Abstract
Actin-like proteins (Alps) are a diverse family of proteins whose genes are abundant in the chromosomes and mobile genetic elements of many bacteria. The low-copy-number staphylococcal multiresistance plasmid pSK41 encodes ParM, an Alp involved in efficient plasmid partitioning. pSK41 ParM has previously been shown to form filaments in vitro that are structurally dissimilar to those formed by other bacterial Alps. The mechanistic implications of these differences are not known. In order to gain insights into the properties and behavior of the pSK41 ParM Alp in vivo, we reconstituted the parMRC system in the ectopic rod-shaped host, E. coli, which is larger and more genetically amenable than the native host, Staphylococcus aureus. Fluorescence microscopy showed a functional fusion protein, ParM-YFP, formed straight filaments in vivo when expressed in isolation. Strikingly, however, in the presence of ParR and parC, ParM-YFP adopted a dramatically different structure, instead forming axial curved filaments. Time-lapse imaging and selective photobleaching experiments revealed that, in the presence of all components of the parMRC system, ParM-YFP filaments were dynamic in nature. Finally, molecular dissection of the parMRC operon revealed that all components of the system are essential for the generation of dynamic filaments.
Introduction
Recent advances in prokaryotic cell biology have challenged the long-held notion that bacterial cells exist merely as casings that contain diffusible chemicals and enzymes. Improved bacterial fluorescent imaging techniques, coupled with the abundance of publically available bacterial genome data, has enabled the spatio-temporal localization of novel proteins to be determined in vivo. Strikingly, bacteria contain an array of proteins which not only adopt specific localizations within cells, but form an integral part of a bacterial subcellular cytoskeleton [1]. For example, the bacterial cytoskeletal protein FtsZ, a distant homologue of eukaryotic tubulin, forms a distinct ring-shape at mid-cell (the ‘Z-ring’) that defines the prokaryotic divisional plane and recruits further proteins involved in bacterial cytokinesis, whereas the actin-like protein MreB that is found in rod-shaped cells forms a discontinuous helical structure involved in controlling the width of a bacterium during cellular growth [2]. While only a few examples of prokaryotic tubulin homologues have been found to date [1], genes encoding actin-like proteins (Alps) are prevalent in the chromosomes and mobile genetic elements (such as plasmids) of many diverse bacterial species [3,4]. Phylogenetic analyses have revealed that chromosomally-encoded Alps, such as MreB, are closely related to each other, whereas Alps present on bacterial mobile genetic elements show vast inter-species sequence divergence [3]. Despite the genetic diversity exhibited by bacterial Alps, crystal structures from distantly related Alps have revealed that they share the basic ‘actin-fold’–the cleft present within all homologues of eukaryotic actin that is required for ATP/GTP binding and hydrolysis–and the ability of the Alp monomer to polymerize into filamentous ultrastructures [5].
The 46 kb Staphylococcus aureus plasmid pSK41 harbors a genetic locus, parMRC (Fig 1A), that encodes an actin-like protein, ParM [6,7]. pSK41 is the prototype of a family of medically important conjugative staphylococcal multiresistance plasmids [8] that have most recently been implicated in the development of vanA-mediated vancomycin resistance in S. aureus [9]. We have previously shown that pSK41 parMRC significantly enhances the segregational stability of an unstable staphylococcal mini-plasmid, and site directed mutagenesis indicated that the NTPase motif of ParM is required for this stability phenotype [6]. The parMRC locus also encodes a DNA binding protein, ParR, which recognizes a series of 10 bp direct repeats, parC, located directly upstream from the parM and parR structural genes. Crystallographic data of ParR bound parC DNA shows that ParR binds as a dimer-of-dimers to the parC repeats, producing an extended macromolecular structure known as the ‘segrosome’ [6]. In the related ParMRC partitioning system of the E. coli multiresistance plasmid R1, ParM interacts with the segrosome to segregate replicated plasmids in a bidirectional fashion. In vitro data indicate that pSK41 ParM adopts a polymeric conformation which is very different to that of actin, MreB and R1 ParM [10]. Whereas pSK41 ParM forms a helical single stranded filament, both R1 ParM and actin adopt a two-start helical conformation, while MreB forms linear protofilaments [10]. Remarkably, database searching using pSK41 ParM crystal structure co-ordinates revealed that this protein is most structurally related to the chromosomally encoded Alp Ta0583 from the archaea Thermoplasma acidophilum, and not the R1 plasmid partitioning protein ParM, underscoring the structural diversity within microbial Alps. Biophysical analyses have suggested that pSK41 ParM filaments undergo a treadmilling-like mechanism of motion in vitro similar to that of F-actin [10]; contrastingly, R1 ParM exhibits a form of dynamic instability similar to that of eukaryotic tubulin [11]. In vivo studies in the native staphylococcal host, using a ParM C-terminal fusion to red fluorescent protein (ParM-RFP), also suggested that pSK41 ParM filaments are not dynamically unstable [12], in agreement with the in vitro observations [10]. Interestingly, the plasmid-partitioning Alp protein Alp7A, from the 55 kb Bacillus subtilis plasmid pLS20, exhibits both treadmilling and dynamic instability [3]. These observations highlight significant diversity in the dynamic properties exhibited by Alps.
Fig 1.
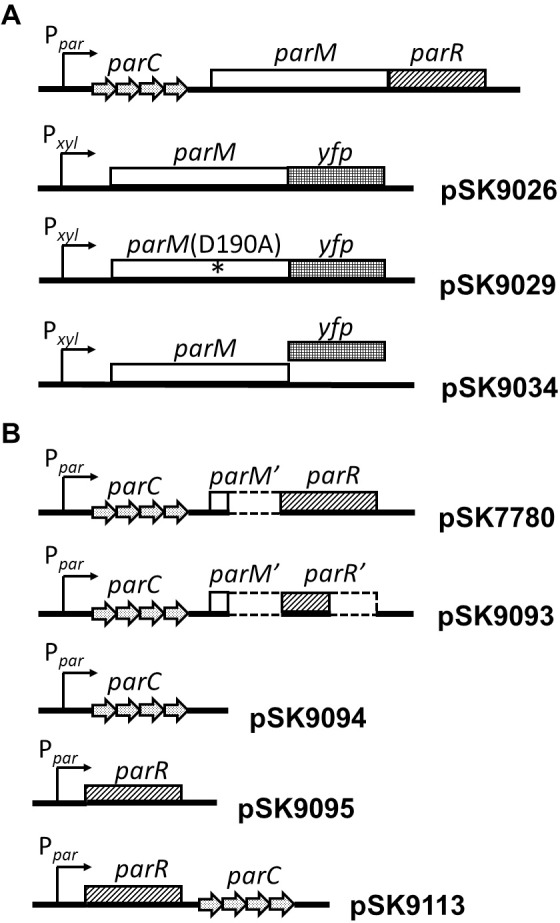
Genetic structures of the pSK41 parMRC operon and parM plasmids (A), and parRC plasmid constructs generated in this study (B). parM is shown as an open box, parR is shown as a cross-hatched box, and yfp is shown as gridded box. Repeats at parC are shown as stippled arrows. Ppar and Pxyl are shown as arrows indicating the direction of transcription. Deleted sequences are denoted by dashed lines. The approximate location of the mutation in parM-yfp of pSK9029 that results in a D190A substitution is indicated by an asterisk. See text for details of plasmid construction. Diagrams are not to scale.
Despite the elucidation of biochemical and biophysical characteristics of the divergent pSK41 ParM Alp [10], little is known about the nature of pSK41 ParM filaments in vivo [12]. Here, we have reconstituted the pSK41 parMRC system in the ectopic rod-shaped host E. coli, and have demonstrated that parMRC is functional in this organism, indicating that pSK41 parMRC is functional as a discrete unit. Using time-lapse fluorescent imaging and selective photobleaching microscopy, we also show that ParM polymers are dynamic in nature, and define the par system components required for this activity. This work enhances the understanding of prokaryotic Alps via the elucidation of the in vivo filament-forming properties of this highly divergent bacterial actin-like ParM protein.
Materials and Methods
Bacterial Strains, Plasmids, Growth Conditions, Media, and Reagents
Plasmid-containing E. coli DH5α cells (Bethesda Research Laboratories) were routinely cultured in Luria Broth (LB) containing 20 μg/ml chloramphenicol or 100 μg/ml ampicillin. Both antibiotics were used at the above concentrations for the selection of co-transformed strains. Liquid broth cultures were grown with agitation using a mechanical orbital shaker (New Brunswick Scientific), set to 220 rpm. Solid media culture was performed using standard non-vented petri plates (Sarstedt, Australia) containing LB supplemented with 1.5% agar and antibiotics where required. All bacterial cell culture was undertaken at 37°C. Reagents for cell culture were purchased from Oxoid (Australia), and chemical reagents were purchased from Sigma-Aldrich (Australia). Bacterial strains and plasmids used in this study are listed in S1 Table.
Plasmid pSK7780 was constructed using the multi-step process detailed below. A ~400 bp fragment encompassing the parC region was amplified from pSK41 plasmid DNA with the primers AB1 and orf346P2HindIII. Concurrent with this process, a second ~400 bp fragment, containing the full-length parR structural gene, was amplified from pSK41 DNA using the primers AB2BamHI and orf346P3HindIII. The purified AB1-orf346P2HindIII PCR product was digested with XbaI and HindIII, and the purified AB2BamHI-orf346P3HindIII product was digested with BamHI and HindIII, before these fragments were co-ligated into pAM401, which had been digested with XbaI and BamHI. The resultant plasmid, pSK7780, contains the pSK41 parC region and the parR open reading frame (ORF), and a deletion derivative of parM (parM‘), which expresses only the first 10 amino acids of the protein. Plasmids pSK9026, pSK9029, pSK9093, pSK9094 and pSK9113 were generated via standard cloning procedures using primers listed in S2 Table. pSK9095 was generated via the multi-step procedure described below. A PCR fragment encompassing Ppar was amplified from pSK7780 using the primers AB1 and AB123. Concurrent with this process, a second PCR fragment harboring the parR ORF was amplified from pSK7780 using the primers AB122 and AB2BamHI. Primers AB122 and AB123 contain regions of complementarity, and these regions were used to join the two amplicons in a PCR reaction which included the primers AB1 and AB2BamHI. The fusion PCR product was digested with XbaI and BamHI and was ligated into similarly prepared pAM401 plasmid DNA, giving rise to pSK9095.
DNA manipulations
DNA manipulations were undertaken using standard protocols detailed in Sambrook et al. [13]. Plasmid DNA extraction from recombinant E. coli cultures was performed using the Bioline Isolate Plasmid Mini-Kit. Polymerase chain reaction (PCR) was done using iProof DNA High-Fidelity DNA Polymerase (Bio-Rad, Australia), using oligonucleotides synthesized by Geneworks, Australia. Primer sequences can be located in S2 Table. Restriction digestion was undertaken using reagents purchased from New England Biolabs (NEB), according to the manufacturer’s instructions. DNA fragments were purified when necessary using the Wizard SV® Gel and PCR Clean-Up system (Promega). Ligation was undertaken at 16°C for 16 hours using T4 DNA ligase purchased from NEB. Recombinant plasmid constructs were sequenced at the Australian Genome Research Facility’s (AGRF) Sydney node.
Plasmid Segregational Stability Assays
Plasmid segregational stability assays were conducted according to a method modified from Schumacher et al. [6]. Briefly, plasmids to be assayed were grown overnight in LB supplemented with both ampicilln and chloramphenicol. The following morning, the stationary phase culture was diluted in 0.1% saline, and viable counts were performed using solid LB media containing ampicillin for the selection of ParM or ParM-Yellow Fluorescent Protein (YFP) fusion expressing plasmids. Using the saline diluted cultures, a 10−4 dilution was made into 10 ml fresh LB containing ampicillin, and was incubated overnight at 37°C with shaking. This process was repeated until approximately 50 generations of growth was achieved (5 days). 50 colonies from the viable count plates were patched onto ampicillin-chloramphenicol double selection media, and the proportion of plasmids remaining in the population was quantified. Stability assays were conducted using three biological replicates, and the standard errors of plasmid-retaining populations were determined using the statistical package available with Microsoft Excel 2007. Differences in plasmid segregational stabilities were evaluated by Fisher’s protected least-significant-difference test after repeated-measures ANOVA, using SPSS Statistics for Macintosh, Version 22.0 (IBM Corporation). A significant difference was defined as a P value of <0.05.
Microscopy
E. coli strains to be assayed were grown with selection overnight. A 1:50 dilution of the saturated culture was made in fresh LB with selection, and the culture was grown to an OD600 nm of 0.6. 0.5 ml of the mid-logarithmic phase culture was harvested by centrifugation, and the pellet was washed once with 0.5 ml phosphate buffered saline (PBS). Cells were collected by centrifugation, and the pellet was resuspended in 50 μl PBS. 3 μl of the cell solution was applied to a 2% agarose pad set within a 65 μl Gene-Frame (Integrated Sciences). Cells were examined using a AxioImager Z1 fluorescence microscope (Carl Zeiss) equipped with a 100 X oil immersion objective lens with a numerical aperture of 1.4. Yellow Fluorescent Protein was excited using light passed through a bandpass 500/20 filter and emitted light was collected through a bandpass 535/30 filter. Images were captured using a Photometrics CoolSNAP HQ camera. Samples for selective photobleaching experiments were prepared as above and then imaged using a LSM 510 Meta confocal microscope (Carl Zeiss) with a 488 nm argon laser at 4% power. A 63 X oil immersion lens with a numerical aperture of 1.4 was used. Regions of interest were photobleached using five iterations of five laser lines (458, 477, 488, 514 and 561 nm), each at 100% power. To monitor recovery of fluorescence after photobleaching, the cells were imaged every 7.6 seconds for three minutes. To capture movies of filament dynamics over time, cells were imaged using a Nikon Eclipse Ti live cell imaging system and a 100 X oil immersion lens with a numerical aperture of 1.45. Excitation light from a LED source passed through a 509/22 filter and emitted light was collected through a 542/25 emission filter. Images were taken every 10 seconds over a 5 minute period in a time series using an Andor iXon Ultra 888 digital camera. To correct for sample drift in the x and y planes over time, each time series was aligned with the Linear Stack Alignment with SIFT plugin within FIJI (http://fiji.sc/Fiji). Movie files were exported at 10 frames per second.
Results and Discussion
Reconstituted trans-acting pSK41 parMRC is functional in the ectopic rod-shaped host, E. coli
Protein components derived from the E. coli plasmid R1 parMRC operon have been shown to retain mechanistic functionality in the presence of parC coated microspheres in vitro [14]. This suggests that parMRC-based partitioning systems are self-contained functional units [14]. In light of this observation, we sought to reconstitute the parMRC system from pSK41 in the heterologous rod-shaped bacterium E. coli, in order to study ParM filament formation and dynamics in vivo. We selected a rod-shaped organism for these studies, rather than the native coccoid host, since the orientation of assembled ParM filaments with respect to the plane of division is easily observable, thereby making it more amenable to functional analyses. Moreover, E. coli cells are larger than S. aureus cells, making them easier to visualize; are easier to genetically transform and manipulate; and have previously been used to study the filament dynamics of partitioning proteins [15]. A diagrammatic representation of the wild-type parMRC operon is shown in Fig 1A.
To reconstitute pSK41 parMRC in E. coli, we constructed a trans-acting system that contains components of the parMRC system distributed across two plasmids. The two-plasmid system described here was necessitated because numerous attempts to clone the entire intact parMRC operon repeatedly resulted in plasmids that accumulate mutations when maintained in E. coli, usually in the par promoter or in the parM gene, indicating that, when expressed in its intact form, the operon is deleterious in E. coli. Plasmids constructed for this system harbor compatible E. coli replication systems, and contain complementary resistance markers. pSK9026 (Fig 1A) is a derivative of the B. subtilis integration plasmid pSG1193 that contains parM cloned as an in-frame fusion to the yfp open reading frame. On pSK9026, transcription of the parM-yfp fusion is controlled by the B. subtilis promoter Pxyl, which is constitutive in E. coli. A site-directed mutagenesis derivative of pSK9026, in which parM was uncoupled from the yfp ORF, was also constructed (pSK9034; Fig 1A)). pSK7780 (Fig 1B) is a moderate copy-number plasmid that replicates using a p15A replicon [16], that contains parC and parR, but with a PCR-generated deletion in parM, so that only the first 10 amino acids of ParM is expressed. To assess the functionality of this reconstituted system, E. coli DH5α cells were co-transformed to ampicillin-chloramphenicol double resistance with pSK7780 and either pSK9026 (parM-yfp), pSK9034 (parM), or pSG9113 (yfp), and the retention of pSK7780 over approximately 50 generations of bacterial growth was determined using segregational stability assays. The assays were performed in the absence of selection for pSK7780, but included ampicillin to ensure the carriage of the other plasmids. These assays (Fig 2) revealed that the parCR plasmid pSK7780 was significantly more stably maintained in the presence of ParM (encoded by co-resident pSK9034; P = 0) or ParM-YFP (from pSK9026; P = 0) than it was in the absence of ParM (vector pSG1193 co-resident), thereby indicating that the reconstituted pSK41 parMRC system was at least partially functional in the ectopic E. coli host. Moreover, ParM-YFP (pSK9026) resulted in pSK7780 stability approximating that mediated by ParM (pSK9034; P = 0.2), suggesting that the YFP tag did not markedly impede ParM function. Although chromosomal segregation systems, which do not encode actin-like NTPases, have been shown to be able to stabilize plasmids in heterologous hosts [17,18], to our knowledge this is the first demonstration of an Alp-based plasmid partitioning system functioning in an ectopic host. Since Gram-positive and Gram-negative bacteria diverged from a common ancestor around 2 billion years ago [19,20], our in vivo data corroborate the in vitro observations of Garner et al. [14] (see above), which indicated that parMRC-like segregation systems are autonomous functional units.
Fig 2. Segregational stability of parC-containing pSK7780 or pSK7780-derived test plasmids in the presence or absence of ParM or ParM-YFP in the E. coli DH5α cells.
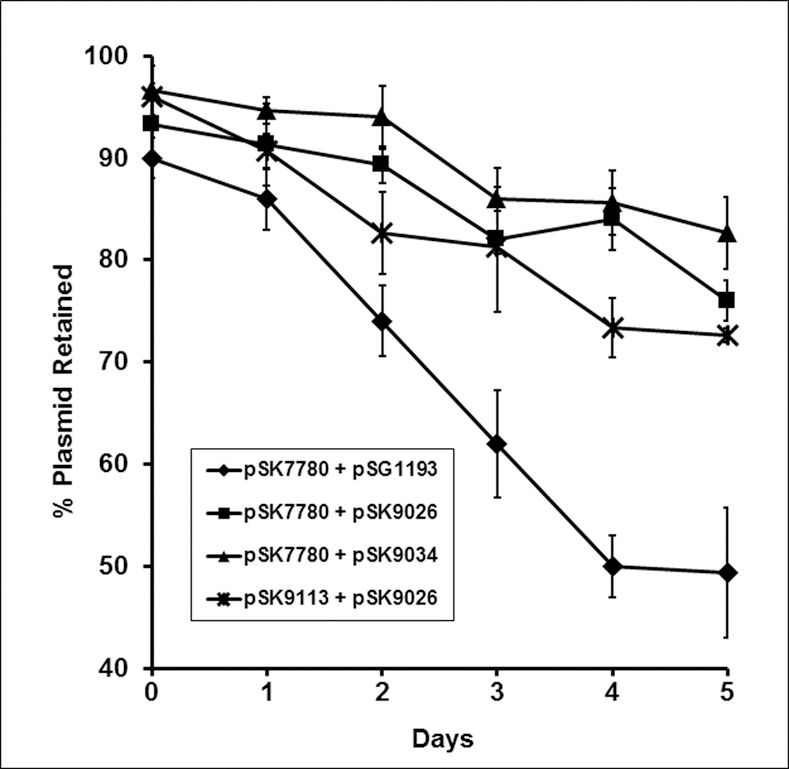
The retention of pSK7780, containing the parC centromere and parR transcribed from Ppar was determined in the presence of pSK9026 (squares; expressing ParM-YFP), pSK9034 (triangles; expressing ParM which had been uncoupled from YFP), or pSG1193 (diamonds; expressing YFP only). The retention of pSK9113 (stars), containing parR transcribed from Ppar, but with parC cloned downstream from the parR ORF, was determined in the presence of pSK9026. Five days of serial subculture represents approximately 50 generations of growth. Each data point is the mean of three biological replicates. Standard error is shown.
ParM-YFP forms filaments in heterologous host cells
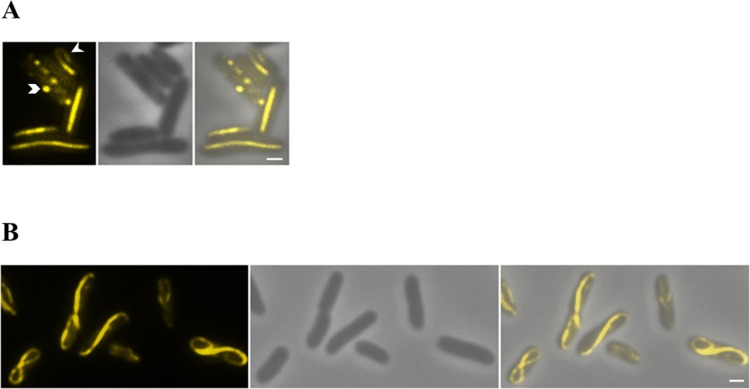
In order to gain insights into the pSK41 parRMC partitioning mechanism in vivo, we conducted fluorescence microscopy using the E. coli strains expressing ParM-YFP described above. In isolation, ParM-YFP (expressed from pSK9026) produced straight pole-to-pole filaments in the majority of cells surveyed (Fig 3A); these are likely to represent filament bundles rather than individual ParM-YFP filaments. Fluorescent foci were present throughout the cytoplasm of occasional cells, possibly representing genesis points for ParM-YFP filaments. In some cells, longer ParM-YFP filaments appeared to curve around the perimeter of the cell, producing a hook-shaped polymer. Most cells contained a single ParM-YFP filament, however, a small number of cells appeared to contain two or three; none contained more. Thus, although longer, the ParM-YFP filaments observed in E. coli closely resembled the straight ParM-RFP filaments seen previously in the smaller coccoid natural host, S. aureus [12].
Fig 3. ParM-YFP filament formation in the presence or absence of parC and ParR.
E. coli cells expressing ParM-YFP in isolation (A) or expressing ParM-YFP in the presence of parC and ParR (B) were visualized by fluorescence microscopy. ParM-YFP filaments exhibit a dramatic shift in morphology in the presence of parC and ParR. Hook shaped filaments are indicated with an arrow-head, and ParM-YFP foci are shown with a chevron. Left to right, both panels: fluorescence image, phase contrast image; merge of phase contrast and fluorescent images. Scale bars represent 1 μm.
Since an intact NTPase motif in ParM is essential for the in vivo partitioning phenotype of the parMRC locus [6], we explored its requirement for ParM filament formation. To do this, we constructed a pSK9026 derivative that contains a parM ORF harboring a D190A mutation in the NTP binding domain [6,7], fused to yfp (pSK9029; Fig 1A; S1 Table). Interestingly, when pSK9029 containing cells were visualized by fluorescence microscopy, only general fluorescence was observed (data not shown). In contrast, equivalent mutations that abolished the ATPase activity of ParM from R1, and the plasmid segregation Alp, AlfA, from B. subtilis plasmid pLS32, did not abolish polymerization of those Alp proteins [7,21]. Although we cannot preclude the possibility that the D190A mutation may prevent proper protein folding, the lack of ParMD190A-YFP filaments might indicate that the D190A mutation in pSK41 ParM disrupts nucleotide binding rather than hydrolysis. However, further experiments are required to investigate the precise effect of the D190A mutation on the ATPase activity of pSK41 ParM. An ATPase deficient mutant of Alp7A was also unable to produce filaments at wild-type cellular concentrations, but was able to produce filaments when the protein was significantly overproduced [22]. In these cells, Alp7A forms large and amorphous polymers that interrupt chromosomal segregation and cell division. Although expression of pSK41 ParM-YFP and ParMD190A-YFP was unregulated in E. coli, cell morphology did not appear to be affected.
In order to assess pSK41 ParM-YFP polypeptide localization with other components of the par system present, E. coli cells harboring both pSK9026 (expressing ParM-YFP) and pSK7780 (Fig 1) were visualized by fluorescence microscopy. Strikingly, cells expressing ParM-YFP and containing the parC centromere-like site and ParR showed a dramatically different ParM-YFP filament morphology compared to cells expressing ParM-YFP alone. Most of the cells within this population contained thinner, curved fluorescent structures that extended along the axis of the E. coli cell, which sometimes appeared lemniscate (figure-eight shaped) in nature (Fig 3B); it is likely that these also represent filament bundles rather than individual filaments (see below). Importantly, since E. coli cells in which all of the components of the pSK41 partitioning system were reconstituted imparted a stable partitioning phenotype to test plasmids (Fig 2), this observation suggests that a shift in ParM filament morphology correlates with an active partitioning phenotype in vivo. Interestingly, the filament morphology of ParM-YFP in the presence of both parC and ParR differs to that of other characterized plasmid Alps; whereas pSK41 ParM-YFP often appeared to be continuous, ParM from R1, AlfA, and Alp7A exhibit a curved, but open-ended, filament shape [3,7,21]. However, as the curved ParM-YFP filaments observed here have been generated in an ectopic host, and since expression of the ParM-YFP fusion protein is unregulated, the observed lemniscate architecture of the polymer may not be representative of its typical conformation in its natural coccoid host at wild-type expression levels.
Curved ParM-YFP filaments are dynamic
The plasmid partitioning Alps ParM from R1, AlfA, and Alp7A form dynamic polymers, and this characteristic is essential for the partitioning function exhibited by these systems [3,7,21]. Mechanistically, dynamic polymerization of plasmid segregation Alps can be achieved in a variety ways: ParM from plasmid R1 exhibits dynamic instability (analogous to that of eukaryotic tubulin); AlfA from pLS32 exhibits treadmilling (analogous to that of eukaryotic actin); and Alp7A from pLS20 exhibits both dynamic instability and treadmilling [3,11,23]. In vitro time-lapse total internal reflection fluorescence (TIRF) imaging using purified pSK41 ParM suggested that ParM polymers do not exhibit dynamic instability, and instead exhibit a treadmilling phenotype [24]. Consistent with this observation, live cell imaging of ParM-RFP filaments in the native S. aureus host indicated that ParM in isolation is not dynamically unstable in vivo [12]. In order to elucidate further mechanistic details of pSK41 ParM filaments in the bacterial cytosol, we conducted live-cell time lapse fluorescent microscopy imaging, using the ParM-YFP expressing strains described above. Images were captured for each ParM-YFP expressing strain every 10 seconds over a time course of five minutes, and images were processed and compiled into a motion picture. This analysis revealed that ParM-YFP filaments formed in the presence of parC and ParR (pSK7780) were dynamic (S1 Movie), whereas ParM-YFP filaments produced in isolation were static (S2 Movie). In all cells visualized, the curved ParM-YFP filaments appeared to undergo active remodeling during the capture period. Filaments did not adopt a preferred distribution within the cell, and polymers of varying lengths were observed forming and redistributing throughout the entire cell cytosol, indicating that ParM filaments are not compartmentalized or confined to particular cellular locales. Importantly, we did not observe any evidence of catastrophic disassembly in vivo, consistent with in vitro and in vivo evidence reported previously [10,12] that indicated that pSK41 ParM filaments do not exhibit dynamic instability [10,12].
To gain further insights into the nature of pSK41 ParM-YFP polymer formation, we conducted selective photobleaching experiments, using the strains described above. Cells containing pSK9026 (ParM-YFP), or pSK9026 and pSK7780 (ParR and parC), were grown to mid-logarithmic phase prior to visualization via fluorescence microscopy. As expected, ParM-YFP filaments expressed in isolation showed no recovery of the photobleached areas over the course of the experiment (Fig 4A; S3 Movie), supporting evidence that ParM-YFP expressed in isolation are static (see above). In contrast, photobleached areas of ParM-YFP filaments formed in the presence of ParR and parC showed a rapid recovery, confirming that ParM-YFP monomers exhibit active turnover within dynamic ParM-YFP polymers (Fig 4B; S4 Movie). Fluorescent recovery to the right of the filament in Fig 4B (S4 Movie) appears to be at the expense of diminishing fluorescence to the left of the bleached region, consistent with dynamic filament turnover. Similar to AlfA, ParM-YFP bleached zones appeared to recover using the same track as observed prior to bleaching [25], which, as suggested by Polka et al., indicates that ParM protofilaments are likely to form a close lateral association with each other [25]. Likewise, cryoelectron tomography-based experiments have revealed that ParM from R1 forms bundles of three to five filaments that are actively involved in plasmid segregation, indicating that filament bundling is likely to be a common mechanism involved in Alp-mediated partitioning [26].
Fig 4. ParM-YFP filaments are dynamic in the presence of parC and ParR.
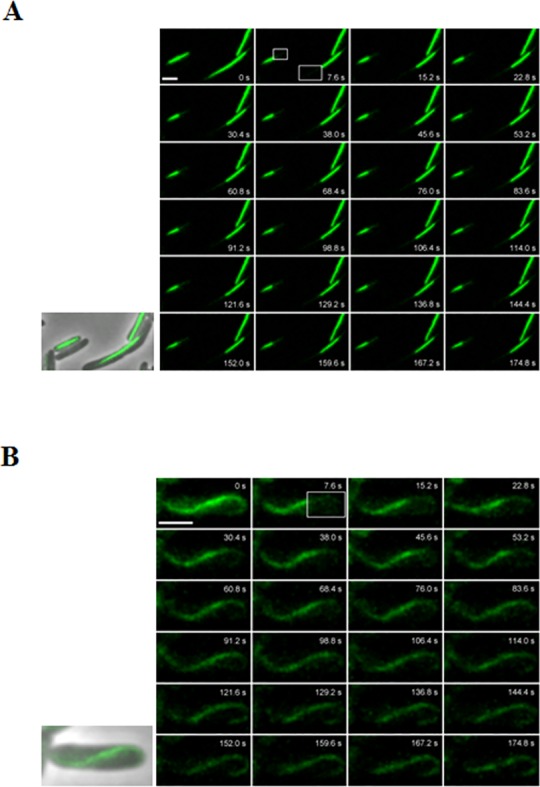
E. coli cells expressing either ParM-YFP in isolation (A) or ParM-YFP in the presence of parC and ParR were grown to mid-logarithmic phase and selective photobleaching experiments were undertaken. Fluorescence recovery was monitored by imaging cells every 7.6 seconds over the course of three minutes. The first image in the series depicts a pre-bleached cell. Phase contrast/fluorescence overlay images of cells expressing ParM-YFP are shown to the left of the photobleaching montage. Boxes indicate regions of laser photobleaching. Time units (seconds; s) are shown. Scale bars represent 2 μm.
ParM-YFP requires parC and ParR for dynamic ParM-YFP polymer turnover
Dynamic ParM-YFP filaments are generated in the presence of all components of the pSK41 parMRC system in vivo (see above). To delineate the requirements for ParM-YFP dynamic filament formation, a series of plasmids containing various components of the pSK41 parMRC system were constructed, in order to determine the contribution of each component individually, or in combination, to filament dynamics. Four constructs were made using the pAM401 parent vector, and separately co-transformed with pSK9026 (expressing ParM-YFP alone) into competent E. coli DH5α cells. The first plasmid generated, pSK9093 (Fig 1B), contains an amplicon encompassing Ppar and parC, and a harbors a fragment of parR (parR’) engineered to express only the first 52 amino acids of the mature ParR polypeptide (ParRN). Our previous work has shown that ParRN is sufficient to be able to bind the pSK41 parC region [6]. pSK9093 was therefore able to be used to determine if the formation of the segrosome, in the absence of the C-terminus of ParR (which is thought to have a role in the recruitment of ParM proteins to the segrosome [6]), is sufficient to be able to generate dynamic ParM-YFP polymers in vivo. pSK9094 (Fig 1B) contains a fragment encompassing only parC and the Ppar promoter, and pSK9095 (Fig 1B) contains the entire parR ORF, transcribed by Ppar, but lacks parC. Since parR transcription is autoregulated via ParR binding to parC [6], any phenotype observed with pSK9095 might be attributable to unregulated parR expression rather than an absence of parC itself (e.g., for assembly of the ParR-parC segrosome). pSK9113 (Fig 1B), containing parC downstream of parR, was therefore constructed to allow these possibilities to be unambiguously differentiated. Thus, any differences in ParM-YFP behavior observed between cells harboring pSK9095 or pSK9113 can be attributed to the presence/absence of parC, and not unregulated parR expression.
Plasmids generated were separately co-transformed with pSK9026 into E. coli DH5α cells, and time-lapse microscopy (as detailed above) was undertaken. The results of these studies showed that ParM-YFP filaments in the presence of pSK9093 (parCR’), pSK9094 (parC), or pSK9095 (parR), were largely static (S5, S6 and S7 Movies). However, in strains harboring pSK9113, which therefore contain the entire reconstituted parMRC system, ParM-YFP active polymer turnover was evident (S8 Movie). Segregational stability assays, which determined the retention of pSK9113 in the presence of pSK9026 (parM-yfp) over the course of approximately 50 generations of growth, showed that pSK9113 is more stable than the parent plasmid pSK7780 co-resident with the yfp vector pSG1193 (P = 0.001), indicating that the reorganized parRC system on pSK9113 retains functionality (Fig 2). Thus, these results indicate that 1) all components of the pSK41 parMRC region are required for dynamic ParM-YFP filament turnover; 2) the C-terminus of ParR is essential for function of the segregation system; and 3) the interaction of parC, ParR, and ParM, to generate the pSK41 segregation complex, activates ParM-YFP dynamic filament formation. These findings are consistent with the assembly mechanism proposed by Gayathri et al. [27], who showed that a 17 amino acid region from the C-terminus of R1 ParR interacts directly with the polymerization interface of the ParM polymer. Based on structural observations, these authors hypothesize that ParR monomers are required to be released from the ParM-ParR complex for ParM polymerization to occur, and that the ParR-parC complex, composed of ten ParR dimers, forms a scaffold that facilitates ParM polymerization via a ‘stair-stepping’ mechanism, analogous to that of eukaryotic formin. It is unknown, at this stage, whether such a ‘stair-stepping’ mechanism for filament polymerization is universally conserved amongst all bacterial Alps.
It should be noted that the parMRC system reconstituted here in E. coli represents a very different context to the native system on pSK41 in S. aureus. In particular, in addition to autoregulation by ParR [6], transcription from the par promoter is controlled by a global regulator of plasmid transcription encoded by pSK41, ArtA [28]. Moreover, pSK41 is a large conjugative plasmid with a tightly-controlled narrow-host-range replication system [29], and at least two other plasmid maintenance determinants; viz., the res multimer resolution system [30] and a fst-like toxin-antitoxin system [31]. Remarkably, isolated from all these extrinsic factors in an unrelated host, the reconstituted minimal pSK41 par system was still able to perform its basic biological function, increasing the segregational stability of a parC-containing plasmid. However, when considering the observations described here it should be remembered that they are, by necessity, of an artificial system involving multiple plasmids, heterologous promoters, a YFP fusion to ParM, and an unrelated host; but they are nonetheless observations of a functional partitioning system. As noted above, the two-plasmid system described here was employed because attempts to clone the entire parMRC operon in E. coli resulted in plasmids that accumulate mutations, indicating that, when expressed in its intact form, the operon is deleterious in E. coli. The basis for this toxicity is not clear but the absence of ArtA control might be a contributing factor. In this regard, it should be noted that Ppar doesn’t drive transcription of parM in the two-plasmid system used in these studies. Additionally, a non wild-type ParM protein (ParM-YFP) was used to track the distribution of ParM proteins within the E. coli cytosol. Caution must be exercised when interpreting the distribution of ParM-YFP filament bundles, since YFP is known to dimerize in in vivo [32,33]. The dimerization properties of fluorescent protein tags have recently been implicated in the aggregation of fused polypeptides in vivo [33,34,35,36]. Nonetheless, it would seem unlikely that YFP dimerization could be responsible for the parCR-dependent morphological shift in ParM-YFP filament bundles observed, particularly in view of the plasmid stabilizing activity of the reconstituted system in the absence of any native ParM.
In summary, we have shown that a YFP-tagged derivative of the staphylococcal pSK41 parMRC system is functional in the heterologous E. coli host, and that the formation of dynamic ParM-YFP filaments correlates with partition function in vivo. All components of the parMRC system were required for the generation of dynamic ParM-YFP filaments. Moreover, the C-terminus of ParR, which facilitates the recruitment of ParM to the segrosome complex, was shown to be required for the conversion of static ParM filaments to a dynamic form proficient for active segregation. This study adds further important information to the suite of growing data elucidating the in vivo properties of the diverse array of recently described bacterial Alps.
Supporting Information
E. coli cells expressing ParM-YFP in the presence of parC and ParR (expressed from pSK7780) were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
E. coli cells expressing ParM-YFP in the absence of ParR and parC were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
E. coli cells expressing ParM-YFP in the absence of ParR and parC were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Regions of interest were photobleached using five iterations of five laser lines (458, 477, 488, 514 and 561 nm), each at 100% power. Recovery of fluorescence of photobleached cells was monitored by imaging every 7.6 seconds for three minutes. Captured images were processed using ImageJ v1.48.
(MOV)
E. coli cells expressing ParM-YFP in the presence of ParR and parC were grown to mid-logarithmic phase fluorescence microscopy was undertaken. Regions of interest were photobleached using five iterations of five laser lines (458, 477, 488, 514 and 561 nm), each at 100% power. Recovery of fluorescence of photobleached cells was monitored by imaging every 7.6 seconds for three minutes. Captured images were processed using ImageJ v1.48.
(MOV)
E. coli cells expressing ParM-YFP in the presence of parC and a truncated ParR protein (ParRN; expressed from pSK9093) were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
E. coli cells expressing ParM-YFP in the presence of parC (on pSK9094), but in the absence of ParR, were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
E. coli cells expressing ParM-YFP in the presence of ParR (expressed from pSK9095), but in the absence of parC, were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
The parMRC system was reconstituted using plasmid pSK9113 so that the expression of ParR is under the control Ppar and parC is present downstream from the parR ORF. pSK9026, expressing ParM-YFP, was co-transformed with pSK9113 and resulting strains were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
(DOC)
(DOC)
Acknowledgments
This work was supported by National Health and Medical Research Council of Australia Project grants 307620 and APP1030003.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Health and Medical Research Council of Australia Project grants 307620 and APP1030003 to NF and RS (https://www.nhmrc.gov.au/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pilhofer M, Jensen GJ (2013) The bacterial cytoskeleton: more than twisted filaments. Curr Opin Cell Biol 25: 125–133. 10.1016/j.ceb.2012.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingerson-Mahar M, Gitai Z (2012) A growing family: the expanding universe of the bacterial cytoskeleton. FEMS Microbiol Rev 36: 256–266. 10.1111/j.1574-6976.2011.00316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, et al. (2009) Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol 73: 534–552. 10.1111/j.1365-2958.2009.06771.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popp D, Narita A, Lee LJ, Ghoshdastider U, Xue B, Srinivasan R, et al. (2012) Novel actin-like filament structure from Clostridium tetani. J Biol Chem 287: 21121–21129. 10.1074/jbc.M112.341016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popp D, Robinson RC (2011) Many ways to build an actin filament. Mol Microbiol 80: 300–308. 10.1111/j.1365-2958.2011.07599.x [DOI] [PubMed] [Google Scholar]
- 6.Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, Skurray RA, et al. (2007) Segrosome structure revealed by a complex of ParR with centromere DNA. Nature 450: 1268–1271. [DOI] [PubMed] [Google Scholar]
- 7.Moller-Jensen J, Jensen RB, Lowe J, Gerdes K (2002) Prokaryotic DNA segregation by an actin-like filament. EMBO J 21: 3119–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA (1998) Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol 180: 4350–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu W, Clark N, Patel JB (2013) pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob Agents Chemother 57: 212–219. 10.1128/AAC.01587-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popp D, Xu W, Narita A, Brzoska AJ, Skurray RA, Firth N, et al. (2010) Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation. J Biol Chem 285: 10130–10140. 10.1074/jbc.M109.071613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garner EC, Campbell CS, Mullins RD (2004) Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306: 1021–1025. [DOI] [PubMed] [Google Scholar]
- 12.Brzoska AJ, Firth N (2013) Two-plasmid vector system for independently controlled expression of green and red fluorescent fusion proteins in Staphylococcus aureus. Appl Environ Microbiol 79: 3133–3136. 10.1128/AEM.00144-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Russell D, editors (2001) Molecular Cloning: A Laboratory Manual. 3rd ed. NY: Cold Spring Harbor Laboratory, Cold Spring Harbor. [Google Scholar]
- 14.Garner EC, Campbell CS, Weibel DB, Mullins RD (2007) Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315: 1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J (2007) Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev 21: 1340–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang AC, Nunberg JH, Kaufman RJ, Erlich HA, Schimke RT, Cohen SN (1978) Phenotypic expression in E. coli of a DNA sequence coding for mouse dihydrofolate reductase. Nature 275: 617–624. [DOI] [PubMed] [Google Scholar]
- 17.Yamaichi Y, Niki H (2000) Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc Natl Acad Sci U S A 97: 14656–14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godfrin-Estevenon AM, Pasta F, Lane D (2002) The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol Microbiol 43: 39–49. [DOI] [PubMed] [Google Scholar]
- 19.Woese CR (1987) Bacterial evolution. Microbiol Rev 51: 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng DF, Cho G, Doolittle RF (1997) Determining divergence times with a protein clock: update and reevaluation. Proc Natl Acad Sci U S A 94: 13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker E, Herrera NC, Gunderson FQ, Derman AI, Dance AL, Sims J, et al. (2006) DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J 25: 5919–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derman AI, Nonejuie P, Michel BC, Truong BD, Fujioka A, Erb ML, et al. (2012) Alp7R regulates expression of the actin-like protein Alp7A in Bacillus subtilis. J Bacteriol 194: 2715–2724. 10.1128/JB.06550-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polka JK, Kollman JM, Mullins RD (2014) Accessory factors promote AlfA-dependent plasmid segregation by regulating filament nucleation, disassembly, and bundling. Proc Natl Acad Sci U S A 111: 2176–2181. 10.1073/pnas.1304127111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popp D, Iwasa M, Narita A, Erickson HP, Maeda Y (2009) FtsZ condensates: an in vitro electron microscopy study. Biopolymers 91: 340–350. 10.1002/bip.21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polka JK, Kollman JM, Agard DA, Mullins RD (2009) The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J Bacteriol 191: 6219–6230. 10.1128/JB.00676-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salje J, Zuber B, Lowe J (2009) Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science 323: 509–512. 10.1126/science.1164346 [DOI] [PubMed] [Google Scholar]
- 27.Gayathri P, Fujii T, Moller-Jensen J, van den Ent F, Namba K, Lowe J (2012) A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science 338: 1334–1337. 10.1126/science.1229091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni L, Jensen SO, Ky Tonthat N, Berg T, Kwong SM, Guan FH, et al. (2009) The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes. Nucleic Acids Res 37: 6970–6983. 10.1093/nar/gkp756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weaver KE, Kwong SM, Firth N, Francia MV (2009) The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61: 94–109. 10.1016/j.plasmid.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeBard RJ, Jensen SO, Arnaiz IA, Skurray RA, Firth N (2008) A multimer resolution system contributes to segregational stability of the prototypical staphylococcal conjugative multiresistance plasmid pSK41. FEMS Microbiol Lett 284: 58–67. 10.1111/j.1574-6968.2008.01190.x [DOI] [PubMed] [Google Scholar]
- 31.Kwong SM, Jensen SO, Firth N (2010) Prevalence of Fst-like toxin-antitoxin systems. Microbiology 156: 975–977; discussion 977. 10.1099/mic.0.038323-0 [DOI] [PubMed] [Google Scholar]
- 32.Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2: 905–909. [DOI] [PubMed] [Google Scholar]
- 33.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J (2012) Segregation of molecules at cell division reveals native protein localization. Nat Methods 9: 480–482. 10.1038/nmeth.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R (2011) Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333: 225–228. 10.1126/science.1203466 [DOI] [PubMed] [Google Scholar]
- 35.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333: 222–225. 10.1126/science.1203285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, et al. (2011) The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A 108: 15822–15827. 10.1073/pnas.1108999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
E. coli cells expressing ParM-YFP in the presence of parC and ParR (expressed from pSK7780) were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
E. coli cells expressing ParM-YFP in the absence of ParR and parC were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
E. coli cells expressing ParM-YFP in the absence of ParR and parC were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Regions of interest were photobleached using five iterations of five laser lines (458, 477, 488, 514 and 561 nm), each at 100% power. Recovery of fluorescence of photobleached cells was monitored by imaging every 7.6 seconds for three minutes. Captured images were processed using ImageJ v1.48.
(MOV)
E. coli cells expressing ParM-YFP in the presence of ParR and parC were grown to mid-logarithmic phase fluorescence microscopy was undertaken. Regions of interest were photobleached using five iterations of five laser lines (458, 477, 488, 514 and 561 nm), each at 100% power. Recovery of fluorescence of photobleached cells was monitored by imaging every 7.6 seconds for three minutes. Captured images were processed using ImageJ v1.48.
(MOV)
E. coli cells expressing ParM-YFP in the presence of parC and a truncated ParR protein (ParRN; expressed from pSK9093) were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
E. coli cells expressing ParM-YFP in the presence of parC (on pSK9094), but in the absence of ParR, were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
E. coli cells expressing ParM-YFP in the presence of ParR (expressed from pSK9095), but in the absence of parC, were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
The parMRC system was reconstituted using plasmid pSK9113 so that the expression of ParR is under the control Ppar and parC is present downstream from the parR ORF. pSK9026, expressing ParM-YFP, was co-transformed with pSK9113 and resulting strains were grown to mid-logarithmic phase and fluorescence microscopy was undertaken. Images were captured every 10 seconds over a time course of 5 minutes. Images were compiled into a motion picture using FIJI.
(AVI)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.