CNCT samples, and showed the CT group had higher cccDNA levels [34]. The reason for this discrepancy is unknown, but may be due to the following reasons. First, PSAD enzyme was used to digest HBV double-stranded linear DNA and single-stranded DNA prior to quantification of cccDNA levels in our study, which would increase the accuracy for cccDNA qualification [35]. Second, the criterion of sample grouping is different for the two studies. HBsAg-negative patients were included in our research while they were excluded in Wong’s report. Several other studies have reported that in HBsAg-negative patients, cccDNA levels were still detectable and were at lower levels than in HBsAg-positive carriers [10,11]. Finally, our study subjects were all Han Chinese, mostly from Chongqing area with typical HBV genotype B and C infections, but we have no information about patients in Wong et al’s study. Therefore, further large scale population analyses need to be done to confirm this. Another issue is HBV cccDNAs do not replicate [36], and it is not clear how cccDNA is distributed into daughter cells during cell division. Meanwhile, it is difficult to quantify the hepatocyte turnover levels in HBV infected patients [37]. Therefore, our study cannot exclude the possibility that some of the loss of cccDNA may be due to proliferation of cancerous cells.
The observation that cccDNA levels in the tumor tissues decreased prompted us seek factors that may be involved in this process. APOBEC3 proteins have been identified as potent inhibitors of HBV, but the molecular mechanism of APOBEC3 inhibition is not completely clear. The mechanism of inhibition for HBV replication has been reported as being both dependent or independent of APOBEC3 enzymatic activity. Bonvin et al found APOBEC3B inhibition of HBV replication was dependent on hypermutation, with G to A accumulating in the plus-polarity DNA strand during reverse transcription [38]. Another report also found APOBEC3G is encapsidated with pgRNA in the core particles and interferes in the process of minus-strand DNA synthesis [39]. Recently, Lucifora et al found APOBEC3A and APOBEC3B are up-regulated by IFN -α and lymphotoxin-β-receptor, respectively, causing cccDNA degradation. These reports demonstrated the importance of APOBEC3 proteins against HBV replication, however, the mechanism still remains unclear, with a number of questions remaining. First, does APOBEC3B edit the HBV RNA or DNA? Although HBV viral RNA were reduced in the presence of APOBEC3B in HepG2.0 cells [40], our data revealed that APOBEC3B does not edit HBV RNA directly (S3 Fig). Another question that is still a matter of debate is whether APOBEC3 deaminates the single strand DNA during reverse transcription in the cytoplasm and/or if it deaminates double strand cccDNA in the nucleus [41] because the rate of deamination for APOBEC3 is 200 or 300 fold higher on ssDNA than dsDNA in vitro [42]. Therefore, further study focusing on how APOBEC3 proteins bind to cccDNA or single-stranded HBV DNA and whether the deamination pattern is similar or different between cccDNA and DNA from core particles would help to clarify this issue.
In our study, we found APOBEC3B was the only APOBEC3 family member significantly up-regulated in HCC patients (Fig 5). Other reports had indicated APOBEC3B was up-regulated in many types of tumor tissues, including ovarian [43], breast and lung carcinomas [44]. Furthermore, as APOBEC3 is mainly located in nucleus [45], which indicates that the reduced cccDNA levels in the CT samples may be in part due to up-regulated APOBEC3B. To clarify this point, we employed RCA and 3D-PCR to analyze the mutations of cccDNA in vivo. We found that more G to A mutations accumulated in the CT group than that in CNCT group (Fig 3), in accordance with the lower levels of cccDNA in the CT group. In addition, dinucleotide context analysis showed GpA motifs were the dominating editing profile for these G to A hypermutations in CT group. These results are consistent with APOBEC3B-mediated deamination contributing to the decline in cccDNA in HCC tissues. Meanwhile, it is reasonable to point out APOBEC3A may also contribute to this effect, but it likely plays a lesser role as it up-regulation did not achieved statistical significance (Fig 6).
In summary, our findings demonstrate that cccDNA levels are significantly lower in HCC tissues, and that the lower levels are likely to stem in part from up-regulation of APOBEC3B, which would target the HBV cccDNA and cause its deamination-dependent degradation. These results provide support for the potential efficacy of inducing APOBEC3B activity as part of an anti-HBV therapeutic regimen.
Supporting Information
A. Specificity of RCA to amplify cccDNA in cell culture or liver tissues. cccDNA purified from HepAD38 or clinical patients was amplified by RCA; DNA from core particles and genomic DNA from HBV- transgenic mice were used as controls. B. RCA product from line 2, line3 and line 4 in Fig A amplified by genomic PCR. C. Southern blot of RCA product amplified by genomic PCR.
(TIF)
CT and CNCT samples from the HCC patients were divided into 2 groups with ratio of cccDNA in the paired samples (CT/CNCT) being either < 1 or > 1. cccDNA was specifically amplified by RCA, then the concatemerized product was analysis by 3D-PCR using a denaturation temperature gradient of 86°C-82°C.
(TIF)
A. Southern blot of HBV core-associated DNA with transfected with a replication-incompetent HBV genomic expression vector (YMHA plasmid). B. 3D-PCR of cDNA reverscribed from RNA in lysates from cells transfected with the YMHA plasmid. Huh7.0 cells were cotransfected with YMHA plasmid plus empty plasmid or APOBEC3B expression plasmid. Total RNA was purified and digested with DNase I, then RT-PCR products served as templates for analysis by 3D-PCR with denaturing temperature ranging from 82–90°C. C. Mutation matrices of RT-PCR products. DNA fragments amplified at 94°C in the presence of empty vector and APOBEC3B were cloned into T vectors, and 15 clones were randomly selected and sequenced.
(TIF)
(DOC)
(DOC)
Acknowledgments
We thank Professor Keyue Ding at Chongqing Medical University for providing liver tissues from the HCC patients. We also thank Dr M. Muramatsu at Kanazawa University in Japan for sharing RCA protocol.
Abbreviations
- HBV
hepatitis B virus
- cccDNA
covalently-closed circular DNA
- RC DNA
relaxed circular DNA
- pgRNA
pregenomic RNA
- HBsAg
hepatitis B surface antigen
- HBcAb
hepatitis B core antibody
- HBeAg
hepatitis B e antigen
- HCC
hepatocellular carcinoma
- CT
cancerous tissues
- CNCT
contiguous noncancerous tissues
- APOBEC3
apolipoprotein B mRNA-editing catalytic polypeptide 3
- RCA
rolling circle amplification
- 3D-PCR
differential DNA denaturation PCR
- UNG
uracil-DNA glycosylase
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a National Sciences Foundation of China award (http://www.nsfc.gov.cn/)(81471945) to Y. Hu. This fund provided the publication. This work was supported by a Natural Science Foundation Project of CQ CSTC (http://www.cstc.gov.cn/)(cstc2014jcyjA10075) to Y. Hu. This fund provided the data collection and analysis. This work was supported by a Major National Science and Technology program (http://www.nhfpc.gov.cn/)(2013ZX10002002) to AL.H. This fund provided study design. This work was supported by a Chongqing Postdoctoral Science Foundation (http://www.cqhrss.gov.cn/u/cqhrss/) (Xm2014006) to Z.T. This fund provided the data collection and analysis. This work was supported by the Scientific and Technological Research Program of Chongqing Municipal Education Commission (http://www.cqedu.cn/) (KJ1400205) to Z.T. This fund provided the data collection and analysis.
References
- 1.Ganem D, Prince AM (2004) Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med 350: 1118–1129. [DOI] [PubMed] [Google Scholar]
- 2.Lin CL, Kao JH (2011) The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol 26 Suppl 1: 123–130. 10.1111/j.1440-1746.2010.06541.x [DOI] [PubMed] [Google Scholar]
- 3.Seeger C, Zoulim F, Mason WS (2007) Hepadnaviruses In: Knipe DM, Howley P, Griffin DE, Lamb RA, Martin MA et al. , editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; pp. 2977–3029. [Google Scholar]
- 4.Seeger C, Mason WS (2015) Molecular biology of hepatitis B virus infection. Virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moraleda G, Saputelli J, Aldrich CE, Averett D, Condreay L, et al. (1997) Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol 71: 9392–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dandri M, Burda MR, Will H, Petersen J (2000) Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology 32: 139–146. [DOI] [PubMed] [Google Scholar]
- 7.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, et al. (2009) Control of cccDNA function in hepatitis B virus infection. J Hepatol 51: 581–592. 10.1016/j.jhep.2009.05.022 [DOI] [PubMed] [Google Scholar]
- 8.Werle–Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, et al. (2004) Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology 126: 1750–1758. [DOI] [PubMed] [Google Scholar]
- 9.Wong DK, Yuen MF, Yuan H, Sum SS, Hui CK, et al. (2004) Quantitation of covalently closed circular hepatitis B virus DNA in chronic hepatitis B patients. Hepatology 40: 727–737. [DOI] [PubMed] [Google Scholar]
- 10.Bai F, Yano Y, Fukumoto T, Takebe A, Tanaka M, et al. (2013) Quantification of pregenomic RNA and covalently closed circular DNA in hepatitis B virus-related hepatocellular carcinoma. Int J Hepatol 2013: 849290 10.1155/2013/849290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Fang M, Gu X, Ji Q, Li D, et al. (2014) The intracellular HBV DNAs as novel and sensitive biomarkers for the clinical diagnosis of occult HBV infection in HBeAg negative hepatocellular carcinoma in China. PLOS One 9: e107162 10.1371/journal.pone.0107162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Zhao J, Zou Z, Liu Y, Li B, et al. (2014) Analysis of hepatitis B virus intrahepatic covalently closed circular DNA and serum viral markers in treatment-naive patients with acute and chronic HBV infection. PLOS One 9: e89046 10.1371/journal.pone.0089046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Fiel MI, Luan W, Blank S, Kadri H, et al. (2013) Impact of intrahepatic hepatitis B DNA and covalently closed circular DNA on survival after hepatectomy in HBV-associated hepatocellular carcinoma patients. Ann Surg Oncol 20: 3761–3770. 10.1245/s10434-013-3046-y [DOI] [PubMed] [Google Scholar]
- 14.Vieira VC, Soares MA (2013) The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int 2013: 683095 10.1155/2013/683095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Y, Du Y, Zhang Q, Han X, Cao G (2014) Human cytidine deaminases facilitate hepatitis B virus evolution and link inflammation and hepatocellular carcinoma. Cancer Lett 343: 161–171. 10.1016/j.canlet.2013.09.041 [DOI] [PubMed] [Google Scholar]
- 16.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM (2012) Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol 23: 258–268. 10.1016/j.semcdb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kock J, Blum HE (2008) Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J Gen Virol 89: 1184–1191. 10.1099/vir.0.83507-0 [DOI] [PubMed] [Google Scholar]
- 18.Suspene R, Guetard D, Henry M, Sommer P, Wain-Hobson S, et al. (2005) Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci U S A 102: 8321–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu R, Zhang X, Zhang W, Fang Y, Zheng S, et al. (2007) Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology 46: 1810–1820. [DOI] [PubMed] [Google Scholar]
- 20.Kitamura K, Wang Z, Chowdhury S, Simadu M, Koura M, et al. (2013) Uracil DNA glycosylase counteracts APOBEC3G-induced hypermutation of hepatitis B viral genomes: excision repair of covalently closed circular DNA. PLOS Pathog 9: e1003361 10.1371/journal.ppat.1003361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, et al. (2014) Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343: 1221–1228. 10.1126/science.1243462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vartanian JP, Henry M, Marchio A, Suspene R, Aynaud MM, et al. (2010) Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLOS Pathog 6: e1000928 10.1371/journal.ppat.1000928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margeridon S, Carrouee-Durantel S, Chemin I, Barraud L, Zoulim F, et al. (2008) Rolling circle amplification, a powerful tool for genetic and functional studies of complete hepatitis B virus genomes from low-level infections and for directly probing covalently closed circular DNA. Antimicrob Agents Chemother 52: 3068–3073. 10.1128/AAC.01318-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohba K, Ichiyama K, Yajima M, Gemma N, Nikaido M, et al. (2014) In vivo and in vitro studies suggest a possible involvement of HPV infection in the early stage of breast carcinogenesis via APOBEC3B induction. PLOS One 9: e97787 10.1371/journal.pone.0097787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao W, Hu J (2007) Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol 81: 6164–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Cheng X, Cao F, Huang A, Tavis JE (2013) beta-Thujaplicinol inhibits hepatitis B virus replication by blocking the viral ribonuclease H activity. Antiviral Res 99: 221–229. 10.1016/j.antiviral.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 27.Tavis JE, Cheng X, Hu Y, Totten M, Cao F, et al. (2013) The hepatitis B virus ribonuclease H is sensitive to inhibitors of the human immunodeficiency virus ribonuclease H and integrase enzymes. PLOS Pathog 9: e1003125 10.1371/journal.ppat.1003125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujii R, Kitaoka M, Hayashi K (2004) One-step random mutagenesis by error-prone rolling circle amplification. Nucleic Acids Res 32: e145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suspene R, Henry M, Guillot S, Wain-Hobson S, Vartanian JP (2005) Recovery of APOBEC3-edited human immunodeficiency virus G->A hypermutants by differential DNA denaturation PCR. J Gen Virol 86: 125–129. [DOI] [PubMed] [Google Scholar]
- 30.Cougot D, Neuveut C, Buendia MA (2005) HBV induced carcinogenesis. J Clin Virol 34 Suppl 1: S75–78. [DOI] [PubMed] [Google Scholar]
- 31.Forner A, Llovet JM, Bruix J (2012) Hepatocellular carcinoma. Lancet 379: 1245–1255. 10.1016/S0140-6736(11)61347-0 [DOI] [PubMed] [Google Scholar]
- 32.Zhang XD, Wang Y, Ye LH (2014) Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol Med 11: 182–190. 10.7497/j.issn.2095-3941.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hai H, Tamori A, Kawada N (2014) Role of hepatitis B virus DNA integration in human hepatocarcinogenesis. World J Gastroenterol 20: 6236–6243. 10.3748/wjg.v20.i20.6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong DK, Yuen MF, Poon RT, Yuen JC, Fung J, et al. (2006) Quantification of hepatitis B virus covalently closed circular DNA in patients with hepatocellular carcinoma. J Hepatol 45: 553–559. [DOI] [PubMed] [Google Scholar]
- 35.Gao YT, Han T, Li Y, Yang B, Wang YJ, et al. (2010) Enhanced specificity of real-time PCR for measurement of hepatitis B virus cccDNA using restriction endonuclease and plasmid-safe ATP-dependent DNase and selective primers. J Virol Methods 169: 181–187. 10.1016/j.jviromet.2010.07.031 [DOI] [PubMed] [Google Scholar]
- 36.Guo JT, Guo H (2015) Metabolism and function of hepatitis B virus cccDNA: Implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res 122: 91–100. 10.1016/j.antiviral.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reaiche-Miller GY, Thorpe M, Low HC, Qiao Q, Scougall CA, et al. (2013) Duck hepatitis B virus covalently closed circular DNA appears to survive hepatocyte mitosis in the growing liver. Virology 446: 357–364. 10.1016/j.virol.2013.08.014 [DOI] [PubMed] [Google Scholar]
- 38.Bonvin M, Greeve J (2007) Effects of point mutations in the cytidine deaminase domains of APOBEC3B on replication and hypermutation of hepatitis B virus in vitro. J Gen Virol 88: 3270–3274. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen DH, Gummuluru S, Hu J (2007) Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J Virol 81: 4465–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T, Cai J, Chang J, Yu D, Wu C, et al. (2013) Evidence of associations of APOBEC3B gene deletion with susceptibility to persistent HBV infection and hepatocellular carcinoma. Hum Mol Genet 22: 1262–1269. 10.1093/hmg/dds513 [DOI] [PubMed] [Google Scholar]
- 41.Chisari FV, Mason WS, Seeger C (2014) Virology. Comment on "Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA". Science 344: 1237. [DOI] [PubMed] [Google Scholar]
- 42.Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709–715. [DOI] [PubMed] [Google Scholar]
- 43.Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, et al. (2013) APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res 73: 7222–7231. 10.1158/0008-5472.CAN-13-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burns MB, Temiz NA, Harris RS (2013) Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45: 977–983. 10.1038/ng.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pak V, Heidecker G, Pathak VK, Derse D (2011) The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B. J Virol 85: 8538–8547. 10.1128/JVI.02645-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Specificity of RCA to amplify cccDNA in cell culture or liver tissues. cccDNA purified from HepAD38 or clinical patients was amplified by RCA; DNA from core particles and genomic DNA from HBV- transgenic mice were used as controls. B. RCA product from line 2, line3 and line 4 in Fig A amplified by genomic PCR. C. Southern blot of RCA product amplified by genomic PCR.
(TIF)
CT and CNCT samples from the HCC patients were divided into 2 groups with ratio of cccDNA in the paired samples (CT/CNCT) being either < 1 or > 1. cccDNA was specifically amplified by RCA, then the concatemerized product was analysis by 3D-PCR using a denaturation temperature gradient of 86°C-82°C.
(TIF)
A. Southern blot of HBV core-associated DNA with transfected with a replication-incompetent HBV genomic expression vector (YMHA plasmid). B. 3D-PCR of cDNA reverscribed from RNA in lysates from cells transfected with the YMHA plasmid. Huh7.0 cells were cotransfected with YMHA plasmid plus empty plasmid or APOBEC3B expression plasmid. Total RNA was purified and digested with DNase I, then RT-PCR products served as templates for analysis by 3D-PCR with denaturing temperature ranging from 82–90°C. C. Mutation matrices of RT-PCR products. DNA fragments amplified at 94°C in the presence of empty vector and APOBEC3B were cloned into T vectors, and 15 clones were randomly selected and sequenced.
(TIF)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.